Background: The lack of cysteine cathepsin inhibitor, stefin B (cystatin B), results in progressive myoclonus epilepsy, type 1.
Results: Stefin B deficiency in macrophages resulted in increased inflammasome activation.
Conclusion: Stefin B-deficient mice are significantly more sensitive to e LPS-induced sepsis due to increased caspase-11 expression and mitochondrial damage.
Significance: Stefin B has an important role in limiting the inflammatory response during LPS-induced sepsis.
Keywords: Caspase, Inflammasome, Mitochondria, Reactive Oxygen Species (ROS), Sepsis
Abstract
Stefin B (cystatin B) is an endogenous cysteine cathepsin inhibitor, and the loss-of-function mutations in the stefin B gene were reported in patients with Unverricht-Lundborg disease (EPM1). In this study we demonstrated that stefin B-deficient (StB KO) mice were significantly more sensitive to the lethal LPS-induced sepsis and secreted higher amounts of pro-inflammatory cytokines IL-1β and IL-18 in the serum. We further showed that increased caspase-11 gene expression and better pro-inflammatory caspase-1 and -11 activation determined in StB KO bone marrow-derived macrophages resulted in enhanced IL-1β processing. Pretreatment of macrophages with the cathepsin inhibitor E-64d did not affect secretion of IL-1β, suggesting that the increased cathepsin activity determined in StB KO bone marrow-derived macrophages is not essential for inflammasome activation. Upon LPS stimulation, stefin B was targeted into the mitochondria, and the lack of stefin B resulted in the increased destabilization of mitochondrial membrane potential and mitochondrial superoxide generation. Collectively, our study demonstrates that the LPS-induced sepsis in StB KO mice is dependent on caspase-11 and mitochondrial reactive oxygen species but is not associated with the lysosomal destabilization and increased cathepsin activity in the cytosol.
Introduction
Sepsis, a systemic inflammatory response syndrome induced by infection, is characterized by multiple organ failures and hemodynamic shock (1). Lipopolysaccharide (LPS), a component on the surface of Gram-negative bacteria, is a potent inducer of sepsis and inflammation and acts by stimulating immune system cells to produce pro-inflammatory cytokines. Upon LPS binding and receptor homodimerization, the signal-transducing adaptor proteins become associated with the receptor and trigger the downstream signaling pathway leading to the activation of NF-κB (2, 3). Members of the NLR family (NLR proteins, Nlrp) and the adaptor ASC form multiprotein complexes, termed inflammasomes, that are required for the activation of the pro-inflammatory caspase-1 that processes pro-interleukin (IL)-1β and pro-IL-18 into mature forms (4, 5). The inflammasome activity should be tightly regulated to defend the host while limiting the excessive inflammation and immunopathology. Nlrp3 inflammasome is the best characterized as a central regulator of inflammation, which regulates caspase-1 and IL-1β production in myeloid cells (4, 5). In this pathway, TLR4 signaling provides the priming step that up-regulates Nlrp3 and pro-IL-1β gene expression (6). Besides the priming step, there are three main cellular processes described in the literature that lead to Nlrp3 inflammasome activation (5, 7). The first is the pannexin-1 membrane pore formation via the ATP-gated P2X7 ion channel and K+ efflux (8). The second way of Nlrp3 activation is the phagocytosis of crystalline or particulate structures. The proposed mechanism suggests that phagocytosis triggers lysosomal destabilization and subsequent release of the lysosomal content, including lysosomal cathepsin proteases, into the cytosol. However, it is not known yet whether the cathepsins interact directly with the inflammasome or whether the process involves molecules activated by the cathepsins (9, 10). The third model proposes that generation of the reactive oxygen species up-regulates activation of the Nlrp3 inflammasome. The role of mitochondria in the inflammasome activation has been recently elucidated by blocking the key enzymes of the respiratory chain, which resulted in the loss of mitochondrial membrane potential, a vast ROS generation and IL-1β secretion (11, 12). Recently, in addition to the canonical (LPS and ATP) Nlrp3 inflammasome signaling, a new noncanonical inflammasome pathway has been described, where caspase-11 is engaged in response to a number of Gram-negative bacteria. Caspase-11 is involved in Nlrp3-dependent activation of caspase-1 leading to IL-1β processing and secretion or in the Nlrp3- and caspase-1-independent pathway leading to cell death (13–16). It is worth noting that the mouse caspase-11 (gene name Casp4) has 46% similarities to caspase-1 and is orthologous to human caspases-4 and -5 (17). Endolysosomal cysteine cathepsins were first described as proteases involved in the protein breakdown within the lysosomes, but many studies revealed their important role in other cellular processes, such as apoptosis, cancer progression, inflammation, cell homeostasis, pathogen recognition, and its elimination (18, 19). Their activity is regulated by the interaction with the endogenous inhibitors, such as cystatins, tyrophins, and some of the serpins (20–23). Stefin B belongs to the type one cystatins and it is up-regulated in the activated macrophages (20, 22). In the nucleus stefin B interacts with the histones and cathepsin L (24). Mutations in the gene of stefin B are associated with the neurodegenerative disease known as the Unverricht-Lundborg disease or progressive myoclonus epilepsy of type 1 (EPM1) (25, 26). EPM1 is characterized by the cerebellar granule neuron apoptosis, progressive ataxia, and myoclonic epilepsy (25, 27). StB KO mice have increased apoptosis in the cerebellum and develop ataxia (28). Houseweart et al. (29) reported that the removal of cathepsin B from StB KO mice reduced the neuronal apoptosis but did not rescue the ataxia and seizure phenotype, suggesting that stefin B has alternative function(s) other than protease inhibition. Stefin B was found as a part of multiprotein complexes in the cerebellum; interestingly, none of the identified proteins interacting with stefin B was a protease (30). The precise mechanism by which stefin B influences the progression of the disease is not yet clear.
In this study, we describe a series of in vivo and in vitro experiments showing that stefin B deficiency profoundly influences the Nlrp3 inflammasome activation and viability of mice in response to LPS. In particular, the data strongly suggest that the lack of stefin B results in an increase of caspase-11 expression, pro-inflammatory cytokine IL-1β and IL-18 serum concentrations, and mitochondrial ROS formation. Thus, the traits observed in StB KO mice result from an increased sensitivity of these mice to inflammation rather than increased enzymatic activity of cathepsin proteases.
EXPERIMENTAL PROCEDURES
General Reagents
LPS from Escherichia coli (E. coli 055:B5, Sigma) was used at a final concentration of 100 ng/ml for the cellular studies. ATP was obtained from Sigma (A7699) and used at a final concentration of 5 mm ATP for 20 min. Recombinant mouse interferon-β (IFN-β) (eBioscience) was used at a final concentration of 100 units/ml. Antibodies used in Western blotting were purchased from Abcam, and anti-caspase-1 (ab108362), anti-caspase-11 (ab10454), anti-IL-18 (ab71495), anti-stefin B (ab53725), anti-STAT1 (ab3987), anti-STAT1 Y 701 (9171S) were from Cell Signaling. 3ZD anti-IL-1β mouse monoclonal antibodies were obtained from the National Cancer Institute-Frederick, MD (31). Anti-mouse IFNAR1 antibody (AF3039) was from R&D Systems. Recombinant mouse macrophage colony-stimulating factor (M-CSF) was purchased from eBioscience and used at a final concentration of 40 ng/ml. The cysteine protease inhibitor E-64d was purchased from the Peptide Research Institute and used at a final concentration of 15 μm.
Mice
Stefin B (cystatin B)-deficient mice were created as described previously (28) and were provided by Dr. R. M. Myers, Stanford University, and bred in our local colony. Mice (8–12 weeks of age) used in this study were wild type (WT) and StB KO, fully backcrossed to FVB/N background. All mouse studies were conducted in accordance with protocols approved by the Veterinary Administration of the Republic of Slovenia and the Government Ethical Committee. Procedures for animal care and use were in accordance with the “PHS Policy on Human Care and Use of Laboratory Animals” and the “Guide for the Care and Use of Laboratory Animals.”
Preparation of Macrophages
Mouse primary bone marrow-derived macrophages (BMDMs) were obtained by ex vivo differentiation from mouse bone marrow progenitors in the presence of M-CSF over 7 days, as described previously (32). Cells were kept in DMEM supplemented with 20% FBS, 1% penicillin/streptomycin, 2 mm l-glutamine, and M-CSF.
Plasmid and Transfections
The cDNA clone for stefin B was obtained from Image (IMAGE, 3453675). It was PCR-amplified, tagged, and cloned into pcDNA3 vector (Invitrogen) at HindIII and XhoI restriction sites (24). Raw-blue cells (InvivoGen) were transiently transfected with pcDNA3 and pcDNA/stefin B-HisTag plasmid using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
NF-κB Reporter Assay
Raw-blue cells (InvivoGen) were derived from the murine RAW 264.7 macrophages with chromosomal integration of a secreted embryonic alkaline phosphatase (SEAP) reporter construct inducible by NF-κB and AP-1. Cells were grown according to the manufacturer's protocol. Upon TLR activation, the signaling pathway leads to activation of the NF-κB-dependent secretion of SEAP. Quanti-blue medium (InvivoGen) was used for detection and quantification of SEAP, according to the manufacturer's protocol. SEAP activity was assessed by reading OD at 655 nm with a microplate reader (Tecan).
Stimulations and Systemic LPS Challenge in Vivo
BMDMs were prepared as described above and seeded in 6-well plates (106 cells/well) for lysate preparation or in 96-well plates (105 cells/well) for supernatant collection. Cell were stimulated as indicated. Briefly, LPS (100 ng/ml) were primed for 4 h, washed with PBS, pH 7.2, and then ATP (5 mm) was applied for 20 min. For cytokine analysis in the serum, StB KO and WT mice were intraperitoneally injected with LPS at 10 mg/kg (E. coli 055:B5, Sigma). Blood was taken before (0 h) and 2, 4, and 6 h after the LPS administration. The serum was prepared by removing the clot by centrifuging at 2000 rpm for 10 min in a refrigerated centrifuge; the resulting supernatants were collected, and the serum levels of the cytokines were determined by flow cytometry. For the viability study, WT and StB KO mice were injected intraperitoneally with a high dose of LPS at 30 mg/kg (E. coli 055:B5, Sigma) and monitored six times daily over the 96 h.
Cell Lysate Preparation and Western Blot Analysis
Monolayer BMDMs at 80% confluence were first washed with ice-cold PBS, followed by the direct addition of the radioimmune precipitation assay lysis buffer supplemented with the Complete protease inhibitor mixture (Sigma) Cell lysates were prepared as described previously (33). The protein concentration of the cell lysates was measured using the Bradford reagent (Bio-Rad). The lysates were subjected to electrophoresis on 12.5% SDS-polyacrylamide gels followed by Western blotting as described previously (33). The proteins were visualized with ECL (Amersham Biosciences) according to the manufacturer's instructions.
Cathepsin Activity Measurements
The cytosolic cathepsin activity assay was performed in a 96-well plate by using sensitive fluorogenic substrates Z-Arg-Arg-AMC (Z-RR-AMC) and Z-Phe-Arg-AMC (Z-FR-AMC) (Bachem). The treated cells were partially lysed with an acetate buffer (50 mm sodium acetate, 150 mm NaCl, 0.5 mm EDTA, pH 5.6) containing 15 μg/ml digitonin (for the cytosolic cathepsin activity) for 12 min on ice as described (34). Immediately after the incubation on ice, the substrate (30 μm)-containing acetate buffer was added, and the liberation of AMC was measured using a fluorimeter (Tecan).
Quantitative Real Time PCR
Total RNA was isolated from the BMDMs after stimulation with LPS using PureLink RNA mini kit (Ambion), followed by DNase treatment using TURBO DNA-free kit (Ambion). RNA concentrations and A260/A230 and A260/A280 ratios were assessed spectrophotometrically using NanoDrop, whereas RNA integrity was confirmed by 1.2% gel electrophoresis. cDNA was synthesized from RNA with Precision nanoScript Reverse Transcription kit (Primer Design Ltd.) using random nanomer primers. The following primers designed by Primer Design were used for the real time quantitative PCR: IL-1β (Il1b) sense, 5′-GCTATGGCAACTGTTCCTGAA-3′, and antisense, 5′-ACAGCCCAGGTCAAAGGTT-3′; IL-18 (Il18) sense, 5′-CCAAGTTCTCTTCGTTGACAAAA-3′, and antisense, 5′-GTCCTCTTACTTCACTGTCTTTG-3′; caspase-1 (Casp1) sense, 5′-CTGCGGTGTAGAAAAGAAACG-3′, and antisense, 5′-TCCATTTATTGTCCCTATACTCACT-3′; caspase-11 (Casp4) sense, 5′-GCTACGATGTGGTGGTGAAA-3′, and antisense, 5′-GGAATGTGCTGTCTGATGTCT-3′. Real time PCR was run on Mx3005P system (Agilent, Stratagene products) using the following thermal profile: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Additionally, a melting curve (55–95 °C) was performed at the end of each run. The mRNA expressions of target genes were normalized to the expression of Gapdh and B2m, which were confirmed to be the most stably expressed reference genes by mouse geNorm reference gene selection kit (Primer Design). The mRNA expression of genes was calculated considering their real time PCR efficiencies using REST 2009 (Relative Expression Software Tool) and presented as a fold increase, relative to the unstimulated cells (control). The mRNA expression levels of genes in control samples were normalized to 1.0.
Chromatin Immunoprecipitation
WT and StB KO BMDMs were grown on six 10-cm culture dishes. The attached cells were washed two times with PBS on the dish and then overlaid with 10 ml of PBS with 1% formaldehyde for fixation at room temperature for 10–15 min with occasional shaking. After fixation, the dishes were washed once with PBS containing 125 mm glycine and two times with cold PBS, and then the cell nuclei were isolated as described (35) and resuspended in 3 mm CaCl2, 50 mm Tris-HCl, pH 8.0. The nuclear preparation was digested with 60 units/ml micrococcal nuclease (Nuclease7, Roche Applied Science) at 37 °C for 30 min with occasional shaking to achieve ∼150-bp-long mononucleosome fragments. The reaction was terminated by 10 mm EDTA. Nuclei were centrifuged for 5 min at 7500 rpm, and the pellet was resuspended in 1 ml of L-CHIP buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.0), 1 mm PMSF, and Complete protease inhibitor mixture (Sigma) and sonicated once at setting 2 for 10 s on Sonic Dismembrator (Fisher, model 100). The lysate was centrifuged at 14,000 rpm for 5 min, and the protein concentration was adjusted to 1 mg/ml with L-CHIP buffer and subjected to immunoprecipitation with antibodies against histone H3 dimethyl K9 (Abcam, ab1220 ChIP grade) as described (36). Before immunoprecipitation, an aliquot of the nuclear lysate was saved for isolation of input DNA. Immunoprecipitated (ChIP) and input chromatin (Input) were treated with 0.5 mg/ml of proteinase K (Roche Applied Science) and RNase A (Roche Applied Science) at 37 °C for 30 min and uncross-linked by incubating at 65 °C overnight. DNA was extracted twice with phenol/chloroform and once with chloroform and ethanol precipitated with glycogen (Roche Applied Science) and sodium acetate. DNA was dissolved in 50 μl of water and used to prepare libraries with NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (E6200). Libraries were sequenced on an Illumina HiSeq 2500 Genome Analyzer at the Pennsylvania State University Genomic Core Facility. ChIP-sequencing reads were mapped to the mouse genome with bowtie 0.12.9 software using the default parameters and the provided index NCBI37/mm9. Sequence reads mapped to multiple genomic locations were excluded from subsequent analysis. All sequence reads and input DNA reads were normalized to genome average, and the ratio of normalized ChIP sequence versus the nonfractionated input DNA was calculated for the promoter-containing region (1 kb upstream of the transcriptional start site) and the open reading frame (1 kb downstream of the transcriptional start site) of the mouse caspase-11 gene (Casp4).
Flow Cytometry
Detection of Cytokines in Cell Supernatants and Serum
The levels of secreted cytokines in the BMDM culture supernatants and mice sera were quantified by bead-based multiplex immunoassay for the flow cytometer (FlowCytomixTM, eBioscience), according to the manufacturer's instructions.
Analysis of Lysosome Integrity
Lysosomes in primed and stimulated BMDMs were stained with the fluorescent acidotropic probe LysoTracker Green probe (Invitrogen) according to the manufacturer's instructions, as described previously (37).
Analysis of Mitochondrial Membrane Potential
The mitochondrial membrane potential (ΔΨm) in BMDMs was determined by the MitoTracker Red CMXRos (Invitrogen) uptake, as described previously (37).
Detection of Mitochondrion-derived ROS (mtROS)
MitoSOX Red (Invitrogen) was used as a mitochondrial superoxide indicator in WT and StB-deficient BMDMs. Upon 4 h of LPS treatment, cells were washed with warm PBS and loaded with 5 μm MitoSOX in PBS for 15 min at 37 °C. ATP was added thereafter for 20 min where indicated. Cells were harvested, pelleted at 1000 rpm for 5 min, immediately resuspended in PBS, and subjected to FACS analysis. According to the manufacturer, MitoSOX Red permeates live cells and is targeted to the mitochondria. The dye was rapidly oxidized by superoxide, which upon binding to nucleic acids emits red fluorescence, which was monitored by flow cytometer using excitation/emission = 488/564–606 nm. All samples were analyzed with the FACSCalibur flow cytometer (BD Biosciences), and fluorescence intensity plots were evaluated using the CellQuest acquisitions software (BD Biosciences), version 3.3. All flow cytometry analyses were performed following live cell gating from forward and side scatter profiles.
Cytotoxicity, Lactate Dehydrogenase Release
Release of the cytoplasmic enzyme lactate dehydrogenase (LDH) into the medium was determined by the cytotoxicity detection kit (Roche Applied Science) following the manufacturer's instruction. BMDMs were lysed with 2% Triton X-100 to obtain total LDH release. The cytotoxicity was expressed as the percent of the total LDH release.
Confocal Microscopy
WT BMDMs were seeded on coverslips, left untreated, or stimulated with LPS. After the indicated treatment, the cells were washed, probed with MitoTracker Red CMXRos (100 nm) in OptiMEM for 45 min at 37 °C, then fixed with 3.7% paraformaldehyde in PBS, pH 7.2, for 15 min, and permeabilized with 0.2% Triton X-100 for 10 min in PBS. Stefin B was labeled with rabbit anti-stefin B antibodies. Fluorescence microscopy was performed using the Carl Zeiss LSM 510 confocal microscope. Secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 546 and rhodamine were excited with an argon (488 nm) or He/Ne (543 nm) laser, and emission was filtered using a narrow band LP 505–530 nm (green fluorescence) and 560 nm (red fluorescence) filter, respectively. Carl Zeiss LSM image software 3.0 (Correlation Plots) was used to evaluate co-localization between the two labeled proteins (i.e. between red and green fluorescence signals).
Statistical Analysis
Average results are presented as the mean ± S.D. from the number of assays indicated. Statistical significance of the results was determined using unpaired Student's t test, assuming unequal variances. Survival rates were analyzed by the Kaplan-Meier method and compared by the log-rank test.
RESULTS
StB KO Mice Are More Sensitive to LPS-induced Lethal Endotoxicosis and Secrete High Levels of IL-1β and IL-18 in the Serum Post-LPS Administration
LPS, known as endotoxin, is found in the outer membrane of various Gram-negative bacteria, yet a small dose of LPS can cause severe inflammation and septic shock (38). Therefore, a high-dose LPS injection into mice is a useful model for the study of endotoxic shock and the associated inflammatory responses. To investigate the overall importance of stefin B during the LPS-induced lethal endotoxicosis, mice were intraperitoneally injected with a high dose of LPS (30 mg/kg weight), and survival was monitored over a 96-h period (Fig. 1A). StB KO mice were found to be significantly more susceptible to the LPS-induced sepsis. All StB KO mice thus succumbed to the high dose of LPS up to the 60-h time point, whereas a higher proportion of the WT mice survived LPS-induced endotoxicosis up to 92 h post-injection.
FIGURE 1.
LPS-induced lethality and LPS-triggered pro-inflammatory cytokine release in vivo is stefin B-dependent. A, Kaplan-Meier plot showing the percentage of survival over time of age-matched wild type (WT) (n = 13) and StB KO mice (n = 11). *, p < 0.05 (log-rank (Mantel-Cox) test). Mice were injected intraperitoneally with 30 mg/kg LPS, and survival was monitored six times daily for a total of 4 days. B, for serum cytokine analysis, mice were injected intraperitoneally with 10 mg/kg LPS, and blood was taken 0, 2, 4, and 6 h after LPS administration. Levels of indicated cytokines in serum were quantified by bead-based multiplex for the flow cytometer. Statistical significance between WT and StB KO mice was determined in serum for IL-1β at 4 and 6 h, for IL-18 at 0, 2, 4, and 6 h, and for TNF-α at 2 h post-LPS injection. Data were obtained from three independent biological experiments performed in triplicate, and the results are presented as means ± S.D. *, p < 0.05; **, p < 0.01.
Furthermore, we challenged StB KO and WT mice (8–12 weeks of age) with a lower dose of LPS (10 mg/kg weight) and analyzed the pro-inflammatory cytokine profile in the serum (Fig. 1B). We determined a dramatic increase in the level of IL-1β and IL-18 in the serum of StB KO mice 4 and 6 h after LPS administration, whereas IL-6 serum levels remained comparable between the WT and StB KO mice. TNF-α release was significantly higher in the serum of StB KO mice only at the 2-h time point, but not at the 4- and 6-h post-LPS administration. These findings suggest that the lack of stefin B results in the greater susceptibility to sepsis, which is at least partly due to the increase of serum cytokines.
Transcription of Caspase-11 Is Up-regulated in LPS-stimulated StB KO BMDMs
The above results suggest that the molecular machinery regulating the IL-1 cytokine family secretion (IL-1β and IL-18), but not the IL-6 and TNF-α, is highly affected by the loss of stefin B. Our question was whether stefin B affects pro-inflammatory cytokine and caspase expression or activation. First, we examined the relative mRNA expression levels of caspases-1 and -11, IL-1β, and IL-18 4 h post-LPS stimulation in the BMDMs derived from StB KO and wild type mice (Fig. 2A). In line with the inducible IL-1β gene expression (39), we detected a dramatic up-regulation of the IL-1β gene expression in both WT and StB KO BMDMs in response to the LPS stimulation (50–90-fold induction). Moreover, the IL-18, caspase-1, and caspase-11 genes were also up-regulated, but to a much lesser extent. Stefin B deficiency had a minimal effect on the caspase-1, IL-1β, and IL-18 mRNA expression, whereas relative caspase-11 expression was significantly higher in StB KO than in WT BMDMs (Fig. 2A). The expression of caspase-11 is regulated by NF-κB and STAT transcription factors (40). In reporter cell line RAW-blue overexpressing stefin B, we confirmed a decreased NF-κB activation upon TLR4 signaling (Fig. 2B). These findings therefore provide evidence that stefin B may be involved in the modulation of NF-κB activity and gene expression.
FIGURE 2.
Transcription of caspase-11 is up-regulated in LPS-stimulated StB KO BMDMs. A, BMDMs were stimulated with LPS and total RNA was isolated 4 h post-stimulation. Relative mRNA expression was determined. RNA expression was normalized to reference genes Gapdh and B2m and presented as fold increase. Data were obtained from three independent experiments performed in triplicate, and the results are presented as means ± S.D. **, p < 0.01. B, increased expression of stefin B diminished NF-κB activation. RAW-blue cells (InvivoGen) were transfected with empty (control) pcDNA3 vector or pcDNA3/Stefin B-HisTag to induce stefin B overexpression. SEAP activity was assessed by reading the OD at 655 nm. C, fold enrichment (ChIP/Input) of H3K9me2 immunoprecipitated nucleosomes at the promoter and the open reading frame (ORF) downstream of the transcriptional start of the mouse caspase-11 gene (Casp4) IL-1β (IL-1β) gene.
Because it has been shown that cathepsin L, an inhibitory target of stefin B, is translocated into the nucleus to process a transcriptional regulator CUX1 (41) and that the nuclear cathepsin L might affect gene expression by altering post-translational histone modifications (42, 43), we hypothesized that the effect of stefin B deficiency on gene expression could be mediated by deregulated cathepsin L. To test this hypothesis, we studied global distribution of histone H3 dimethylated at lysine 9 (H3K9me2) and histone H3 trimethylated at lysine 9 (H3K9me3), two repressive chromatin marks strongly affected by inhibition of nuclear cathepsin L by MENT (44) and by cathepsin L knock-out (43). Immunofluorescence experiments with chromosome spreads showed that global arrangement of H3K9me3 and H3K9me2 was not significantly changed in either WT or StB KO cells (data not shown). To precisely identify gene-specific distribution of H3K9me2, a repressive histone modification playing an important role in gene-specific silencing (45), we applied chromatin immunoprecipitation followed by sequencing (Chip-seq). We were particularly interested in the caspase-11 gene (Casp4), as it was highly up-regulated in StB KO BMDMs. As expected, we observed a decreased level of H3K9me2 at the promoter of caspase-11 in StB KO BMDMs (Fig. 2C). Moreover, there were essentially no differences in the distribution of the H3K4me2 variant at promoter regions in the IL-1β gene of WT and StB KO BMDMs (Fig. 2C). We concluded that the lack of the H3K9me2 mark at the caspase-11 promoter could contribute to the increased caspase-11 expression.
Increased Expression of Caspase 11 Is Not Because of Activation of TRIF Pathway and Type I IFN Signaling
Rathinam et al. (13) reported that TLR4-TRIF-IFN-β signaling pathway is essential for caspase-11 activation. To assess the role of IFN-β in increased caspase-11 expression in StB KO BMDMs, we pretreated BMDMs with interferon α/β receptor (IFNAR1)-blocking antibody prior to LPS stimulation. Anti-IFNAR1 antibody treatment did not diminish caspase-11 expression in StB KO BMDMs (Fig. 3A). Next, we examined the relative mRNA expression levels of IFN-β 2 h after LPS stimulation in the BMDMs derived from StB KO and WT mice, and we determined that relative IFN-β mRNA expression was lower in StB KO than in WT BMDMs (Fig. 3B). Fang et al. (46) showed that epigenetic mark H3K9me2 is a suppressor of IFN and IFN-inducible antiviral gene expression. We applied Chip-seq and determined increased levels of H3K9me2 at the promoter of IFN-β in StB KO BMDMs in comparison with WT BMDMs in normalized samples (Fig. 3D). This result suggested that the increased H3K9me2 mark at the IFN-β promoter could contribute to the lower IFN-β expression. The biological effects of IFN-β are mediated largely through activation of STAT1 and STAT2. We show that IFN-β-induced phosphorylation of STAT1 was significantly augmented in the absence of stefin B (Fig. 3E). Because IFN-β expression and signaling are decreased in StB KO BMDMs, we reasoned that that TRIF signaling is not essential for the increased caspase-11 expression determined in StB KO BMDMs.
FIGURE 3.
Decreased IFN-β expression and signaling in StB KO BMDMs. A, BMDMs from WT and StB KO mice were treated with inhibitory anti IFNAR1 antibody and stimulated for 4 h with LPS (100 ng/ml). Cell lysates were immunoblotted with anti-caspase-11 antibodies recognizing pro-caspase (casp)-11 (inactive zymogen) and subunits generated by proteolytic cleavages. Lysates were probed with anti-β-actin antibody as a loading control. Data shown are representative of three independent experiments. B, BMDMs were stimulated with LPS, and total RNA was isolated 2 h post-stimulation. Relative mRNA expression was determined. RNA expression was normalized to reference gene B2m and presented as fold increase. Data were obtained from three independent experiments performed in triplicate. C, fold enrichment (ChIP/Input) of H3K9me2-immunoprecipitated nucleosomes at the promoter and the open reading frame (ORF) downstream of the transcriptional start of the mouse IFN-β (IFN-β). D, input DNA normalized to genome average. E, immunoblotting of p-STAT1 (pY-701) and total STAT1 in WT and StB KO BMDM at the indicated times following stimulation with IFN-β. Data shown are representative of three independent experiments.
Caspase-11 and Caspase-1 Activation Is Enhanced in StB KO BMDMs
Because the levels of IL-1β and IL-18 were highly up-regulated in the serum of LPS-injected mice, we examined the effects of stefin B deficiency on the activation of the Nlrp3 inflammasome in the BMDMs derived from StB KO and wild type mice. Pro-inflammatory caspase-11 is essential for caspase-1-independent macrophage death and caspase-1-dependent IL-1β and IL-18 production in response to inflammasome activators (15). Upon LPS priming and ATP stimulation, the formation of the p26 subunit of caspase-11 and the p10 subunit of caspase-1 was more prominent in StB KO BMDMs, suggesting a more efficient pro-inflammatory caspase proteolytic activation in StB KO BMDMs (Fig. 4). Pro-IL-1β in control samples was found at undetectable levels, which increased significantly at 4 h after LPS priming (Fig. 4), which correlates well with the inducible IL-1β gene expression (Fig. 2A). A high extracellular ATP concentration provides the second signal for the NLRP3 inflammasome assembly and the caspase-1-mediated processing of pro-inflammatory cytokines. The levels of IL-1β and IL-18 pro-form decreased in both genotypes upon addition of ATP; however, the drop was more evident in StB KO samples that is in line with a higher caspase-1 activation (Fig. 3). IL-1β and IL-18 mature forms were determined by Western blots of cell culture supernatants (Fig. 3), additionally secreted IL-1β was quantified in media by a flow cytometer (Fig. 4D). In line with better processing of pro-form, we determined significantly increased levels of mature IL-1β in StB KO BMDM's media (Fig. 4D).
FIGURE 4.
Enhanced processing and secretion of IL-1β and IL-18 in stefin B-deficient BMDMs is caspase-1-dependent. BMDMs from WT and StB KO mice were primed for 4 h with LPS (100 ng/ml), washed, and stimulated with ATP for 20 min (5 mm) to activate the Nlrp3 inflammasome. Cell lysates and culture supernatants were immunoblotted with the indicated antibodies. Anti-caspase-1 and anti-caspase-11 antibodies recognized pro-caspase (inactive zymogen) and subunits generated by proteolytic cleavages. Anti-IL-1β and anti-IL-18 antibodies recognized inactive pro-form in cell extracts and mature secreted cytokines in cell supernatants. Lysates were immunoblotted with anti-β-actin as a loading control. Data shown are representative of three independent experiments.
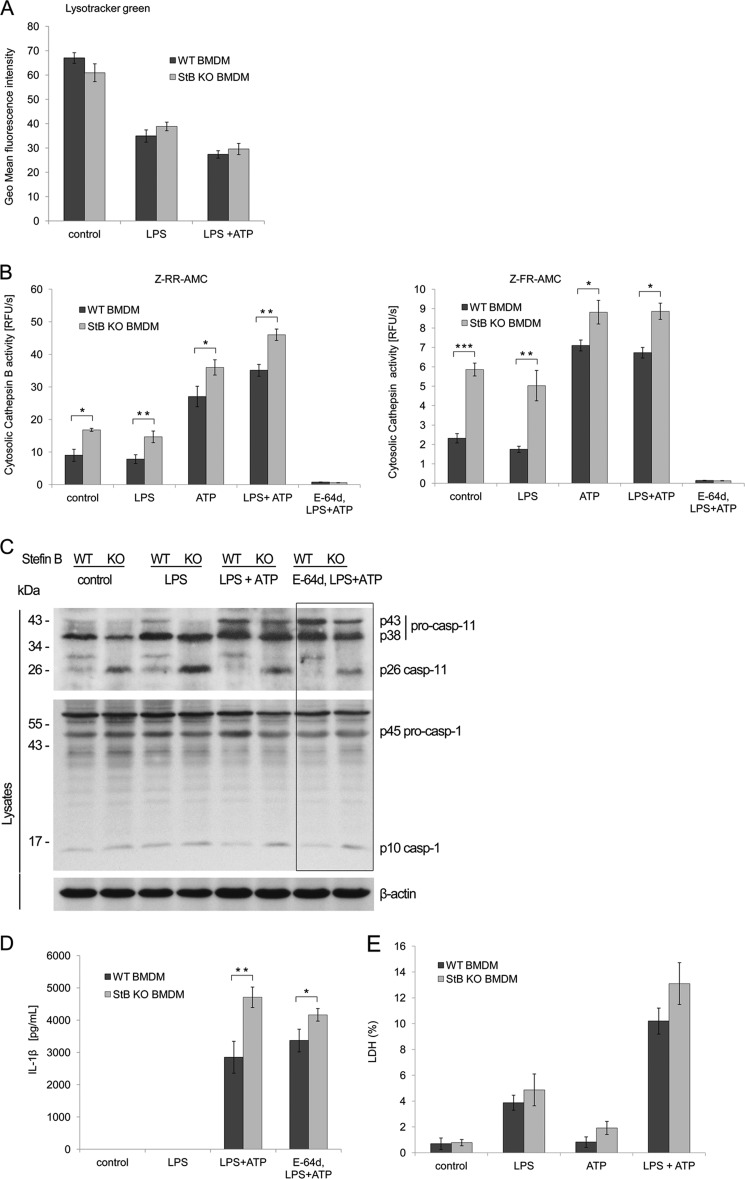
Inhibition of Lysosomal Cathepsin Activity Has No Effect on Nlrp3 Inflammasome Activation in StB KO BMDMs
Because it was reported that endolysosomal cathepsins participate in the NLRP3 inflammasome activation and IL-1β release (9, 10, 47), we hypothesized that the increased cathepsin activity, determined in the cells of EPM1 patients (48), is responsible for the Nlrp3 inflammasome activation in our model. A previous study showed that the loss of the lysosomal integrity resulted in the increase of cysteine cathepsin activity in the cytosol (37). First, we tested whether lysosomes get destabilized upon LPS and ATP stimulation. Lysosomal membrane integrity was determined with LysoTracker Green. LPS evoked slight attenuation in the green fluorescence, but addition of ATP increased lysosomal destabilization (Fig. 5A). However, stefin B deficiency had no effect on the lysosomal integrity (Fig. 5A). To quantify the extent of the lysosomal destabilization, we determined the cytosolic cathepsin activity by the fluorogenic broad cathepsin substrates Z-FR-AMC and Z-RR-AMC that are preferentially cleaved by cathepsin L and B (Fig. 5B). Cells were lysed with the 15 μg/ml digitonin extraction buffer under experimental conditions that did not disrupt lysosomes and then incubated with the substrates. We determined the highest levels of cathepsin activity in ATP and LPS/ATP-stimulated StB KO BMDMs, whereas LPS itself had no effect on the cathepsin lysosomal release and activation in the cytosol (Fig. 5B). This finding is in line with recent studies showing that ATP-mediated lysosomal breakdown and cathepsin release were LPS-independent (49). Contrary to the hypothesis concerning the cathepsin involvement in the inflammasome activation, we found that preincubation of BMDMs with the membrane-permeable broad spectrum cathepsin inhibitor E-64d before the Nlrp3 inflammasome activation did not alter activation of pro-inflammatory caspases-1 and caspase-11 (Fig. 5C), and consequently, it had no effect on IL-1β release (Fig. 4D).
FIGURE 5.
Increased inflammasome activation in StB KO BMDMs is independent of lysosomal cysteine cathepsins. A, biosynthesis and integrity of lysosomes were studied by LysoTracker Green probe, as described under “Experimental Procedures.” The lysosomal fluorescence intensity was quantified upon LPS and ATP stimulations. Lysosomal destabilization was increased by ATP addition and was comparable between genotypes. Data were obtained from three independent biological experiments performed in triplicate, and the results are presented as means of geometric mean fluorescence intensity ± S.D. B, release of cathepsins into the cytosol and their activity were measured after indicated stimulations using fluorogenic substrates specific for cathepsins. Incubation of BMDMs with E-64d completely prevented cathepsin activity. C, BMDMs from WT and StB KO mice were primed for 4 h with LPS (100 ng/ml), washed, and stimulated with ATP for 20 min (5 mm) or primed with broad spectrum cysteine cathepsin inhibitor E-64d for 1 h (15 μm), followed by LPS and ATP stimulation. Cell lysates were immunoblotted with indicated antibodies. Lysates were immunoblotted with anti-β-actin as a loading control. Incubation of BMDMs with E-64d did not affect the processing of caspase-1 and pro-inflammatory cytokine release. Data shown are representative of three independent experiments. D, BMDMs were seeded into 96-well plates and stimulated as above. IL-1β release was quantified in the media by FlowCytomixTM. StB KO BMDMs secreted higher amounts of IL-1β into the media compared with the WT; however, priming with E-64d did not affect IL-1β release (n.s., nonsignificant difference). Data were obtained from four independent experiments performed in triplicate, and the results are presented as means ± S.D. *, p < 0.05; **, p < 0.01. E, viability of BMDMs was assessed by LDH release into the cell culture media upon inflammasome activation. The cytotoxicity was expressed as the percent of the total LDH release. Data were obtained from three independent experiments performed in triplicate, and the results are presented as means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Besides IL-1β maturation and secretion, activation of caspase-1 can result in caspase-1-dependent inflammatory cell death termed pyroptosis (50). To investigate whether pro-inflammatory cytokines and cathepsins are released due to the cytolysis, we quantified the release of the cytolytic marker LDH in media (Fig. 5E). The difference in cell death between StB KO and WT BMDMs was not statistically significant. In addition, the levels of LDH in media were quite low (<15%), suggesting that the cell death was not the cause of cathepsin release.
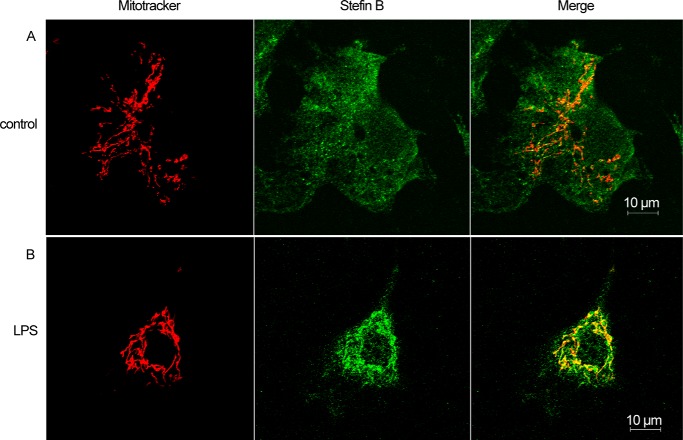
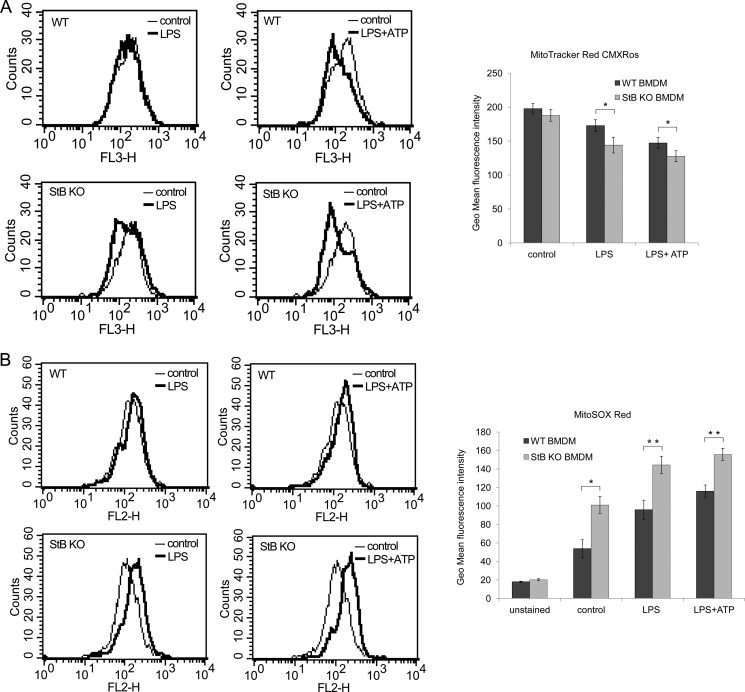
LPS Induced Translocation of Stefin B into Mitochondria Where It Prevented Excessive Superoxide Generation
Recently, several studies examined the involvement of the mitochondrion-derived reactive oxygen species (mtROS) and mitochondrial DNA (mtDNA) in the inflammasome activation; however, the precise mechanism of how cellular stress induces mitochondria breakdown and ROS release is still unknown (11, 51, 52). Because we determined that the lysosomal leakage was not essential for the Nlrp3 inflammasome activation, we next investigated whether mtROS are implicated in an increased IL-1β secretion in the mice lacking stefin B. To demonstrate the intracellular localization of stefin B upon inflammasome activation, WT BMDMs were probed with the MitoTracker Red. Unstimulated cells showed no co-localization between stefin B and the MitoTracker probe, whereas in the LPS-treated WT BMDMs we determined co-localized regions (Fig. 6). Furthermore, we monitored the loss of mitochondrial membrane potential (ΔΨm) by flow cytometry. As the mitochondrial membrane potential collapses, the MitoTracker Red CMXRos probe decreases its fluorescence. Unlike lysosomal integrity that was comparable between the WT and StB KO BMDMs (Fig. 4A), we detected a larger degree of damaged mitochondria in the LPS and LPS/ATP-stimulated StB KO BMDMs, suggesting a potentially protective role of stefin B in preserving the mitochondrial membrane integrity (Fig. 7A). Dysfunctional mitochondria are a key source of mtROS generation, and paralleled with the detected breakdown of ΔΨm, LPS and LPS/ATP stimulations highly increased superoxide generation in StB KO and to a lesser extent in WT BMDMs (Fig. 6B). The above results show that in the mitochondria stefin B protects against the loss of ΔΨm and prevents the production of mtROS.
FIGURE 6.
Stefin B is localized into mitochondria upon LPS stimulation. WT BMDMs were left untreated (A) or LPS (100 ng/ml)-primed (B). The localization of stefin B was determined by immunofluorescence. The images were merged to visualize co-localization of stefin B and mitochondria. Scale bar, 10 μm.
FIGURE 7.
Mitochondrial disruption resulted in increased superoxide generation in StB KO BMDMs. A, MitoTracker Red CMXRos uptake was quantified by flow cytometry. The decrease in red fluorescence correlates with the loss of mitochondrial membrane potential. Mitochondria of StB KO BMDMs turned to be more susceptible to LPS and ATP stimulation. Results are representative of three independent experiments performed in duplicate and presented as histogram plots and as columns of geometric mean fluorescence intensity of three independent experiments performed in duplicate and presented as means ± S.D. *, p < 0.05. B, BMDMs were stimulated and probed with 5 μm MitoSOX Red fluorogenic dye as indicated under “Experimental Procedures.” Red fluorescence was measured by flow cytometry. LPS and LPS/ATP stimulations induced significant increase in superoxide (mtROS) generation in StB KO compared with WT BMDMs. Results are representative of three independent experiments performed in duplicate and presented as histogram plots and as columns of geometric mean fluorescence intensity of three independent experiments performed in duplicate and presented as means ± S.D. *, p < 0.05; **, p < 0.01.
DISCUSSION
In our study, we show that the StB KO mice are significantly more sensitive to the LPS-induced sepsis (Fig. 1A), suggesting an important role of stefin B during inflammatory responses. We have determined significantly increased caspase-11 mRNA expression in the LPS-stimulated StB KO BMDMs, although the differences in caspase-1, IL-1β, or IL-18 mRNA expression between StB KO and WT BMDMs were not significant (Fig. 2A). Kayagaki et al. (15) showed that the loss of caspase-11 rather than caspase-1 protected the mice from a lethal dose of LPS, and we propose that the increased lethality upon LPS challenge could be attributed to the increased caspase-11 expression in StB KO BMDMs.
The cathepsin L deficiency and cathepsin L inhibition by a nuclear protease inhibitor, MENT, were shown to cause a global rearrangement of specifically modified histones H3K9me3 and H3K9me2 from euchromatin to heterochromatin (43, 44). However, our observation that the global distribution of methylated histone 3 (H3K9me2 and H3K9me3) was not dramatically changed between stefin B-deficient and WT BMDMs suggests that the increased caspase-11 expression is unlikely to result from a global epigenetic rearrangement, but could be due to some gene-specific mechanism. Stefin B was shown to interact with cathepsin L and histones H3, H2B, and H2A.Z in the nucleus (24). The cathepsin L deficiency and cathepsin L inhibition by the nuclear protease inhibitor MENT were shown to cause a global rearrangement of specifically modified histones H3K9me3 and H3K9me2 from euchromatin to heterochromatin (43, 44). The decreased level of histone variant H3K9me2 at the promoter of caspase-11 (Casp4) but not Il1b in StB KO BMDMs (Fig. 2C) is in accordance with the increased expression of caspase-11 in those cells. Furthermore, increased expression of stefin B in the nucleus delayed cell cycle progression, due to decreased cleavage of CUX1 transcription factor by cathepsin L (24), suggesting that the processing of CUX1 or some other gene-specific mechanism could be responsible for the up-regulation of caspase-11. Recently, it was reported that caspase-11 expression upon LPS-induced TLR4 signaling is dependent on MyD88-dependent and TRIF-dependent pathways that both lead to the activation of NF-κB (16).
The activation of TRIF pathway leads to type-I interferon expression (13). Engagement of IFNAR1 then stimulates STAT1 and IRF9 signaling pathways that also contribute to the up-regulation of caspase-11 expression (13, 14). In our work, we showed that blocking of IFNAR1 with inhibitory antibody did not prevent caspase-11 up-regulation in StB KO BMDMs. In addition, we determined decreased IFN-β expression, as well STAT1 activation in StB KO BMDMs in comparison with WT BMDMs. Previous studies reported that stefin B interacted with the STAT-1 in monocyte-derived and placental macrophages (53). However, the recent study from the same group could not confirm the direct interaction of stefin B with STAT1 (54). They reported that stefin B interacted with several different proteins in HIV-1-infected macrophages; among others the two proteins were involved in JAK/STAT signaling pathway (54). In our recent work, we showed significantly lower STAT3 signaling in LPS-stimulated StB KO BMDMs, which resulted in decreased IL-10 expression (55). Therefore, we propose that stefin B interferes with the signaling pathway upstream of STAT1 and STAT3. The precise mechanism by which stefin B influences JAK/STAT signaling has yet to be elucidated.
Because stefin B overexpression was found to down-regulate the NF-κB signaling (Fig. 2B), we propose that stefin B could modulate the caspase-11 gene expression in an NF-κB-dependent manner. Stefin B deficiency resulted in significantly higher serum levels of IL-1β and IL-18 (Fig. 1B), as well as in higher levels of IL-1β (Fig. 4D) in BMDM media compared with the WT controls. The maturation and release of the pro-inflammatory cytokines IL-1β and IL-18 are dependent on the caspase-1 activation on inflammasome (56). In noncanonical inflammasome activation, triggered by some of the bacteria, caspase-11 can interact with caspase-1 and forms a heterodimeric complex (15, 57). Both pro-caspase-11 p43 and its cleaved p26 subunit interacted with caspase-1 in the BMDMs, when exposed to cholera toxin B and E. coli; however, a much weaker interaction was detected upon canonical LPS/ATP stimulation (15). The differences in caspase-1 activation (Fig. 3) that we observed in StB KO and WT BMDMs upon canonical inflammasome activation with LPS/ATP could be at least partially attributed to the increased caspase-11 expression and activation. Because stefin B is essentially a cathepsin inhibitor, our first hypothesis was that the increased cathepsin activity reported before in the lymphocytes of EPM1 patients (48) and confirmed in our experiments in BMDMs (Fig. 4B) is responsible for the increased Nlrp3 inflammasome activation. We have shown that under our experimental conditions lysosomes get destabilized, and the released cathepsins are more active in StB KO BMDMs. However, the addition of broad spectrum inhibitor E-64d did not alter caspase-1 and -11 processing. Although the cathepsin activity was higher in StB KO BMDMs, the effect of stefin B deficiency on ATP-mediated caspase-1 activation and consequently on IL-1β processing was not reversed by the addition of the cathepsin inhibitor E-64d, a result showing that the cysteine proteinase activity is not essential for the differences in inflammasome activation in StB KO and WT BMDMs. The above results are in line with other studies, showing that ATP-induced IL-1β release was unaltered in the presence of cathepsin inhibitors (9, 49). The generation of ROS is another evolutionarily well conserved mechanism, triggered in response to infection or injury, and emerging studies strongly support the ROS pathway in the inflammasome activation (11, 58). Recent studies have shown that the mitochondrial dysfunction and relocation of the Nlrp3 complex to the mitochondrion-associated endoplasmic reticulum membranes is essential for the inflammasome activation (11, 59). Furthermore, Nakahira et al. (12) reported that depletion of the autophagy proteins beclin-1 or LC3B in macrophages leads to the activation of caspase-1 and the release of IL-1β and IL-18 by promoting the accumulation of dysfunctional mitochondria. These data suggested that the mitochondrial dysfunction represents an endogenous stimulus for inflammasome activation. Our findings revealed that stefin B is localized in the mitochondria upon LPS stimulation (Fig. 5), and with flow cytometry experiments, we confirmed its role in the protection of mitochondrial membrane integrity (Fig. 6A). Recently two independent studies demonstrated that mtROS and oxidized mtDNA released into the cytosol are key players in the Nlrp3 inflammasome activation (12, 51). Our results show that the lack of stefin B in BMDMs resulted in increased mtROS production (Fig. 6B). Interestingly, Lehtinen et al. (60) showed that stefin B protects cerebellar granule neurons from the oxidative stress, and the stefin B deficiency resulted in the increased apoptosis in the cerebellum. In addition, Butinar et al. (61) reported that stefin B-deficient tumor cells were significantly more sensitive to cell death upon the increased oxidative stress. Furthermore, Shimada et al. (51) showed that staurosporin (STS), a pro-apoptotic protein kinase inhibitor, was sufficient to act as a second signal for the NLRP3 inflammasome activation. In our previous study, we showed that STS-treated StB KO thymocytes were more sensitive to STS-induced apoptosis (33). Although we did not measure ROS in that study, other studies reported the disruption of electron transport, loss of Δψm, and the production of ROS in the STS-induced apoptosis (62).
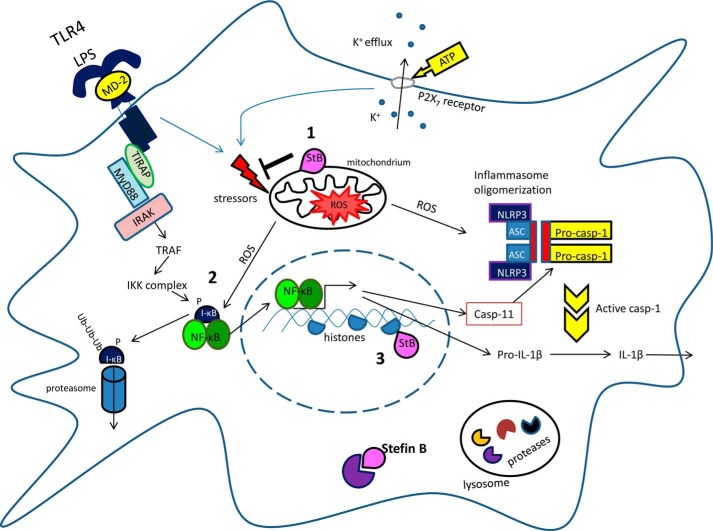
Based on these results, the following model is proposed for the protective role of stefin B in inflammation (Fig. 8). Upon LPS stimulation, stefin B is translocated into mitochondria and protects mitochondrial integrity. Stefin B deficiency resulted in the breakdown of mitochondrial membrane potential and increased mtROS generation. It was reported that NF-κB could act as a sensor for oxidative stress (63). ROS were shown to activate NF-κB directly (64) or indirectly by other stimuli such as TNF-α and IL-1β (65). The increased mtROS detected in StB KO BMDMs upon LPS/ATP stimulation could therefore lead to the increased NF-κB activation and consequently caspase-11 expression. In the nucleus, stefin B deficiency resulted in a decreased level of H3K9me2 repressive histone variant at the caspase-11 promoter, and this could contribute to the increased level of caspase-11 mRNA expression.
FIGURE 8.
Schematic of proposed model for the role of stefin B in Nlrp3 inflammasome activation. Step 1, upon LPS stimulation stefin B translocates into mitochondria and protects mitochondrial integrity. Step 2, mitochondrial ROS up-regulate NF-κB activation. Step 3, in the nucleus, stefin B deficiency resulted in a decreased level of H3K9me2-repressive histone variant at caspase-11 promoter and increased level of caspase-11 mRNA expression.
Halle et al. (9) reported NLRP3 inflammasome activation by amyloid-β in microglia in the case of Alzheimer disease resulted in the maturation and secretion of IL-1β. Because in StB KO mice early microglial activation preceded neuronal loss in the brain (66), it is tempting to speculate that Nlrp3 inflammasome activation is an important stage of the disease progression. In addition, Penacchio et al. (28) reported that StB KO mice develop corneal lesions as they age. TLR and NLRP3 expression was reported also in corneal tissue (67, 68). This might suggest that the increased incidence of the corneal inflammation in StB KO mice is a consequence of the increased inflammasome activation in the cornea.
In conclusion, in this study we report for the first time the mitochondrial localization of stefin B and the increased mtROS formation in StB KO BMDMs. The increased mortality of StB KO mice upon LPS challenge could be a consequence of an increased mtROS formation that leads to an increased NF-κB activation and expression of NF-κB target genes. Our in vivo data indicate that stefin B may be the critical effector of deleterious inflammatory responses. Therefore, elucidating the precise mechanism by which stefin B up-regulates caspase-11 (caspase-4 in humans) expression may be essential for the treatment of sepsis.
Acknowledgments
We thank Janja Završnik for help with mouse experiments and discussions and Mojca Trstenjak-Prebanda and Loulou Kroon-Žitko for expert technical assistance. We are grateful to R. M. Myers (Stanford University, Stanford, CA) for kindly providing StB KO mice. Confocal images were taken at the Carl Zeiss Reference Center for Confocal Microscopy (LN-MCP, Institute of Pathophysiology, School of Medicine, Ljubljana, Slovenia). We thank Phillip I. Bird (Monash University, Australia) and Roman Jerala (National Institute of Chemistry, Slovenia) for the critical reading of the manuscript and Marko Kreft for advice in confocal microscopy.
This work was supported in part by Slovenian Research Agency Grants J3-0612 (to N. K. J.) and P-0140 (to B. T.) and Commissariat a l'Énergie Atomique et aux Énergies Alternatives France-Slovenia grant (to N. K. J.).
- TLR
- toll-like receptor
- BMDM
- bone marrow-derived macrophage
- mtROS
- mitochondrial reactive oxygen species
- StB
- stefin B-deficient
- SEAP
- secreted embryonic alkaline phosphatase
- STS
- staurosporin
- Z
- benzyloxycarbonyl
- AMC
- 7-amino-4-methylcoumarin hydrochloride
- LDH
- lactate dehydrogenase
- TRIF
- TIR domain-containing adapter-inducing IFN-β.
REFERENCES
- 1. Cohen J. (2002) The immunopathogenesis of sepsis. Nature 420, 885–891 [DOI] [PubMed] [Google Scholar]
- 2. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 3. Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 4. Martinon F., Tschopp J. (2007) Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 14, 10–22 [DOI] [PubMed] [Google Scholar]
- 5. Latz E., Xiao T. S., Stutz A. (2013) Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., Latz E. (2009) Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rathinam V. A., Vanaja S. K., Fitzgerald K. A. (2012) Regulation of inflammasome signaling. Nat. Immunol. 13, 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kahlenberg J. M., Dubyak G. R. (2004) Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+ release. Am. J. Physiol. Cell Physiol. 286, C1100–C1108 [DOI] [PubMed] [Google Scholar]
- 9. Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 12. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rathinam V. A., Vanaja S. K., Waggoner L., Sokolovska A., Becker C., Stuart L. M., Leong J. M., Fitzgerald K. A. (2012) TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell 150, 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broz P., Monack D. M. (2013) Noncanonical inflammasomes: caspase-11 activation and effector mechanisms. PLoS Pathog. 9, e1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W. P., Roose-Girma M., Dixit V. M. (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 [DOI] [PubMed] [Google Scholar]
- 16. Broz P., Ruby T., Belhocine K., Bouley D. M., Kayagaki N., Dixit V. M., Monack D. M. (2012) Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490, 288–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S., Miura M., Jung Yk, Zhu H., Gagliardini V., Shi L., Greenberg A. H., Yuan J. (1996) Identification and characterization of Ich-3, a member of the interleukin-1β converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J. Biol. Chem. 271, 20580–20587 [DOI] [PubMed] [Google Scholar]
- 18. Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bird P. I., Trapani J. A., Villadangos J. A. (2009) Endolysosomal proteases and their inhibitors in immunity. Nat. Rev. Immunol. 9, 871–882 [DOI] [PubMed] [Google Scholar]
- 20. Kopitar-Jerala N. (2006) The role of cystatins in cells of the immune system. FEBS Lett. 580, 6295–6301 [DOI] [PubMed] [Google Scholar]
- 21. Lenarcic B., Bevec T. (1998) Thyropins–new structurally related proteinase inhibitors. Biol. Chem. 379, 105–111 [PubMed] [Google Scholar]
- 22. Turk V., Stoka V., Turk D. (2008) Cystatins: biochemical and structural properties, and medical relevance. Front. Biosci. 13, 5406–5420 [DOI] [PubMed] [Google Scholar]
- 23. Turk B., Turk D., Salvesen G. S. (2002) Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Des. 8, 1623–1637 [DOI] [PubMed] [Google Scholar]
- 24. Ceru S., Konjar S., Maher K., Repnik U., Krizaj I., Bencina M., Renko M., Nepveu A., Zerovnik E., Turk B., Kopitar-Jerala N. (2010) Stefin B interacts with histones and cathepsin L in the nucleus. J. Biol. Chem. 285, 10078–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pennacchio L. A., Lehesjoki A. E., Stone N. E., Willour V. L., Virtaneva K., Miao J., D'Amato E., Ramirez L., Faham M., Koskiniemi M., Warrington J. A., Norio R., de la Chapelle A., Cox D. R., Myers R. M. (1996) Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1). Science 271, 1731–1734 [DOI] [PubMed] [Google Scholar]
- 26. Lalioti M. D., Mirotsou M., Buresi C., Peitsch M. C., Rossier C., Ouazzani R., Baldy-Moulinier M., Bottani A., Malafosse A., Antonarakis S. E. (1997) Identification of mutations in cystatin B, the gene responsible for the Unverricht-Lundborg type of progressive myoclonus epilepsy (EPM1). Am. J. Hum. Genet. 60, 342–351 [PMC free article] [PubMed] [Google Scholar]
- 27. Joensuu T., Lehesjoki A. E., Kopra O. (2008) Molecular background of EPM1-Unverricht-Lundborg disease. Epilepsia 49, 557–563 [DOI] [PubMed] [Google Scholar]
- 28. Pennacchio L. A., Bouley D. M., Higgins K. M., Scott M. P., Noebels J. L., Myers R. M. (1998) Progressive ataxia, myoclonic epilepsy, and cerebellar apoptosis in cystatin B-deficient mice. Nat. Genet. 20, 251–258 [DOI] [PubMed] [Google Scholar]
- 29. Houseweart M. K., Pennacchio L. A., Vilaythong A., Peters C., Noebels J. L., Myers R. M. (2003) Cathepsin B but not cathepsins L or S contributes to the pathogenesis of Unverricht-Lundborg progressive myoclonus epilepsy (EPM1). J. Neurobiol. 56, 315–327 [DOI] [PubMed] [Google Scholar]
- 30. Di Giaimo R., Riccio M., Santi S., Galeotti C., Ambrosetti D. C., Melli M. (2002) New insights into the molecular basis of progressive myoclonus epilepsy: a multiprotein complex with cystatin B. Hum. Mol. Genet. 11, 2941–2950 [DOI] [PubMed] [Google Scholar]
- 31. Lew W., Oppenheim J. J., Matsushima K. (1988) Analysis of the suppression of IL-1α and IL-1β production in human peripheral blood mononuclear adherent cells by a glucocorticoid hormone. J. Immunol. 140, 1895–1902 [PubMed] [Google Scholar]
- 32. Weischenfeldt J., Porse B. (2008) Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. 10.1101/pdb.prot5080 [DOI] [PubMed] [Google Scholar]
- 33. Kopitar-Jerala N., Schweiger A., Myers R. M., Turk V., Turk B. (2005) Sensitization of stefin B-deficient thymocytes towards staurosporin-induced apoptosis is independent of cysteine cathepsins. FEBS Lett. 579, 2149–2155 [DOI] [PubMed] [Google Scholar]
- 34. Zhang H., Zhong C., Shi L., Guo Y., Fan Z. (2009) Granulysin induces cathepsin B release from lysosomes of target tumor cells to attack mitochondria through processing of bid leading to necroptosis. J. Immunol. 182, 6993–7000 [DOI] [PubMed] [Google Scholar]
- 35. Nekrasov M., Amrichova J., Parker B. J., Soboleva T. A., Jack C., Williams R., Huttley G. A., Tremethick D. J. (2012) Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 19, 1076–1083 [DOI] [PubMed] [Google Scholar]
- 36. Popova E. Y., Xu X., DeWan A. T., Salzberg A. C., Berg A., Hoh J., Zhang S. S., Barnstable C. J. (2012) Stage- and gene-specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One 7, e46867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. špes A., Sobotic B., Turk V., Turk B. (2012) Cysteine cathepsins are not critical for TRAIL- and CD95-induced apoptosis in several human cancer cell lines. Biol. Chem. 393, 1417–1431 [DOI] [PubMed] [Google Scholar]
- 38. Morrison D. C., Ryan J. L. (1987) Endotoxins and disease mechanisms. Annu. Rev. Med. 38, 417–432 [DOI] [PubMed] [Google Scholar]
- 39. Puren A. J., Fantuzzi G., Dinarello C. A. (1999) Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1β are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc. Natl. Acad. Sci. U.S.A. 96, 2256–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schauvliege R., Vanrobaeys J., Schotte P., Beyaert R. (2002) Caspase-11 gene expression in response to lipopolysaccharide and interferon-gamma requires nuclear factor-κB and signal transducer and activator of transcription (STAT) 1. J. Biol. Chem. 277, 41624–41630 [DOI] [PubMed] [Google Scholar]
- 41. Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. (2004) A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14, 207–219 [DOI] [PubMed] [Google Scholar]
- 42. Duncan E. M., Muratore-Schroeder T. L., Cook R. G., Garcia B. A., Shabanowitz J., Hunt D. F., Allis C. D. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bulynko Y. A., Hsing L. C., Mason R. W., Tremethick D. J., Grigoryev S. A. (2006) Cathepsin L stabilizes the histone modification landscape on the Y chromosome and pericentromeric heterochromatin. Mol. Cell. Biol. 26, 4172–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Istomina N. E., Shushanov S. S., Springhetti E. M., Karpov V. L., Krasheninnikov I. A., Stevens K., Zaret K. S., Singh P. B., Grigoryev S. A. (2003) Insulation of the chicken β-globin chromosomal domain from a chromatin-condensing protein, MENT. Mol. Cell. Biol. 23, 6455–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shinkai Y., Tachibana M. (2011) H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fang T. C., Schaefer U., Mecklenbrauker I., Stienen A., Dewell S., Chen M. S., Rioja I., Parravicini V., Prinjha R. K., Chandwani R., MacDonald M. R., Lee K., Rice C. M., Tarakhovsky A. (2012) Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J. Exp. Med. 209, 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoegen T., Tremel N., Klein M., Angele B., Wagner H., Kirschning C., Pfister H. W., Fontana A., Hammerschmidt S., Koedel U. (2011) The NLRP3 inflammasome contributes to brain injury in pneumococcal meningitis and is activated through ATP-dependent lysosomal cathepsin B release. J. Immunol. 187, 5440–5451 [DOI] [PubMed] [Google Scholar]
- 48. Rinne R., Saukko P., Järvinen M., Lehesjoki A. E. (2002) Reduced cystatin B activity correlates with enhanced cathepsin activity in progressive myoclonus epilepsy. Ann. Med. 34, 380–385 [DOI] [PubMed] [Google Scholar]
- 49. Lopez-Castejon G., Theaker J., Pelegrin P., Clifton A. D., Braddock M., Surprenant A. (2010) P2X(7) receptor-mediated release of cathepsins from macrophages is a cytokine-independent mechanism potentially involved in joint diseases. J. Immunol. 185, 2611–2619 [DOI] [PubMed] [Google Scholar]
- 50. Bergsbaken T., Fink S. L., Cookson B. T. (2009) Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimada K., Crother T. R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V. K., Wolf A. J., Vergnes L., Ojcius D. M., Rentsendorj A., Vargas M., Guerrero C., Wang Y., Fitzgerald K. A., Underhill D. M., Town T., Arditi M. (2012) Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murakami T., Ockinger J., Yu J., Byles V., McColl A., Hofer A. M., Horng T. (2012) Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 109, 11282–11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luciano-Montalvo C., Meléndez L. M. (2009) Cystatin B associates with signal transducer and activator of transcription 1 in monocyte-derived and placental macrophages. Placenta 30, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rivera-Rivera L., Perez-Laspiur J., Colón K., Meléndez L. M. (2012) Inhibition of interferon response by cystatin B: implication in HIV replication of macrophage reservoirs. J. Neurovirol. 18, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maher K., Završnik J., Jerič-Kokelj B., Vasiljeva O., Turk B., Kopitar-Jerala N. (2014) Decreased IL-10 expression in stefin B-deficient macrophages is regulated by the MAP kinase and STAT-3 signaling pathways. FEBS Lett. 588, 720–726 [DOI] [PubMed] [Google Scholar]
- 56. Schroder K., Tschopp J. (2010) The inflammasomes. Cell 140, 821–832 [DOI] [PubMed] [Google Scholar]
- 57. Wang S., Miura M., Jung Y. K., Zhu H., Li E., Yuan J. (1998) Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92, 501–509 [DOI] [PubMed] [Google Scholar]
- 58. Jabaut J., Ather J. L., Taracanova A., Poynter M. E., Ckless K. (2013) Mitochondria-targeted drugs enhance Nlrp3 inflammasome-dependent IL-1β secretion in association with alterations in cellular redox and energy status. Free Radic. Biol. Med. 60, 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tschopp J. (2011) Mitochondria: Sovereign of inflammation? Eur. J. Immunol. 41, 1196–1202 [DOI] [PubMed] [Google Scholar]
- 60. Lehtinen M. K., Tegelberg S., Schipper H., Su H., Zukor H., Manninen O., Kopra O., Joensuu T., Hakala P., Bonni A., Lehesjoki A. E. (2009) Cystatin B deficiency sensitizes neurons to oxidative stress in progressive myoclonus epilepsy, EPM1. J. Neurosci. 29, 5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Butinar M., Prebanda M. T., Rajkovič J., Jerič B., Stoka V., Peters C., Reinheckel T., Krüger A., Turk V., Turk B., Vasiljeva O. (2014) Stefin B deficiency reduces tumor growth via sensitization of tumor cells to oxidative stress in a breast cancer model. Oncogene 33, 3392–3400 [DOI] [PubMed] [Google Scholar]
- 62. Ricci J. E., Muñoz-Pinedo C., Fitzgerald P., Bailly-Maitre B., Perkins G. A., Yadava N., Scheffler I. E., Ellisman M. H., Green D. R. (2004) Disruption of mitochondrial function during apoptosis is mediated by caspase cleavage of the p75 subunit of complex I of the electron transport chain. Cell 117, 773–786 [DOI] [PubMed] [Google Scholar]
- 63. Li N., Karin M. (1999) Is NF-κB the sensor of oxidative stress? FASEB J. 13, 1137–1143 [PubMed] [Google Scholar]
- 64. Schreck R., Rieber P., Baeuerle P. A. (1991) Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-κB transcription factor and HIV-1. EMBO J. 10, 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schreck R., Grassmann R., Fleckenstein B., Baeuerle P. A. (1992) Antioxidants selectively suppress activation of NF-κB by human T-cell leukemia virus type I Tax protein. J. Virol. 66, 6288–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tegelberg S., Kopra O., Joensuu T., Cooper J. D., Lehesjoki A. E. (2012) Early microglial activation precedes neuronal loss in the brain of the Cstb−/− mouse model of progressive myoclonus epilepsy, EPM1. J. Neuropathol. Exp. Neurol. 71, 40–53 [DOI] [PubMed] [Google Scholar]
- 67. Karthikeyan R. S., Leal S. M., Jr., Prajna N. V., Dharmalingam K., Geiser D. M., Pearlman E., Lalitha P. (2011) Expression of innate and adaptive immune mediators in human corneal tissue infected with Aspergillus or Fusarium. J. Infect. Dis. 204, 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu X. Y., Gao J. L., Ren M. Y. (2007) Expression profiles and function of Toll-like receptors in human corneal epithelia. Chin. Med. J. 120, 893–897 [PubMed] [Google Scholar]