Background: GILT is known to reduce disulfide bonds in endosomes, lysosomes, and phagosomes.
Results: GILT, in addition to reducing disulfide bonds, maintains phagosomal proteolytic activity, particularly in alternatively activated macrophages.
Conclusion: GILT maintains activity of cysteine proteases in phagosomes.
Significance: These results reveal a novel role for GILT that may affect antigen processing and efficiency of hydrolysis of phagocytosed protein.
Keywords: Cysteine Protease, Lysosome, Macrophage, Oxidation-Reduction (Redox), Proteolysis, Reductase, Phagosome
Abstract
Although it is known that lysosomal cysteine cathepsins require a reducing environment for optimal activity, it is not firmly established how these enzymes are maintained in their reduced-active state in the acidic and occasionally oxidative environment within phagosomes and lysosomes. γ-Interferon-inducible lysosomal thiol reductase (GILT) has been the only enzyme described in the endosomes, lysosomes, and phagosomes with the potential to catalyze the reduction of cysteine cathepsins. Our goal in the current study was to assess the effect of GILT on major phagosomal functions with an emphasis on proteolytic efficiency in murine bone marrow-derived macrophages. Assessment of phagosomal disulfide reduction upon internalization of IgG-opsonized experimental particles confirmed a major role for GILT in phagosomal disulfide reduction in both resting and interferon-γ-activated macrophages. Furthermore we observed a decrease in early phagosomal proteolytic efficiency in GILT-deficient macrophages, specifically in the absence of an NADPH oxidase-mediated respiratory burst. This deficiency was more prominent in IL-4-activated macrophages that inherently possess lower levels of NADPH oxidase activity. Finally, we provide evidence that GILT is required for optimal activity of the lysosomal cysteine protease, cathepsin S. In summary, our results suggest a role for GILT in maintaining cysteine cathepsin proteolytic efficiency in phagosomes, particularly in the absence of high NADPH oxidase activity, which is characteristic of alternatively activated macrophages.
Introduction
The phagosome is a versatile organelle that performs a variety of functions including the clearance of cellular debris, destruction of microorganisms and generation of antigenic peptides. These functions are mediated by the highly orchestrated fusion events that drive the maturation of nascent phagosomes to highly degradative, mature phagolysosomes. During this transition, the lumens of maturing phagosomes undergo a rapid transformation characterized by decreasing pH, changes in ion composition, the addition of various hydrolases and antimicrobial peptides, and often, dramatic changes to the redox microenvironment (1–4). Although the individual chemistries within the phagosome are for the most part well characterized, how these chemistries affect one another and alter overall phagosome function has recently garnered great attention. Microenvironmental conditions that inhibit the activity of lysosomal proteases within the phagosomal lumen have undergone recent scrutiny (5, 6). Of particular relevance to the current study, an oxidative microenvironment within the phagosome has been shown to inhibit lysosomal cysteine cathepsins within the phagosome of macrophages and dendritic cells (7, 8). Because the proteolytic activity of cysteine cathepsins relies on the active site cysteine (Cys-25, papain numbering) being in a thiol (reduced) state, the oxidation of this residue renders the enzyme inactive. This is particularly evident during a respiratory burst when the reactive oxygen species (ROS)2 produced by the phagosomal NADPH oxidase (NOX2) complex lead to a dramatic decrease in phagosome-specific proteolytic efficiency. Because NOX2 activity is induced by the exposure of macrophages to proinflammatory stimuli such as interferon γ (IFNγ) and lipopolysaccharide, the effect of NOX2 on phagosomal proteolysis is more pronounced in classically activated macrophages (M1). In contrast, alternatively activated macrophages (M2) show little inhibition of cysteine cathepsins after phagocytosis due to a down-regulation of NOX2 in these cells. The lowered level of oxidative inactivation of cysteine cathepsins combined with induced expression of several lysosomal proteases enables alternatively activated macrophages to become extremely proficient at hydrolyzing phagocytosed protein (9).
Not only does the regulation of the lysosomal cysteine cathepsins (including cathepsin B, L, Z, and S) influence the efficiency of protein hydrolysis within the phagosome, it is also critical to the efficient antigen and invariant chain processing necessary for antigenic presentation (10, 11). Unsurprisingly, the activity of these enzymes is tightly regulated at multiple levels, including expression, trafficking, zymogen processing, and pH (12). However, although it has been recently shown that the oxidative inactivation of these enzymes during a respiratory burst additionally influences their phagosomal activity, how these enzymes are returned and maintained in their reduced, active state is poorly defined (13). In other cellular compartments, reductive machinery consisting of small molecule reductants (such as glutathione and cysteine) and enzymes (such as thioredoxin and glutaredoxin) can maintain or restore the activity of redox-sensitive proteins by reducing thiols in cysteine residues that have been oxidized (14). For example, thioredoxin, the prototypical cellular thiol reductase, is involved in maintaining the activity of certain peroxidases that are oxidized after reaction with hydrogen peroxide (15). Whether or not reductive machinery present in the endo/lysosomal system can maintain cysteine cathepsin activity in a mechanistically similar fashion is currently unknown. In reconstituted systems cathepsin B activity can be maintained with glutathione as well as thioredoxin (16, 17) and papain by NADPH through the thioredoxin reductase/thioredoxin system (18). However, glutathione and thioredoxin have not been shown to be active in the lysosome. To date, γ-interferon-inducible lysosomal thiol reductase (GILT) has been the only reductase described within endosomes, lysosomes, and phagosomes with the potential to maintain cysteine cathepsins in their reduced and active states (19).
Our objective in the current study was to evaluate the role of GILT on major phagosomal chemistries in macrophages under various activation states, with an emphasis on proteolysis. Using fluorometric-based assays that evaluate phagosomal parameters in live macrophages in real time, we observed a role for GILT in maintaining early phagosomal proteolytic efficiency, particularly in the absence of NOX2 activity. This role was most apparent in alternatively activated macrophages, which have decreased phagosomal NOX2 activity and increased cysteine cathepsin expression.
EXPERIMENTAL PROCEDURES
Cells and Animals
C57BL/6 (wild type (WT)) and the congenic mouse strain B6.129S6-Cybb−/− (Cybb−/−) were purchased from The Jackson Laboratory (20). C57BL/6 GILT−/− (GILT−/−) mice were provided by Dr. Peter Cresswell (Yale University School of Medicine, New Haven, CT). C57BL/6 CtsS−/− (CatS−/−) mice were provided by Dr. Kenneth Rock (University of Massachusetts Medical School, Worcester, MA). C57BL/6 GILT−/−/CatS−/− and GILT−/−/Cybb−/− double knock-out mice were generated at the University of Calgary using the GILT−/−, CatS−/−, and Cybb−/− mice as a part of this study. All animal experiments were conducted according to protocols approved by the University of Calgary Animal Care and Use Committee. Bone marrow-derived macrophages (BMMØs) were derived from 8–12-week-old mice using L929-conditioned medium as previously described (21). Alternative activation was achieved by treating BMMØs with 10 ng/ml recombinant murine interleukin-4 (IL-4; Peprotech) for 40 h before phagosomal assessment (9). IFNγ activation was achieved by treating BMMØs with 100 units/ml recombinant murine IFNγ (Peprotech) for 18 h before phagosomal assessment. Pharmacologic inhibition of NOX2 was achieved with treatment of BMMØs with 0.5 μm diphenyleneiodonium (DPI; EMD Chemicals) for 10 min preceding phagocytosis of experimental particles (7). For fluorometric phagosomal analysis, 1.5 × 105 BMMØs were plated in every well of a μ-clear 96-well plate (Greiner Bio-One) to achieve confluency.
Analysis of Phagosomal Hydrolytic Activities
3-μm silica experimental particles bearing various fluorescent substrates/reporters that allow for phagosomal lumenal characterization in a population of live BMMØs were prepared as previously described (21–23). Briefly, the hydrolytic activities of phagosomal β-galactosidase, bulk protease, cathepsin B/S/L, and cathepsin D/E were measured by recording the rates of substrate-liberated fluorescence relative to a calibration fluor (Alexa Fluor 594 succinimidyl ester (SE); Invitrogen) using the particle-bound fluorogenic substrates 5-dodecanoylaminofluorescein di-β-d-galactopyranoside (Invitrogen), DQ Green BODIPY albumin (DQ-albumin; Invitrogen), (biotin-LC-Phe-Arg)2-rhodamine 110 (kindly provided by David Russell, Cornell University, Ithaca, NY), and methoxycoumarin-GKPILFFRLK(dinitrophenol)-r-NH2 (Anaspec), respectively. Reduced DQ-albumin was prepared by incubating experimental particles in 1 mm dithiothreitol followed by alkylation with 2 mm iodoacetic acid as previously described (24). β-Galactosidase activity was measured using a Fluostar OPTIMA microplate reader (BMG Labtech). Bulk proteolysis was measured using a Flex Station microplate reader (Molecular Devices). Cathepsin B/S/L and cathepsin D/E activity was monitored using an Envision multilabel reader (PerkinElmer Life Sciences). Measurements were taken every 120 s. All assays were performed at 37 °C in assay buffer (tissue culture grade PBS supplemented with 1 mm CaCl2, 2.7 mm KCl, 0.5 mm MgCl2, 5 mm dextrose, and 0.25% gelatin).
Analysis of Phagosomal Acidification and Lysosomal Fusion
Phagosomal pH was calculated by recording the ratio of the fluorescence emissions at 520 nm of particle-bound carboxyfluorescein SE (Invitrogen) excited at 490 and 450 nm followed by polynomial regression to a standard curve using a modified nigericin-based technique as previously described (8). The rate and extent of phagosome-lysosome fusion (PL fusion) were monitored by measuring the fluorescence resonance energy transfer (FRET) efficiency between the donor fluor Alexa Fluor 488 SE (Invitrogen) conjugated to albumin-coated experimental particles and the fluid-phase acceptor fluor Alexa Fluor 594 hydrazide (Invitrogen) in lysosomes as previously described (22). Loading of the lysosomes with acceptor fluor was achieved by incubating macrophages in BMMØ medium containing 50 μg/ml Alexa Fluor 594 hydrazide for 18 h followed by a chase period of 4 h before analysis. Phagosomal pH and PL fusion were monitored using a Fluostar OPTIMA microplate reader. Measurements were taken every 120 s.
Analysis of Phagosomal Redox Chemistries
Intraphagosomal reductase activity was evaluated by measuring the dequenching of particle-bound BODIPY FL l-cystine (Invitrogen) relative to the Alexa Fluor 594 SE calibration fluor using a Fluostar OPTIMA microplate reader. Measurements were taken every 120 s. BODIPY FL l-cystine particle conjugation was achieved by coupling the free carboxyl group present within cystine to amine-modified dextran (Invitrogen)-coated 3-μm silica particles. Briefly, 0.1 mg of BODIPY FL l-cystine was reacted with 3.2 mg of 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride and 6.4 mg of N-hydroxysulfosuccinimide in a buffer containing 100 mm MES and 500 mm NaCl, pH 6 (25, 26). The amine-reactive BODIPY FL l-cystine-(NHS ester)2 product was directly coupled to amine-modified 70,000 Mr dextran (Invitrogen)-coated silica particles in MES buffer (total reaction volume of 0.2 ml) for 2 h at room temperature. Beads were washed and resuspended in 0.1 m borate buffer, pH 8. Biotin SE (Invitrogen) was then conjugated to the beads through the free amines present on the modified dextran. Particles were washed in PBS then opsonized with Alexa Fluor 594 SE-labeled anti-biotin IgG before use. The generation of reactive oxygen species by NOX2 within the phagosomal lumen was evaluated by measuring the fluorescence after oxidation of the experimental particle-restricted H2HFF-OxyBURST substrate (Invitrogen) as previously described using an Envision multilabel reader (23). Measurements were taken every 120 s. For H2O2 production, plated BMMØs were washed then incubated for 1 h in assay buffer containing 10 mg/ml zymosan (Sigma). Amplex UltraRed (Invitrogen) at a final concentration of 10 ng/ml plus 1 unit of horseradish peroxidase (Sigma) were added to each well supernatant and incubated for 15 min. Amplex UltraRed fluorescence was monitored using a GENios Pro microplate reader (Tecan Group Ltd.).
Statistical Analysis for Phagosomal Fluorescence Kinetic Measurements
Relative rates of phagosomal activity were determined by calculation of the slope between the times indicated of the real-time trace (as described by y = mx + c, where y = relative fluorescence, m = slope, and x = time). Error bars denote S.E. p values were determined by one-way analysis of variance with the Tukey post hoc test or paired t test where indicated using GraphPad Prism software.
Real-time Polymerase Chain Reaction (QPCR)
To measure the GILT mRNA levels present in BMMØs, RNA was extracted from BMMØs plated for 18 h with or without IFNγ or for 48 h with or without IL-4 according to the instructions outlined in the Aurum Total mRNA Mini kit (Bio-Rad). cDNA was made from 0.4 μg of RNA using iScript Reverse Transcriptase Supermix for RT-QPCR (Bio-Rad). All primers for QPCR were at 300 nm, had a single melting curve, had efficiencies between 90 and 100%, and were designed or verified using Primer 3 (NCBI). QPCR was conducted using the following primers: 18 S (Fwd, 5′-AGTCGGCATCGTTTATGGTC-3′; Rev, 5′-CGCGGTTCTATTTTGTTGGT-3′); GILT (Fwd-5′-GCTTGTCGCTACTTCCTCGT-3′; Rev (5′-ATGGTTAGGAACGCTGCCTC-3′). 18 S was used as an internal control and did not vary across treatments. All reactions were used under the following PCR conditions (in a Bio-Rad iQ5 thermocycler): 95°C for 5 min, 40 cycles of 95 °C for 30 s, and 58 °C for 30 s and ran using the iQ SYBER Green Supermix (Bio-Rad) protocol. mRNA levels are presented relative to 18 S expression and made relative to unactivated BMMØs. Experimental groups were compared by paired t test using GraphPad Prism software.
SDS-Polyacrylamide Gel Electrophoresis and Western Blotting
Analysis of cathepsin expression in whole cell lysates derived from unactivated or IL-4-activated BMMØs was performed by Western blotting with the following antibodies: cathepsin B (Biovision), cathepsin S, cathepsin D, (Santa Cruz Biotechnology), cathepsin L (R&D Systems), and GAPDH (Cell Signaling Technology) as previously described (9). Volume of pixels was determined using Quantity One 1-D analysis software (Bio-Rad). Each calculated pixel volume was normalized to the calculated pixel volume of GAPDH expression from the same sample. Relative band densities of pro and mature forms were determined by calculation of normalized pixel volume relative to WT controls. The pro-form of cathepsin B was not detectable. Experimental groups were compared by paired t test where appropriate using GraphPad Prism software. Images of bands in the unsaturated range of exposure were used to quantify pixel volume. Representative images presented in the figures may not be at the exposure used for data quantification.
Reconstituted Cathepsin S Activity Assays
The cathepsin S substrate Ac-KQKLR-AMC (Anaspec) was diluted in assay buffer (20 mm sodium acetate, pH 5.5, containing 0.675 mm KCl, 0.25 mm CaCl2, 0.125 mm MgCl2) supplemented with 500 μm l-cysteine:cystine (600:1 molar ratio) (12). 0.5 μg of cathepsin S purified from human spleen (Sigma) and 0.625 μg of human recombinant GILT (rGILT) (Acris Antibodies Inc.) were each diluted in assay buffer then sequentially added to each well in a total reaction volume of 50 μl. Heat inactivation of rGILT was performed at 90 °C for 10 min before the addition to cathepsin S. Experiments were performed in ½ area μ-clear 96-well plates (Greiner), and cleavage of the cathepsin S substrate was monitored using a Fluostar OPTIMA microplate reader.
RESULTS
Assessment of Phagosomal Redox Functions in GILT−/− BMMØs
The reduction of disulfide bonds plays a critical role in the processing of endocytosed/phagocytosed antigens (27). To date, GILT has been the only thiol reductase described in endosomes, lysosomes, and phagosomes (19); however, its contribution to overall reductive activity has not been directly assessed in live cells in real time. We, therefore, evaluated phagosomal relative rates of disulfide reduction between 40 and 60 min in GILT−/− BMMØs using experimental particles bearing the self-quenched BODIPY FL l-cystine substrate in unactivated and IFNγ-activated BMMØs. Although GILT is constitutively active in antigen-presenting cells, we measured GILT mRNA levels in IFNγ-activated cells. In IFNγ-activated WT BMMØs we observed a modest but significant increase in GILT mRNA levels (1.31 ± 0.07-fold increase; Fig. 1A). After phagocytosis of the experimental particle, disulfide reduction was monitored by measuring fluorescence dequenching of the BODIPY FL l-cystine substrate relative to the calibration fluor. Because the generation of ROS by NOX2 has been shown to significantly decrease rates of disulfide reduction in phagosomes, these assays were performed in the presence or absence of the respiratory burst using the NOX2 inhibitor DPI. As expected, we found that rates between 40 and 60 min of phagosomal disulfide reduction were significantly lower in GILT−/− BMMØs compared with WT controls in both untreated and DPI-treated cells (54.4 ± 7.7% and 51.0 ± 7.1% decrease respectively; Fig. 1, B and D). Although overall rates of phagosomal disulfide reduction were decreased after BMMØ activation with IFNγ, the relative differences between WT and GILT−/− BMMØs were more notable (68.1 ± 9.7% and 62.7 ± 5.7% decrease for untreated and DPI-treated samples respectively; Fig. 1, C and D), possibly indicating a greater role for GILT in reducing phagosomal disulfides after classical activation. The observed negative values seen in the real-time traces may represent photobleaching of the substrate. This was most apparent in experimental groups, which exhibit low disulfide reductase activity. Because the absence or inhibition of certain cellular reductive systems are known to affect the ROS scavenging and antioxidant functions in other cellular compartments (28–30), we set out to determine whether GILT impacted the NOX2 respiratory burst by measuring the rates of oxidation of cargo within the phagosome and hydrogen peroxide (H2O2) liberated from BMMØs after phagocytosis. No significant differences in the oxidation rates of particle-restricted OxyBURST Green H2HFF BSA were observed within the phagosomes of WT and GILT−/− BMMØs in the presence or absence of IFNγ (Fig. 1, E and F). Furthermore, no significant differences in extracellular H2O2 levels between WT and GILT−/− BMMØs (with or without prior IFNγ exposure) were noted after phagocytosis of serum-opsonized zymosan (as measured by Amplex UltraRed; Fig. 1G). Together these results confirm a major role for GILT in phagosomal disulfide reduction. However, GILT does not appear to impact potential antioxidant functions in BMMØ phagosomes at a level that would impact the effects of the phagosomal respiratory burst.
FIGURE 1.
Phagosomal reductive and oxidative processes in GILT−/− BMMØs. BMMØs derived from WT and GILT−/− mice were incubated for 18 h in the presence/absence of IFNγ. A, total mRNA levels of GILT in unactivated and IFNγ-activated BMMØs were determined by QPCR. Averaged relative mRNA levels from four independent QPCR experiments are shown. Relative expression was expressed as mRNA levels relative to 18 S and presented relative to unactivated BMMØs. Error bars denote S.E. *, p < 0.05. 10 min before addition and subsequent phagocytosis of experimental particles, BMMØs were treated with the NOX2 inhibitor DPI (0.5 μm) where indicated. B, C, and D, phagosomal disulfide reduction of the BODIPY FL l-cystine substrate conjugated to IgG-opsonized dextran-coated experimental particles. E and F, phagosomal oxidation of the OxyBURST Green H2HFF BSA substrate conjugated to IgG-opsonized experimental particles. G, extracellular H2O2 production relative to unactivated/untreated WT control after phagocytosis of serum opsonized zymosan particles as measured by oxidation of the Amplex UltraRed reagent. B, C, and E, representative real-time traces. Relative fluorescence units (RFU) values are proportional to the degree of substrate reduction/oxidation. D and F, averaged rates relative to unactivated/untreated WT controls of four independent experiments are shown for each phagosomal measurement. Average relative rates of disulfide reduction were calculated between 40 and 60 min after particle internalization. Relatives rate of OxyBURST Green H2HFF BSA oxidation were calculated between 30 and 80 min after particle internalization. A, D, F, and G, error bars denote S.E. of four independent experiments. *, p < 0.05.
GILT−/− Macrophages Display Lower Levels of Phagosomal Proteolysis during NOX2 Inhibition
It is well established that cysteine cathepsins require a reducing environment for optimal activity (31). Due to the acidic environment of lysosomes and its effect on the pKa of thiols, it is likely that the reductive maintenance of thiols within lysosomes would be facilitated by an enzyme such as GILT with a low pH optimum as opposed to small molecule reductants such as cysteine or glutathione alone (12). We, therefore, evaluated bulk phagosomal proteolysis in GILT−/− BMMØs to test the hypothesis that the reductive activity of GILT could sustain activity. Bulk phagosomal proteolysis was measured in both unactivated and IFNγ-activated BMMØs using IgG-opsonized experimental particles bearing the proteolytic substrate DQ-albumin. Because cysteine cathepsin activities have been shown to be overwhelmingly inhibited in the phagosome during the oxidative burst, we evaluated the effect of GILT on phagosomal proteolysis in the presence and absence of the NOX2 inhibitor DPI (7). In the absence of NOX2 activity, we found a modest but significant decrease (27.6 ± 4.0% decrease) in the rates of bulk phagosomal proteolysis between 40 and 60 min between GILT−/− and WT unactivated BMMØs, whereas no statistical significance was observed in the absence of NOX2 inhibition or in IFNγ-activated BMMØs (Fig. 2, A–C). Similar results were obtained using non-opsonized mannosylated particles targeted for phagocytic uptake through the mannose receptor (Fig. 2, D–F). To rule out the potential effect of differential cathepsin expression in GILT−/− BMMØs, we evaluated the expression of a selection of major phagosomal proteases. Semiquantitative Western blotting revealed no significant differences in the expression levels of cathepsin B (mature form), S, L (pro/mature forms), and cathepsin D (pro/mature form) between unactivated WT and GILT−/− BMMØs, indicating that the lower phagosomal proteolysis in GILT−/− BMMØs was not due to decreased cathepsin expression (Fig. 2G). Because GILT did not significantly impact proteolytic efficiency of phagosomes in the absence of NOX2 inhibition and given that GILT is also likely to be susceptible to oxidative inactivation, we reasoned that phagosomal ROS may be sufficient to prevent the activity of GILT. Thus, in this scenario the effect of GILT on phagosomal proteolytic efficiency would be greatest in macrophage states with low NOX2 activity.
FIGURE 2.
GILT−/− BMMØs display lower rates of phagosomal proteolysis in the absence of NOX2 activity. BMMØs derived from WT and GILT−/− mice were incubated for 18 h in the presence/absence of IFNγ. 10 min before addition and subsequent phagocytosis of experimental particles, BMMØs were treated with the NOX2 inhibitor DPI (0.5 μm) where indicated. A, B, and C, bulk phagosomal proteolysis as measured by hydrolysis of DQ-albumin conjugated to IgG-opsonized experimental particles in unactivated/IFNγ-activated BMMØs in the presence/absence of DPI. D, E, and F, bulk phagosomal proteolysis as measured by hydrolysis of DQ-albumin conjugated to mannosylated experimental particles in unactivated/IFNγ-activated BMMØs in the presence/absence of DPI. A, B, D, and E, representative real-time traces. Relative fluorescence units (RFU) values are proportional to the degree of substrate hydrolysis. C and F, averaged rates relative to unactivated/untreated WT controls from four independent experiments. Relative rates were calculated between 40 and 60 min after particle internalization. G, relative abundance of the lysosomal proteases in whole cell lysates of WT and GILT−/− BMMØs as detected by semiquantitative Western blotting. Mature forms of cathepsins B, S, L, and D and the pro-forms for cathepsins S, L, and D are shown. Representative images and average of band relative density from 4–6 independent experiments are shown. Each calculated pixel volume was normalized to the calculated pixel volume of GAPDH expression from the same sample. Relative density was determined by calculation of normalized pixel volume relative to WT sample using Quantity One analysis software. ns, not significant. Error bars denote S.E. *, p < 0.05. H, phagosomal oxidation of the OxyBURST Green H2HFF BSA substrate conjugated to IgG-opsonized experimental particles in unactivated, IL-4-activated (40 h), and IFNγ-activated (40 h) WT BMMØs. Averaged rates relative to unactivated control are shown. I, relative extracellular H2O2 production in unactivated, IL-4-activated (40 h), and IFNγ-activated (40 h) WT BMMØs after phagocytosis of serum-opsonized zymosan particles as measured by oxidation of the Amplex UltraRed reagent. Error bars denote S.E. *, p < 0.05.
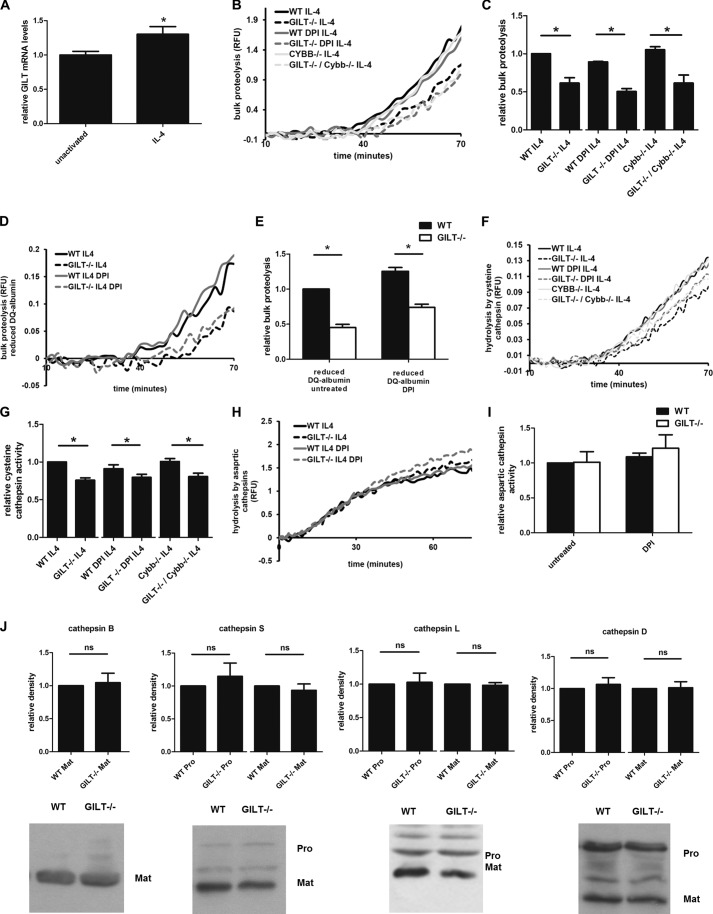
The Decrease in Phagosomal Proteolysis Observed in GILT−/− Macrophages Is More Pronounced in an Alternatively Activated State
The production of ROS, even at low levels, may promote an oxidizing environment that could limit the ability of GILT to maintain proteolytic activity. As IL-4 has been shown to decrease NOX2 activity in macrophages (Fig. 2, H and I) (9) as well as increase GILT expression (32), we hypothesized that the role of GILT in maintaining cysteine cathepsin activities would be greater in alternatively activated macrophages. Furthermore, IL-4 specifically increases the expression of cysteine cathepsins S and L, both of which could be potential targets of GILT (9, 33). Because the mannose receptor is highly expressed in alternatively activated macrophages (34) and to avoid Fc receptor-enhanced NOX2 assembly (35), we used mannosylated experimental particles in these experiments. QPCR analysis of GILT mRNA in IL-4-activated wild type BMMØs revealed a modest but significant increase (1.31 ± 0.10-fold increase; Fig. 3A). In IL-4-activated BMMØs, proteolytic rates measured between 40 and 60 min were significantly lower in GILT−/− BMMØs in the presence and absence of NOX2 inhibition by DPI (decreases of 38.5 ± 7.1% and 43.5 ± 4.4%, respectively; Fig. 3, B and C). Although DPI is a commonly used inhibitor of NOX2 activity (36, 37), DPI has been shown to inhibit flavoenzymes other than NOX2, such as those involved in mitochondrial ROS production (38, 39). To omit any off-target effects of DPI that may affect phagosomal function, we generated a GILT−/−/Cybb−/− double knock-out mouse to further observe the effect of GILT on phagosomal proteolysis in BMMØs without the gp91phox subunit of NOX2. As anticipated, the decrease in bulk phagosomal proteolysis in GILT−/−/Cybb−/− BMMØs compared with Cybb−/− BMMØs was similar to the decrease seen in DPI-treated samples (42.4 ± 8.5%; Fig. 3, B and C). Interestingly, the differences in proteolytic rates at later time points were not significant, suggesting a role for GILT in supporting earlier phagosomal proteolytic events.
FIGURE 3.
GILT−/− BMMØs display significantly lower rates of phagosomal proteolysis after IL-4-activation. BMMØs derived from WT, GILT−/−, Cybb−/−, and GILT−/−/Cybb−/− mice were incubated for 40 h in the presence 10 ng/ml IL-4. 10 min before the addition and subsequent phagocytosis of experimental particles, BMMØs were treated with the NOX2 inhibitor DPI (0.5 μm) where indicated. A, total mRNA levels of GILT in unactivated and IL-4-activated BMMØs were determined by QPCR. Averaged relative mRNA levels from four independent QPCR experiments are shown. Relative expression was expressed as mRNA levels relative to 18 S and presented relative to unactivated BMMØs. Error bars denote S.E. *, p < 0.05. RFU, relative fluorescence units. B and C, bulk proteolysis as measured by hydrolysis of the DQ-albumin substrate conjugated to experimental particles in IL-4-activated BMMØs (four independent experiments). D and E, bulk proteolysis as measured by hydrolysis of the prereduced and alkylated DQ-albumin substrate (four independent experiments). Reduced DQ-albumin was prepared by incubating experimental particles in 1 mm dithiothreitol followed by alkylation with 2 mm iodoacetic acid. F and G, phagosomal hydrolysis of the cysteine cathepsin-specific substrate (biotin-LC-Phe-Arg)2-rhodamine 110 conjugated to experimental particles in IL-4-activated BMMØs (seven independent experiments). H, and I, phagosomal hydrolysis of the aspartic cathepsin specific substrate in IL-4-activated BMMØs (four independent experiments). B, D, F, and H, representative real-time traces. Relative fluorescence values are proportional to the degree of substrate hydrolysis. C, E, G, and I, averaged rates relative to unactivated/untreated WT controls. Relative rates were calculated between 40–60 min after particle internalization. Error bars denote S.E. *, p < 0.05 as determined by paired t test of the indicated experimental groups. J, relative abundance of the lysosomal proteases in whole cell lysates of WT and GILT−/− IL-4-activated BMMØs as detected by semi-quantitative Western blotting. Mature (Mat) forms of cathepsins B, S, L, and D and the pro-forms (Pro) for cathepsins S, L, and D are shown. Representative images and averages of band relative density from four (cathepsins B and D), six (cathepsin S), and seven (cathepsin L) independent experiments are shown. Each calculated pixel volume was normalized to the calculated pixel volume of GAPDH expression from the same sample. Relative density was determined by calculation of normalized pixel volume relative to WT sample using Quantity One analysis software. Error bars denote S.E. ns, not significant.
In the early phagosome, GILT may directly support the proteolytic machinery or, conversely, may indirectly support early phagosomal proteolysis through the reduction of intramolecular disulfides of phagocytosed protein facilitating its denaturation. To examine whether or not the observed increase in phagosomal proteolysis in the presence of GILT was a result of GILT-mediated reduction of disulfides within the DQ-albumin substrate itself, we evaluated proteolysis of a pre-reduced/alkylated DQ-albumin substrate in WT and GILT−/− phagosomes. The decrease in phagosomal bulk proteolysis of the reduced DQ-albumin substrate was maintained in untreated and DPI-treated GILT−/− BMMØs relative to wild type controls (46.9 ± 3.7% and 41.0 ± 3.7%, respectively; Fig. 3, D and E). These data support a role for GILT in directly maintaining activities of proteolytic enzymes within the phagosome.
As DQ-albumin measures total proteolysis and thus the combined activity of phagosomal proteases other than cysteine cathepsins (9, 21) and to further rule out the enhancement of proteolytic activity through GILT-mediated reduction of disulfide bonds within the DQ-albumin substrate, we evaluated the efficiency of proteolysis of the cysteine cathepsin-specific substrate (biotin-LC-Phe-Arg)2-rhodamine 110 (cathepsin B > S > L) in IL-4-activated BMMØs. Indeed, cysteine cathepsin-specific activity was significantly decreased in IL-4-activated GILT−/− BMMØs in the absence and presence of DPI compared with wild type controls (24.3. ± 3.1% and 12.4 ± 2.3%, respectively; Fig. 3, F and G). In addition, a significant decrease in the cysteine cathepsin-specific substrate hydrolysis was observed in GILT−/−/Cybb−/− double knock-out BMMØs compared with the Cybb−/− single knock-out control (19.7 ± 3.3%; Fig. 3, F and G). No significant differences in aspartic protease activity between WT and GILT−/− BMMØs were observed in the presence or absence of DPI (Fig. 3, H and I). We also evaluated any differences in major protease expression between IL-4-activated WT and GILT−/− BMMØs. No significant differences in the expression of cathepsins B, S, L, and D were noted (Fig. 3J).
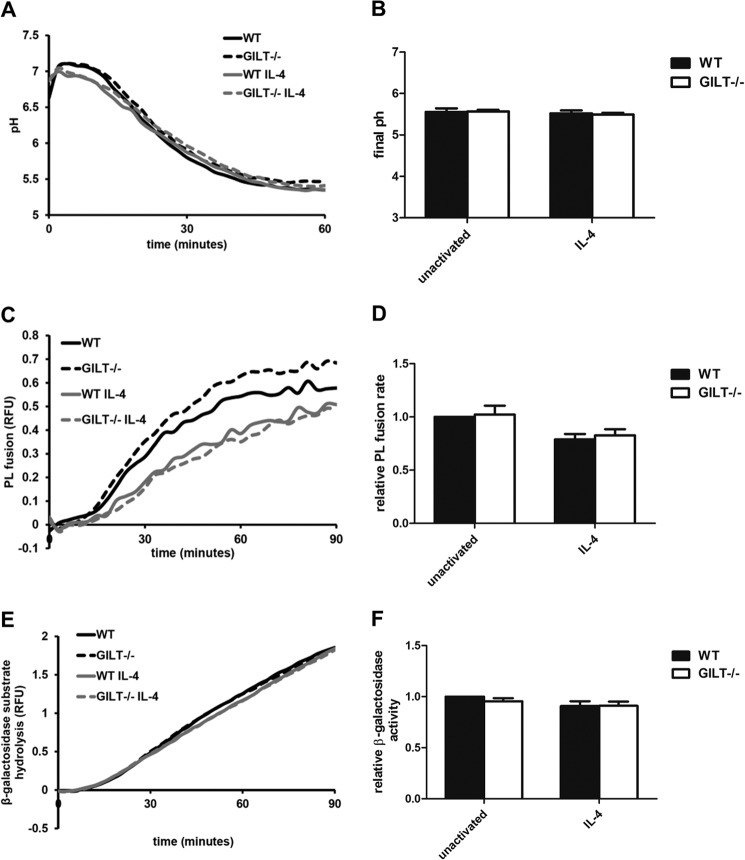
GILT−/− Macrophages Display Normal Rates of PL Fusion and Phagosomal Acidification
To determine whether the differences in disulfide reduction and bulk proteolysis were due to any changes in membrane trafficking events during phagosomal maturation, we monitored phagosomal acidification and PL fusion of engulfed mannosylated particles in WT and GILT−/− BMMØs with/without IL-4 activation. No significant differences were observed in either phagosomal acidification or the delivery of lysosomal content (Fig. 4, A–D). Furthermore, because delivery of β-galactosidase (a specific lysosomal enzyme) to the phagosome is known to correlate with the degree of PL fusion, we measured β-galactosidase activity in unactivated and IL-4-activated macrophages. Similarly, no significant differences were found in β-galactosidase substrate hydrolysis in any of the experimental groups (Fig. 4, E and F). These results indicate that the defects in disulfide reduction and decreased proteolytic activity observed in phagosomes of GILT−/− BMMØs are not a result of altered acidification or membrane trafficking during phagosomal maturation and thus are likely due to direct enzymatic activity of GILT within the phagosome.
FIGURE 4.
Phagosomal maturation is not altered in the absence of GILT. BMMØs derived from WT and GILT−/− mice were incubated for 40 h in the presence/absence of 10 ng/ml IL-4. A, phagosomal pH calculated from excitation ratios of the pH-sensitive carboxyfluorescein-SE conjugated to experimental particles by regression to a standard curve. B, average final pH as measured 60 min post experimental particle internalization over 3 independent experiments. C and D, PL fusion as measured by FRET between phagocytosed Alexa Fluor 488-conjugated experimental particles and Alexa Fluor 594 hydrazide-pulsed lysosomes. E and F, phagosomal hydrolysis of a fluorogenic β-galactosidase substrate relative to a calibration fluor. A, C, and E, representative real-time traces. Relative fluorescence units (RFU) values are proportional to the degree of phagosome/lysosome fusion/β-galactosidase substrate hydrolysis. D and F, averaged rates relative to unactivated/untreated WT controls from three independent experiments. Relative rates were calculated between 40 and 60 min after particle internalization. Error bars denote S.E.
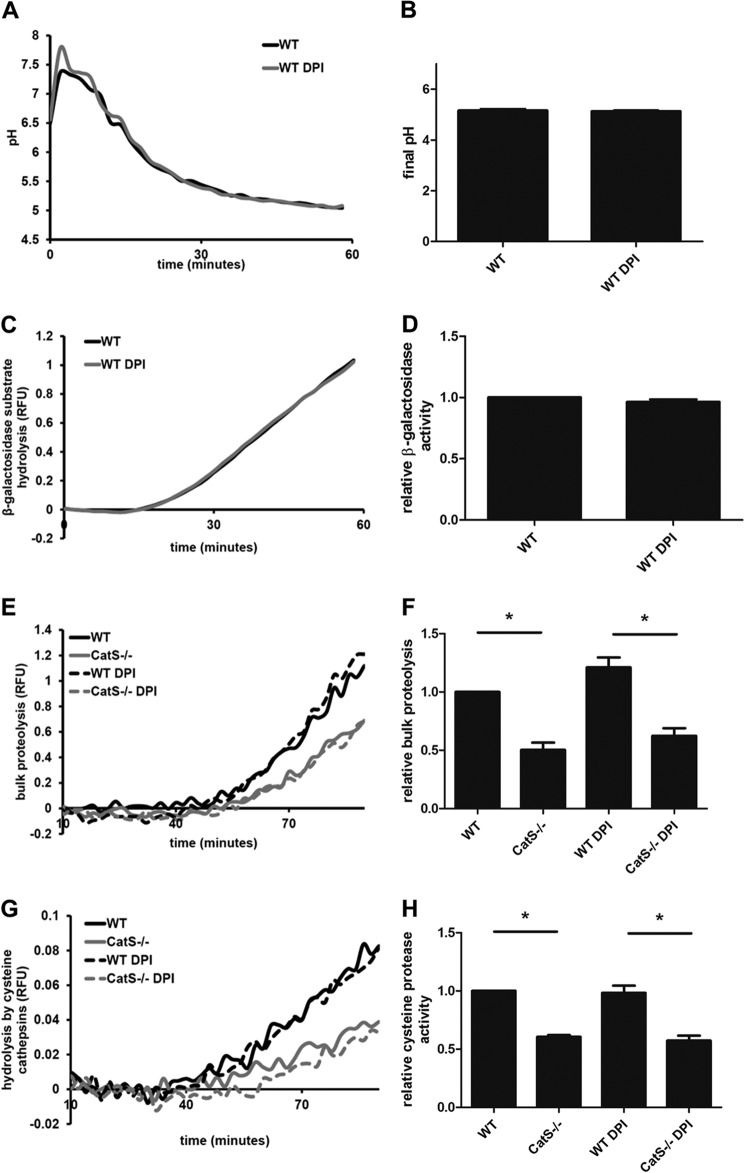
Validation of the Use of DPI and Proteolytic Substrates in IL-4-activated BMMØs
In IL-4-activated BMMØs, the largest difference in DQ-albumin proteolytic efficiency between GILT and WT BMMØs was observed in the presence of DPI. To ensure that DPI was not non-specifically altering general phagosomal function, we measured both phagosomal acidification and β-galactosidase activity in IL-4-activated wild type BMMØs in the presence of DPI. No significant differences in phagosomal acidification or β-galactosidase activity were observed in the presence of DPI (Fig. 5, A–D). Next, to ensure that the proteolytic substrates used in this study were suitable to assess phagosome-specific cathepsin-mediated proteolysis in IL-4-activated BMMØs, we evaluated DQ-albumin and (biotin-LC-Phe-Arg)2-rhodamine 110 hydrolysis in cathepsin S-deficient (CtsS−/−) BMMØs. In the presence and absence of DPI, we observed significant decreases in DQ-albumin (49.7 ± 6.3%; DPI, 48.8 ± 2.0%) and (biotin-LC-Phe-Arg)2-rhodamine 110 (39.4 ± 1.4%; DPI, 41.5 ± 4.1%) hydrolysis in CtsS−/− BMMØs compared with wild type controls (Fig. 5, E and F).
FIGURE 5.
Assessing the use of DPI and proteolytic substrates in IL-4-activated BMMØs. BMMØs derived from WT mice were incubated for 40 h in the presence/absence of 10 ng/ml IL-4. 10 min before the addition and subsequent phagocytosis of experimental particles, BMMØs were treated with the NOX2 inhibitor DPI (0.5 μm). A, phagosomal pH calculated from excitation ratios of the pH-sensitive carboxyfluorescein-SE conjugated to mannosylated experimental particles by regression to a standard curve. B, average final pH as measured 60 min post experimental particle internalization over four independent experiments. C and D, phagosomal hydrolysis of a fluorogenic β-galactosidase substrate relative to a calibration fluor. RFU, relative fluorescence units. E and F, bulk proteolysis as measured by hydrolysis of the DQ-albumin substrate conjugated to mannosylated experimental particles in IL-4-activated BMMØs in the presence/absence of DPI. G and H, phagosomal hydrolysis of the cysteine cathepsin-specific substrate (biotin-LC-Phe-Arg)2-rhodamine 110 conjugated to mannosylated experimental particles in IL-4-activated BMMØs in the presence/absence of DPI. A, C, E, and G, representative real-time traces. Relative fluorescence values are proportional to the degree of substrate hydrolysis. D, F, and H, averaged rates relative to untreated WT controls from three independent experiments. Relative rates were calculated between 40–60 min after particle internalization. Error bars denote S.E. *, p < 0.05.
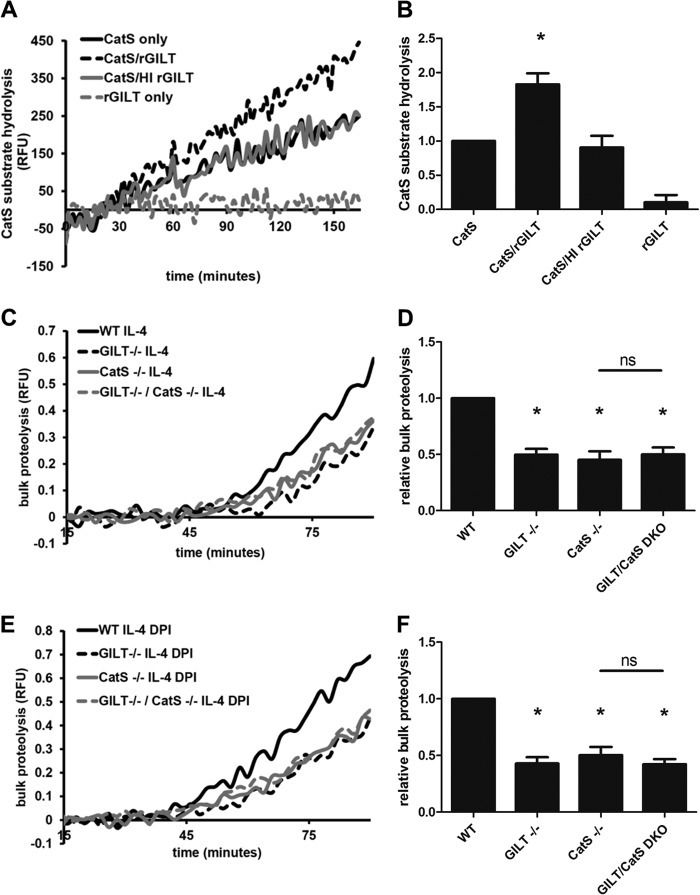
GILT Maintains Cathepsin S Activity in a Reconstituted System
As GILT exhibited the greatest effect on early phagosomal proteolytic activity, where cathepsin S is a dominant protease (40), we hypothesized that GILT may specifically or disproportionately act to maintain cathepsin S activity. We, therefore, evaluated the ability of GILT to alter cathepsin S activity in a reconstituted system. Enzymatic activity of purified human cathepsin S on the cathepsin S-specific substrate Ac-KQKLR-AMC was measured in the presence/absence of human rGILT. rGILT increased the rate of Ac-KQKLR-AMC hydrolysis by purified cathepsin S nearly 2-fold (1.83 ± 0.16-fold increase) as compared with cathepsin S alone (Fig. 6, A and B). No increase in Ac-KQKLR-AMC hydrolysis was observed when rGILT was heat-inactivated before the addition to cathepsin S. rGILT alone did not display any Ac-KQKLR-AMC hydrolysis. To determine whether GILT influences cathepsin S activity in BMMØs, we compared bulk proteolysis in IL-4-activated macrophages derived from WT, GILT−/−, CatS−/−, and GILT−/−/CatS−/− double knock-out mice. GILT−/− BMMØs displayed similar levels of proteolysis as CatS−/− single knock-out BMMØs compared with WT BMMØs in the presence/absence of DPI (untreated, 50.2 ± 5.2%, 54.8 ± 7.6%, respectively; DPI, 57.1 ± 5.5%, 49.7 ± 7.2%, respectively; Fig. 6, C–F). Furthermore, BMMØs derived from the GILT−/−/CatS−/− double knock-out had no significant changes in DQ-albumin proteolysis compared with the CatS−/− single knock-out control. This suggests that the majority of the decrease in total proteolysis observed in GILT−/− phagosomes is due to decreased cathepsin S activity in these compartments. Collectively these results demonstrate the involvement of GILT in the maintenance of cathepsin S activity within the phagosome of macrophages.
FIGURE 6.
GILT maintains cathepsin S activity in a reconstituted system and in the early phagosome of IL-4-activated BMMØs. A and B, purified cathepsin S (CatS) activity on the cathepsin S-specific substrate Ac-KQKLR-AMC in the presence/absence of rGILT in a reconstituted system. Where indicated, heat inactivation (HI) of rGILT was performed by subjecting the enzyme to 95 °C for 10 min before addition. *, p < 0.05 compared with CatS alone and CatS/HI rGILT samples. A, representative real-time traces. RFU, relative fluorescence units. B, averaged rates relative to CatS only controls from four independent experiments. Relative rates were calculated between 90 and 150 min. Error bars denote S.E. C and D, bulk phagosomal proteolysis as measured by hydrolysis of the DQ-albumin substrate conjugated to experimental particles in IL-4-activated BMMØs derived from WT, GILT−/−, CatS−/−, and GILT−/−/CatS−/− mice. E and F, bulk proteolysis as measured by hydrolysis of the DQ-albumin substrate conjugated to experimental particles in IL-4-activated BMMØs treated with the NOX2 inhibitor DPI (0.5 μm) 10 min before the addition and subsequent phagocytosis of experimental particles. C, and E, representative real-time traces. Relative fluorescence units values are proportional to the degree of substrate hydrolysis. D, and F, averaged rates relative to unactivated/untreated WT controls from five independent experiments. Relative rates were calculated between 40 and 60 min after particle internalization. Error bars denote S.E. *, p < 0.05 compared with WT control. ns, not significant.
DISCUSSION
This body of work in addition to demonstrating the disulfide reductive function of GILT within phagosomes in real time, reports on the involvement of GILT in maintaining the proteolytic efficiency of phagosomes in the macrophage. This novel role of GILT in supporting cysteine cathepsin activity is most apparent during states of low or no NOX2 activity, as observed in alternatively activated macrophages. Thus GILT may function to additionally increase the proteolytic proficiency of phagosomes in alternatively activated macrophages that are implicated in the efficient clearance of cellular debris and apoptotic bodies during the resolution of inflammation (41–43). In addition, this may have relevance in other scenarios where NOX2 activity is down-regulated, such as after macrophage exposure to IL-10 or apoptotic cells (44–46).
One interesting finding was that the role for GILT in proteolysis was diminished in the presence of ROS, particularly in unactivated macrophages. Conversely, the role for GILT in substrate disulfide reduction was maintained in the presence of ROS. The susceptibility of GILT-mediated activation of cysteine cathepsins to the presence of ROS could be due to the fact that certain forms of thiol-oxidation may not be reversible by GILT. For example thiol oxidation to the sulfonic state is irreversible and cannot be reduced enzymatically (47). In the presence of ROS, the degree of irreversible thiol oxidation of the cysteine cathepsin active site thiol may increase, potentially limiting the effect of GILT. In the case of disulfide reduction, the target substrate is a disulfide that is less affected by the presence of ROS as it is already oxidized. Thus the effect of GILT on disulfide-containing substrates would not be influenced by the degree of ROS production.
Although the ability of GILT to reduce disulfide bonds within endosomes, lysosomes, and phagosomes has been well documented (19), by using a real-time fluorescence assay we were able to visualize and quantify the reductase activity of GILT within phagosomes in both resting and classically activated macrophages in the presence and absence of NOX2 activity. GILT-catalyzed disulfide reduction was evident within 15 min after phagocytosis and continued within the mature phagolysosome. Although the absence of GILT significantly compromised phagosomal disulfide reductive efficiency, it was not completely abolished in unactivated cells. The residual disulfide reduction noted in these phagosomes supports the finding that the reduction of certain model antigens is independent of GILT (48). It was also interesting to note that phagosomes of BMMØs classically activated with IFNγ did not possess significantly higher levels of disulfide reduction compared with unactivated controls even in the presence of DPI despite increased expression of GILT. This may indicate that disulfide reduction by GILT is not rate-limiting in these phagosomes. In the absence of GILT, however, the reductive capacity of these phagosomes was near-abolished. This could suggest that the primary role of the induction of GILT by IFN-γ is not to increase disulfide reduction in activated macrophages but is to maintain levels of disulfide reduction in the more oxidative endolysosomal system of these cells.
Historically, GILT has been shown to play an important role in antigen processing in a variety of in vivo and in vitro models (49–51). It is clear that this function of GILT is primarily mediated through its ability to reduce inter- and intramolecular disulfides of antigens. However, our data suggest that in addition to reducing disulfide bonds of antigens directly, GILT may impact antigen processing through maintenance of cysteine cathepsin activity under particular redox conditions.
Although this study found that GILT is required for optimal cathepsin S activity, a role for GILT in supporting other cysteine cathepsins either directly or indirectly cannot be ruled out. Indeed, expression of GILT in melanoma and B cell lines has been shown to increase the processing of pro-cathepsin B, S, L, and D to their mature forms or to specifically destabilize cathepsin S (52, 53). Although we found no evidence of either altered processing or destabilization of cysteine or aspartic cathepsins mediated by native levels of GILT in macrophages (GILT−/− BMMØs exhibited similar levels of mature cathepsin B, S, L, and D), increased cysteine cathepsin activities mediated by GILT could indeed enhance the proteolytic processing of these pro-enzymes as well as accelerate their decay (54, 55).
The current study does not describe the nature of the interaction between GILT and cathepsin S. GILT may directly reduce oxidized Cys-25 in the active site of cathepsin S. Alternatively, GILT may support proteolysis indirectly by maintaining a high concentration of reducing equivalents (such as cysteine) required for optimal cysteine cathepsin activity, before, or shortly after phagocytosis. GILT has been shown to alter cellular glutathione levels in murine fibroblasts, although glutathione is unlikely to be a major reducing equivalent in the endolysosomal system (12, 56). Elucidating mechanisms by which GILT supports cathepsin S activity is a focus of ongoing research.
In addition to cathepsin S, GILT may be involved in maintaining the activity of other enzymes present in these compartments, such as thiol-dependent peroxidases. In the current study we surprisingly did not see evidence of GILT supporting antioxidant functions during a respiratory burst as we noted no changes in ROS levels in GILT−/− BMMØs over the period of phagosomal maturation measured. It is possible that GILT could still be involved with reestablishing a reductive environment after NOX2 activity by activating currently uncharacterized ROS scavenging functions in the phagosome similar to the role of thioredoxin in the activation of certain peroxidases (57). These issues are difficult to address as the presence of other thiol reductases and the identities of endosomal/lysosomal reducing equivalents are currently unknown (12, 13). For instance, the mechanisms that maintain the activity of GILT itself (as it is also reliant on reduced active-site cysteines) are uncharacterized. Although earlier reports suggested l-cysteine as a source of reducing equivalents for both GILT and cysteine cathepsins (19, 58), direct experimental evidence for this is still lacking. The full extent of the role of GILT as a reductive enzyme within the endo/lysosomal system will be more defined as we uncover the exact mechanisms by which redox conditions are established and maintained within these dynamic compartments.
This work was supported by the Canadian Institutes of Health Research and Natural Sciences and Engineering Research Council of Canada.
- ROS
- reactive oxygen species
- GILT
- γ-interferon-inducible lysosomal thiol reductase
- rGILT
- recombinant GILT
- NOX2
- NADPH oxidase 2
- BMMØ
- bone marrow-derived macrophage
- DPI
- diphenyleneiodonium
- SE
- succinimidyl ester
- DQ-albumin
- DQ green BODIPY albumin
- PL fusion
- phagosome-lysosome fusion
- QPCR
- real-time PCR
- AMC
- amidomethylcoumarin.
REFERENCES
- 1. Huynh K. K., Grinstein S. (2007) Regulation of vacuolar pH and its modulation by some microbial species. Microbiol. Mol. Biol. Rev. 71, 452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nunes P., Demaurex N., Dinauer M. C. (2013) Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic 14, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 3. Flannagan R. S., Cosío G., Grinstein S. (2009) Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366 [DOI] [PubMed] [Google Scholar]
- 4. Claus V., Jahraus A., Tjelle T., Berg T., Kirschke H., Faulstich H., Griffiths G. (1998) Lysosomal enzyme trafficking between phagosomes, endosomes, and lysosomes in J774 macrophages. Enrichment of cathepsin H in early endosomes. J. Biol. Chem. 273, 9842–9851 [DOI] [PubMed] [Google Scholar]
- 5. Kotsias F., Hoffmann E., Amigorena S., Savina A. (2013) Reactive oxygen species production in the phagosome: impact on antigen presentation in dendritic cells. Antioxid. Redox Signal. 18, 714–729 [DOI] [PubMed] [Google Scholar]
- 6. Yates R. M. (2013) Redox considerations in the phagosome: current concepts, controversies, and future challenges. Antioxid. Redox Signal. 18, 628–629 [DOI] [PubMed] [Google Scholar]
- 7. Rybicka J. M., Balce D. R., Khan M. F., Krohn R. M., Yates R. M. (2010) NADPH oxidase activity controls phagosomal proteolysis in macrophages through modulation of the lumenal redox environment of phagosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 10496–10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rybicka J. M., Balce D. R., Chaudhuri S., Allan E. R., Yates R. M. (2012) Phagosomal proteolysis in dendritic cells is modulated by NADPH oxidase in a pH-independent manner. EMBO J. 31, 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balce D. R., Li B., Allan E. R., Rybicka J. M., Krohn R. M., Yates R. M. (2011) Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood 118, 4199–4208 [DOI] [PubMed] [Google Scholar]
- 10. Hsing L. C., Rudensky A. Y. (2005) The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol. Rev. 207, 229–241 [DOI] [PubMed] [Google Scholar]
- 11. Lennon-Duménil A. M., Bakker A. H., Maehr R., Fiebiger E., Overkleeft H. S., Rosemblatt M., Ploegh H. L., Lagaudrière-Gesbert C. (2002) Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J. Exp. Med. 196, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pillay C. S., Elliott E., Dennison C. (2002) Endolysosomal proteolysis and its regulation. Biochem. J. 363, 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lockwood T. D. (2013) Lysosomal metal, redox, and proton cycles influencing the CysHis cathepsin reaction. Metallomics 5, 110–124 [DOI] [PubMed] [Google Scholar]
- 14. Go Y. M., Jones D. P. (2008) Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780, 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chae H. Z., Chung S. J., Rhee S. G. (1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670–27678 [PubMed] [Google Scholar]
- 16. Lockwood T. D. (2002) Cathepsin B responsiveness to glutathione and lipoic acid redox. Antioxid. Redox Signal. 4, 681–691 [DOI] [PubMed] [Google Scholar]
- 17. Kerblat I., Drouet C., Chesne S., Marche P. N. (1999) Importance of thioredoxin in the proteolysis of an immunoglobulin G as antigen by lysosomal Cys-proteases. Immunology 97, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stephen A. G., Powls R., Beynon R. J. (1993) Activation of oxidized cysteine proteinases by thioredoxin-mediated reduction in vitro. Biochem. J. 291, 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phan U. T., Arunachalam B., Cresswell P. (2000) γ-Interferon-inducible lysosomal thiol reductase (GILT). Maturation, activity, and mechanism of action. J. Biol. Chem. 275, 25907–25914 [DOI] [PubMed] [Google Scholar]
- 20. Pollock J. D., Williams D. A., Gifford M. A., Li L. L., Du X., Fisherman J., Orkin S. H., Doerschuk C. M., Dinauer M. C. (1995) Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9, 202–209 [DOI] [PubMed] [Google Scholar]
- 21. Yates R. M., Hermetter A., Russell D. G. (2005) The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic 6, 413–420 [DOI] [PubMed] [Google Scholar]
- 22. Yates R. M., Russell D. G. (2008) Real-time spectrofluorometric assays for the lumenal environment of the maturing phagosome. Methods Mol. Biol. 445, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. VanderVen B. C., Yates R. M., Russell D. G. (2009) Intraphagosomal measurement of the magnitude and duration of the oxidative burst. Traffic 10, 372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Voss E. W., Jr., Workman C. J., Mummert M. E. (1996) Detection of protease activity using a fluorescence-enhancement globular substrate. Biotechniques 20, 286–291 [DOI] [PubMed] [Google Scholar]
- 25. Anjaneyulu P. S., Staros J. V. (1987) Reactions of N-hydroxysulfosuccinimide active esters. Int. J. Pept. Protein Res. 30, 117–124 [DOI] [PubMed] [Google Scholar]
- 26. Carraway K. L., Triplett R. B. (1970) Reaction of carbodiimides with protein sulfhydryl groups. Biochim. Biophys. Acta 200, 564–566 [DOI] [PubMed] [Google Scholar]
- 27. Collins D. S., Unanue E. R., Harding C. V. (1991) Reduction of disulfide bonds within lysosomes is a key step in antigen processing. J. Immunol. 147, 4054–4059 [PubMed] [Google Scholar]
- 28. Shrimali R. K., Irons R. D., Carlson B. A., Sano Y., Gladyshev V. N., Park J. M., Hatfield D. L. (2008) Selenoproteins mediate T cell immunity through an antioxidant mechanism. J. Biol. Chem. 283, 20181–20185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carlson B. A., Yoo M. H., Tsuji P. A., Gladyshev V. N., Hatfield D. L. (2009) Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules 14, 3509–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamamoto M., Yang G., Hong C., Liu J., Holle E., Yu X., Wagner T., Vatner S. F., Sadoshima J. (2003) Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J. Clin. Invest. 112, 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lockwood T. D. (2000) Redox control of protein degradation. Antioxid. Redox Signal. 2, 851–878 [DOI] [PubMed] [Google Scholar]
- 32. Welch J. S., Escoubet-Lozach L., Sykes D. B., Liddiard K., Greaves D. R., Glass C. K. (2002) TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J. Biol. Chem. 277, 42821–42829 [DOI] [PubMed] [Google Scholar]
- 33. Gocheva V., Wang H. W., Gadea B. B., Shree T., Hunter K. E., Garfall A. L., Berman T., Joyce J. A. (2010) IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 24, 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stein M., Keshav S., Harris N., Gordon S. (1992) Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 176, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tian W., Li X. J., Stull N. D., Ming W., Suh C. I., Bissonnette S. A., Yaffe M. B., Grinstein S., Atkinson S. J., Dinauer M. C. (2008) Fc γ R-stimulated activation of the NADPH oxidase: phosphoinositide-binding protein p40phox regulates NADPH oxidase activity after enzyme assembly on the phagosome. Blood 112, 3867–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I. C., Lennon-Duménil A. M., Seabra M. C., Raposo G., Amigorena S. (2006) NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218 [DOI] [PubMed] [Google Scholar]
- 37. Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., Brumell J. H. (2009) Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Y., Trush M. A. (1998) Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 253, 295–299 [DOI] [PubMed] [Google Scholar]
- 39. Bulua A. C., Simon A., Maddipati R., Pelletier M., Park H., Kim K. Y., Sack M. N., Kastner D. L., Siegel R. M. (2011) Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 208, 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Riese R. J., Wolf P. R., Brömme D., Natkin L. R., Villadangos J. A., Ploegh H. L., Chapman H. A. (1996) Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity 4, 357–366 [DOI] [PubMed] [Google Scholar]
- 41. Wang Y., Wang Y. P., Zheng G., Lee V. W., Ouyang L., Chang D. H., Mahajan D., Coombs J., Wang Y. M., Alexander S. I., Harris D. C. (2007) Ex vivo programmed macrophages ameliorate experimental chronic inflammatory renal disease. Kidney Int. 72, 290–299 [DOI] [PubMed] [Google Scholar]
- 42. Zeng M. Y., Pham D., Bagaitkar J., Liu J., Otero K., Shan M., Wynn T. A., Brombacher F., Brutkiewicz R. R., Kaplan M. H., Dinauer M. C. (2013) An efferocytosis-induced, IL-4-dependent macrophage-iNKT cell circuit suppresses sterile inflammation and is defective in murine CGD. Blood 121, 3473–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fernandez-Boyanapalli R. F., Frasch S. C., McPhillips K., Vandivier R. W., Harry B. L., Riches D. W., Henson P. M., Bratton D. L. (2009) Impaired apoptotic cell clearance in CGD due to altered macrophage programming is reversed by phosphatidylserine-dependent production of IL-4. Blood 113, 2047–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bogdan C., Vodovotz Y., Nathan C. (1991) Macrophage deactivation by interleukin 10. J. Exp. Med. 174, 1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roilides E., Dimitriadou A., Kadiltsoglou I., Sein T., Karpouzas J., Pizzo P. A., Walsh T. J. (1997) IL-10 exerts suppressive and enhancing effects on antifungal activity of mononuclear phagocytes against Aspergillus fumigatus. J. Immunol. 158, 322–329 [PubMed] [Google Scholar]
- 46. Serinkan B. F., Gambelli F., Potapovich A. I., Babu H., Di Giuseppe M., Ortiz L. A., Fabisiak J. P., Kagan V. E. (2005) Apoptotic cells quench reactive oxygen and nitrogen species and modulate TNF-α/TGF-β1 balance in activated macrophages: involvement of phosphatidylserine-dependent and -independent pathways. Cell Death Differ. 12, 1141–1144 [DOI] [PubMed] [Google Scholar]
- 47. Lim J. C., Choi H. I., Park Y. S., Nam H. W., Woo H. A., Kwon K. S., Kim Y. S., Rhee S. G., Kim K., Chae H. Z. (2008) Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 283, 28873–28880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sinnathamby G., Maric M., Cresswell P., Eisenlohr L. C. (2004) Differential requirements for endosomal reduction in the presentation of two H2-E(d)-restricted epitopes from influenza hemagglutinin. J. Immunol. 172, 6607–6614 [DOI] [PubMed] [Google Scholar]
- 49. Maric M., Arunachalam B., Phan U. T., Dong C., Garrett W. S., Cannon K. S., Alfonso C., Karlsson L., Flavell R. A., Cresswell P. (2001) Defective antigen processing in GILT-free mice. Science 294, 1361–1365 [DOI] [PubMed] [Google Scholar]
- 50. Haque M. A., Li P., Jackson S. K., Zarour H. M., Hawes J. W., Phan U. T., Maric M., Cresswell P., Blum J. S. (2002) Absence of γ-interferon-inducible lysosomal thiol reductase in melanomas disrupts T cell recognition of select immunodominant epitopes. J. Exp. Med. 195, 1267–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singh R., Cresswell P. (2010) Defective cross-presentation of viral antigens in GILT-free mice. Science 328, 1394–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goldstein O. G., Hajiaghamohseni L. M., Amria S., Sundaram K., Reddy S. V., Haque A. (2008) γ-IFN-inducible-lysosomal thiol reductase modulates acidic proteases and HLA class II antigen processing in melanoma. Cancer Immunol. Immunother. 57, 1461–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Phipps-Yonas H., Semik V., Hastings K. T. (2013) GILT expression in B cells diminishes cathepsin S steady-state protein expression and activity. Eur. J. Immunol. 43, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turk B., Turk D., Turk V. (2000) Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta 1477, 98–111 [DOI] [PubMed] [Google Scholar]
- 55. Barry Z. T., Platt M. O. (2012) Cathepsin S cannibalism of cathepsin K as a mechanism to reduce type I collagen degradation. J. Biol. Chem. 287, 27723–27730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chiang H. S., Maric M. (2011) Lysosomal thiol reductase negatively regulates autophagy by altering glutathione synthesis and oxidation. Free Radic. Biol. Med. 51, 688–699 [DOI] [PubMed] [Google Scholar]
- 57. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 58. Pisoni R. L., Acker T. L., Lisowski K. M., Lemons R. M., Thoene J. G. (1990) A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. J. Cell Biol. 110, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]