Abstract
Acoustic startle responses have been studied extensively in relation to individual differences and psychopathology. We examined three indices of the blink response in a picture-viewing paradigm—overall startle magnitude across all picture types, and aversive and pleasant modulation scores—in 3,323 twins and parents. Biometric models and molecular genetic analyses showed that half the variance in overall startle was due to additive genetic effects. No single nucleotide polymorphism was genome-wide significant, but GRIK3 did produce a significant effect when examined as part of a candidate gene set. In contrast, emotion modulation scores showed little evidence of heritability in either biometric or molecular genetic analyses. However, in a genome-wide scan, PARP14 did produce a significant effect for aversive modulation. We conclude that, although overall startle retains potential as an endophenotype, emotion-modulated startle does not.
Descriptors: Endophenotypes, Startle, Heritability, Genome-wide association study, Molecular genetics, Gene-based tests, GCTA
Presented with a startling stimulus, many animal species will show a whole body response that facilitates initial orienting and attention, and if necessary, escape and protective responses. One component of the startle reaction is the eye blink, a reflexive response that protects this key organ. The startle blink response is one of the most widely used indices of emotional, especially defensive, reactivity in neurobiology. Extensive work using rodents has shown that it is mediated by two circuits. One involves a basic reflex response that is initiated by the nucleus reticularis pontis caudalis after input from a startling stimulus. The other receives input from the amygdala, and can modulate the basic reflex initiated by the first circuit. This latter system, in turn, is also divided into a “fear” system that is directly linked to the central nucleus of the amygdala, and a more tonic system associated with longer-lasting stressors (presumably anxiety-related responses) that is modulated by the extended amygdala or the bed nucleus of the stria terminalis (Davis, 1998; Davis, Walker, & Lee, 1997). These circuits are distinct in that they can be selectively disabled, and consequently affect different parameters of the blink response (Davis, Walker, Miles, & Grillon, 2010). They are also interrelated, which permits individual differences in experiences of stressors to influence the basic reflex via the second circuit. There is also some evidence that inhibition of startle is linked to the nucleus accumbens (Koch, Schmid, & Schnitzler, 1996), though the precise mechanism by which this occurs is relatively less explored.
In psychopathology research, startle blinks are sometimes elicited using air puffs delivered to the eye or, more commonly, through the abrupt presentation of a loud acoustic stimulus. The strength of the eye blink response can be indexed from electrodes placed around the eye to record electromyographic activity from the orbicularis oculi muscle. Of interest are individual differences in the strength of the basic or general reflexive response and its modulation. Attentional modulation can be achieved by preceding the startling stimulus with a less intense auditory stimulus. In healthy subjects using this prepulse inhibition (PPI) procedure, preservation of attention to the first auditory stimulus (and relative inhibition of the blink response to the subsequent, startling stimulus) is thought to facilitate attention to the information represented in the initial stimulus by preventing a strong response to subsequent, possibly conflicting stimuli. However, the normal reduction in startle responses to noises preceded by a prepulse may not occur in individuals with mental disorders (e.g., schizophrenia, see Braff, 2010, for a review), presumably because they lack the ability to filter—or “gate”—such stimuli.
Emotional modulation of the startle response can be achieved using various methods, but one common approach is to present unwarned acoustic probes while subjects view affectively arousing pictures with neutral, pleasant, or unpleasant content. When startled against a pleasant picture foreground, healthy individuals show a reduced eye blink response compared to their response in the presence of a neutral scene. When the picture is unpleasant, potentiation of the eye blink is observed. These bidirectional effects result in a linear pattern of startle response modulation across picture valence categories (i.e., pleasant < neutral < unpleasant startle response magnitude). Deviations from this pattern are associated with psychopathology, especially internalizing disorders and psychopathy (see Vaidyanathan, Patrick, & Cuthbert, 2009, for a review).
In this article, we examine the molecular genetic basis of the general startle response and its emotional modulation using the affect-picture startle paradigm (cf. Vrana, Spence, & Lang, 1988). Although we did not use the PPI startle procedure, our results as they pertain to the general startle response are likely relevant at least in part to understanding the genetics of PPI startle as well.
Psychopathology and the Affect-Picture Startle Paradigm
Numerous studies have found that the blink reflex is altered among individuals with phobic disorders where potentiation of the response in the presence of an unpleasant emotional foreground (especially involving fear) is exaggerated (Grillon & Baas, 2003; Lang & McTeague, 2008; Vaidyanathan et al., 2009). Results have been less consistent for other anxiety disorders and depression (Vaidyanathan et al., 2009). In a recent study involving a subset of 515 female twins from the Minnesota Twin Family Study (MTFS) who were also included in the current report, Vaidyanathan et al. (2014) found that major depression was associated with an abnormal startle pattern only in those with recurrent depression when compared to those with single episode depression and nondepressed controls. Those with multiple episodes showed a potentiated response to both pleasant and unpleasant pictures.
Quite different startle patterns have been observed for psychopathy. Psychopaths have been found to show a decreased startle response, but only to unpleasant pictures generally (cf. Patrick, Bradley, & Lang, 1993). A similar finding has been reported in 355 MTFS male twins who also comprise part of the sample for the present investigation. Twenty-year-old males with psychopathic dispositions, as reflected in high scores on a scale derived from a personality questionnaire to predict psychopathic traits, showed a similar pattern of reduced potentiation of the startle response by unpleasant pictures (Benning, Patrick, & Iacono, 2005). These different patterns of startle found for phobia, depression, and psychopathy presumably indicate the degree to which the functioning of fear circuitry, indexed by the modulation of the startle response and mediated by the amygdala, is differentially implicated in these disorders.
In addition to assessing patterns of emotion startle modulation, the general or baseline startle reflex (referred to as overall startle in the current report) has also been of interest. The general startle response has been measured in various ways, including during a period preceding emotional manipulation (Morgan, Grillon, Southwick, Davis, & Charney, 1995), during intertrial intervals (Cuthbert et al., 2003), or across all trial types in an affect picture context (Kaviani et al., 2004). The general response thus provides an index of startle reactivity absent any emotional modification—presumably a direct index of the functioning of the obligatory circuit mentioned above. However, the general startle response has been less consistently linked to psychopathology, although there is evidence that heightened general startle is linked to anxiety disorders (Vaidyanathan et al., 2009), including posttraumatic stress disorder (Pole, 2007).
Startle as an Endophenotype
As we noted in the introductory paper outlining the rationale and methodology used in the articles comprising this special issue (Iacono, Malone, Vaidyanathan, & Vrieze, 2014), the affect picture startle paradigm was added to MTFS in the 1990s because of the well-developed literature and theory supporting its relevance to understanding fundamental psychological and neurobiological processes relevant to psychopathology (Lang, Bradley, & Cuthbert, 1990). At that time, relatively little research had been carried out that could be used to evaluate the endophenotypic potential of startle. Of particular interest is research emerging since the adoption of this paradigm by the MTFS that shows that it is heritable and present in well relatives of those with psychiatric disorder, two fundamental characteristics of an endophenotype (Gottesman & Gould, 2003; Iacono & Malone, 2011).
For the general startle response, substantial heritable individual differences (about .67 across studies) in overall magnitude have been reported (Anokhin, Golosheykin, & Heath, 2007; Anokhin, Heath, Myers, Ralano, & Wood, 2003; Carlson, Katsanis, Iacono, & McGue, 1997; Hasenkamp et al., 2010). To date, two studies (Anokhin et al., 2007; Carlson et al., 1997) have examined the heritability of the emotion-modulated blink response in twin samples using behavior genetic methodology. One found that startle modulation scores were also heritable (Carlson et al., 1997), whereas the second, larger study did not (Anokhin et al., 2007). Family studies have indicated that abnormal startle responses appear to be inherited with anxiety disorders and depression, even if individuals are only at risk for them and do not show evidence of the disorders themselves. For example, both children and grandchildren of subjects with anxiety disorders and depression showed varying patterns of startle abnormality in either threatening context or for overall startle, compared to relatives of controls who did not have any psychiatric disorders (Grillon, Dierker, & Merikangas, 1998; Grillon et al., 2005). These family findings provide the strongest support available for startle modulation as an endophenotype, in this case, for anxiety and depression.
Individual differences in PPI startle measures have also been shown to be heritable (Anokhin et al., 2003; Greenwood et al., 2007). Moreover, reduced PPI is observed in relatives of schizophrenia probands, and there is a relatively strong body of research supporting this measure as an endophenotype for schizophrenia (Braff, 2010).
To date, studies of candidate genes in relation to the emotion-modulated startle response have found that single nucleotide polymorphisms (SNPs) or other variants in several genes are related to aspects of the blink response: the serotonin transporter (5-hydroxytryptamine transporter gene-linked region—5-HTTLPR; Brocke et al., 2006), dopamine transporter (DAT; Pauli et al., 2010), catechol-O-methyltransferase (COMT; Montag et al., 2008), and monoamine oxidase A (MAOA; Larson, Taubitz, & Robinson, 2010). However, to our knowledge, successful replications are lacking, and, in general, molecular genetic studies of the overall startle response and affect modulated startle are few in number. In particular, there have not been any genome-wide association studies (GWAS) investigating how genetic variants spread across the genome might be linked to startle.
Aims of the Current Study
The current study was undertaken to fill this gap in the literature. We conducted a GWAS in a large sample of more than 3,000 individuals, including MTFS twins and their parents, from the Minnesota Center for Twin and Family Research (MCTFR). We pursued several lines of investigation. First, because it is not clear to what extent the emotion-modulated startle response is heritable, using our large, family-based twin sample, we examined the biometric heritability of three affect picture startle paradigm measures: general overall startle, emotion-potentiated startle to aversive picture stimuli, and emotion-inhibited startle to pleasant picture stimuli. Second, these analyses were complemented by an examination of SNP heritability using genome-wide complex trait analysis (GCTA; Yang, Lee, Goddard, & Visscher, 2011). GCTA estimates the proportion of variance in startle due to the aggregate, additive effect of all SNPS on our genotyping array.
Third, we conducted a GWAS of each of more than 527,000 SNPs on our genotyping array, followed by a more focused analysis of 1,180 SNPs identified through meta- and “mega-” analyses of psychiatric disorders and traits that are plausibly relevant to understanding the molecular genetic basis of endophenotypes generally, including the startle response and startle modulation. Fourth, we conducted gene-based analyses examining the degree of association between each of 17,601 autosomal genes available through the versatile gene-based association study method (VEGAS; Liu et al., 2010), in a genome-wide scan analogous to our scan of all individual SNPs. This was followed by a more targeted analysis of 204 candidate genes of potential relevance to psychophysiological endophenotypes broadly considered, and 92 schizophrenia-endophenotype candidate genes selected by the Consortium for the Genetics of Schizophrenia (COGS; Greenwood et al., 2011). In an even more focused gene-based analysis, we examined those specific candidate genes tentatively linked to the startle response by previous studies. Thus, the present study provides a particularly comprehensive assessment of the molecular genetic basis of the emotion-modulated startle response.
Method
Details of the method, including the rationale that guided the development of this project and information regarding how the three startle indices were related to each other and to the other 14 endophenotypes assessed in these same participants, can be found in Iacono et al. (2014).
Participants
Subjects for the current study consisted of same-sex male and female twin pairs and their parents from the older and younger cohorts and the enrichment sample (ES) of the MTFS. Briefly, the MTFS uses a cohort-sequential longitudinal design, with twins seen at approximately 3-year intervals. The samples are population based and broadly representative of the state of Minnesota at the time twins were born, and thus the participants are primarily Caucasian. Because differences among ethnic and racial groups in allele frequencies can confound genetic analyses, we limited the present investigation to subjects of European descent. This was based on self-reported ethnicity corroborated by analysis of the whole genome (Miller et al., 2012).
As described in Iacono et al. (2014), the affective acoustic startle modulation task was not part of the original MTFS psychophysiology protocol. Unlike the other endophenotypes examined in this special issue, therefore, twins in the present investigation were not all assessed using startle at their age-17 assessment. We nevertheless tried to maximize uniformity of twin ages as well as sample size. Data from the age-20 assessment were used for most twins. However, there were two exceptions: data for older-cohort females were from their age-17 assessment and data for ES twins were from their intake assessment, when twins were approximately 11 years old, because this was the only age at which startle was assessed for this sample. Because parents did not accompany twins at age 20, fewer parents completed this task than the tasks used in the other papers in this special issue. Thus, the sample comprised data from both parents and twins assessed at different ages. In all, the sample for this investigation comprised 3,323 subjects, 2,195 of them twins (1,176 females) and 1,128 parents (496 mothers). Subjects were from 1,473 families, 939 of them monozygotic (MZ) twin families (63.7%) and 534 of them dizygotic (DZ) twin families. Twins ranged in age from 10.9 to 24.2, with a mean of 17.8, while parents ranged from 28.4 to 63.7 (M = 43.7).
Startle Blink Responses
Experimental Paradigm
The experimental paradigm consisted of an affective-picture startle task in which subjects viewed a series of affective pictures, during which auditory startle probes were delivered through earphones at varying times, between 2–5 s after the onset of each picture. Auditory probes were 105 dB bursts of white noise lasting 50 ms, with near instantaneous rise and fall times. All subjects viewed a series of 27 affective pictures chosen from the International Affective Picture System (IAPS; Lang, Bradley, & Cuthbert, 1999), which lasted 6 s each; viewing distance was approximately 95 cm. Pictures were divided into pleasant, neutral, and unpleasant valence categories. Normative ratings of valence and arousal were matched across valence categories. Probes were delivered during 18 of these pictures (6 of each picture type). Six more probes were heard between intertrial intervals (ITI) lasting 10–15 s with probes delivered 5–10 s before the following picture’s onset. Three pictures in each category were associated with no probes either during the picture or ITI.
Pictures were presented in semirandom order such that no more than two pictures from the same valence category occurred consecutively. However, given the longitudinal nature of the study and the fact that we were collecting data from parents, twin adults, and twin children, counterbalancing pictures across all participants presented logistical constraints. Thus, all female subjects saw the same gender-appropriate slides in the same counterbalanced order. Fathers and 11-year-old male subjects also saw the same gender-appropriate slide set (see online supporting information Table S11 for IAPS pictures used for each cohort and for valence ordering of pictures). Male twins from the age-20 assessment viewed images that tended to be more intense in nature, including erotic images; everyone else viewed stimuli that were less intense in nature. Thus, parents, because of potential concerns about the content of images presented to their children, viewed the same less intense stimuli as the majority of 11- and 17-year-old children. Overall, 2,135 participants viewed the less intense picture set, while 1,188 viewed the more mature images. Stimuli were counterbalanced for valence in the identical way for all participants. Hardware and software constraints present at the time startle data collection was initiated in the early 1990s precluded data collection during ITI trials; to maintain consistency in data collection procedures and data file formats, this practice was continued in later waves of data collection as well.
Data Collection and Processing
Eye blink responses were collected using two electrodes placed on the orbicularis oculi muscle, one below the right eye and the other right next to it. Impedances for the sensors were kept below 20 kΩ. Stimulus delivery and data collection were performed using custom software. A Grass Neurodata 12 acquisition system was used for data acquisition, with data filtered at low and high frequency settings of 100 and 1000 Hz (6 dB down), and digitized to 12 bits resolution at 1000 Hz.
Data were processed further using in-house scripts written in MATLAB. We started out with an initial participant sample of 3,927 Caucasian subjects with genetic data. As a first step, trials in which the amplifier clipped the recorded signal were identified and excluded (10.5% of all trials). Next, electromyographic (EMG) data were rectified and smoothed forward and backward to prevent any phase shift using a third-order low-pass filter with a cutoff frequency of 40 Hz. At this point, trial epochs were defined from −50 ms to 150 ms with startle blink magnitude as the highest value of the signal occurring between 20 to 150 ms after the startle probe, relative to a 50-ms preprobe baseline. As part of the data reduction process, trials in which the variance of the baseline (-50 ms to stimulus onset) or poststimulus (stimulus onset to 150 ms) periods exceeded three standard deviations from the sample mean were also identified and excluded from analyses (3.7% of nonclipped trials). In addition, subjects who had less than a third of their trials for analyses (i.e., < 7 out of 18 trials) were also excluded. Following this, 254 subjects were eliminated for a variety of reasons such as hearing problems; falling asleep during the experiment; head trauma that led to loss of consciousness for more than 24 hours, hospitalization, or residual cognitive effects; neurological disorders; or being under the influence of medication, alcohol or illicit substances on the day of testing.
Molecular Genetic Data
Genetic data were primarily obtained via blood draws, with saliva samples collected in a small minority of cases. Samples were genotyped using the Illumina Human660W-Quad array (Illumina, Inc., San Diego, CA). The genotyping pipeline and quality control filters are described thoroughly elsewhere (Iacono et al., 2014; Miller et al., 2012). Principal component analysis (PCA) of genotypes, which was used to confirm self-reported ethnicity, was also applied using EIGENSTRAT (Price et al., 2006). The first 10 components identified from this PCA were used in all analyses reported here in order to account for unknown effects on our endophenotypes of subtle residual ethnic variation (Price et al., 2006).
Phenotypes
The startle blink response was operationalized as follows:
Overall (general) startle—the average magnitude of log-transformed startle response across all 18 trials.
-
Emotion modulation scores: Because the response to the first trial tended to be disproportionately large compared with subsequent responses, it was excluded. To eliminate interindividual differences in general magnitude, responses following the first trial were transformed to z scores for each subject, yielding a mean of zero across the 17 probed trials of the task and a standard deviation of 1 (cf. Bradley, Codispoti, Cuthbert, & Lang, 2001). Following this within-subject score transformation, trials were averaged for each individual within each valence category. Emotion modulation difference scores were then computed as follows:
Aversive difference modulation = mean (startle to aversive pictures) – mean (startle to neutral pictures).
Pleasant difference modulation = mean (startle to pleasant pictures) – mean (startle to neutral pictures).
All analyses used the same covariates: age, gender, generation (parent vs. twin), and a dummy variable coding for the ES sample, which was identified by means of a different sampling frame than the older and younger cohorts in order to enrich the sample for substance abuse risk. In addition, as indicated in the previous section, we included the first 10 PCs from EIGENSTRAT to eliminate any remaining sources of ethnic variation in the data (see supporting information for more details on effect of covariates).
Statistical Analyses
The following analyses were undertaken in a comprehensive evaluation of the heritable and molecular genetic basis of the different startle indices.
Biometric heritability
We used biometric models to estimate the degree to which the amount of variance in each startle index was accounted for by genetic and environmental factors. Biometric approaches model observed data in terms of the influence of three latent variables: additive genes (A), shared or common environment (C), and unique or unshared environment (E). Broadly speaking, and given a few assumptions, biometric models compare observed covariances for MZ and DZ twin families with expected covariances based on the known magnitude of genetic and environmental correlation within families. For instance, MZ twins share all of their genes, making the genetic correlation between them exactly 1, whereas DZ twins and parent-offspring pairs share approximately half of their genes, although by different mechanisms, making the genetic correlation between them 0.5. We fit biometric models to data from four-member families as well as to twins only. The latter allows comparison of our results with published findings derived from twin studies. In addition to the above, we also fit ADE models using the twin data, with D estimating a dominance term.
SNP heritability
We used GCTA (Yang et al., 2011) to estimate the total amount of variance accounted for in each putative endophenotype by the combined effect of all SNPs simultaneously. GCTA does this by using a linear mixed model, treating the SNPs as random effects and estimating the total variance due to all SNPs. The covariance matrix of random effects consists of normalized pairwise genetic relationships among all participants (the genetic relationship matrix, or GRM). When the sample comprises families, as in the present investigation, GCTA estimates of SNP heritability are driven by phenotypic relationships. Yang and colleagues recommend filtering the sample using different thresholds of genetic relatedness to produce genetically unrelated subsamples (Yang, Lee, Goddard, & Visscher, 2013). Ideally, these different subsamples produce relatively stable estimates. We used thresholds of .025, .05, and .10 for this purpose, with the most stringent of them corresponding to approximately third or fourth cousins. We repeated these analyses using GRMs based on SNPs that were weighted to adjust for local linkage disequlibrium (LD) patterns, which can inflate heritability estimates (Speed, Hemani, Johnson, & Balding, 2012).
Yang and colleagues have recently also recommended an approach whereby one uses the full sample of closely related subjects while modeling the environmental influence shared by family members (Yang et al., 2013), which is equivalent to the C latent variable in biometric models. This constituted our third variant of GCTA. We conducted GCTA on the whole sample as well but without modeling C, which allowed us to compare estimates from the two family-based approaches and thus estimate C from the molecular genetic data.
SNP effects: Genome-wide scan
Our GWAS consisted of examining the impact on the different startle indices of each of the 527,829 genotyped SNPs that survived quality control. This was done in a generalized least squares (GLS) regression framework in which each startle index was regressed against the allele count of each SNP (coded 0, 1, or 2), which constitutes an additive model of inheritance. GLS extends ordinary regression to account for correlations within clustered units, such as families. We used the R package rapid feasible generalized least squares (RFGLS; Li, Basu, Miller, Iacono, & McGue, 2011), developed to account for the presence of families in a computationally efficient manner. RFGLS estimates the variance-covariance matrix separately for each family type (MZ or DZ twin families or the 39 stepparents in the present investigation, who are treated as families of one). Each SNP was evaluated for statistical significance at the conventional GWAS threshold of p < 5 × 10−8.
SNP effects: Candidate SNPs
We also investigated a specific set of 1,180 endophenotype-general candidate SNPs selected from meta-analyses and related studies of interest examining the molecular genetic bases of disorders and traits likely related to the startle response, such as depression, schizophrenia, and antisocial behavior (see notes for Tables S1–S3 in supporting information for a list of the sources used). SNPs in this candidate set that were not on the Illumina array were imputed. Those that could be imputed accurately (imputation r2 > .30) were used in additional RFGLS analyses with the allele dosage, a count of the minor allele weighted by the posterior probability of each genotype (0, 1, or 2 minor alleles), as independent variables. A Bonferroni-corrected threshold of p ≤ 4.24 × 10−5 was used to determine statistical significance.
Gene effects: Genome-wide scan
As a complement to analyses of individual SNPs, we conducted gene-based tests using VEGAS (Liu et al., 2010) software. Such tests can be powerful when there are multiple causal variants within a gene, which may essentially deflate the associated p values, causing them to be indistinguishable from statistical noise. VEGAS aggregates all SNPs in a particular gene into a single score by converting p values for each SNP into a chi-squared statistic and summing them. The resulting test statistic is adjusted for LD patterns among SNPs in the gene. Because the p values were derived from RFGLS analyses, they are corrected for family correlations. We examined all autosomal genes available, which amounted to a total of 17,601, using a Bonferroni-corrected criterion of p ≤ 2.84 × 10−6.
Gene effects: Candidate genes
In addition to this genome-wide scan, analogous to GWAS, we evaluated three sets of candidate genes. First, we examined three startle-specific candidate genes—the serotonin transporter, dopamine transporter, and COMT genes—that have been found to be related to the startle blink response in prior studies, at a threshold of p < .05.1 Next, we examined a list of 204 endophenotype-general candidate genes determined a priori as of interest to all the endophenotypes examined in this special issue because they are involved in the major neurotransmitter systems; they code for receptors for nicotine, cocaine, and alcohol or are involved in metabolizing these substances; or they are part of the endogeneous cannabinoid and opioid systems. We used a Bonferroni-corrected threshold of p ≤ 2.45 × 10−4. As a final test, we examined 92 autosomal genes identified by COGS (Greenwood et al., 2011) as related to different endophenotypes for schizophrenia. Genes in this COGS endophenotype candidate gene set were evaluated a Bonferroni-corrected threshold of 5.43 × 10−4.
Results
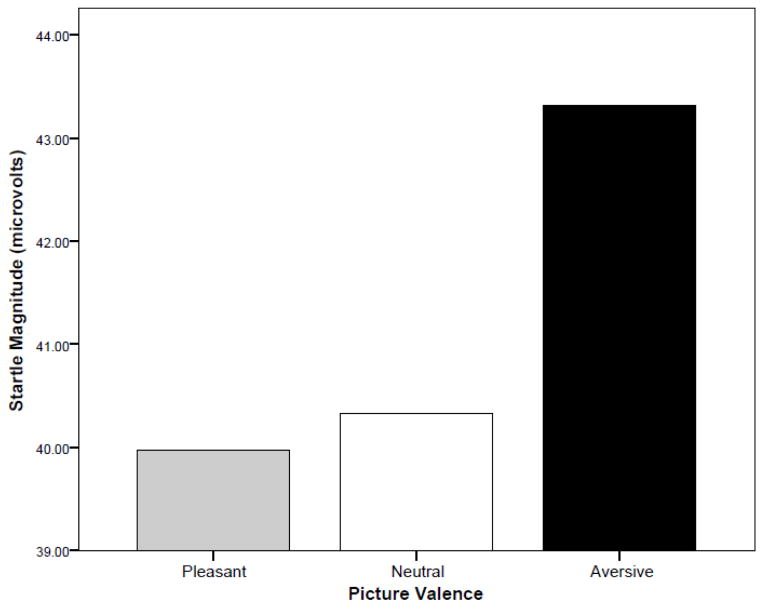
Figure 1 depicts the pattern of startle modulation (in microvolts), confirming that the startle task produced the expected results in the sample as a whole, with subjects startling less to pleasant and more to unpleasant pictures. Figures S1 through S3 depict the distribution of scores for overall startle magnitude, aversive modulation, and pleasant modulation scores, respectively. As can be seen, while overall startle magnitude was somewhat positively skewed, the z-transformed aversive and pleasant modulation scores tended to be more normally distributed. Table 2 in Iacono et al. (2014) in this issue provides descriptive statistics for each participant type (male twin, female twin, mothers, and fathers).
Figure 1.
Startle response in relation to picture valence. Mean response magnitudes in microvolts (mV) for the entire sample are plotted separately by picture valence condition.
Heritability from Biometric Models
Table 1 shows correlations between family members for the various startle indices, adjusted for model covariates. Correlations for overall startle are consistent with moderate heritability: the MZ twin correlation (.54) was somewhat less than twice the magnitude of the DZ twin correlation (.34). For the modulation scores, MZ twin values did not exceed DZ values, and correlations overall were little different from zero, especially for pleasant difference scores, suggesting little familial resemblance or heritability for modulated startle measures.
Table 1.
Correlations for Various Startle Indices Among Family Members
| Overall startle | Aversive difference | Pleasant difference | |
|---|---|---|---|
| Mom-dad | .053 | −.129 | .007 |
| Mothers-twins | .179 | .019 | −.007 |
| Fathers-twins | .238 | .043 | −.036 |
| MZ twins | .535 | .101 | .096 |
| DZ twins | .335 | .171 | .088 |
Note. All correlations are from RFGLS, and they are therefore adjusted for model covariates: gender, chronological age, generation cohort (adult or twin), ES (enrichment sample), and the first 10 PCs from EIGENSTRAT.
Results from biometric models were in accordance (see Table 2) and indicated that, while as much as half of the variance in overall startle was due to genetic factors, modulation scores were influenced very little or not at all by genetic factors, with the small heritability estimates all showing confidence intervals that overlapped with zero. Shared environmental effects were small for all measures (C in Table 2), although only in twin models and again with confidence intervals overlapping zero. For modulated startle, effects for nonshared environmental influences, which include measurement error, approached unity, with the confidence interval for one effect overlapping 1.0.
Table 2.
Biometric Model Estimates for Startle Indices
| A | C | E | |
|---|---|---|---|
| Overall startle | |||
| Twin-family | .518 (.420–.567) | .000 (.000–.072) | .482 (.433–.535) |
| Twin | .367 (.158–.570) | .164 (.000–.352) | .467 (.418–.526) |
| Aversive difference | |||
| Twin-family | .109 (.000–.172) | .000 (.000–.063) | .891 (.828–.955) |
| Twin | .000 (.000–.157) | .128 (.000–.192) | .872 (.807–.937) |
| Pleasant difference | |||
| Twin-family | .052 (.000–.116) | .000 (.000–.055) | .947 (.884–1.000) |
| Twin | .014 (.000–.176) | .079 (.000–.156) | .907 (.824–.976) |
Note. Entries are point estimates of the variance components from an ACE biometric model (with 95% confidence intervals). These have been standardized by the total variance so that they sum to 1 across the three latent variables, A, C, and E. Twin-family models are fit to the whole four-member families, while twin models are fit only to the twin data. ADE models were also fit using the twin data, but none produced a significant effect for D. Confidence intervals included 0 and D could be constrained to 0 without significantly degrading model fit (all likelihood ratio test p values > .05). A = Additive genetic effects; C = Shared environmental effects; E = unique environment/measurement error.
SNP Heritability
Estimates from GCTA results generally paralleled those found in the biometric models (see Table 3). Although standard errors are necessarily large, point estimates were relatively consistent across three thresholds used for genetic unrelatedness and the two GCTA methods (using weighted or unweighted SNPs). The median estimate for overall startle was .49, which almost exactly matched the method modeling shared environment in the family (Family C in Table 3) and was somewhat less than the biometric heritability estimate. This pattern suggests that the vast majority of variance in this index is due to additive genes.
Table 3.
GCTA Estimates of SNP Heritability for Startle Indices
| Threshold | ||||
|---|---|---|---|---|
| .025 | .050 | .100 | None | |
| Overall startle | ||||
| Unweighted | .493 (.223) | .429 (.219) | .373 (.218) | .526 (.025) |
| Weighted | .493 (.283) | .526 (.279) | .505 (.279) | — |
| Family C | — | — | — | .498 (.052) |
| Aversive difference | ||||
| Unweighted | .000 (.213) | .000 (.208) | .000 (.207) | .108 (.031) |
| Weighted | .008 (.278) | .000 (.271) | .000 (.269) | — |
| Family C | — | — | — | .118 (.061) |
| Pleasant difference | ||||
| Unweighted | .001 (.213) | .000 (.209) | .000 (.206) | .047 (.031) |
| Weighted | .062 (.274) | .000 (.270) | .000 (.267) | — |
| Family C | — | — | — | .048 (.062) |
Note. Threshold refers to the genetic relatedness threshold used for selecting unrelated individuals. None indicates that no threshold was imposed and all subjects were included. Unweighted GRM is the raw GRM, whereas Weighted GRM uses weights based on LD patterns to discount those SNPs in high LD (Speed et al., 2012). This is not used in the full sample, because the method was designed for samples of unrelated individuals or samples containing a small number of large pedigrees (Doug Speed, e-mail communication, May 4, 2014). Family C uses all subjects while simultaneously modeling shared environmental influences. Sample sizes for the different GRM cutoffs for unrelated people ranged from 1,640 to 1,677 for the unweighted estimates and from 1,353 to 1,396 for the weighted estimates. For the full sample, it was 3,321 to 3,323. GCTA = genome-wise complex trait analysis; SNP = single nucleotide polymorphism; GRM = genetic relationship matrix; LD = linkage disequilibrium
By contrast, point estimates for the modulation scores across the three thresholds of genetic unrelatedness and two methods were essentially 0. Estimates for both aversive and pleasant difference scores obtained by modeling shared environmental influence within families (Family C) largely matched estimates using all of the data (i.e., without a threshold), suggesting that C is essentially 0, an inference supported by the biometric models. That SNP heritability estimates based on unrelated individuals were largely 0 and those based on an approach that models shared environmental influences in family data were small but nonzero (approximately .11 and .05 for aversive and pleasant difference scores, respectively) raise the possibility that nonadditive genetic effects and rare variants may contribute to these modulation scores. However, the large standard errors require caution when interpreting GCTA point estimates.
SNP Effects: Genome-Wide Scan
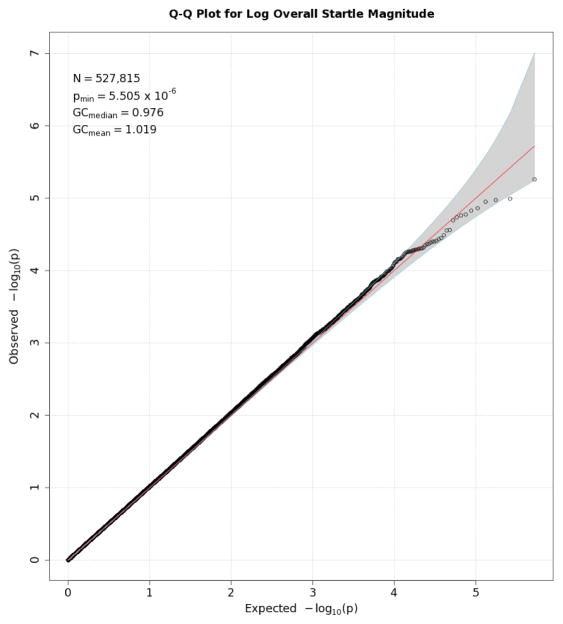
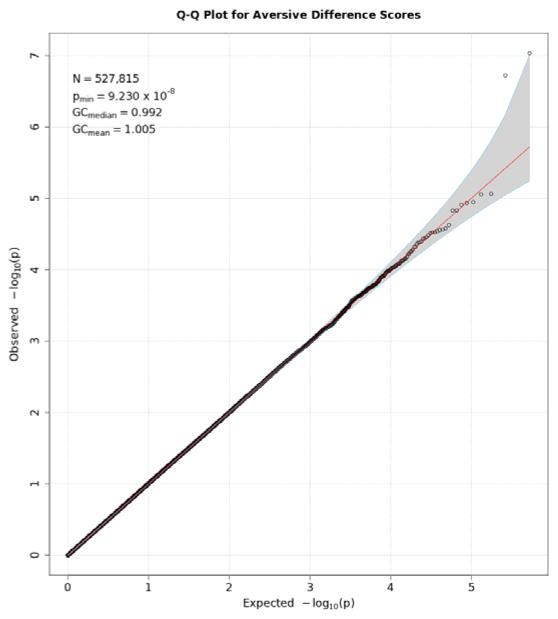
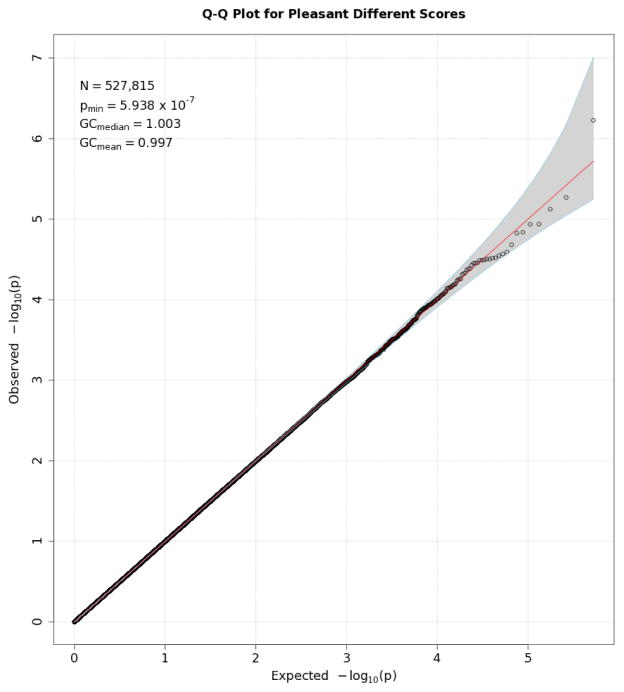
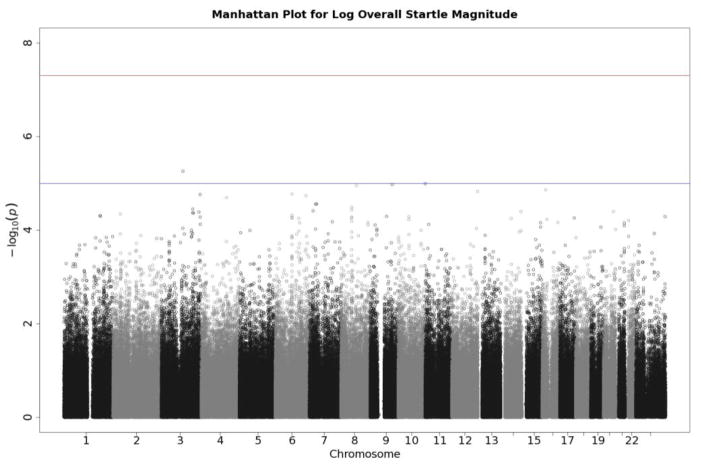
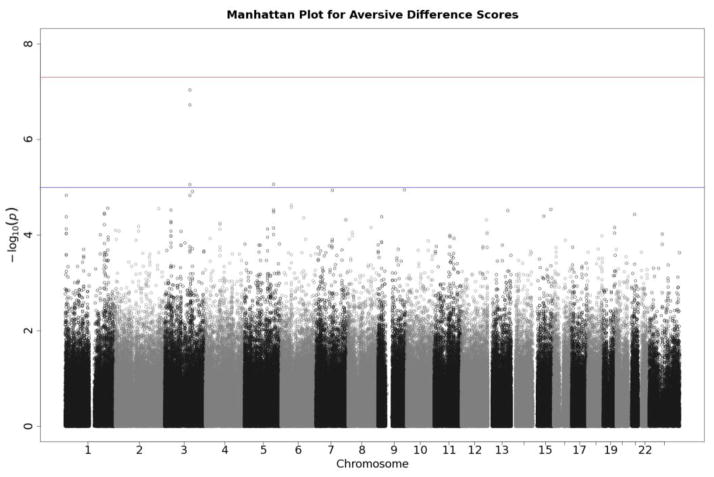
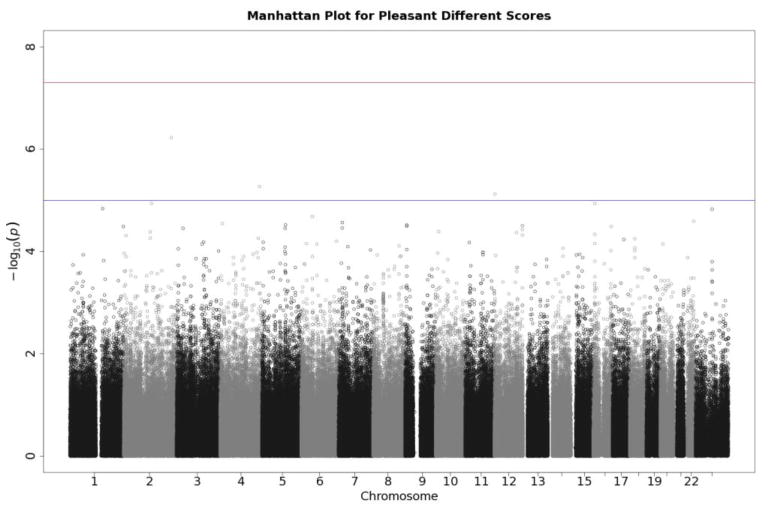
Q-Q plots for the three startle indices are presented in Figures 2, 3, and 4. These depict the relationship between observed p values and those expected under the null distribution. They generally reflect what would be expected—that is, observed and expected p values agreed closely, falling along a 45-degree line, except for a few SNPs with smaller than expected p values (larger -log(p values) which might reflect true associations, albeit subthreshold ones). There is no evidence of non-normality and the genomic inflation statistics are close to 1 (see figure insets); thus, our results are likely not due to artifacts such as unaccounted-for ethnic stratification that can produce spuriously small p values. Figures 5, 6, and 7 depict Manhattan plots, showing the negative log of the observed p values for all SNPs tested in our GWAS on the y axis ordered by their respective location on each chromosome in the genome. The red line depicts the conventional statistical threshold for significance of p ≤ 5 × 10−8. As can be seen from these, no individual SNP across any of the startle indices exceeded this threshold.
Figure 2.
Q-Q plot for SNP associations with overall startle. The line bisecting the graph gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 14 fewer than the number of SNPs on the array because there was no variation for 14 SNPs in this sample. Q-Q plots in GWAS give the observed p values against the expected p values under the null distribution of no association, although the additive inverse of the common log of p values (-log10[p value]) is used in order to emphasize small p values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p values should conform closely to their expected values, falling on or very close to a 45° line, which is plotted in the center. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 3.
Q-Q plot for SNP associations with aversive modulation scores. See Figure 2 for further details.
Figure 4.
Q-Q plot for SNP associations with pleasant modulation scores. See Figure 2 for further details.
Figure 5.
Manhattan plot of individual SNP associations with overall startle. Manhattan plots also depict the distribution of –log10(p values) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p values. The horizontal upper line indicates the genome-wide significance level (5E-08). The horizontal lower line indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 6.
Manhattan plot of individual SNP associations with aversive modulation scores. See Figure 3 for further details.
Figure 7.
Manhattan plot of individual SNP associations with pleasant modulation scores. See Figure 3 for further details.
Tables 4–6 list SNPs with p values less than 10−4 for each of the indices from these analyses. While no SNP was quite statistically significant by the genome-wide significance threshold, two SNPs on Chromosome 3 came close to the 5 × 10−8 threshold for aversive difference scores (see Table 5 and Figure 4).2
Table 4.
SNPs with p Values < 10−4 for Overall Startle Scores
| SNP | Chr | Position | Allele1 | Allele2 | Beta | SE | t | p | Gene | Class |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2399126 | 3 | 106734131 | T | C | −0.057 | 0.013 | −4.552 | 5.51E-06 | ||
| rs2056082 | 11 | 1369465 | A | G | −1.063 | 0.240 | −4.420 | 1.02E-05 | ||
| rs12115792 | 9 | 112108334 | G | A | −0.090 | 0.020 | −4.411 | 1.06E-05 | ||
| rs16939284 | 8 | 77417193 | T | G | −0.085 | 0.019 | −4.400 | 1.12E-05 | LINC01111 | intron |
| rs12933411 | 16 | 17826882 | C | T | −0.072 | 0.016 | −4.355 | 1.37E-05 | ||
| rs12317869 | 12 | 131051506 | A | C | −0.085 | 0.020 | −4.337 | 1.49E-05 | LOC102724399 | intron |
| rs1170348 | 6 | 84020324 | C | T | −0.055 | 0.013 | −4.310 | 1.68E-05 | ME1 | intron |
| rs13068219 | 3 | 193407650 | A | G | −0.053 | 0.012 | −4.303 | 1.73E-05 | OPA1 | intron |
| rs11966566 | 6 | 154607017 | G | A | 0.129 | 0.030 | 4.291 | 1.83E-05 | IPCEF1 | intron |
| rs1511138 | 4 | 127516859 | C | T | 0.103 | 0.024 | 4.269 | 2.02E-05 | ||
| rs17170881 | 7 | 37142574 | G | A | 0.122 | 0.029 | 4.200 | 2.75E-05 | ELMO1 | intron |
| rs4723113 | 7 | 31965003 | G | A | 0.051 | 0.012 | 4.197 | 2.78E-05 | PDE1C | intron |
| rs7816250 | 8 | 53749559 | A | G | 0.058 | 0.014 | 4.161 | 3.24E-05 | ||
| rs391833 | 3 | 156480263 | C | T | −0.080 | 0.019 | −4.140 | 3.55E-05 | LINC00886 | intron |
| rs2884946 | 8 | 53745838 | G | T | 0.057 | 0.014 | 4.132 | 3.68E-05 | ||
| rs6971743 | 7 | 19478992 | T | C | −0.065 | 0.016 | −4.121 | 3.86E-05 | ||
| rs10148201 | 14 | 101959581 | C | A | −0.050 | 0.012 | −4.114 | 3.98E-05 | ||
| rs4811276 | 20 | 50632282 | C | T | −0.064 | 0.016 | −4.114 | 3.99E-05 | ||
| rs1568293 | 3 | 187101435 | A | G | −0.050 | 0.012 | −4.108 | 4.10E-05 | ||
| rs9847692 | 3 | 156531322 | T | C | −0.057 | 0.014 | −4.096 | 4.30E-05 | LINC00886 PA2G4P4 | multiple |
| rs2171846 | 3 | 159155504 | G | T | −0.059 | 0.015 | −4.095 | 4.32E-05 | IQCJ-SCHIP1 SCHIP1 | intron |
| rs3845785 | 2 | 37402955 | A | G | 0.053 | 0.013 | 4.085 | 4.51E-05 | SULT6B1 | Intron |
| rs1535588 | 6 | 84051050 | C | A | −0.053 | 0.013 | −4.071 | 4.79E-05 | ME1 | intron |
| rs10911676 | 1 | 185071831 | C | T | −0.108 | 0.027 | −4.064 | 4.94E-05 | RNF2 | downstream |
| rs12035073 | 1 | 185103113 | G | A | −0.110 | 0.027 | −4.063 | 4.97E-05 | TRMT1L | intron |
| rs12043495 | 1 | 185116727 | G | A | −0.110 | 0.027 | −4.063 | 4.97E-05 | TRMT1L | intron |
| rs1330209 | 9 | 83950796 | G | T | −0.164 | 0.040 | −4.057 | 5.08E-05 | ||
| rs10482211 | 23 | 150833344 | C | A | −0.043 | 0.010 | −4.055 | 5.12E-05 | PASD1 | intron |
| rs16937445 | 10 | 55227588 | A | G | 0.066 | 0.016 | 4.054 | 5.14E-05 | ||
| rs2108 | 8 | 53557226 | C | T | 0.052 | 0.013 | 4.050 | 5.25E-05 | RB1CC1 | intron |
| rs7624750 | 3 | 193334991 | A | G | −0.050 | 0.012 | −4.049 | 5.25E-05 | OPA1 | multiple |
| rs3798886 | 6 | 84131526 | A | G | −0.052 | 0.013 | −4.044 | 5.38E-05 | ME1 | intron |
| rs2119395 | 8 | 53491159 | G | A | 0.055 | 0.014 | 4.040 | 5.47E-05 | ||
| rs1170484 | 6 | 84038522 | G | A | −0.052 | 0.013 | −4.039 | 5.48E-05 | ME1 | intron |
| rs6937193 | 6 | 84095331 | T | C | −0.052 | 0.013 | −4.039 | 5.48E-05 | ME1 | intron |
| rs425815 | 17 | 75392712 | A | G | −1.251 | 0.310 | −4.039 | 5.48E-05 | MIR4316 SEPT9 | multiple |
| rs492132 | 6 | 117643433 | C | T | 0.076 | 0.019 | 4.035 | 5.58E-05 | ROS1 | intron |
| rs1253704 | 14 | 52400054 | T | C | 0.097 | 0.024 | 4.034 | 5.62E-05 | GNG2 | intron |
| rs271015 | 16 | 24488497 | T | C | 0.054 | 0.013 | 4.023 | 5.87E-05 | LOC101927875 | noncoding RNA |
| rs351833 | 10 | 55227059 | G | T | 0.065 | 0.016 | 4.019 | 5.97E-05 | ||
| rs1340958 | 22 | 19253332 | A | G | −0.050 | 0.012 | −4.009 | 6.24E-05 | CLTCL1 | intron |
| rs7843825 | 8 | 53494271 | T | C | 0.055 | 0.014 | 4.001 | 6.44E-05 | ||
| rs10235210 | 7 | 116003445 | C | T | −0.049 | 0.012 | −3.995 | 6.62E-05 | ||
| rs3739425 | 8 | 135602581 | G | A | 0.063 | 0.016 | 3.988 | 6.82E-05 | ZFAT | multiple |
| rs7206316 | 16 | 80354117 | G | T | −0.053 | 0.013 | −3.986 | 6.86E-05 | ||
| rs1573086 | 15 | 94391572 | C | A | −0.069 | 0.017 | −3.984 | 6.91E-05 | LOC283682 | intron |
| rs11969110 | 6 | 122433229 | C | T | −0.086 | 0.022 | −3.984 | 6.94E-05 | ||
| rs3788013 | 21 | 43841328 | A | C | −0.049 | 0.012 | −3.981 | 7.02E-05 | UBASH3A | intron |
| rs1061648 | 3 | 193412672 | A | G | 0.050 | 0.012 | 3.966 | 7.47E-05 | OPA1 | 3′ UTR |
| rs4757676 | 11 | 18640477 | C | T | −0.055 | 0.014 | −3.964 | 7.51E-05 | SPTY2D1 | intron |
| rs1412515 | 9 | 25321111 | A | G | −0.068 | 0.017 | −3.957 | 7.73E-05 | ||
| rs11988811 | 8 | 135598675 | A | G | 0.062 | 0.016 | 3.957 | 7.75E-05 | ZFAT | intron |
| rs9976767 | 21 | 43836390 | G | A | −0.048 | 0.012 | −3.949 | 8.02E-05 | UBASH3A | intron |
| rs1342525 | 15 | 94399786 | G | A | −0.069 | 0.017 | −3.933 | 8.56E-05 | LOC283682 | intron |
| rs16959242 | 19 | 46560693 | A | G | −0.064 | 0.016 | −3.931 | 8.64E-05 | ||
| rs11914364 | 3 | 106722004 | G | A | 0.054 | 0.014 | 3.918 | 9.10E-05 | ||
| rs11057555 | 12 | 124756292 | T | C | −0.065 | 0.017 | −3.917 | 9.15E-05 | ZNF664-FAM101A | intron |
| rs2890545 | 9 | 34277053 | C | T | 0.057 | 0.015 | 3.909 | 9.46E-05 | KIF24 | intron |
| rs12216600 | 7 | 31965969 | C | T | −0.047 | 0.012 | −3.909 | 9.46E-05 | PDE1C | intron |
| rs6122316 | 20 | 61447759 | T | C | −0.048 | 0.012 | −3.904 | 9.63E-05 | COL9A3 | upstream |
Note. Chr is the chromosome on which each SNP is located. Position gives its location in assembly GRCh37 (hg19). The two alleles at each locus are listed, with the minor allele listed first. Alleles are aligned to the forward strand of assembly GRCh37 (hg19). Beta is the RFGLS regression coefficient, and SE its standard error. df range from 3,289 to 3,307. Gene indicates the nearest gene based on the NCBI database. If none is listed, then the SNP is located in an intergenic region. Class indicates the SNP’s NCBI function class, where “intron” is a noncoding variant, “upstream” indicates that the SNP is located within 2 kilobases of the 5′ UTR, “downstream” indicates that it is located within 500 bases of the 3′ UTR, “synon codon” is a synonymous codon, “noncoding RNA” is a transcript of noncoding RNA, and “multiple” indicates that the SNP is contained in multiple transcripts and has multiple functions. SNP = single nucleotide polymorphism.
Table 6.
SNPs with p Values < 10−4 for Pleasant Difference Startle Scores
| SNP | Chr | Position | Allele1 | Allele2 | Beta | SE | t | p | Gene | Class |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1002353 | 2 | 218395145 | C | T | 0.093 | 0.019 | 5.003 | 5.9380E-07 | DIRC3 | intron |
| rs13125519 | 4 | 180678226 | C | A | −0.127 | 0.028 | −4.557 | 5.3670E-06 | ||
| rs956451 | 12 | 2684702 | T | C | −0.105 | 0.023 | −4.486 | 7.5057E-06 | CACNA1C | intron |
| rs17562548 | 16 | 7397851 | C | T | 0.117 | 0.027 | 4.394 | 1.1498E-05 | RBFOX1 | intron |
| rs3768857 | 2 | 127829282 | T | C | 0.085 | 0.019 | 4.393 | 1.1545E-05 | BIN1 | intron |
| rs7516108 | 1 | 152542229 | T | C | −0.068 | 0.016 | −4.342 | 1.4514E-05 | ||
| rs1005295 | 23 | 79342388 | G | A | 0.063 | 0.015 | 4.337 | 1.4893E-05 | ||
| rs1984813 | 6 | 51119277 | G | A | 0.070 | 0.016 | 4.263 | 2.0736E-05 | ||
| rs6519866 | 22 | 46007623 | T | C | 0.064 | 0.015 | 4.215 | 2.5631E-05 | ||
| rs10259109 | 7 | 17540774 | A | C | 0.071 | 0.017 | 4.202 | 2.7133E-05 | LOC101927630 | intron |
| rs4697760 | 4 | 10601918 | T | C | −0.081 | 0.019 | −4.192 | 2.8343E-05 | CLNK | intron |
| rs7728213 | 5 | 108872146 | G | T | 0.063 | 0.015 | 4.178 | 3.0114E-05 | ||
| rs7858815 | 9 | 8494221 | A | G | −0.074 | 0.018 | −4.178 | 3.0231E-05 | PTPRD | intron |
| rs7309727 | 12 | 129955359 | T | C | −0.074 | 0.018 | −4.170 | 3.1303E-05 | TMEM132D | intron |
| rs1873727 | 12 | 129955902 | A | C | −0.074 | 0.018 | −4.170 | 3.1303E-05 | TMEM132D | intron |
| rs7862199 | 9 | 8470572 | A | G | −0.087 | 0.021 | −4.165 | 3.1927E-05 | PTPRD | intron |
| rs11862535 | 16 | 83661466 | A | G | −0.063 | 0.015 | −4.162 | 3.2403E-05 | CDH13 | intron |
| rs6671837 | 1 | 247778888 | G | A | 0.107 | 0.026 | 4.161 | 3.2514E-05 | LOC102724435 | intron |
| rs13228338 | 7 | 17559252 | A | G | 0.099 | 0.024 | 4.145 | 3.4785E-05 | LOC101927630 | intron |
| rs9882137 | 3 | 29494147 | T | C | −0.220 | 0.053 | −4.143 | 3.5182E-05 | RBMS3 | intron |
| rs6894788 | 5 | 106847274 | G | A | 0.079 | 0.019 | 4.142 | 3.5267E-05 | EFNA5 | intron |
| rs10847881 | 12 | 129946148 | G | A | −0.073 | 0.018 | −4.130 | 3.7230E-05 | TMEM132D | intron |
| rs1218486 | 10 | 14385265 | G | T | −0.070 | 0.017 | −4.107 | 4.1051E-05 | ||
| rs4622731 | 2 | 121948307 | G | A | −0.063 | 0.015 | −4.105 | 4.1413E-05 | ||
| rs7306268 | 12 | 103036915 | G | A | 0.136 | 0.033 | 4.098 | 4.2766E-05 | ||
| rs11866524 | 16 | 7405782 | C | A | 0.103 | 0.025 | 4.079 | 4.6247E-05 | RBFOX1 | intron |
| rs7485201 | 12 | 129940298 | G | A | −0.073 | 0.018 | −4.072 | 4.7622E-05 | TMEM132D | intron |
| rs4669685 | 2 | 11213936 | A | G | −0.069 | 0.017 | −4.067 | 4.8726E-05 | ||
| rs17006117 | 2 | 121948846 | A | G | 0.062 | 0.015 | 4.039 | 5.4916E-05 | ||
| rs10520276 | 4 | 175183493 | G | A | −0.078 | 0.019 | −4.038 | 5.5232E-05 | FBXO8 | intron |
| rs9962891 | 18 | 25282724 | T | C | 0.177 | 0.044 | 4.033 | 5.6223E-05 | ||
| rs11654392 | 17 | 54696980 | A | G | −0.120 | 0.030 | −4.025 | 5.8254E-05 | ||
| rs13211741 | 6 | 161846654 | G | A | −0.061 | 0.015 | −4.008 | 6.2710E-05 | PARK2 | intron |
| rs9832424 | 3 | 122365136 | T | C | −0.146 | 0.036 | −3.999 | 6.4895E-05 | ||
| rs555851 | 5 | 6559575 | G | A | 0.060 | 0.015 | 3.996 | 6.5850E-05 | ||
| rs10833162 | 11 | 19770798 | T | C | 0.096 | 0.024 | 3.994 | 6.6455E-05 | NAV2 | intron |
| rs13203985 | 6 | 23918178 | A | C | −0.082 | 0.021 | −3.982 | 6.9716E-05 | ||
| rs8045750 | 16 | 7515902 | G | A | −0.061 | 0.015 | −3.981 | 6.9965E-05 | RBFOX1 | intron |
| rs4611819 | 3 | 115713604 | G | A | 0.062 | 0.016 | 3.976 | 7.1717E-05 | LSAMP | intron |
| rs3177118 | 20 | 11905642 | A | G | −0.061 | 0.015 | −3.975 | 7.1742E-05 | BTBD3 | 3′ UTR |
| rs9442693 | 6 | 72386001 | A | G | −0.158 | 0.040 | −3.975 | 7.1928E-05 | ||
| rs11562750 | 8 | 119175763 | A | G | −0.091 | 0.023 | −3.955 | 7.8053E-05 | ||
| rs11763488 | 7 | 43299455 | G | A | 0.069 | 0.017 | 3.949 | 8.0157E-05 | HECW1 | intron |
| rs10491394 | 5 | 106844951 | C | T | 0.077 | 0.019 | 3.943 | 8.2228E-05 | EFNA5 | intron |
| rs9954254 | 18 | 25299829 | A | G | 0.169 | 0.043 | 3.936 | 8.4732E-05 | ||
| rs2158358 | 14 | 69011972 | C | T | 0.068 | 0.017 | 3.932 | 8.6113E-05 | RAD51B | intron |
| rs567431 | 5 | 6560041 | T | C | 0.059 | 0.015 | 3.927 | 8.7642E-05 | ||
| rs578665 | 3 | 8076372 | G | A | 0.064 | 0.016 | 3.926 | 8.8295E-05 | ||
| rs17650044 | 9 | 8482778 | T | C | −0.069 | 0.018 | −3.917 | 9.1302E-05 | PTPRD | intron |
| rs2710126 | 7 | 147753897 | C | T | −0.061 | 0.016 | −3.912 | 9.3405E-05 | CNTNAP2 | intron |
| rs4783277 | 16 | 82829401 | T | G | 0.061 | 0.016 | 3.905 | 9.6168E-05 | CDH13 LOC101928446 | intron |
| rs6508497 | 18 | 25264341 | T | C | 0.066 | 0.017 | 3.904 | 9.6672E-05 | ||
| rs9816545 | 3 | 190978515 | T | C | −0.063 | 0.016 | −3.901 | 9.7832E-05 | OSTN | intron |
| rs430123 | 5 | 106081733 | C | T | −0.094 | 0.024 | −3.896 | 9.9639E-05 |
Note. Chr is the chromosome on which each SNP is located. Position gives its location in assembly GRCh37 (hg19). The two alleles at each locus are listed, with the minor allele listed first. Alleles are aligned to the forward strand of assembly GRCh37 (hg19). Beta is the RFGLS regression coefficient, and SE its standard error. df range from 3,298 to 3,306. Gene indicates the nearest gene based on the NCBI database. If none is listed, then the SNP is located in an intergenic region. Class indicates the SNP’s NCBI function class, where “intron” is a noncoding variant, “upstream” indicates that the SNP is located within 2 kilobases of the 5′ UTR, “downstream” indicates that it is located within 500 bases of the 3′ UTR, “synon codon” is a synonymous codon, “noncoding RNA” is a transcript of noncoding RNA, and “multiple” indicates that the SNP is contained in multiple transcripts and has multiple functions. SNP = single nucleotide polymorphism.
Table 5.
SNPs with p Values < 10−4 Aversive Difference Startle Scores
| SNP | Chr | Position | Allele1 | Allele2 | Beta | SE | t | p | Gene | Class |
|---|---|---|---|---|---|---|---|---|---|---|
| rs790110 | 3 | 122384020 | G | T | −0.182 | 0.034 | −5.353 | 9.23E-08 | ||
| rs2650951 | 3 | 122368347 | T | C | −0.177 | 0.034 | −5.221 | 1.89E-07 | ||
| rs2112743 | 5 | 144736245 | A | G | 0.087 | 0.020 | 4.455 | 8.65E-06 | ||
| rs9832424 | 3 | 122365136 | T | C | −0.174 | 0.039 | −4.453 | 8.76E-06 | ||
| rs10988301 | 9 | 132051298 | G | A | −0.143 | 0.032 | −4.397 | 1.13E-05 | LOC101929331 | intron |
| rs731160 | 7 | 82059917 | T | C | −0.071 | 0.016 | −4.393 | 1.16E-05 | CACNA2D1 | intron |
| rs3772637 | 3 | 134913950 | A | G | 0.078 | 0.018 | 4.379 | 1.23E-05 | EPHB1 | intron |
| rs278020 | 1 | 6764643 | T | C | −0.070 | 0.016 | −4.339 | 1.47E-05 | ||
| rs2280070 | 3 | 122397717 | G | A | −0.139 | 0.032 | −4.338 | 1.48E-05 | PARP14 | upstream |
| rs1984813 | 6 | 51119277 | G | A | 0.074 | 0.018 | 4.234 | 2.36E-05 | ||
| rs2505263 | 6 | 51102772 | G | T | 0.072 | 0.017 | 4.209 | 2.63E-05 | ||
| rs9804001 | 1 | 212300741 | C | T | −0.110 | 0.026 | −4.201 | 2.73E-05 | ||
| rs7604632 | 2 | 213680645 | A | G | 0.572 | 0.136 | 4.196 | 2.78E-05 | ||
| rs8032463 | 15 | 89440745 | G | A | 0.098 | 0.023 | 4.187 | 2.90E-05 | HAPLN3 | upstream |
| rs6863915 | 5 | 144744479 | G | T | 0.077 | 0.018 | 4.180 | 2.99E-05 | ||
| rs17433029 | 3 | 31012087 | A | G | 0.092 | 0.022 | 4.180 | 3.00E-05 | ||
| rs1924575 | 13 | 97463114 | A | G | −0.139 | 0.033 | −4.174 | 3.07E-05 | HS6ST3 | intron |
| rs1450332 | 5 | 144735126 | A | G | 0.077 | 0.018 | 4.159 | 3.28E-05 | ||
| rs1339556 | 1 | 195487796 | A | C | −0.068 | 0.016 | −4.144 | 3.50E-05 | ||
| rs10801428 | 1 | 195483327 | G | A | −0.068 | 0.016 | −4.135 | 3.63E-05 | ||
| rs2831672 | 21 | 29613132 | C | T | −0.071 | 0.017 | −4.132 | 3.68E-05 | ||
| rs4774727 | 15 | 55196173 | C | T | −0.117 | 0.028 | −4.112 | 4.02E-05 | ||
| rs10811051 | 9 | 18861021 | C | T | 0.068 | 0.017 | 4.106 | 4.12E-05 | ADAMTSL1 | intron |
| rs277672 | 1 | 6771607 | G | T | −0.066 | 0.016 | −4.105 | 4.14E-05 | ||
| rs9487538 | 6 | 111157593 | A | G | 0.170 | 0.042 | 4.094 | 4.34E-05 | AMD1 | intron |
| rs2014450 | 7 | 148265863 | G | A | −0.079 | 0.019 | −4.071 | 4.80E-05 | ||
| rs11057929 | 12 | 125440331 | G | A | 0.091 | 0.022 | 4.070 | 4.80E-05 | DHX37 | intron |
| rs6788647 | 3 | 31007847 | C | T | 0.088 | 0.022 | 4.051 | 5.21E-05 | ||
| rs6550032 | 3 | 31008073 | A | G | 0.088 | 0.022 | 4.039 | 5.49E-05 | ||
| rs7656234 | 4 | 72356146 | C | T | −0.074 | 0.018 | −4.033 | 5.63E-05 | SLC4A4 | intron |
| rs10938141 | 4 | 72356928 | C | A | −0.073 | 0.018 | −4.020 | 5.94E-05 | SLC4A4 | intron |
| rs12094330 | 1 | 195444102 | C | T | −0.067 | 0.017 | −4.017 | 6.03E-05 | ||
| rs7559909 | 2 | 115253288 | A | G | −0.070 | 0.018 | −3.997 | 6.56E-05 | DPP10 | intron |
| rs4802869 | 19 | 52314013 | T | C | −0.079 | 0.020 | −3.984 | 6.93E-05 | FPR3 | intron |
| rs10955602 | 8 | 112825223 | A | C | 0.117 | 0.029 | 3.984 | 6.94E-05 | ||
| rs1450333 | 5 | 144729692 | T | C | 0.073 | 0.018 | 3.977 | 7.13E-05 | ||
| rs546126 | 1 | 6776772 | C | T | −0.064 | 0.016 | −3.968 | 7.40E-05 | ||
| rs6888963 | 5 | 114799354 | A | G | −0.066 | 0.017 | −3.966 | 7.45E-05 | ||
| rs6858376 | 4 | 72361951 | A | G | −0.073 | 0.018 | −3.964 | 7.52E-05 | SLC4A4 | intron |
| rs11127542 | 2 | 1004430 | C | T | −0.124 | 0.031 | −3.952 | 7.93E-05 | SNTG2 | intron |
| rs2201439 | 3 | 31019269 | A | G | 0.087 | 0.022 | 3.943 | 8.23E-05 | ||
| rs17712682 | 2 | 115219528 | A | G | −0.069 | 0.018 | −3.941 | 8.28E-05 | DPP10 | multiple |
| rs7560092 | 2 | 18280641 | C | T | −0.139 | 0.035 | −3.941 | 8.28E-05 | ||
| rs1155947 | 3 | 80377469 | T | C | 0.072 | 0.018 | 3.938 | 8.38E-05 | ||
| rs12677314 | 8 | 20984753 | A | G | −0.090 | 0.023 | −3.927 | 8.79E-05 | ||
| rs2130777 | 12 | 128172301 | T | G | 0.076 | 0.019 | 3.926 | 8.82E-05 | ||
| rs10421380 | 19 | 52304146 | A | G | −0.084 | 0.021 | −3.920 | 9.05E-05 | FPR3 | intron |
| rs277679 | 1 | 6783548 | T | C | −0.063 | 0.016 | −3.914 | 9.26E-05 | LOC100505887 | upstream |
| rs202577 | 1 | 6784053 | A | G | −0.063 | 0.016 | −3.914 | 9.26E-05 | LOC100505887 | upstream |
| rs10847435 | 12 | 128183164 | G | T | 0.085 | 0.022 | 3.911 | 9.36E-05 | ||
| rs5980803 | 23 | 68458859 | G | A | 0.055 | 0.014 | 3.908 | 9.50E-05 | ||
| rs10515549 | 5 | 144697489 | G | A | 0.063 | 0.016 | 3.902 | 9.71E-05 | ||
| rs6691883 | 1 | 175364423 | G | A | 0.078 | 0.020 | 3.900 | 9.80E-05 | TNR | intron |
Note. Chr is the chromosome on which each SNP is located. Position gives its location in assembly GRCh37 (hg19). The two alleles at each locus are listed, with the minor allele listed first. Alleles are aligned to the forward strand of assembly GRCh37 (hg19). Beta is the RFGLS regression coefficient, and SE its standard error. df range from 3,301 to 3,305. Gene indicates the nearest gene based on the NCBI database. If none is listed, then the SNP is located in an intergenic region. Class indicates the SNP’s NCBI function class, where “intron” is a noncoding variant, “upstream” indicates that the SNP is located within 2 kilobases of the 5′ UTR, “downstream” indicates that it is located within 500 bases of the 3′ UTR, “synon codon” is a synonymous codon, “noncoding RNA” is a transcript of noncoding RNA, and “multiple” indicates that the SNP is contained in multiple transcripts and has multiple functions. SNP = single nucleotide polymorphism.
SNP Effects: Candidate SNPs
Examining our list of 1,180 endophenotype-general candidate SNPs revealed a result similar to the results of the broader RFGLS analyses: no SNP crossed the Bonferroni-corrected statistical threshold (see supporting information Tables S1–S3).
Gene Effects: Genome-Wide Scan
Analysis of all 17,601 genes available in the VEGAS program revealed that, while no gene was statistically significant for overall startle or pleasant difference startle scores, one gene—PARP14 on Chromosome 3—was associated with aversive difference startle scores (p = 1 × 10−6). The next most significant gene for aversive difference startle (although not statistically so) was PARP15, approximately 45 kilobases from the 3′ UTR (untranslated region) of PARP14 on Chromosome 3 (p = 1.4 × 10−5). Similar to the GWAS results discussed above, such findings suggest that an area on Chromosome 3 may be implicated in fear-potentiated startle responses. Moreover, although no gene was significant for pleasant differences startle scores, PARP14 had the second smallest p value out of all 17,601 genes that were tested (p = 5.44 × 10−4). This pattern of results is at best suggestive but implies a common genetic basis to both affective modulated startle responses.
Gene Effects: Candidate Genes
Examination of the three startle-specific candidate genes implicated in prior work on the startle reflex revealed that none was statistically significant (see Table S4). Indeed, the smallest p value among all three indices that any of the genes attained was .42. One of the 204 endophenotype-general candidate genes exceeded the Bonferroni-corrected threshold for significance: GRIK3 on Chromosome 1 was associated with overall startle responses. Results for all genes and the three startle indices are provided in Tables S5–S7. GRIK3 is also one of the 92 COGS endophenotype candidate genes that have been related to schizophrenia endophenotypes in prior work. None of the other genes in this set of candidate genes approached significance for any of the indices (see Tables S8–S10).
Discussion
The current study comprised an investigation of the behavioral and molecular genetic bases of three differing parameters of the startle blink reflex—overall startle magnitude, and pleasant and aversive modulation (difference) scores. The startle reflex represents a fundamental defensive mechanism of the mammalian nervous system and the neural circuitry of the startle response and startle modulation has been well characterized (see Davis et al., 2010, for a recent overview). Understanding molecular genetic influences on these measures is of broad scientific interest.
To determine whether the indices we examined were heritable, we first examined biometric model-fitting estimates. Overall startle was clearly heritable, with the biometric analyses suggesting that perhaps as much as half the variance in this index was due to additive genetic influences. In contrast, biometric modeling of the emotion modulation scores provided little support for their heritability, a finding that is consistent with the results from one other twin study (Anokhin et al., 2007). Our results add to the finding of Anokhin et al. (2007), who studied only young women, by showing that the same results apply in a large sample that includes males and extends over a broad age range.
For overall startle, measures of SNP heritability derived from GCTA, which estimated the heritability in each index from the measured SNPs on our genotyping array (and those in LD with them), was approximately .49 to .50 whether based on subsamples of largely unrelated subjects or on the whole sample in an analysis that modeled shared environmental influence. These estimates largely corroborated the biometric model-fitting results for overall startle using the family data, which, when considered together with the SNP heritability, find no support for a shared environmental effect. Thus, the vast majority of the additive genetic variance in this index (at least judging by point estimates) seems to be accounted for by the SNPs on the Illumina array. In contrast, SNP heritability of the modulation scores was effectively 0 in subsamples of unrelated subjects. As these point estimates are necessarily imprecise, it is unwise to make too much of them, but it seems clear that heritability of the modulation scores due to measured SNPs was very small, which is consistent with the biometric estimates, especially those based on twin data.
GWAS results failed to yield a single association that was genome-wide significant, whether for overall startle response or modulation scores. Examination of endophenotype general candidate SNPs paralleled the genome-wide results in indicating that there were no SNPs with statistically significant associations with any of the startle indices.
Gene-based tests, by contrast, produced two significant associations. VEGAS gene-based tests aggregate the effects of all individual SNPs in a gene and within 50 kilobases of either end of the gene into a single score. Such tests can be particularly powerful if there are a number of causal variants in the gene, in which case none of them will likely surpass the stringent genome-wide significance threshold. In an unbiased analysis of 17,601 genes, we obtained a significant association between aversive difference scores and PARP14 (poly [ADP-ribose] polymerase family, member 14) on Chromosome 3. This parallels the GWAS results, in which a similar region appeared to contain SNPs related to the same index, although not surpassing the threshold for genome-wide significance. PARP14 codes for a protein that helps in the survival of injured cells, including histones and other nuclear proteins (Amé, Spenlehauer, & de Murcia, 2004). However, it has not been implicated in prior work on the startle response. Tests of 204 endophenotype general candidate genes yielded evidence that GRIK3 (glutamate receptor, ionotropic, kainate 3) on Chromosome 1 was significantly related to overall startle response. This gene codes for the kainite family of glutamate receptors, which function as ligand-gated ion channels and play a role in synaptic plasticity. Although associations between a polymorphism in GRIK3 and schizophrenia have been reported (Begni et al., 2002; Djurovic et al., 2009; Kilic et al., 2010), as well as between the gene and depression (Luciano et al., 2010; Schiffer & Heinemann, 2007), it is unclear how it might be involved in the startle response. Of course, these findings require independent replication. We also did not corroborate previous findings concerning a number of emotion-modulated startle specific candidate genes (serotonin and dopamine transporters and COMT).
Implications for Startle as an Endophenotype
These results raise an important question regarding the use of startle, especially modulation scores, and even more so aversive difference scores, as an endophenotype. Overall startle, which has not received the degree of attention that emotion-modulated startle has received for the study of individual differences, appears to have potential as an endophenotype. It was heritable in this sample, and was associated with one of our candidate genes, GRIK3, one of the genes in the endophenotype schizophrenia-relevant set adopted by COGS. Of interest would be whether, in studies of schizophrenia, GRIK3 could be confirmed as related to the overall startle component that is part of PPI.
As we have noted, patterns of modulation are frequently investigated in various disorders. Aversive modulation is thought to be a reliable indicator of defensive reactivity, individual variation in which is linked to disorders including psychopathy, depression, and phobias (see Vaidyanathan et al., 2009; also see NIMH Research Domain Criteria [RDoC]; NIMH, 2011). It is intriguing then that, even though such disorders appear to be heritable, and these modulation scores are reliably linked to them, the modulation scores themselves showed no evidence of additive heritability. Emotion-modulated startle thus appears to fail as an endophenotype, and it appears reasonable to posit that individual differences in modulated startle are acquired largely by experience. Were this the case, the biological system governing startle could be viewed as shaped by learning and environmental exposure over the course of development that is unique to each individual. A similar conclusion was reached in the female twin study of Anokhin et al. (2007) who hypothesized that plasticity in a functional system governing emotion may prove adaptive in evolutionary perspective when considering ever-changing environmental demands.
However, it is worth noting that our results here and in the exome chip companion article in this special issue also provide some support for a genetic contribution to modulated startle. Although point estimates of biometric heritability were small or even 0, confidence intervals indicated that as much as 17% of the variance in modulated startle could be due to additive genetic influence. In the present report, PARP14 was associated with the aversive difference score. Although this could be a false positive result, it could also be contributing to the weak genetic signal suggested by the GCTA results, which yielded point estimates of 0 in genetically unrelated subsamples and small but nonzero estimates in families. This GCTA pattern suggests that additive influence on modulated startle is virtually nonexistent but that nonadditive effects or rare variants not well tagged by the Illumina array might account for a small proportion of variance, as implied by the difference between the two SNP heritability estimates. In addition, it is worth noting that the absence of strong evidence of heritability does not preclude finding genetic variants associated with a trait. For example, twin studies show limited support for genetic influence on Parkinson’s disease, yet molecular genetic investigation of this medical disorder has proved fruitful (Maraganore et al., 2005; Pankratz et al., 2009).
Limitations
Our conclusions are necessarily tempered by the fact that our sample consisted of approximately 3,000 people—exceptionally large by the average psychophysiology study standards, but currently considered on the smaller end for GWAS. Our sample also consisted of a mix of subjects evaluated for startle responses at different ages. In addition, different but gender-appropriate slide sets were used for males and females, and only twins assessed at age 20 were exposed to pictorial stimuli that could be considered intense in nature. These differences in procedure were dictated in part by the inappropriateness of exposing minor children to unpleasant imagery considered disturbing and pleasant images considered to be age inappropriate (e.g., erotic nudes). Although we made statistical adjustments designed to mitigate against the possible effects contributed by the use of these different slides and different subject age groupings, it is possible our results would have been different had everyone 17 and older received the intense slide stimuli. Stronger startle probes, such as electric shocks, might have yielded different (i.e., possibly stronger) results, but the use of such stimuli with minor children would also raise ethical concerns. Moreover, it can be argued that aversive and pleasant modulation scores are somewhat crude measures in the sense that they literally involve subtracting one psychophysiological response from another. However, this procedure is used widely in the field and has yielded interesting results at the group level in many studies of psychopathology. In addition, in our own studies using these slide sets in different subsets of male and female twins, we obtained effects that point to the construct validity of the procedures we used (Benning et al., 2005; Vaidyanathan, Welo, Malone, Burwell, & Iacono, 2014). Given current practices, the existing literature, and our own success using difference scores, we elected to use them rather than invent a new way of refining measurement of modulation scores. Although some of these same concerns apply to the overall startle measure, the results were comparatively quite different. Overall startle was heritable, and the MZ twin correlation of .54 supports the measurement reliability of this index (if you consider MZ twins to, in effect, constitute parallel forms of the same person). However, that we obtained so few significant findings in relation to the different facets of startle is striking and raises some intriguing questions about how best to understand the mechanisms linking startle psychophysiology to psychiatric disorders.
Conclusions
Although we obtained a significant association between the PARP14 gene and aversive difference scores, this report provides little evidence to suggest that individual differences in startle modulation are heritable or that it could be an endophenotype. Modulation scores nevertheless fit RDoC (NIMH, 2011) as an example of a physiological variable related to negative valence systems that can be used to better understand the neurobiology of constructs such as fear, anxiety, and loss. As such, it may better be conceptualized as an environmentally mediated biological system than one governed primarily by genetic influences.
Unlike the modulation scores, overall startle resembles the other psychophysiological indices we examined in this special issue in that approximately half the variance in it is attributable to genetic factors, a finding supported by SNP heritability estimates as well. In fact, SNP heritability estimates suggest that the vast majority of genetic influence on the overall startle response is tagged by SNPs on the Illumina array. Given these biometric and SNP heritability results, we might have expected that the genetic signal related to overall startle magnitude would yield more associations with individual SNPs and genes than the modulation scores, which was not the case. Overall magnitude might be highly polygenic, reflecting the influence of a large number of variants, each with very small effect. As a result, we would have been underpowered to detect all but the very largest. The variants influencing startle modulation, by contrast, may be fewer in number and have larger effects on average. In the absence of replication studies and meta-analyses based on the work of large consortia, it is difficult to know. However, it is also possible that the GWAS approach, which is limited to examining individual variants in isolation, whether SNPs or genes, is not ideally suited to detect the major sources of genetic influence on complex traits such as the startle response.
Supplementary Material
Acknowledgments
This research was supported by NIH grants DA 024417, DA 05147, DA 036216, AA 09367, and DA 13240. We would also like to thank Scott Burwell for his help in developing the MATLAB scoring algorithm for the startle blink data.
Footnotes
One other study exists that has examined MAOA (Larson et al., 2010) in relation to startle. However, MAOA is on the X chromosome, which we did not analyze.
Further inspection of Table 5 reveals that 4 out of the top 10 SNPs for aversive difference scores were in the same region of Chromosome 3. To ensure that we were not missing a stronger signal from another SNP in the region but not on the Illumina array, we conducted a finer-grained analysis of that area on Chromosome 3 using RFGLS analyses of imputed SNPs. The 1000 Genomes (2/2012) CEU reference samples were used for imputation. None of the imputed SNPs in the area produced a smaller p value than rs790110. To test whether these four SNPs were acting in concert or in LD and thus reflect one signal, the SNP with the smallest p value, rs790110, was entered as a covariate in RFGLS, and the GWAS undertaken again. Doing so increased the p values of the 3 additional SNPs on Chromosome 3 quite substantially (to the point that all were greater than .05), suggesting that they had no effect on aversive differences scores that was statistically independent of the rs790110 effect.
Additional supporting information may be found in the online version of this article:
Figure S1: Distribution of overall startle magnitude residuals.
Figure S2: Distribution of aversive startle modulation difference scores residuals.
Figure S3: Distribution of pleasant startle modulation difference scores residuals.
Table S1: SNP associations for endophenotype-general candidate SNPs and overall startle magnitude.
Table S2: SNP associations for endophenotype-general candidate SNPs and aversive startle modulation scores.
Table S3: SNP associations for endophenotype-general candidate SNPs for pleasant startle modulation scores.
Table S4: Results of VEGAS gene-based tests of startle specific candidate genes and the three startle indices.
Table S5: Results of VEGAS gene-based tests of endophenotype-general candidate genes and overall startle magnitude.
Table S6: Results of VEGAS gene-based tests of endophenotype-general candidate genes and aversive startle modulation scores.
Table S7: Results of VEGAS gene-based tests of endophenotype-general candidate genes and pleasant startle modulation scores.
Table S8: Results of VEGAS gene-based tests of COGS endophenotype candidate genes and overall startle magnitude.
Table S9: Results of VEGAS gene-based tests of COGS endophenotype candidate genes and aversive startle modulation scores.
Table S10: Results of VEGAS gene-based tests of COGS endophenotype candidate genes and pleasant startle modulation scores.
Table S11: IAPS pictures used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Anokhin AP, Golosheykin S, Heath AC. Genetic and environmental influences on emotion-modulated startle reflex: A twin study. Psychophysiology. 2007;44:106112. doi: 10.1111/j.1469-8986.2006.00486.x. [DOI] [PubMed] [Google Scholar]
- Anokhin AP, Heath AC, Myers E, Ralano A, Wood S. Genetic influences on prepulse inhibition of startle reflex in humans. Neuroscience Letters. 2003;353:45–48. doi: 10.1016/j.neulet.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Begni S, Popoli M, Moraschi S, Bignotti S, Tura GB, Gennarelli M. Association between the ionotropic glutamate receptor kainate 3 (GRIK3) ser310ala polymorphism and schizophrenia. Molecular Psychiatry. 2002;7:416–418. doi: 10.1038/sj.mp.4000987. [DOI] [PubMed] [Google Scholar]
- Benning SD, Patrick CJ, Iacono WG. Psychopathy, startle blink modulation, and electrodermal reactivity in twin men. Psychophysiology. 2005;42:753–762. doi: 10.1111/j.1469-8986.2005.00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley MM, Codispoti M, Cuthbert BN, Lang PJ. Emotion and motivation I: defensive and appetitive reactions in picture processing. Emotion. 2001;1:276–298. [PubMed] [Google Scholar]
- Braff DL. Prepulse inhibition of the startle reflex: A window on the brain in schizophrenia. Current Topics in Behavioral Neurosciences. 2010;4:349–371. doi: 10.1007/7854_2010_61. [DOI] [PubMed] [Google Scholar]
- Brocke B, Armbruster D, Muller J, Hensch T, Jacob CP, Lesch KP, Strobel A. Serotonin transporter gene variation impacts innate fear processing: Acoustic startle response and emotional startle. Molecular Psychiatry. 2006;11:1106–1112. doi: 10.1038/sj.mp.4001908. [DOI] [PubMed] [Google Scholar]
- Carlson SR, Katsanis J, Iacono WG, McGue M. Emotional modulation of the startle reflex in twins: Preliminary findings. Biological Psychology. 1997;46:235–246. doi: 10.1016/s0301-0511(97)00014-8. [DOI] [PubMed] [Google Scholar]
- Cuthbert BN, Lang PJ, Strauss C, Drobes D, Patrick CJ, Bradley MM. The psychophysiology of anxiety disorder: Fear memory imagery. Psychophysiology. 2003;40:407–422. doi: 10.1111/1469-8986.00043. [DOI] [PubMed] [Google Scholar]
- Davis M. Are different parts of the extended amygdala involved in fear versus anxiety? Biological Psychiatry. 1998;44:1239–1247. doi: 10.1016/s0006-3223(98)00288-1. [DOI] [PubMed] [Google Scholar]
- Davis M, Walker DL, Lee Y. Roles of the amygdala and bed nucleus of the stria terminalis in fear and anxiety measured with the acoustic startle reflex. Annals of the New York Academy of Sciences. 1997;821:305–331. doi: 10.1111/j.1749-6632.1997.tb48289.x. [DOI] [PubMed] [Google Scholar]
- Davis M, Walker DL, Miles L, Grillon C. Phasic vs sustained fear in rats and humans: Role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology. 2010;35:105–135. doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djurovic S, Kahler AK, Kulle B, Jonsson EG, Agartz I, Le Hellard S, Andreassen OA. A possible association between schizophrenia and GRIK3 polymorphisms in a multicenter sample of Scandinavian origin (SCOPE) Schizophrenia Research. 2009;107:242–248. doi: 10.1016/j.schres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, Schork NJ. Initial heritability analyses of endophenotypic measures for schizophrenia: The consortium on the genetics of schizophrenia. Archives of General Psychiatry. 2007;64:1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Hardiman G. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2011;168:930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillon C, Baas JM. A review of the modulation of the startle reflex by affective states and its application in psychiatry. Clinical Neurophysiology. 2003;114:1557–1579. doi: 10.1016/s1388-2457(03)00202-5. [DOI] [PubMed] [Google Scholar]
- Grillon C, Dierker L, Merikangas KR. Fear-potentiated startle in adolescent offspring of parents with anxiety disorders. Biological Psychiatry. 1998;44:990–997. doi: 10.1016/s0006-3223(98)00188-7. [DOI] [PubMed] [Google Scholar]
- Grillon C, Warner V, Hille J, Merikangas KR, Bruder GE, Tenke CE, Weissman MM. Families at high and low risk for depression: A three-generation startle study. Biological Psychiatry. 2005;57:953–960. doi: 10.1016/j.biopsych.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Hasenkamp W, Epstein MP, Green A, Wilcox L, Boshoven W, Lewison B, Duncan E. Heritability of acoustic startle magnitude, prepulse inhibition, and startle latency in schizophrenia and control families. Psychiatry Research. 2010;178:236–243. doi: 10.1016/j.psychres.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM. Developmental endophenotypes: Indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Development Perspectives. 2011;5:239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology. 2014 doi: 10.1111/psyp.12343. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaviani H, Gray JA, Checkley SA, Raven PW, Wilson GD, Kumari V. Affective modulation of the startle response in depression: Influence of the severity of depression, anhedonia, and anxiety. Journal of Affective Disorders. 2004;83:21–31. doi: 10.1016/j.jad.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Kilic G, Ismail Kucukali C, Orhan N, Ozkok E, Zengin A, Aydin M, Kara I. Are GRIK3 (T928G) gene variants in schizophrenia patients different from those in their first-degree relatives? Psychiatry Research. 2010;175:43–46. doi: 10.1016/j.psychres.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Koch M, Schmid A, Schnitzler HU. Pleasure-attenuation of startle is disrupted by lesions of the nucleus accumbens. NeuroReport. 1996;7:1442–1446. doi: 10.1097/00001756-199605310-00024. [DOI] [PubMed] [Google Scholar]
- Lang PJ, Bradley MM, Cuthbert BN. Emotion, attention, and the startle reflex. Psychological Review. 1990;97:377–395. [PubMed] [Google Scholar]
- Lang PJ, Bradley MM, Cuthbert BN. International affective picture system (IAPS): Technical manual and affective ratings. Gainesville, FL: The Center for Research in Psychophysiology, University of Florida; 1999. [Google Scholar]
- Lang PJ, McTeague LM. The anxiety disorder spectrum: Fear imagery, physiological reactivity, and differential diagnosis. Anxiety, Stress & Coping. 2008;22:5–25. doi: 10.1080/10615800802478247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Taubitz LE, Robinson JS. MAOA T941G polymorphism and the time course of emotional recovery following unpleasant pictures. Psychophysiology. 2010;47:857–862. doi: 10.1111/j.1469-8986.2010.01005.x. [DOI] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono WG, McGue M. A rapid generalized least squares model for a genome-wide quantitative trait association analysis in families. Human Heredity. 2011;71:67–82. doi: 10.1159/000324839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Houlihan LM, Harris SE, Gow AJ, Hayward C, Starr JM, Deary IJ. Association of existing and new candidate genes for anxiety, depression and personality traits in older people. Behavior Genetics. 2010;40:518–532. doi: 10.1007/s10519-009-9326-4. [DOI] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. American Journal of Human Genetics. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag C, Buckholtz JW, Hartmann P, Merz M, Burk C, Hennig J, Reuter M. COMT genetic variation affects fear processing: Psychophysiological evidence. Behavioral Neuroscience. 2008;122:901–909. doi: 10.1037/0735-7044.122.4.901. [DOI] [PubMed] [Google Scholar]
- Morgan CA, III, Grillon C, Southwick SM, Davis M, Charney DS. Fear-potentiated startle in posttraumatic stress disorder. Biological Psychiatry. 1995;38:378–385. doi: 10.1016/0006-3223(94)00321-S. [DOI] [PubMed] [Google Scholar]
- NIMH. NIMH Research Domain Criteria (RDoC) 2011 Retrieved from http://www.nimh.nih.gov/research-funding/rdoc/nimh-research-domain-criteria-rdoc.shtml.
- Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, Pugh EW, Foroud T. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Human Genetics. 2009;124:593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick CJ, Bradley MM, Lang PJ. Emotion in the criminal psychopath: Startle reflex modulation. Journal of Abnormal Psychology. 1993;102:82–92. doi: 10.1037/0021-843x.102.1.82. [DOI] [PubMed] [Google Scholar]
- Pauli P, Conzelmann A, Mucha RF, Weyers P, Baehne CG, Fallgatter AJ, Lesch KP. Affect-modulated startle reflex and dopamine D4 receptor gene variation. Psychophysiology. 2010;47:25–33. doi: 10.1111/j.1469-8986.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- Pole N. The psychophysiology of posttraumatic stress disorder: A meta-analysis. Psychological Bulletin. 2007;133:725–746. doi: 10.1037/0033-2909.133.5.725. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Schiffer HH, Heinemann SF. Association of the human kainate receptor GluR7 gene (GRIK3) with recurrent major depressive disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2007;144:20–26. doi: 10.1002/ajmg.b.30374. [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Patrick CJ, Cuthbert BN. Linking dimensional models of internalizing psychopathology to neurobiological systems: Affect-modulated startle as an indicator of fear and distress disorders and affiliated traits. Psychological Bulletin. 2009;135:909–942. doi: 10.1037/a0017222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Welo EJ, Malone SM, Burwell SJ, Iacono WG. The effects of recurrent episodes of depression on startle responses. Psychophysiology. 2014;51:103–109. doi: 10.1111/psyp.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana SR, Spence EL, Lang PJ. The startle probe response: A new measure of emotion? Journal of Abnormal Psychology. 1988;97:487–491. doi: 10.1037/0021-843x.97.4.487. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis (GCTA): Methods, data analyses, and interpretations. In: Gondro C, van der Werf J, Hayes B, editors. Genome-wide association studies and genomic prediction. Vol. 1019. New York, NY: Humana Press; 2013. pp. 215–236. 2013/06/13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.