Abstract
The high-altitude hypoxic environment represents one of the most extreme challenges for mammals. Previous studies of humans on the Tibetan plateau and in the Andes Mountains have identified statistical signatures of selection in different sets of loci. Here, we first measured the hemoglobin levels in village dogs from Tibet and those from Chinese lowlands. We found that the hemoglobin levels are very similar between the two groups, suggesting that Tibetan dogs might share similar adaptive strategies as the Tibetan people. Through a whole-genome sequencing approach, we have identified EPAS1 and HBB as candidate genes for the hypoxic adaptation on the Tibetan plateau. The population genetic analysis shows a significant convergence between humans and dogs in Tibet. The similarities in the sets of loci that exhibit putative signatures of selection and the hemoglobin levels between humans and dogs of the same environment, but not between human populations in different regions, suggests an extraordinary landscape of convergent evolution between human beings and their best friend on the Tibetan plateau.
Keywords: convergence, high-altitude adaptation, EPAS1, Tibetan village dog
Introduction
High-altitude plateaus (>2,500 m) are one of the most extreme living conditions on earth (Zhao et al. 2009; Li and Zhang 2012). A formidable physiological challenge for animals is the low oxygen level. Genome-wide scans for detecting molecular signatures of high-altitude adaptation have been conducted across a wide range of geographic locations (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Peng et al. 2011; Xu et al. 2011; Ji et al. 2012). Interestingly, studies of signatures of selection among Tibetan, Andean, and high-altitude Ethiopian human populations have revealed distinct genetic components and different physical characteristics for hypoxia adaptation (Bigham et al. 2010; Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012). For example, Tibetan people are found to have similar hemoglobin levels (Hb) to people from lower areas, whereas people from Andes or Ethiopia have much elevated Hb levels (Beall 2007; Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012), suggesting that diverse strategies that can be employed for high-altitude adaptation.
As the first domesticated animal and man’s closest friend, domestic dog (canis familiaris) has followed human beings and travelled across a wide variety of ecological niches with us (Clutton-Brock 1995; Savolainen et al. 2002; Pang et al. 2009). Previous research found that convergent evolution driven by similar environments seems to have shaped both of our genomes at multiple phenotypes (Wang et al. 2013). How the two species response to the high-altitude environment poses an interesting and important question.
Using a single nucleotide polymorphisms (SNPs) genotyping strategy, one previous research provided the first glimpse on the genetic mechanisms for high-altitude adaptation of Tibetan Mastiffs. (Li et al. 2014). However, considering the sparse spacing in the canid SNP array (average distance is 70 kb), the survey of the adaptive mechanisms might be quite incomplete. In the present study, we want to take a whole-genome sequencing approach and wish to learn the adaptive strategy of Tibetan village dogs at both the phenotypic and genic level and to compare the high-altitude adaptation between humans and dogs.
Materials and Methods
Measuring Hb Level
The peripheral bloods were collected for measuring Hb level using in four populations at the stated elevations: 9 Tibetan dogs in Yushu (∼3,500-m altitude), 56 Tibetan dogs in Lijiang (∼2,700-m altitude), 52 breeds dogs in Beijing (∼50-m altitude), and 24 breed dogs in Guangzhou (∼50-m altitude), respectively. The dog populations that measured Hb level are different from those used in the population genomic survey.
Populations and Resequencing
Four dog populations were collected for this study: 1) Tibetan dogs living in Tibet and Qinghai province at elevations of about 3,500 m; 2) population of indigenous dogs gathered from other provinces of China with elevations lower than 2,000 m; and 3) a population of modern dog breeds (DB) mixed by Chow Chow, Rottweiler, Chihuahua, and Saint Bernard. The genomic DNA was extracted using the Phenol/Chloroform method from peripheral venous blood. For each population, genomic DNA for ten individuals was pooled together equally. Note that 3 chow chows, 3 Rottweilers, 3 Chihuahuas, and 1 Saint Bernard were mixed as one population to represent modern DB. The Illumina Genome Analyzer GAII was used to generate paired-end reads for each population pool according to standard protocols specified by the manufacturer. The read length of raw sequence data was shown on table 1.
Table 1.
Whole-Genome Resequencing Results for Four Dog Populations
| Populations | Reads Length | Number of Reads (M) | Total Bases | Matching Reads (M) | Matching bp | Effective Depth (Fold) |
|---|---|---|---|---|---|---|
| TID1 | 171 | 317.26 | 54.25 | 292.89 | 38.01 | 15.94 |
| TID2 | 125 | 361.45 | 45.18 | 321.83 | 36.76 | 15.41 |
| Indigenous dog from low-altitude area (CID) | 121 | 248.52 | 30.07 | 209.27 | 21.27 | 8.92 |
| Breed dogs (DB) | 125 | 296.68 | 37.08 | 269.25 | 36.62 | 15.35 |
Reads Mapping and SNP Calling
After trimming low-quality bases, paired-end reads were aligned to the dog genome assembly (CanFam2) (Lindblad-Toh and Wade 2005) using Burrows–Wheeler algorithm implemented in BWA-short module with default parameters (Li and Durbin 2009). SAMtools was subsequently used to call raw SNPs (Li et al. 2009) with these four conditions: 1) The base quality is of no less than 20; 2) the mapping quality is no less than 20; 3) the coverage is no less than 8; and 4) the coverage is no more than 100.
Population Differentiation
We estimated the allele frequency of each population by counts of different alleles in the sequencing pool. Fst is calculated using the method from Weir and Cockerham (1984) (Weir and Cockerham 1984; Akey et al. 2002). The rms of Fst was calculated as
Fisher’s exact test was carried out by testing each position against their population grouping (high or low elevation). Because the number of founding chromosomes in each pool in the association studies is 40 (20 diploid genomes), in order to avoid the situation where positions with very high coverage will generate too small P-values, we used the hypergeometric distribution to project down the total coverage to 40. The average P values across multiple replicates (1,000) were used as the P value for that site.
Genotyping Candidate SNPs on Large Population
Polymerase chain reaction (PCR) primers were designed for sequencing the three SNPs in the EPAS1 and HBB showing the most significant frequency differences between dogs groups. Three regions surrounding EPAS1 and six regions surrounding HBB were selected to measure the accuracy of site frequencies surveyed using the pooled strategy. After PCR amplification, Sanger sequence technology was employed to sequence the target regions in two population samples. The PCR primers were given in the supplementary table S1, Supplementary Material online.
Enrichment Analysis
Gene orthologous relationship between human and dog was downloaded from Ensembl database (www.ensembl.org, last accessed August 12, 2014). Whole-genome alignment between human and dogs was downloaded from UCSC (genome.ucsc.edu, last accessed August 12, 2014). The proportions of positively selected candidates (>100 kb) in two species were both calculated. The P value was calculated as the proportion of simulated data sets that have equal or higher number of overlapped genes and segments than the observed count (1,000,000 replications).
Results
Hemoglobin Concentrations
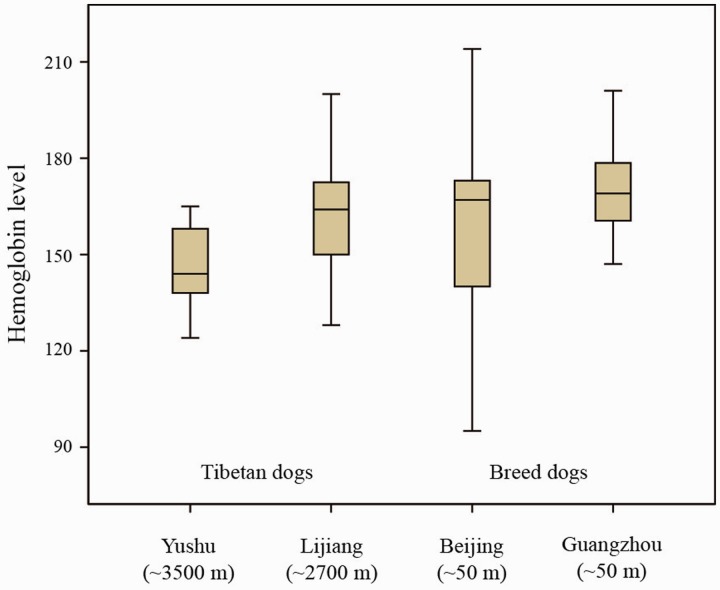
Firstly, we measured the Hb level in the peripheral blood of 141 dogs from two high-altitude dog populations: Tibetan dogs in Lijiang (∼2,700-m altitude) and Yushu (∼3,500-m altitude), and two low-altitude dog populations: Breed dogs in Beijing (∼50-m altitude) and Guangzhou (∼50-m altitude). Statistical analysis reveals that the Hb levels of Tibetan dogs from high-altitude areas are similar to breed dogs from low-altitude area (fig. 1). The average Hb concentrations are 160 and 161 g/l for Tibetan dogs and breed dogs, respectively. These concentrations are quite close to the canine hematology reference value (137.7–203.8) (Moritz et al. 2004). The observation suggests Tibetan dogs appear to share the same strategy as Tibetan people.
Fig. 1.—
Hb levels over different HBB genotypes in different dog populations. Boxplot of Hb levels measured in the Tibetan dogs in Yushu (∼3,500-m altitude) and Lijiang (∼2,700-m altitude) and breed dogs in Beijing (∼50-m altitude) and Guangzhou (∼50-m altitude).
Population Sampling and Sequencing
In order to identify possible signatures of positive selection in the genomes of high-altitude dogs, we carried out whole-genome sequencing on ten individuals from four different populations of dogs. They are 1) two Tibetan indigenous dog populations (TID1 and TID2), both being from the Tibetan plateau and living in an elevations of more than 3,500 m; 2) chinese indigenous dogs (CID), and 3) a mixture of modern DB; the latter two being from provinces across China at altitudes below 2,000 m. Genomic DNA samples of each population were pooled equally and sequenced with Illumina GAII platform, resulting in 248–317 million raw reads for each population. After mapping the short reads to the reference genome (version CanFam2, May 2005) (Lindblad-Toh and Wade 2005), we were able to cover the dog genome for about 8.9- to 15.9-fold per population (table 1). By a set of stringent criteria, 2.4 million high-quality SNPs were extracted. Adjacent SNPs were separated by a median distance of 272 bp (mean distance of 907 bp) that facilitates evolutionary analyses. In order to measure the accuracy of site frequency on the pooled strategy, 39 SNPs were selected and individually sequenced using Sanger technology in all 40 individuals (supplementary tables S1 and S2, Supplementary Material online). Correlation analysis found that allele frequencies calculated from the pooled sample match with the results from the individual sequencing (supplementary fig. S1, Supplementary Material online). The significant correlation observed in the individual and pooled sequencing suggests that pooled strategy provides a cost-effective approach for extracting information about allele frequencies from a population (supplementary note S1, Supplementary Material online).
Genomic Regions with High Population Differentiation
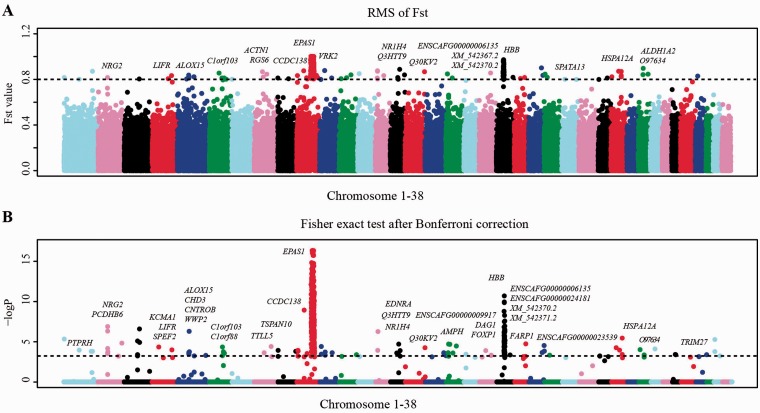
We employed the F statistic (Fst) to look for genomic regions with high population differentiation (Weir and Cockerham 1984; Akey et al. 2002). Although population demographic histories affect gene frequencies at all loci similarly, positive selection on the other hand will increase allele frequency at some loci in some populations only, leading to much elevated Fst value (Barreiro et al. 2008). Using allele frequencies extracted from sequence reads, we calculated Fst values in all four high–low population comparisons (Weir and Cockerham 1984; Akey et al. 2002). Fst(TID1, CID), Fst(TID1, DB), Fst(TID2, CID), and Fst(TID2, DB) across all 2.4 million SNPs distributed over 38 autosomes were calculated using the Weir and Cockerham (1984) method (supplementary fig. S2, Supplementary Material online) (Weir and Cockerham 1984; Akey et al. 2002). Chromosome X was not included because of a different effective population size (Akey et al. 2002). In order to combine multiple values of Fst, we calculated the root mean square of Fst: Fst(rms) based on all four comparisons. Using a stringent cutoff of 0.8, we identified 437 high Fst SNPs (∼0.018%, supplementary table S3, Supplementary Material online), of which 257 of them located in genic regions over a total of 20 genes (fig. 2A and supplementary table S4, Supplementary Material online).
Fig. 2.—
Genome-wide association mapping of high-altitude adaptation in dogs. (A) The distribution of RMS of Fst along 38 autosomes. The horizontal dashed line indicates the threshold of Fst(rms) (0.8). (B) The distribution of -log(P value) from the Fisher’s exact tests after a Bonferroni correction procedure along 38 autosomes. The dashed black line indicates the cutoff significance level (0.1%).
In an alternative measure, we performed a Fisher’s exact test, as a second empirical outlier analysis, combining the Tibetan dogs (TID1 + TID2) as the case population and treating the other lower altitude dogs (CID + DB) as a control group. This provides a complementary measure of allele frequency differentiation between the two phenotypically distinct groups. Using the approach, we identified 469 polymorphic sites showing statistical evidence of positive selection (supplementary table S5, Supplementary Material online). Among them, 277 were found in genic regions distributed over 34 genes (fig. 2B and supplementary table S6, Supplementary Material online).
In summary, 14 genes in 12 genomic regions pass the threshold using both the Fst value and Fisher’s exact test. Among them, endothelial per-ARNT-Sim (PAS) domain protein 1 (EPAS1) and hemoglobin beta (HBB) harbor nonsynonymous mutations as shown in supplementary table S7, Supplementary Material online. The most prominent one is EPAS1, with 225 SNPs in a region of 145 kb that significantly differentiate Tibetan and lowland dogs. Four nonsynonymous mutations were founded on EPAS1: G305S (Fst(rms) = 0.95; PFET = 4.47E-07), D494E (Fst(rms) = 0.93; PFET = 8.83E-11), V500M (Fst(rms) = 0.95; PFET = 1.06E-09), and P750S (Fst(rms) = 0.92; PFET = 2.77E-07). We chose G305S for a more extensive survey on a larger sample of 61 Tibetan dogs from Tibet and 149 indigenous dogs from low-altitude areas. The frequency of the mutant allele at the EPAS1 locus is 95% in Tibet but only 6% among the lowland dogs (P = 1.14E-70). A second noteworthy gene, HBB, is the HBB gene, which functions in the oxygen transport from the lung to various peripheral tissues (Wang 1961; Storz and Moriyama 2008; Mairbaurl and Weber 2012). Two nonsynonymous mutations were found in HBB: G14S (Fst(rms) = 0.87; PFET = 3.19E-08) and L15M (Fst(rms) = 0.92; PFET = 1.94E-11), which was also strongly altitude dependent by the extensive survey (86.4% in Tibet vs. 17.5% on the lowlands, P = 3.27E-41). Interestingly, the L15M mutation is predicted to be probably damaging with a score of 0.996 based on polyphen-2 (Adzhubei et al. 2010), which could influence Hb-O2 affinity (Natarajan et al. 2013).
The EPAS1 Gene and its Convergent Evolution between Humans and Dogs
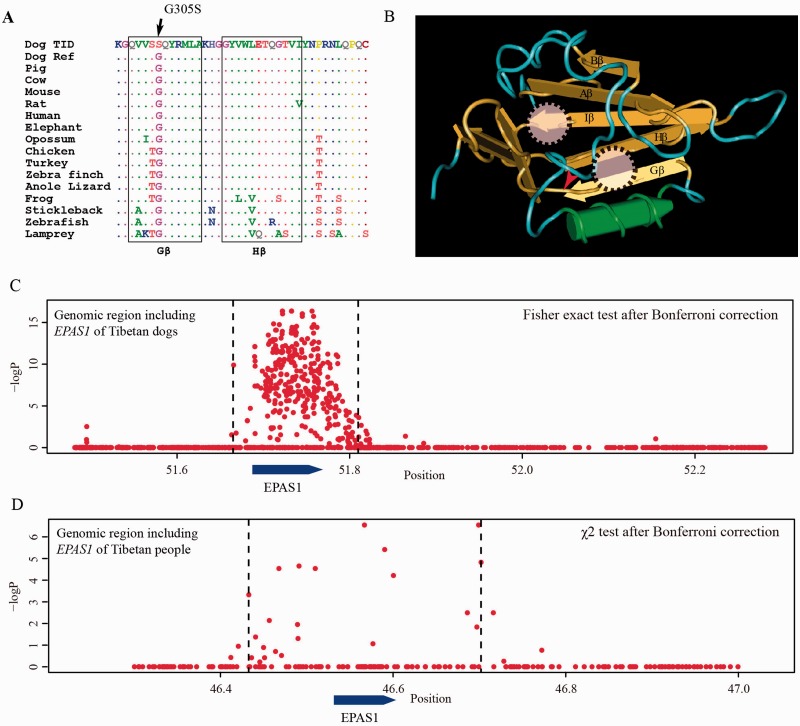
Several earlier studies have identified noncoding SNPs near EPAS1 as extreme outlier loci in genome scans in the high-altitude adaptation in Tibetan human populations (Beall et al. 2010; Yi et al. 2010; Peng et al. 2011; Xu et al. 2011). This gene is a member of the basic helix–loop-helix/PAS family of transcription factors and play a crucial role in systemic responses to hypoxia (Lee and Percy 2011). It encodes a transcription factor involved in the induction of genes regulated by oxygen, which is induced as oxygen levels fall. Sequence comparisons found that the glycine at codon 305 is conserved across all known vertebrates as shown in figure 3A. Interestingly, the G305S, indicated by a red arrow in figure 3B, is in the Gβ-sheet of the PAS-B domain (Key et al. 2009; Scheuermann et al. 2009). This domain is located in a preformed solvent-inaccessible cavity and is the major functional domain binding to its ligands (Key et al. 2009). The G305S mutation with a predicted functional change in the binding domain (a score of 0.998 based on polyphen-2 [Adzhubei et al. 2010]) might be the major contributing factor for the high-altitude hypoxic adaptation.
Fig. 3.—
Population differentiation at the EPAS1 gene together with its evolution and structure. (A) Sequence alignment across vertebrates surrounding the G305S mutation at the EPAS1 protein. The two black boxes indicate two conserved β sheet regions in the PAS-B domain. (B) Structural model of the C-Terminal PAS-B domain of EPAS1 (PDB ID: 1P97) plotted using the Cn3D software (Wang et al. 2000). The PAS-B domain (Erbel et al. 2003) adopts a typical α/β PAS domain fold with several α-helices flanking a five-stranded anticonvergent β sheet (Erbel et al. 2003). Circles indicate the locations of the ligand-binding sites (Erbel et al. 2003; Scheuermann et al. 2009) and the mutation G305S is marked with a red tick. (C) Genetic association across a 800-kb region surrounding the EPAS1 locus in Tibetan dogs. (D) Genetic association for a 800-kb region surrounding the EPAS1 locus in Tibetan people from human genotype data (Beall et al. 2010).
The observation that EPAS1 emerged as a Fst outlier of Tibetan dogs is surprising because human adaptation to the Andes does not appear to involve this locus. Note that the mutation frequencies in EPAS1 are comparable in humans and dogs. The mutant allele in Tibet is 94% for dogs and 87% in humans. In lowlands, the mutant frequency is low in dogs and humans, at 4.9% and 8.75%, respectively (Yi et al. 2010). The population genetic signature shows a strong convergence between humans and dogs in Tibet as well. Figure 3C and D depict the regions of hitchhiking that differentiate Tibetans from lowlanders. Both regions in two species center around the EPAS1 locus. Although the sizes differ somewhat in the two species (145 kb in dogs and 220 kb in humans) and level of frequency difference is higher in dogs and than in humans, the modest differences probably reflect the general differences in the genomes of the two species. For example, the haplotype blocks in dogs are typically much lower than in humans (<10 kb for indigenous dogs (Wang et al. 2013), approximately 60 kb in non-Africans humans [Reich et al. 2001]). Statistical analysis revealed that the overlapping of selected segments between dogs and humans was significantly higher than expected (P < 10E-7, chi-square test). Meanwhile, when we compared our list of selected genes in dogs with that from humans presented in Yi et al, both the EPAS1 and HBB exist in this overlapping set between two species. This level of overlapping is highly significant (P = 4E-4, χ2 test). This is also true when we try to overlap our list of selected genes with Simonson et al (EPAS1 exists in both sets with a P value of 0.009) (supplementary note S2, Supplementary Material online). Taken together, the data present a strongly convergent pattern between humans and dogs in their adaptation to the Tibetan environment.
Discussion
Whole-genome resequencing strategy offers more dense genetic markers for the population genetic analysis compared with the microarray approach (Li et al. 2014). The Tibetan village dogs are characterized by a rapid rate of decay of linkage disequilibrium as a function of physical distance, which allowed us to perform genome-wide scans for recent positive selection for traits that are adaptive at high altitude (Lindblad-Toh and Wade 2005; Karlsson et al. 2007). Using more than 2.4 million SNPs extracted from populations of village dogs that both lived at high altitude on the Plateau and from lower altitudes, we successfully mapped the two most evident genes, EPAS1 and HBB, that could have contributed to high-altitude hypoxic adaptation in dogs. Interestingly, a recent work also identified these two genes as candidates showing signs of high-elevation adaptation based on a set of independent populations and approaches (Gou et al. 2014), which also supports the conclusion of the present study.
Furthermore, we found that a nonsynonymous mutation on EPAS1, G305S, might be the major contributing factor for the high-altitude hypoxic adaptation, as the codon 305 is not only conserved across all known vertebrates but also located in the PAS-B domain that is the major functional domain binding to its ligands (Key et al. 2009). Meanwhile, HBB also carries a nonsynonymous mutation, L15M, that could potentially contribute to the hypoxic adaptation. Several previous studies in other high-altitude species showed that amino acid mutations in the alpha- and/or beta-globin genes can alter Hb-O2 affinity, for example, Andean hummingbirds (Projecto-Garcia et al. 2013) and deer mice (Storz et al. 2009, 2010; Natarajan et al. 2013). If we intersected the results of Fst analysis and Fisher’s exact test approaches, the EDNRA gene (Endothelin-1 receptor Precursor) shows up as an another candidate selected gene involved in the hypoxia-inducible-factor (HIF) pathway and is also found in Tibetans presented in Simonson et al. (2010).
Interestingly, several previous studies suggested that EPAS1 aided hypoxic adaptation in Tibetan human populations (Beall et al. 2010; Simonson et al. 2010). Surprisingly, studies looking at high-altitude adaptation in people living around the Andes mountains revealed rather different genetic components in the hypoxia response (Bigham et al. 2010) with several differences in physiological traits from the Tibetan regions (Beall 2007). These conclusions collectively seem to suggest that humans and dogs in the Tibetan plateau have adapted to the high-altitude environment by modifying a set of similar genes and pathways. Through evolutionary analysis, we found that there are significant convergence between human and dogs at genetic level on the Tibetan plateau. The convergent adaptive evolution between dogs and humans in the same environment in the absence of such convergence between different human populations in different regions suggests that the shared environment may have played a pivotal role (Wang et al. 2013). For example, sharing much of the diet and complex interactions between human and dogs might have shaped both species adapting to the common environment. Similarly, the differences between Andeans and Tibetans might be due to dissimilarities between local ecological pressures in their respective geographic locations. Differences in genetic background in the two human populations might have also contributed to this observed disparity. As we look further into many organisms living in Tibetan regions and other high-altitude environments, we might unveil a very intriguing array of successful adaptations with diverse mechanisms as well as a new understanding of the convergent evolution between human beings and their closest animal companions. These results shed new light on the field of convergent evolution and offers opportunities to gain new insights into high-altitude settlement shared by dogs and humans.
Supplementary Material
Supplementary notes S1 and S2, tables S1–S8, and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Masatoshi Nei of Pennsylvania State University for helpful comments and suggestions to the manuscript and Andrew Willden of the Kunming Institute of Zoology for assistance in editing. The authors thank Chang-Qing Zeng and Peter A. Robbins for help with previously published human genotype data. They also thank Bing Liu of CAS Tibetan Mastiff CO, LTD for helping to measure Hb level in Tibetan dogs in Lijiang. This work was supported by grants from the National Natural Science Foundation of China (91231108), the 973 program (2013CB835200), and the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L07).
Literature Cited
- Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkorta-Aranburu G, et al. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 2007;104(Suppl. 1):8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J. Origins of the dog: domestication and early history. In: Serpell J, editor. The domestic dog: its evolution, behaviour, and interactions with people. New York: Cambridge University Press; 1995. pp. 7–20. [Google Scholar]
- Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X, et al. Whole genome sequencing of six dog breeds from continuous altitudes reveals adaption to high-altitude hypoxia. Genome Res. 2014;24:1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji LD, et al. Genetic adaptation of the hypoxia-inducible factor pathway to oxygen pressure among Eurasian human populations. Mol Biol Evol. 2012;29:3359–3370. doi: 10.1093/molbev/mss144. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Pathol. 2011;6:165–192. doi: 10.1146/annurev-pathol-011110-130321. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Population variation revealed high altitude adaptation of Tibetan Mastiffs. Mol Biol Evol. 2014;31:1200–1205. doi: 10.1093/molbev/msu070. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang YP. High genetic diversity of Tibetan Mastiffs revealed by mtDNA sequences. Chin Sci Bull. 2012;57:1483–1487. [Google Scholar]
- Lindblad-Toh K, Wade CM. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Mairbaurl H, Weber RE. Oxygen transport by hemoglobin. Compr Physiol. 2012;2:1463–1489. doi: 10.1002/cphy.c080113. [DOI] [PubMed] [Google Scholar]
- Moritz A, Fickenscher Y, Meyer K, Failing K, Weiss DJ. Canine and feline hematology reference values for the ADVIA 120 hematology system. Vet Clin Pathol. 2004;33:32–38. doi: 10.1111/j.1939-165x.2004.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Natarajan C, et al. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JF, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- Projecto-Garcia J, et al. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 2013;110:20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann TH, et al. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Storz JF, Moriyama H. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol. 2008;9:148–157. doi: 10.1089/ham.2007.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- Wang JH. Transport of oxygen through hemoglobin solutions. Science. 1961;133:1770–1771. doi: 10.1126/science.133.3466.1770. [DOI] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci U S A. 2009;106:21230–21235. doi: 10.1073/pnas.0907844106. [DOI] [PMC free article] [PubMed] [Google Scholar]