Background: Late embryogenesis-abundant proteins accumulate under water-deficit and are widely distributed in plants and anhydrobiotic organisms.
Results: A common bean group-6 LEA protein shows structural disorder, adopts helicity under water deficit or high molecular crowding, and presents oligomeric forms.
Conclusion: PvLEA6 protein adopts different conformations and/or a quaternary structure depending on its environment.
Significance: Similar characteristics might be present in different LEA proteins, which could be relevant to their function.
Keywords: Intrinsically Disordered Protein, Isothermal Titration Calorimetry (ITC), Protein Conformation, Protein Self-assembly, Protein Structure, Stress Response, Plant Biology
Abstract
Late embryogenesis-abundant proteins accumulate to high levels in dry seeds. Some of them also accumulate in response to water deficit in vegetative tissues, which leads to a remarkable association between their presence and low water availability conditions. A major sub-group of these proteins, also known as typical LEA proteins, shows high hydrophilicity and a high percentage of glycine and other small amino acid residues, distinctive physicochemical properties that predict a high content of structural disorder. Although all typical LEA proteins share these characteristics, seven groups can be distinguished by sequence similarity, indicating structural and functional diversity among them. Some of these groups have been extensively studied; however, others require a more detailed analysis to advance in their functional understanding. In this work, we report the structural characterization of a group 6 LEA protein from a common bean (Phaseolus vulgaris L.) (PvLEA6) by circular dichroism and nuclear magnetic resonance showing that it is a disordered protein in aqueous solution. Using the same techniques, we show that despite its unstructured nature, the addition of trifluoroethanol exhibited an intrinsic potential in this protein to gain helicity. This property was also promoted by high osmotic potentials or molecular crowding. Furthermore, we demonstrate that PvLEA6 protein is able to form soluble homo-oligomeric complexes that also show high levels of structural disorder. The association between PvLEA6 monomers to form dimers was shown to occur in plant cells by bimolecular fluorescence complementation, pointing to the in vivo functional relevance of this association.
Introduction
Most spermatophyte species have seeds able to tolerate high levels of dehydration (1). This trait is acquired late in plant embryogenesis during a stage where, in addition to the synthesis of storage products, abscisic acid, and the establishment of dormancy, the seed developmental program induces the production of the so-called Late embryogenesis-abundant (LEA)3 proteins and the loss of water that leads to a severe desiccation state (2, 3). To survive, plant embryos need to endure this condition, an outstanding feature of mature orthodox seeds. Many studies have reported various mechanisms that allow plants to withstand desiccation and/or low water availability, in which a diversity of molecules play fundamental roles in diverse processes throughout plant development under different intensities of water deficit (2, 4). Among these molecules, LEA proteins have been described as a set of proteins, identified in vascular and nonvascular plants, whose synthesis and accumulation is highly associated with low water availability in the different plant tissues (5). As members of hydrophilins, LEA proteins share particular physicochemical properties such as high hydrophilicity, high content of glycine and other small amino acids (Ala, Ser, and Thr), and lack or low content of Trp and Cys residues (5, 6). This amino acid composition is similar to that found in intrinsically disordered proteins (IDPs) (7), such that prediction programs allowed the classification of most LEA proteins as IDPs, a characteristic that has been validated for some of them when they are in aqueous solution (5, 8, 9). IDPs are now recognized as essential for many biological processes and are widely distributed in viruses and organisms from the three domains of life (10), where their occurrence seems to increase with the complexity of the organisms (7). The absence of structural constraints in these proteins allows plasticity and binding promiscuity, in agreement with their involvement in diverse processes, including signaling, regulation, and responses to various developmental and environmental cues (7, 11). Accordingly, transient or stable binding to different partner molecules has been shown for some IDPs, suggesting their potential multifunctionality (7, 12, 13).
Even though LEA proteins share a number of physicochemical characteristics, they show great diversity in their amino acid sequences such that they can be grouped in seven families, without significant sequence similarity between them and showing distinctive motifs in each group. Some of the groups of LEA proteins have been detected beyond the plant kingdom (5, 14).
In this study, we present some of the structural features of a group 6 LEA protein from common bean (Phaseolus vulgaris L.), PvLEA6, a representative of one of the most conserved LEA protein families, where four distinctive motifs have been identified (5, 15). Group 6 LEA proteins are highly hydrophilic, do not contain Cys and/or Trp residues, and do not coagulate when exposed to high temperatures. PvLEA6 protein, previously called PvLEA18, was the first protein identified from this group (11). Previous data showed that this protein and its transcript not only accumulate to high levels in dry seeds and pollen grains but also in vegetative tissues upon water deficit and abscisic acid treatments (16–18). PvLEA6 protein also accumulates during normal development in the expansion region of bean seedling hypocotyls of well irrigated plants, a region that shows lower water potentials than those from nongrowing regions, and also in the vascular cylinder and in meristematic zones such as the apical meristem and root primordia (17). Although its participation in the plant response to water deficit is well documented, PvLEA6 protein function is still unknown.
As in other LEA proteins, group 6 LEA proteins have been predicted to be unstructured (16); however, their structural properties have been scarcely studied (19) in contrast with LEA proteins from other groups (5, 8, 20, 21). In this report, analysis by circular dichroism (CD) and nuclear magnetic resonance (NMR) shows the disordered structure of the PvLEA6 protein in aqueous solution, as well as the potential of this protein to acquire up to 40% α-helix. In addition, we show that low osmotic potential induced with glycerol or molecular crowding induced with polyethylene glycol (PEG) promotes, although to a limited extent, the formation of the α-helical structure. We also demonstrate that PvLEA6 protein is able to form homo-oligomeric complexes in solution, which maintain the monomer structural disorder. Furthermore, we show the formation of PvLEA6 dimers in planta by bimolecular fluorescence complementation (BiFC), suggesting their biological relevance.
EXPERIMENTAL PROCEDURES
Cloning of PvLEA6 Protein
The PvLEA6 cDNA coding region (249 bp; GenBankTM accession number AF240774 (18)) was amplified by PCR using gene-specific primers containing SapI (5′-GGTGGTTGCTCTTCCAACATGGAGAAGGAGAAAAAGACAG-3′) and PstI (5′-CCCCAAGCTTGGGCTGCAGTCACTTGTGATTAGTGGCACC-3′) restriction sites. The amplified fragment was cloned in the pCR®4-TOPO plasmid (Invitrogen); the resulting plasmid was digested with SapI and PstI restriction enzymes, and the PvLEA6-containing fragment was ligated into the SapI/PstI sites of the pTYB11 expression vector (New England Biolabs Inc.).
Expression and Purification of Recombinant PvLEA6 Protein
To obtain the recombinant PvLEA6 (rPvLEA6) protein, the IMPACTTM-CN expression system (New England Biolabs Inc.) was used. In this system the protein is fused to an intein-chitin binding domain tag that allows for its purification in one step using a chitin resin. For this purpose, Escherichia coli strain ER2566 carrying a chromosomal copy of the T7 RNA polymerase gene under the control of the lac promoter was used as host of the pTYB11/PvLEA6 plasmid, where the fusion protein is expressed from T7/lac promoter. Overexpression of the intein-PvLEA6 protein was induced with a final concentration of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 0.5–0.8 and incubated at 25 °C during 6 h. At this point, bacterial cells were centrifuged, and the pellet was resuspended in lysis buffer (20 mm sodium phosphate, pH 8, 500 mm NaCl, 0.1% Triton X-100, 20 μm phenylmethylsulfonyl fluoride (PMSF)) (12.5 ml/g cells). Cells were lysed by sonication in ice, and the extract was clarified by centrifugation at 20,000 × g for 30 min at 4 °C. To obtain the protein without the tag, the clarified extract was loaded onto a chitin column following the procedure described by the manufacturer (IMPACTTM-CN kit). Protein was quantified considering the PvLEA6 molar extinction coefficient at 274.6 nm (ϵ = 6,186 m−1 cm−1).
Plant Material
Embryos were extracted from dry seeds of P. vulgaris L. cv Negro Jamapa, kindly provided by Dr. J. Acosta (National Institute for Forest and Agricultural Research, Celaya, México). After separation from seeds, embryos were frozen in liquid nitrogen and stored at −80 °C until their use for protein extraction.
Preparation of Cell Extracts
Plant embryos were homogenized in the presence of liquid nitrogen and extracted with 1.3 volumes of extraction buffer (20 mm TES, pH 8.0, 0.5 m NaCl, protease inhibitor mixture (Complete, Roche Applied Science)) at 4 °C. The homogenate was centrifuged to remove insoluble materials, and the supernatant was boiled for 10 min in a water bath, and coagulated material was removed by centrifugation. Thereafter, the soluble supernatant was separated and dialyzed against 50 mm potassium phosphate buffer, pH 8.0, 150 mm NaCl. When needed, protein was concentrated by acetone precipitation and dissolved in the same buffer. Total protein was quantified using Bradford assay with Bio-Rad dye reagent.
Gel Filtration Chromatography
Recombinant PvLEA6 (rPvLEA6) protein (100 μg), dialyzed against 50 mm potassium phosphate buffer, pH 8.0, 150 mm NaCl, was applied onto a Superdex-75 (10/300 GL) gel filtration high performance column in an FPLC system (ÄKTA FPLC Amersham Biosciences) at a flow rate of 0.5 ml min−1. The eluate was monitored with a 280-nm filter, and the resulting fractions were analyzed by SDS-PAGE and Western blot. Calibration was carried out using as molecular mass markers the following: bovine serum albumin 66 kDa; egg albumin 45 kDa; carbonic anhydrase 29 kDa; trypsinogen 24 kDa; and trypsin inhibitor 20.1 kDa.
Antibodies and Immunoblotting
PvLEA6 antibodies were produced using the purified GST-PvLEA6 fusion protein expressed in E. coli. Polyclonal antibody production was performed as described by Colmenero-Flores et al. (17). The specificity of the antibody was verified by competition assays using anti-PvLEA6 polyclonal antibody previously incubated with 50 μg of the purified rPvLEA6 protein. Proteins were separated in SDS-PAGE using 15% polyacrylamide gels and transferred to nitrocellulose membranes (Hybond C, GE Healthcare), washed with TBST (150 mm NaCl, 20 mm Tris-HCl and 0.1% Tween 20), blocked with 5% skimmed milk (in TBST), and probed with anti-PvLEA6 (1:1000 or 1:2000) antibodies. Membranes were washed and incubated with secondary antibody (1:30,000) (anti-rabbit horseradish peroxidase, Zymed Laboratories Inc.) for 1 h at room temperature, washed with TBST, and developed with peroxidase substrates from Supersignal West Pico chemiluminescent substrate kit (Pierce and Thermo Fisher Scientific Inc.). Membranes were exposed to x-ray films (Kodak, México).
Protein-Protein Cross-linking
Cross-linking experiments were carried out using Tris-bipyridylruthenium (II) di-cation Ru(II)bpy32+ (TdClRu, Sigma), a photoactivatable reagent able to induce cross-linking of proteins without any previous chemical modification. In this procedure, also known as photoinduced cross-linking of unmodified proteins (PICUP), cross-linking occurs by an efficient reaction that is induced by a very fast photolysis of a tris-bipyridyl Ru(II) complex triggered by visible light in the presence of an electron acceptor. Cross-linking reaction conditions were as described by Fancy and Kodadek (22) with minor modifications. Briefly, purified rPvLEA6 was diluted to a concentration of 20 μm in cross-linking buffer (150 mm NaCl, 15 mm Na2HPO4, pH 6.5); the protein mixture was brought to 10 μl containing 1.25 mm Ru(II)bpy32+ and 0.025 or 2.5 mm ammonium persulfate, as indicated, and it was flashed for 0–150 s through a 2.5-cm water filter using a white light bulb as source (75 watts). Samples were subsequently quenched with an equal volume of 2× Laemmli sample buffer containing 4% SDS and 10% β-mercaptoethanol and resolved by SDS-PAGE. Protein bands were visualized by chemiluminescence from Western blots using anti-PvLEA6 antibody and anti-rabbit IgG peroxidase.
CD Spectroscopy
CD spectra were recorded on a CD spectropolarimeter (model J-715, Jasco Analytical Instruments) using a 0.1-cm path length cell over the 190–260-nm range. Protein concentration used was 0.3 mg/ml. The temperature was regulated by a Peltier temperature-controlled cell holder (model PTC-4235, Jasco). CD spectra were acquired every 1 nm with a 2-s averaging time per point and a 1-nm band pass. The average of four spectra were recorded to reduce noise and smoothed before structure analysis was performed.
Estimation of Secondary Structure
The secondary structure prediction was performed using the CDSSTR algorithm, and 4, 7, and SP175 data sets, accessed through the Dichroweb server, which requires data from 190 to 240 nm (19).
Fluorometry
Measurements were carried out in a luminescence spectrometer (LS50B, PerkinElmer Life Sciences) using a 1-cm path length cell over the 280–500-nm range. The temperature was controlled by a circulating water bath (Thermo Scientific HAAKE, model K10) and determined directly into the cell using a thermocouple.
Near-UV CD Spectroscopy
Spectra were recorded on a CD spectropolarimeter (model J-715, Jasco Analytical Instruments) using a 1-cm path length cell over the 250–350-nm range. The temperature was regulated by a Peltier temperature-controlled cell holder (model PTC-4235, Jasco). CD spectra were acquired every 1 nm with 8 s averaging time per point and a 1-nm band pass. The averages of four spectra were recorded to reduce noise.
NMR Spectroscopy
All NMR spectra were recorded on a 700 MHz Agilent VNMR-S spectrometer equipped with a cryogenically cooled triple-resonance pulsed field gradient probe at the Laboratorio Nacional de Estructura de Macromoléculas at Centro de Investigaciones Químicas (Universidad Autónoma del Estado de Morelos). Proteins were isotopically labeled (15N) according to Marley et al. (23), reducing the volume of the bacterial culture 4-fold and inducing the protein expression with a final concentration of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Two-dimensional 1H-15N correlated NMR spectra of rPvLEA6 (50–125 μm) were recorded in 10 mm potassium phosphate buffer, pH 8.0, and 62.5% trifluoroethanol (TFE) at 298 K. Data were processed and analyzed with NMRPipe (24) and CARA software (25).
Isothermal Dilution Calorimetry
The dissociation of the rPvLEA6 homodimer was characterized using isothermal dilution calorimetry (IDC) with a MicroCalTM iTC200 system (GE Healthcare). Before the experiment, the protein solution was extensively dialyzed against 10 mm potassium phosphate buffer, pH 8. Stepwise additions of small aliquots of a solution with a high protein concentration (0.47–0.63 mm of monomer equivalent) were applied into the calorimetric reaction cell loaded with buffer solution. All experiments were performed at 30 °C. According to a simple dimer dissociation model (26), the heat measured upon addition of the ith injection of volume dVi into the cell calorimeter is shown in Equation 1,
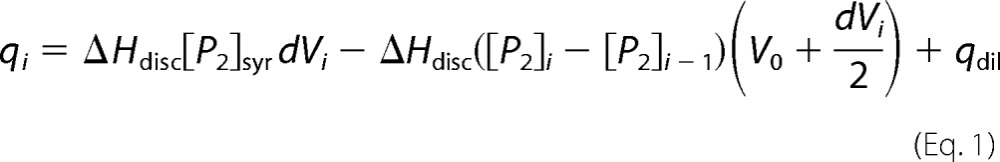 |
where ΔHdisc is the dissociation enthalpy of the dimer, [P] and [P2] are the molar concentrations of the free monomer and dimer, respectively, and qdil is the dilution heat of the protein. The dimer concentration in the reaction cell, [P2]i, and the syringe, [P2]syr, are in turn related to the corresponding equivalent monomer concentration, [PT], through the dissociation constant shown in Equation 2,
where Kdisc is the equilibrium dissociation constant. ΔHdisc and Kdisc were determined using nonlinear regression routines implemented in Origin 7.0 (OriginLab, Co., Northampton, MA). Fittings with higher order dissociation models were performed by using a private version of the AFFINImeter software.
Mass Spectrometry
Mass spectrometry analyses were carried out by the Proteomic Facility at the Instituto de Biotecnología-Universidad Nacional Autónoma de México. Samples to be sequenced were previously reduced with dithiothreitol (DTT), alkylated with iodoacetamide, and digested in gel with trypsin. Resultant peptides were applied into an LC-MS system composed of a micro-flux liquid chromatograph with splitter (1:20) (Accela, Thermo-Fisher Scientific Inc.) and a mass spectrometer (LTQ-Orbitrap Velos; Thermo-Fisher Scientific Inc.) with a nano-spray ionization system (ESI). Liquid chromatography was performed with a 10–100% gradient of solvent B (acetonitrile, 0.1% acetic acid) for 120 min in a capillary column (PicoFrit ProteoPepTM 2 C18 75 μm inner diameter × 50 mm; New Objective Inc., Woburn, MA) with a flow of 0.4 μl/min.
To determine the molecular mass of the PvLEA6 monomer, the sample was purified by HPLC (Agilent Technologies 1200) equipped with Vydac C18 column using a 0–60% acetonitrile, 0.1% trifluoroacetic acid linear gradient. Subsequently, the sample was injected into the same LC-MS system described above. Molecular mass was calculated using Xcalibur software (Thermo-Fisher Scientific Inc.).
Molecular masses of PvLEA6 oligomeric forms were determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) using a 4800 MALDI-TOF/TOFTM analyzer (Applied Biosystems/MDS SCIEX). For this analysis, samples were co-crystallized with an organic matrix of sinapinic acid (50% acetonitrile, 0.1% trifluoroacetic acid). This study was performed using a linear mode, 4000 laser shots, and a delayed time of 1500 ns. Mass resolution was 12,000; signal/noise, 20. The internal and external calibrations were performed using calibration mixtures 3 and 5, respectively (Applied Biosystems/MDS SCIEX).
BiFC Experiments
To assess the formation and localization of PvLEA6 protein dimer in vivo, BiFC experiments were carried out using Nicotiana benthamiana leaves. YFPN43-PvLEA6 and YFPC43-PvLEA6 plasmids were constructed by recombination of pENTR-PvLEA6 ORF with pYFPN43 or pYFPC43 vectors (27), using Gateway® recombination cloning system. Resulting plasmids were sequenced to verify the correct reading frames. Plasmids were introduced into Agrobacterium tumefaciens GV3101 strain and used for plant transformation. Four-week-old N. benthamiana plants were used for transient transformation as described by Marion et al. (28). Briefly, Agrobacterium cells from an overnight culture (A600 = 3.0) were centrifuged. and the resultant pellet was resuspended in 5% sucrose, 200 μm acetosyringone (3′,5′-dimethoxy-4′-hidroxyacetophenone) to an A600 of 2. After 2 h at room temperature, cultures were mixed for the corresponding co-expression. To increase transient transformation efficiency, A. tumefaciens C58 strain carrying p19 suppressor from tomato bushy stunt virus grown to an A600 of 2.0 was added to the cell mixture containing the constructions of interest (29). Cell suspensions were infiltrated on the abaxial side of the leaf, and the infiltrated zone was demarcated for expression analysis. Plants were maintained under optimal conditions for 3 to 4 days. At this time, expression in leaves was analyzed by confocal microscopy.
Confocal Microscopy
Leaves co-transformed with PvLEA6-YFPN43 and PvLEA6-YFPC43 fusion proteins (targets) or with the corresponding empty vectors pYFPN43 and pYFPC43 were examined under a confocal microscope (Olympus FluoViewTM FV1000, Inverted IX81, ×60 S/1.3 objective). Saturation level was verified for each image. The settings for image capture were established with target (YFPN43-PvLEA6 and YFPC43-PvLEA6) expression and used to capture control images (empty vectors or one target and empty vector, respectively). Images were processed using Fiji image processing package (30).
RESULTS
PvLEA6 Protein Is Disordered in Aqueous Solution
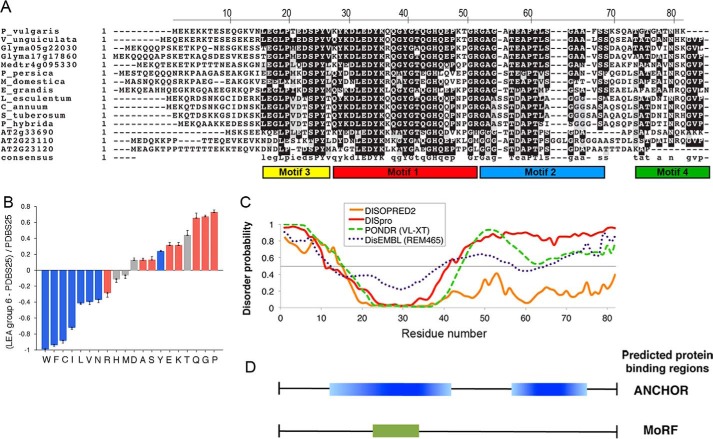
Among LEA proteins, group 6 is one of the most conserved, where four distinctive motifs have been identified (Fig. 1A) (5, 15). Previous analysis of PvLEA6 protein amino acid sequence indicated that this protein is highly hydrophilic with a high content of polar amino acids such as Lys (12%), Glu (11%), and Thr (11%). In addition, it contains a high percentage of Gly residues (13%), and thus it is considered a hydrophilin (6). A comparison of the amino acid composition of group 6 LEA proteins against Protein Data Base (PDBS25) using a Composition Profiler software (31) confirmed bias in the amino acid composition of this protein family, showing a conspicuously higher representation of disorder promoting amino acids (Fig. 1B). Consistent with its physicochemical characteristics, structure predictors suggested that PvLEA6 is a disordered protein; however, it shows a 20-amino acid region for which some structural order is predicted where the four Tyr residues of the protein are concentrated (Fig. 1C) (16). Computational analyses using ANCHOR (32) and MoRFpred (33) predict that this segment may correspond to a protein binding region (Fig. 1D). Interestingly, this region is highly conserved among the members of this family, with minimal variations in the Tyr residues (Fig. 1A) (5).
FIGURE 1.
In silico analysis of the PvLEA6 protein sequence. A, alignment of group 6 LEA proteins showing the conserved regions and distinctive motifs of the family. B, comparative analysis of amino acid composition between group 6 LEA proteins and a subset of ordered proteins amenable to crystallization studies (PDBS25) using Composition Profiler software (31). Red, blue and gray bars refer to amino acid residues promoting disorder, order, or neutrality, respectively. C, prediction of ordered/disordered regions in PvLEA6 protein using different disorder predictors as indicated. Numbers in the x axis correspond to those in the upper line in A. D, putative protein binding regions in PvLEA6 protein predicted using ANCHOR (32) and MoRFpred (33).
The first indications of the unstructured nature of PvLEA6 were observed during initial characterization of the protein. rPvLEA6 protein was produced and purified from E. coli, from which a highly pure native protein was obtained that migrated in SDS-PAGE with an apparent molecular mass of 14 kDa (Fig. 2A; see “Experimental Procedures” for details). The molecular mass and amino acid sequence of the purified rPvLEA protein were proven by LC-MS, showing a molecular mass of 8.76 kDa (data not shown), in agreement with informatic predictions, and the same amino acid sequence as that deduced from its open reading frame, as reported previously (18). Therefore, indicating that the difference in its molecular mass obtained by SDS-PAGE migration or by LC-MS is likely due to the absence of hydrophobic clusters and therefore a low interaction with SDS (Fig. 1), and it is consistent with its predicted structural disorder, which usually gives rise to large protein hydrodynamic dimensions (34).
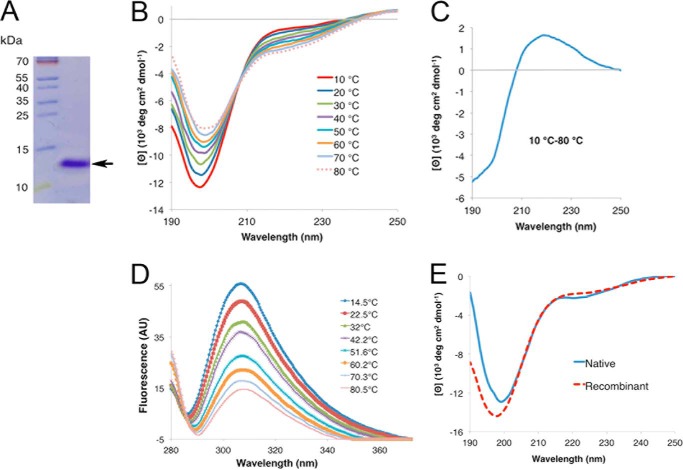
FIGURE 2.
Circular dichroism spectroscopy analysis of rPvLEA6 protein. A, separation by SDS-PAGE of the purified rPvLEA6 protein used in these analyses. Molecular mass markers are shown to the left of the figure in kDa. B, CD spectra showing the effect of temperature on the secondary structure of rPvLEA6. C, CD difference spectrum obtained subtracting the CD spectrum at 80 °C from the CD spectrum at 10 °C (10–80 °C). D, rPvLEA6 protein analyzed by UV fluorescence showing the Tyr residue emission spectra obtained at different temperatures. The maximum emission fluorescence of Tyr residues occurs at 307 nm. AU, arbitrary units. E, CD spectrum of the native PvLEA6 protein obtained from dry common bean embryos compared with that from rPvLEA6 protein. Proteins were in 10 mm potassium phosphate buffer, pH 8.0.
To test the disordered nature predicted for PvLEA6 protein, its secondary structure in aqueous solution at pH 8.0 (10 mm potassium phosphate) was explored by CD spectroscopy in the far-UV light. The spectrum showed a strong minimum ellipticity around 200 nm, indicative of a largely unordered conformation (Fig. 2B). However, there is experimental and theoretical information suggesting that similar spectra can be obtained from polypeptides containing some local order such as extended helix or β-like structures (35–40). To explore whether the amino acid charges could favor a particular conformation, CD spectra were obtained from the protein at different pH values. The results showed similar CD spectra profiles indicating that changes in the protein amino acid charges between pH 5.7 and 8.0 do not produce detectable structural modifications (data not shown). The effect of temperature on rPvLEA6 protein conformation was also investigated by CD analysis in a temperature range from 10 to 80 °C (Fig. 2B). In this case, the temperature rise led to a significant and progressive decrease in the intensity of the negative band at 197–200 nm and to a gradual increment in the intensity of the negative signal between 210 and 235 nm, corresponding to a wavelength band, where dichroic signals have been detected for polyproline II (PPII) or β-like structures (41, 42). The intensity changes in these two negative signals suggested that, in contrast to pH, temperature was promoting protein conformational modifications, which, as has been reported for other IDPs, are also consistent with the formation of secondary structure (43, 44). Furthermore, the analysis of these data uncovered the presence of a clearly defined isodichroic point at 208 nm and −4.7 × 103 degrees cm2 dmol−1, in the temperature range analyzed, suggesting that the rPvLEA6 protein in aqueous solution undergoes a transition between two structural conformations (Fig. 2B). One conformation corresponded to a random coil unordered structure and PPII-like extended helices, this last favored at low temperatures, and a second one represented by unordered and β-like structures, as revealed by the increase in the negative signal between 215 and 220 nm that occurs with the gradual temperature increase (Fig. 2B) (36, 37, 39–42). Consistent with this, the CD difference spectrum (Δ10–80 °C) showed a negative signal near 200 nm and a positive band with a maximum at 219 nm (Fig. 2C), characteristics that have been reported for peptides where PPII-like conformations are common (41), and in group 2 and group 1 LEA proteins from soybean, where this extended helix conformation was also detected at low temperatures (45, 46). This phenomenon has also been observed in other intrinsically disordered proteins (40) and in small unstructured model peptides (36, 37, 39). It is worth mentioning that similar CD spectra were obtained for the rPvLEA6 protein and from that purified from common bean embryos (Fig. 2E), suggesting that this kind of structure is also predominant in vivo.
To monitor changes in the local environment around aromatic residues, UV fluorescence spectroscopy was used. PvLEA6 protein contains only five aromatic residues, one Phe at position 67 and four Tyr at positions 26, 29, 35 and 40. As shown in Fig. 2D, upon excitation at 274 nm, the Tyr fluorescence emission occurred at 308 nm. An increase in fluorescence intensity would be indicative of Tyr residues being in a more rigid and hydrophobic environment (47); however, for this protein temperature induced a decrease in fluorescence intensity, a general phenomenon due to a diminish quantum yield of the Tyr residue fluorescence at high temperatures (48, 49). This effect could counteract any change in intensity due to formation of some tertiary interactions.
Although far UV-CD signal is reflecting the general behavior of the polypeptide chain, near UV-CD shows changes in the environment around the fluorescent probes in the protein. Hence, as an alternative attempt to determine whether Tyr residues could be sensing some conformational changes, the near-UV CD spectrum at different temperatures was obtained. The spectrum profile showed a small but sharp peak around 278 nm indicative of some rigidity of at least one of the Tyr residues present in the protein (Fig. 3) (50, 51). The intensity of this band did not change significantly with temperature; however, the band seems to broaden as temperature increases. In addition, three isodichroic points were detected at 254, 271, and 282 nm, which indicate changes in the structure of this protein (Fig. 3). A negative band around 294 nm was also detected, which cannot be attributed to Trp residues given their absence in this protein; therefore, a likely explanation is that this signal originates from the phenolate state of Tyr. Interestingly, as temperature increases, this negative signal decreases monotonically and shows a slight red shift up to 296 nm at 80 °C (Fig. 3), indicative of a more hydrophobic environment for the chromophore responsible for this signal.
FIGURE 3.
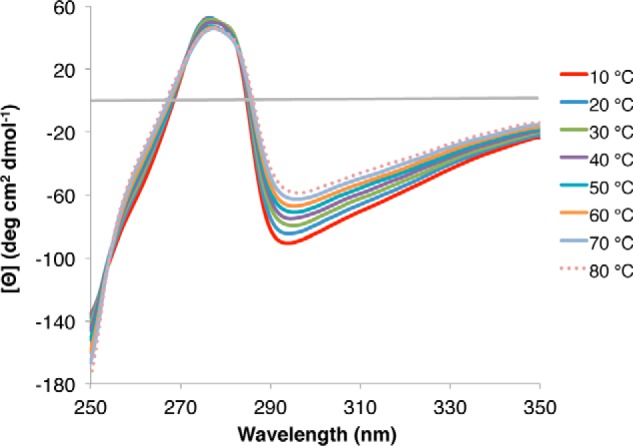
Near-UV circular dichroism spectroscopy analysis of rPvLEA6 protein. Near-UV CD spectra showing the effect of different temperatures (10–80 °C) on rPvLEA6 secondary structure. CD spectrum in this UV region is attributable to phenylalanine (250–270 nm), tyrosine (270–290 nm), and tryptophan residues (280–300 nm), and thus changes in the local environment of these amino acids could be detected.
PvLEA6 Protein Has an Intrinsic Propensity to Fold in α-Helical Conformation
TFE has been widely used in the study of the conformational properties of proteins. Although no single mechanism accounts for all TFE effects on protein conformation, it is well known that this co-solvent promotes the helicity of peptides and proteins depending on their amino acid sequence, such that it actually increases the proportion of helices in those protein regions with an intrinsic proclivity to form α-helical secondary structure (52–54).
Even though the CD analysis showed that rPvLEA6 protein is unstructured in aqueous solution, it is possible that this protein has an intrinsic propensity to form secondary structure when exposed to particular environments. To investigate such a possibility, we analyzed the effect of different TFE concentrations on rPvLEA6 protein structural organization by CD spectroscopy. This study showed that the presence of increasing TFE concentrations promotes and stabilizes concomitantly larger levels of α-helices, as revealed by the progressive appearance of a positive band at ∼190 nm and two negative bands at 208 and 222 nm (Fig. 4A). In the presence of 20% (v/v) TFE, rPvLEA6 did not show a significant change in its helicity; however, with 60% (v/v) TFE, rPvLEA6 acquired a significant calculated value of α-helical structure (∼37%), which increased further (∼41%) in the presence of 90% (v/v) TFE (Fig. 4A). This effect is clearer in the difference spectrum, where the typical signals attributed to α-helix conformations are evident as follows: a positive band at 190 nm and negative bands between 208 and 222 nm (Fig. 4B). The lack of an isodichroic point in this case indicates that more than two structural species participate in this transition.
FIGURE 4.
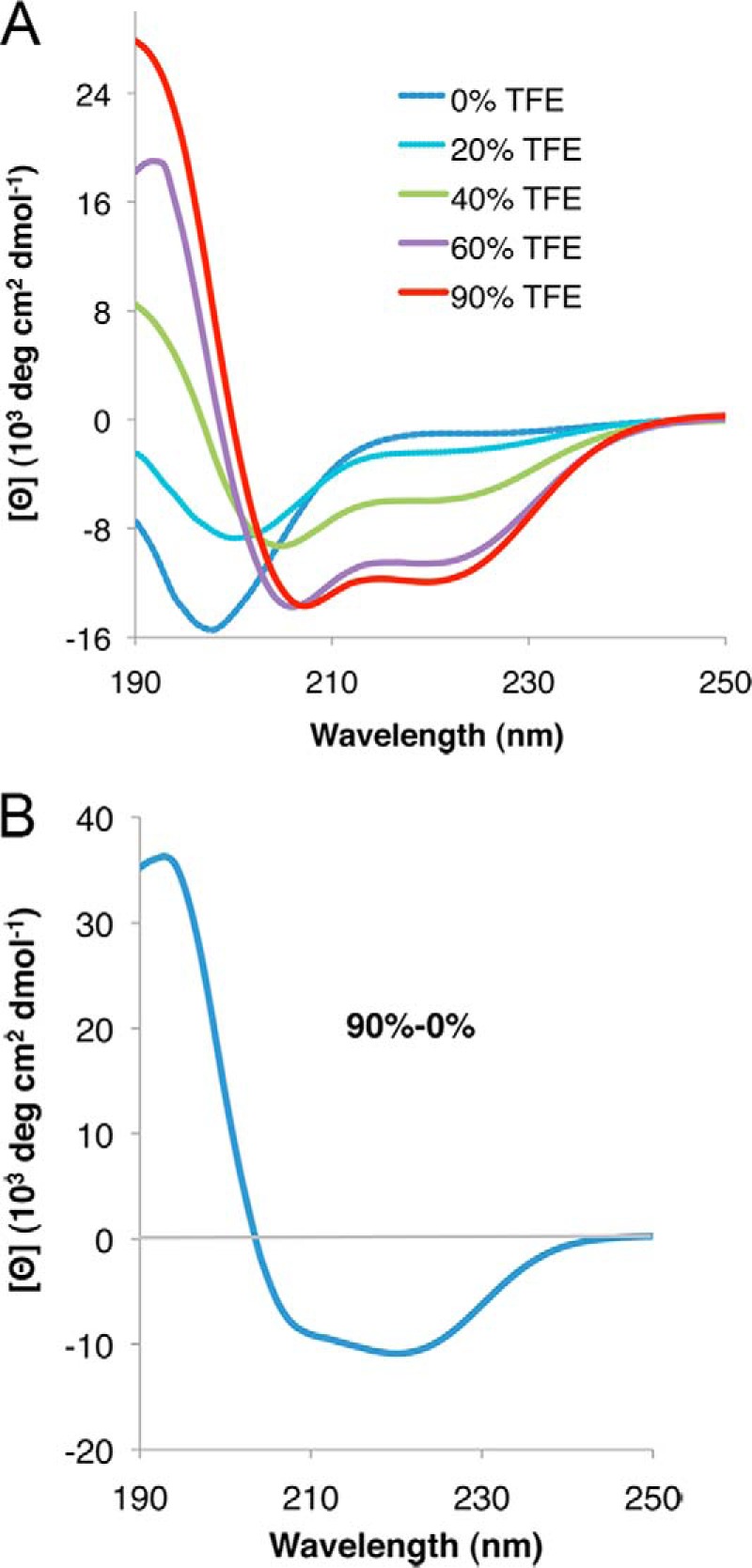
rPvLEA6 protein analyzed by CD spectroscopy in the presence of TFE. A, CD spectra of rPvLEA6 protein in the presence of different concentrations of TFE (0–90%). B, CD difference spectrum obtained by subtracting the CD spectrum obtained at 0% TFE from the CD spectrum obtained at 90% TFE (90 to 0%).
To obtain a deeper insight into the global PvLEA6 structural organization, two-dimensional NMR spectra were recorded using a 15N-labeled protein. As a first approach, a two-dimensional 1H-15N heteronuclear single quantum coherence correlation spectrum was obtained, which shows the correlation between the 15N and 1H resonance frequencies (Fig. 5). The low level of chemical shift dispersion apparent in the 1H dimension clustered between 7.7 and 8.5 ppm (Fig. 5A) was indicative of structural disordered state for this protein under native conditions in aqueous solution. A two-dimensional 1H-15N heteronuclear single quantum coherence correlation spectrum was also obtained for the protein in the presence of 62.5% TFE, a condition where gain of structural order was previously observed in the CD spectroscopy analysis. As shown in Fig. 5B, TFE induced larger spreading of the proton chemical shift spectrum when compared with the protein in the absence of the co-solvent indicating structural changes in the protein. The dispersal of some N-H correlation peaks toward lower parts/million values than 8.0 ppm and higher than 8.5 ppm suggested the attainment of structure (Fig. 5B), in agreement with CD data.
FIGURE 5.
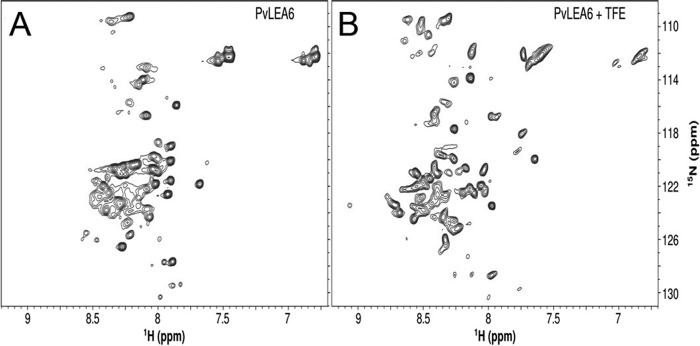
Two-dimensional 1H-15N correlated NMR spectra of rPvLEA6. A, NMR spectrum of rPvLEA6 in 10 mm KHPO4 buffer, pH 8. B, NMR spectrum of rPvLEA6 in 62.5% TFE. The spectra were recorded on a 700 MHz spectrometer at 298 K.
High Osmolarity and/or Molecular Crowding Induce Changes in the Structural Organization of the PvLEA6 Protein
The delicate balance in the interaction between the intrinsic physicochemical properties of a protein and its environment guides its structure and stability, as reflected in the interactions that stabilize and/or destabilize a folded or unfolded polypeptide. The structural flexibility of disordered proteins implies a relevant role of the solvent environment such that some proteins might exhibit significantly different folding behavior depending on the solvent properties and in response to changes in their milieu.
Given the fact that PvLEA6 protein accumulates under low water availability in plant tissues and its intrinsic propensity to acquire some helicity as shown by the TFE treatments, it is expected that environments simulating this physiological condition could induce a different folding behavior than that observed in the aqueous solutions tested. To simulate this condition we used glycerol, a natural osmolyte that accumulates in diverse organisms upon water deficit treatments. CD spectra of rPvLEA6 showed that the addition of different concentrations of glycerol (0–87%) induces various changes as follows: the appearance of a positive ellipticity signal close to 190 nm; a slight migration of the minimum in the negative band from 198 to 204 nm with a decrease in the negative signal upon addition of higher glycerol concentrations; and an increase of the negative signal between 215 and 235 nm (Fig. 6A). The difference spectrum in Fig. 6A (inset) shows the more evident structural changes taking place in the presence of this osmolyte, which according to Dichroweb analysis represent 16–20% formation of α-helix, indicating that despite the slight modifications, environmental variations might promote a differential folding behavior in the protein.
FIGURE 6.
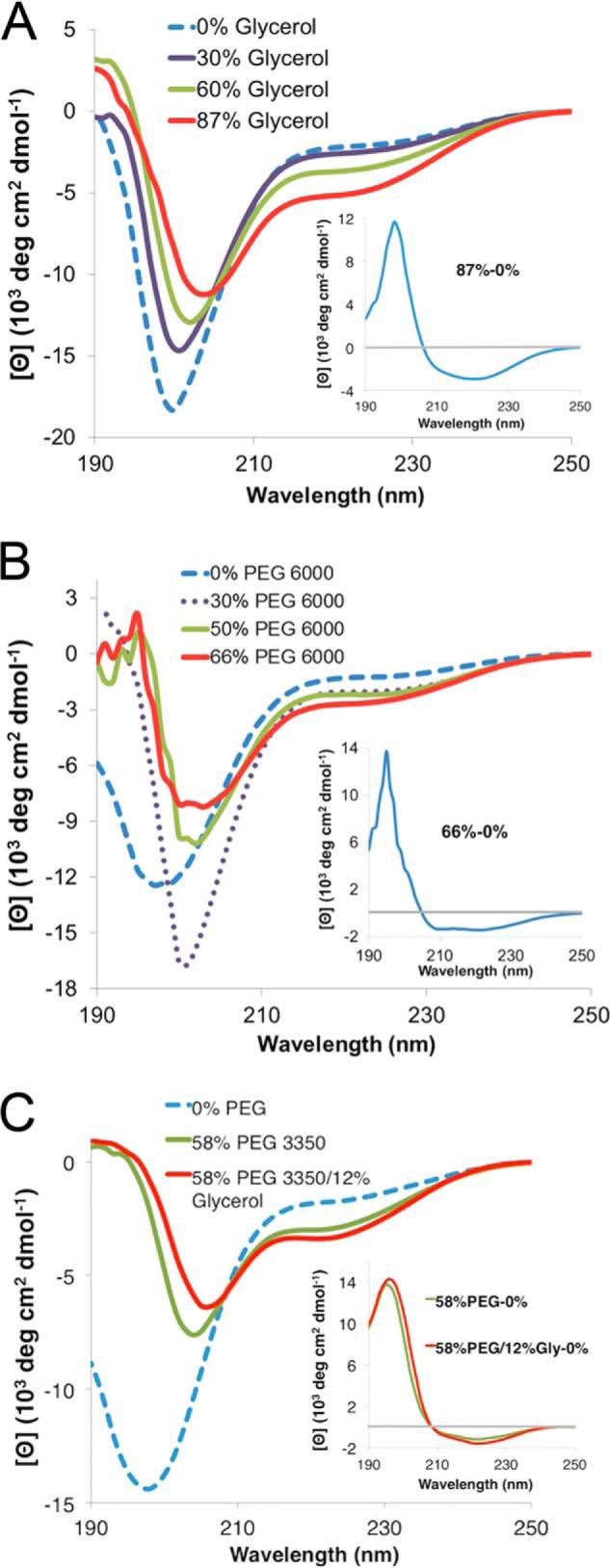
Effect of high osmolarity and molecular crowding on rPvLEA6 protein secondary structure. A, CD spectra in the presence of different glycerol concentrations (0–87%). Inset, CD difference spectrum obtained by subtracting the CD spectrum obtained at 0% glycerol from the CD spectrum obtained at 87% glycerol (87 to 0%). B, CD spectra in the presence of different PEG-6000 concentrations (0–66%). Inset, CD difference spectrum obtained by subtracting the CD spectrum obtained at 0% PEG-6000 from the CD spectrum obtained at 66% PEG-6000 (66 to 0%). C, CD spectra in the presence of PEG-3350 (58%) or PEG-3350 (58%) plus glycerol (12%). Inset, CD difference spectra obtained by subtracting the CD spectrum at 0% PEG-3350 from the CD spectrum obtained at 58% PEG-3350 (58 to 0%) or by subtracting the CD spectrum at 0% solutes from the CD spectrum obtained at 58% PEG-3350 plus 12% glycerol (58/12 to 0%).
An additional factor that will be also present in the cellular environment under low water availability is an increase in the molecular crowding present inside the cell, which represents a complex situation, where the concentration of not only ions and osmolytes rises but also that of different macromolecules such as carbohydrates, nucleic acids, and proteins; a situation frequently neglected in most of the in vitro characterization analyses of proteins. To simulate, at least, a simple molecular crowding condition, model crowding agents such as PEG and Ficoll were used, and their effect on PvLEA6 structural properties was analyzed by CD spectroscopy. Different concentrations of PEG-6000 (0–66%) also induced structural changes in the PvLEA6 protein (Fig. 6B); however, in this case a trend to form α-helices is more evident given the presence of a positive band near 190 nm and the characteristic negative bands in 208 and 222 nm as revealed by the difference spectrum (Fig. 6B, inset); thereby indicating that these crowding conditions promote modifications in the structural properties of this protein, although to a small extent. For this case, Dichroweb analysis was estimated between 12 and 16% of α-helix formation. As for TFE, there is no isodichroic point indicating the inter-conversion of more than two species in a more complex process. A comparable effect was observed using PEG-3350, PEG-4000, or PEG-8000; however, when Ficoll-70 (0–58%) was used, no effect was detected (data not shown).
To analyze the combined effect of molecular crowding and low water availability, we obtained the CD spectrum of rPvLEA6 protein in the presence of PEG-3350 and glycerol using the highest solute concentration possible, 58% PEG-3350 and 12% glycerol. However, no further significant changes were detected in addition to those observed with only 58% PEG-3350 (Fig. 6C).
PvLEA6 Protein Forms Oligomers
During the characterization of the purified rPvLEA6, we realized that the loading of large amounts of this protein in SDS-PAGE allowed for the detection of additional bands with an apparent molecular mass higher than that of the monomer (data not shown). These bands were detected by the PvLEA6-specific antibody and were not apparent when the antibody was previously incubated with the purified protein indicating that they corresponded to monomer ensembles that match with possible dimers and trimers according to their apparent molecular mass (∼24.6 and 37.8 kDa, respectively) (Fig. 7A).
FIGURE 7.
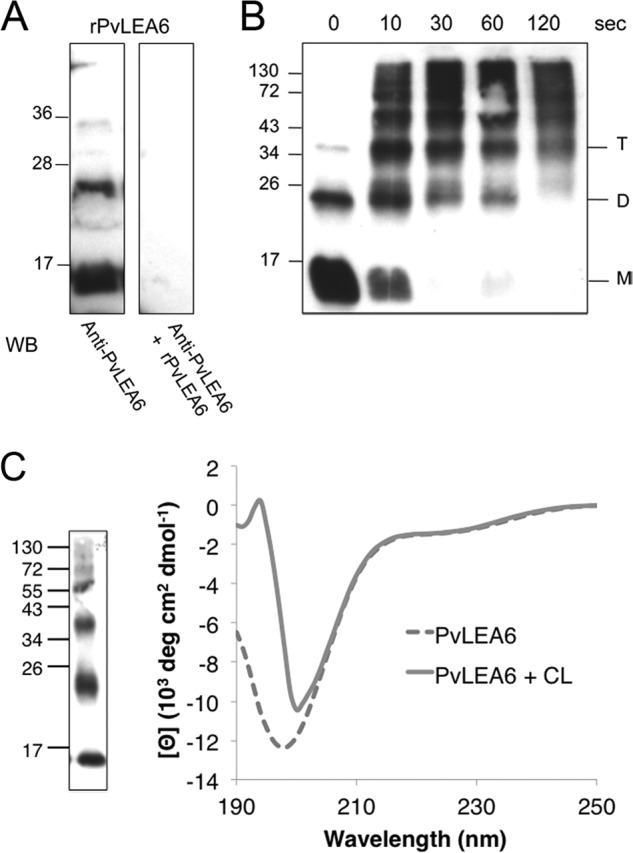
Analyses of the rPvLEA6 protein oligomeric forms. A, Western blot (WB) assay using anti-PvLEA6-specific antibody or anti-PvLEA6-specific antibody previously incubated with the rPvLEA6 protein. Purified rPvLEA6 protein was separated in 15% SDS-PAGE. B, Western blot assay of rPvLEA6 protein cross-linked by photoinduction using Ru(II)bpy32+ during different times as indicated. Cross-linked or noncross-linked purified rPvLEA6 protein was separated in 15% SDS-PAGE. Molecular mass markers are indicated in kDa at the left of gel images. M, monomer; D, dimer; and T, trimer. C, CD spectra of cross-linked (CL) and noncross-linked rPvLEA6 proteins. At the left of the figure, a Western blot of the cross-linked rPvLEA6 protein used to obtain this CD spectrum is shown. Protein was separated in 15% SDS-PAGE and probed with specific anti-PvLEA6 protein antibody.
To further investigate this possibility and add information on its structural organization, PICUP was applied to purified rPvLEA6 protein, a method that does not require previous protein modifications nor the inclusion of the cross-linking agent, thus enabling covalent bond formation between closely interacting polypeptides (see “Experimental Procedures” for details). Several time points were taken upon photoinduction as indicated, and small aliquots were resolved by SDS-PAGE, and the products were detected with anti-PvLEA6 antibody (Fig. 7B). The results showed the formation of various oligomeric products of monomer addition ranging from dimer through hexamer, and even higher molecular mass complexes, which increase with the cross-linking time together with the decreasing of the monomer signal, supporting the idea that rPvLEA6 is able to form oligomers.
To verify this observation, an rPvLEA6 sample was separated by gel filtration chromatography. Fractions were collected and analyzed by immunoblotting using anti-PvLEA6 antibody. Although the larger oligomers were barely detected due to the amount of the loaded sample and probably to the buffer composition (see below), chromatograms allowed the detection of monomers, dimers, and scarcely trimers, which were separated as follows: rPvLEA6 monomer between fractions 20 and 24, dimers between fractions 16 and 20, and trimers overlapping with some dimers in fractions 16–18 (indicated by M, D, T, respectively in Fig. 8, A and B). The molecular mass deduced from this separation method for each of the species obtained was 23.6, 45.8, and 57.8 kDa, respectively. To have a more precise determination of the molecular mass of these rPvLEA6 oligomers, a cross-linked sample was subjected to MALDI-TOF-MS analysis. The spectrum, obtained only for protein molecules smaller than 50,000 Da, showed three peaks at 8760, 17,520, and 26,280 Da, corresponding to monomer, dimer, and trimer, respectively (Fig. 8C). The differences between the molecular masses estimated by SDS-PAGE (see above) and gel filtration chromatography with those obtained by mass spectrometry suggested that oligomeric forms maintain the monomer structural disorder. In agreement with this, the apparent high molecular mass deduced from gel filtration chromatography (size exclusion) reflects a large hydrodynamic radius as expected for the extended structures in a disordered protein. To validate this inference, a similarly cross-linked rPvLEA6 sample was analyzed by CD. Results showed that indeed the association between rPvLEA6 monomers does not lead to significant structural modifications (Fig. 7C).
FIGURE 8.
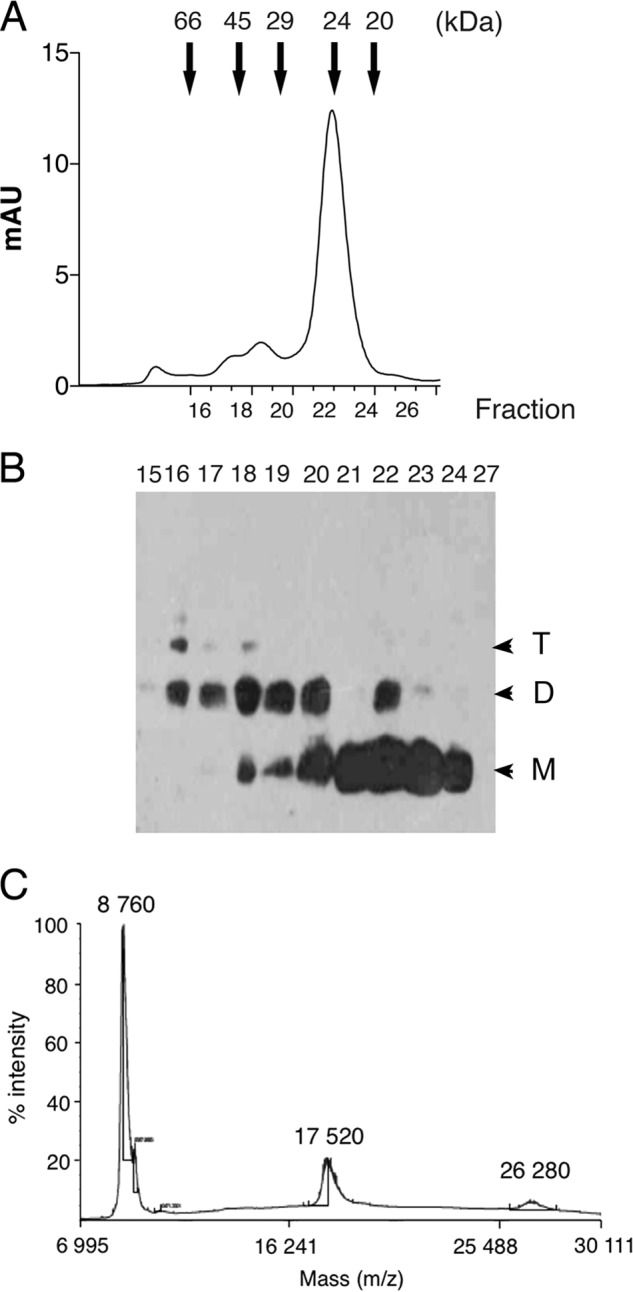
Characterization of PvLEA6 oligomers. A, elution profile from the FPLC Superdex-75 (10/300 GL) gel filtration high performance column. Arrows indicate the molecular mass markers used for the corresponding calibration curve as follows: bovine serum albumin 66 kDa; egg albumin 45 kDa; carbonic anhydrase 29 kDa; trypsinogen 24 kDa; and trypsin inhibitor 20.1 kDa. B, Western blot using the specific antibody anti-PvLEA6 protein of relevant fractions obtained from FPLC and separated in a 15% SDS-PAGE. M, monomer; D, dimer; and T, trimer. C, MALDI-TOF mass spectrometry analysis of cross-linked rPvLEA6 protein showing their corresponding molecular mass. mAU, milli-absorbance units.
The detection of oligomeric forms when rPvLEA6 was separated by SDS-PAGE suggested that these intermolecular interactions were SDS-resistant. The composition of this protein shows a high percentage of charged residues, the absence of Cys or Trp, and the low representation of other hydrophobic residues such as Phe (1), Tyr (4), Leu (4), or Val (2). Given the presence of Tyr, the possibility that covalent di-tyrosine bonds were accountable for oligomer formation was investigated. To rule out this possibility, the gel band corresponding to dimeric forms was obtained and subjected to MS analysis, which did not detect di-tyrosine covalently bonded peptides (data not shown), allowing us to conclude that SDS-resistant oligomers were not the result of covalent associations between tyrosine residues. SDS-resistant intermolecular interactions have been identified in a number of proteins leading to the recognition of some of the structural determinants involved (55), suggesting that the interaction between the negatively charged SDS and the polar PvLEA6 monomer interface is not a favorable process, an idea also supported by the anomalous migration observed in SDS-PAGE. Moreover, the abundance of charged residues in the PvLEA6 protein suggested that its homo-oligomers could be stabilized by a network of polar interactions between monomer interfaces, consequently resistant to dissociation by SDS. To explore this possibility, photo-induced cross-linking reactions were carried out under different ionic strengths, using different NaCl concentrations. Results in Fig. 9 showed that salt interferes with monomeric interface interactions, preventing an efficient formation of the oligomeric forms and indicating that salt bridges and other ionic interactions participate in the oligomerization process.
FIGURE 9.
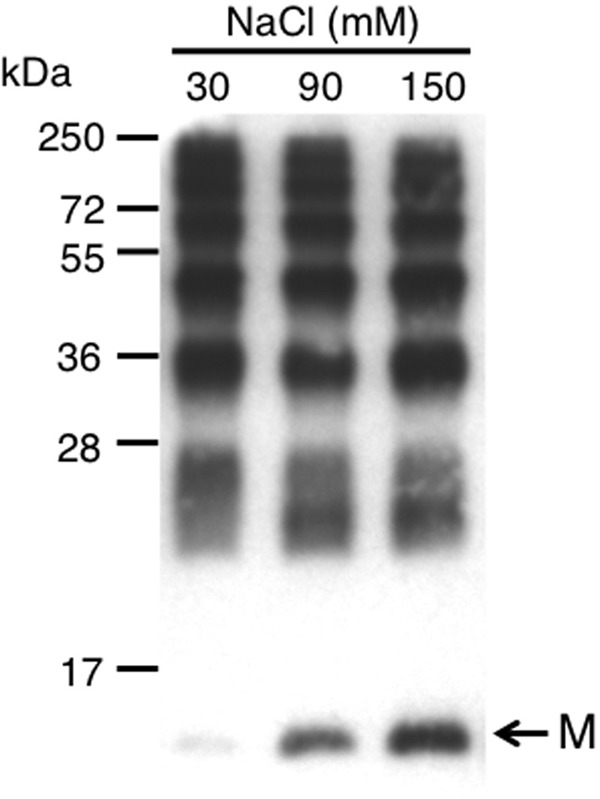
Effect of different NaCl concentrations on the formation of rPvLEA6 oligomers. Purified rPvLEA6 protein was cross-linked by photo-induction using Tris-bipyridylruthenium(II) di-cation Ru(II)bpy32+ in the presence of different NaCl concentrations (30, 90, and 150 mm). Protein was separated in 15% SDS-PAGE and probed with specific anti-PvLEA6 protein antibody. M, monomer. The migration position of the molecular mass markers is shown to the left.
Calorimetric Determination of PvLEA6 Dimer Dissociation Constant
To evaluate the self-association strength of rPvLEA6 protein, the dissociation constant (Kd) was determined by IDC. The stepwise addition of a high concentration protein solution into the calorimetric cell, initially loaded with buffer solution, evolved endothermic heat indicative of homo-oligomer dissociation. The solid line in the dissociation isotherm corresponded to the best fit of a dimer dissociation model (Fig. 10). The use of higher order dissociation models yielded no fitting convergence and/or too large standard errors of the fitting parameters (data not shown). Hence, the good fitting observed with the dimer model is consistent with rPvLEA6 monomer-monomer interaction mostly at a 1:1 ratio, indicating the predominant formation of dimers (Fig. 10). The measurements (carried out in triplicate) yielded a Kd = 60 ± 20 μm and a dissociation enthalpy, ΔHd, = 1.0 ± 0.3 kcal/mol. This information allowed estimation of the following thermodynamic parameters for dimer formation (30 °C): ΔGa = −5.9 kcal/mol; ΔHa = −1.0 kcal/mol; and TΔSa = 4.9 kcal/mol, consistent with a low affinity, mainly entropically driven interaction between rPvLEA6 monomers under these conditions.
FIGURE 10.
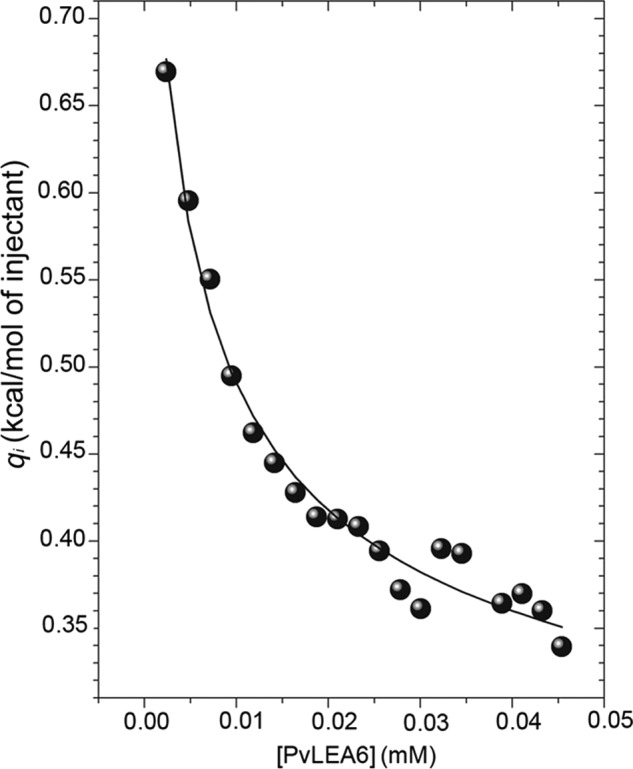
Calorimetric dissociation isotherm of rPvLEA6 protein. Twenty aliquots of 1 μl of LEA protein (0.47 mm, monomer equivalent concentration) were added stepwise to the calorimeter cell loaded with buffer solution. Isotherm was obtained at 30 °C in 10 mm KHPO4 buffer, pH 8.0. The solid line represents the best fitting of a simple dimer dissociation model to the calorimetric data.
PvLEA6 Dimer Is Formed in Vivo
To determine whether PvLEA6 monomer-monomer interaction was occurring in living plant cells, the BiFC system was used. For this assay, PvLEA6 ORF was independently fused to two nonfluorescent but complementary fragments of YFP and introduced into N. benthamiana leaf cells by Agro-infiltration (see “Experimental Procedures” for details) for fluorescence analysis. As shown in Fig. 11, fluorescence was detected only when the mixture containing the YFPN43-PvLEA6 and YFPC43-PvLEA6 was infiltrated but not when any of these were individually introduced into plant cells, indicating that the two nonfluorescent YFP fragments were brought together by the interaction between PvLEA6 monomers, hence forming a fluorescent complex. This experiment also allowed us to observe the localization of the protein complex in the cytosol and in the nucleus, in agreement with previous protein immunolocalization data of the endogenous protein in the common bean (17).
FIGURE 11.
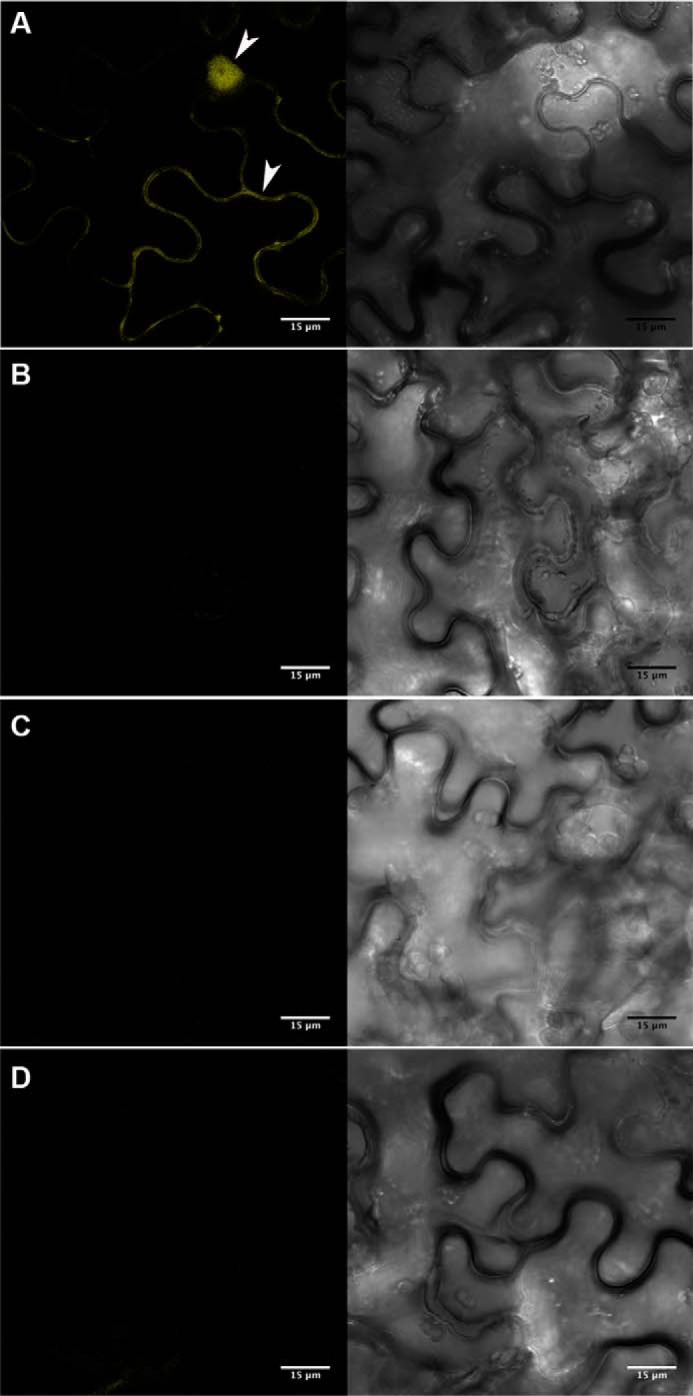
Visualization of PvLEA6 dimer using BiFC assay in N. benthamiana epidermis cells. Complete PvLEA6 ORF was cloned in BiFC vectors (27). Transient expression was analyzed by confocal microscopy. A, PvLEA6 dimers were detected in cytoplasm and nuclei of epidermal cells co-transformed with YFPN43-PvLEA6 and YFPC43-PvLEA6. White arrowheads indicate cytosol and nuclear signals. Cytosol is pressed by a large central vacuole against cell membrane. No fluorescence was detected in controls. B, pYFPN43 and pYFPC43, empty vectors. C, YFPN43-PvLEA6 and pYFPC43. D, pYFPN43 and YFPC43-PvLEA6. Scale bar corresponds to 15 μm. Both images within A–D correspond to the same field. Images obtained with UV light are shown at the left side and with white light at the right side.
DISCUSSION
Structural disorder in proteins is now recognized as a relevant property involved in many important functions in organisms from the different kingdoms of life (9). From the many IDPs predicted in plants (95 in Arabidopsis), very few have been structurally and/or functionally characterized (11, 56). By prediction, most typical LEA proteins constitute one of the sets of proteins considered as IDPs, whose accumulation is remarkably associated with water deficit conditions (5, 6, 11). Despite their common response to similar developmental and stress cues, the lack of significant sequence similarity between the different groups of LEA proteins indicates functional diversity between them. Although LEA proteins from some particular groups, such as group 2 or dehydrins, have been extensively studied, the structural and functional properties of some others are rather poorly characterized. This is the case of group 6 LEA proteins, one of the most conserved LEA protein families. Previously, we have studied the transcript and protein accumulation patterns of PvLEA6 gene, the only member of this family in common bean (15, 18), and shown its close association with a reduction in water availability, even with that present during normal development under optimal growth conditions (17). In this work, we investigated the structural characteristics of this protein, showing that, as predicted, it contains a high level of disorder, similar to that obtained for the Arabidopsis group 6 LEA proteins (19), a characteristic that was also shown for the native PvLEA6 protein obtained from common bean dry embryos (this work). The analysis of the temperature effect on the CD spectra showed that rPvLEA6 protein undergoes a structural transition that, given the presence of an isodichroic point, indicated that this protein in aqueous solution is in equilibrium between two conformational states, a phenomenon also observed in other LEA proteins from group 1 (GmD-19) and group 2 (GmDHN1) (45, 46). According to the changes observed in the spectrum signals, the rPvLEA6 protein could transit between unordered and PPII- or extended β-like structures. The negative CD band at 200 nm and the positive band with a maximum between 215 and 220 nm shown in the difference spectrum are consistent with the presence of left-handed extended helices of PPII-like conformations at low temperatures (41). The presence of the PPII helical structure in polypeptides with the abundance of highly soluble charged amino acid residues is an energetically favorable alternative, which allows an extensive formation of hydrogen bonds with the solvent, and seems to be a common property of protein unfolded states, hence suggesting an important component of structural elements in proteins considered as structurally disordered (35, 57). An alternative and likely explanation for the temperature-induced changes in the rPvLEA6 far-UV CD spectra is the formation of secondary structure promoted by the increasing temperatures, which has been proposed to be driven by hydrophobic interactions and has also been reported for various IDPs (43, 44).
Because the detection by UV fluorescence spectroscopy of possible tertiary interactions could be hindered by a reduction in the quantum yield of tyrosine fluorescence induced by temperature, a near-UV CD spectroscopy at different temperatures was carried out. The three isodichroic points exhibited by this analysis suggested changes in the local environment of more than one amino acid residue in this protein, possibly those corresponding to Phe and to one or more of the Tyr residues. The isodichroic points at 271 and 282 nm are in a region corresponding to Tyr signals (50), suggesting that some of the Tyr residues may be in equilibrium between three conformational stages or, alternatively, two Tyr in equilibrium with two conformations. These profiles are consistent with temperature-induced changes in the structure of this protein as suggested by far-UV CD spectroscopy. Moreover, the progressive reduction in the signal of the band around 294 nm with increasing temperature together with its migration up to 296 nm at 80 °C was also suggestive of modifications in the environment of another chromophore, possibly the phenolate form of Tyr. Albeit more experiments would be needed to attribute the behavior observed in the near-UV CD for the negative signal at 294 nm to a particular residue, it is possible that the region responsible for these changes is the sequence from residues 25 to 32 (PYVKYKDL), a region for which some secondary structure was predicted, and that shows a small stretch of hydrophobic residues that could be forming some local structure. This stretch is followed by another Tyr residue flanked by two Lys that could be affecting its pKa value (favoring the phenolate form), and then an aspartic acid that could be interacting with one of the Lys through a salt bridge. The decrease of the band at 294 nm is consistent with a reduction in the phenolate fraction expected to occur as electrostatic interactions decrease upon heating. Alternatively, the decrement in this band could be reflecting a less constrained environment for the residue responsible for the signal.
Proteins in the cell are under conditions where water availability is lower and molecular crowding is higher than those commonly present during their in vitro characterization. These conditions are even more extreme when cells are under water deficit. Structural changes upon severe dehydration intending to simulate the dryness of the seed or the anhydrobiosis of ferns, mosses, and their spores (58) have been reported for LEA proteins from various groups, including Arabidopsis group 6 LEA proteins (19, 59, 60); however, LEA proteins also accumulate under conditions where water limitation is not so extreme, even where stress is rather mild (water potentials of approximately −6.5 bars in common bean hypocotyl tissues) (5, 16, 17). In this work, we showed that PvLEA6 proteins possess an intrinsic ability to attain some structural order, given the progressive gain of α-helices upon treatments with increasing TFE concentrations. This capacity was also detected under low water availability induced with increasing glycerol concentrations and/or molecular crowding simulated with different PEG concentrations. This attainment of structure agrees with the prediction of some structural order (up to 40%) obtained from the PvLEA6 amino acid sequence analysis, order that seems to occur in the most conserved region, containing motifs 1 and 3, which in turn cluster the four highly conserved Tyr residues present in this protein. Although the observed changes were between 12 and 16% of α-helix formation, upon low osmotic potentials and/or molecular crowding induced with PEG, these slight structural modifications were consistent and reproducible in independent experiments, indicating that these low levels of helicity could play a significant role in vivo. The conformational dynamics that individual proteins might experience, particularly those with such a high degree of flexibility as that expected for structurally disordered proteins (9, 61–63), lead us to consider small structural modifications as relevant for their function, so as to attain the proper conformational adaptation for a functional interaction with, for example, their corresponding ligands under specific environmental conditions.
Many proteins in the cell modulate their function(s) by the association of their monomeric units. Some of them are active as oligomers, and others exhibit their activity only as monomers (64). In this work, we demonstrate that PvLEA6 protein can form oligomers, dimers being the most conspicuous association, but with the capacity to organize itself into higher order homo-oligomeric complexes, as shown by PICUP, a method that allows the detection of not only high affinity complexes but also metastable protein ensembles because of its capacity to cross-link proteins within very short time intervals (1 s) (22). The abundance of polar and charged amino acid residues in PvLEA6 protein suggested that polar interactions and salt bridges played a major role in the establishment of PvLEA6 monomer associations, which was consistent with the inhibition of oligomerization by increasing the ionic strength in the solution. The interaction between two highly flexible PvLEA6 monomers presumes a dynamic process, where transient protein-protein interactions might be responsible for PvLEA6 oligomer formation. The determined Kd of 60 ± 20 μm for dimer formation supports this statement, implying a low binding affinity and therefore a weak transient interaction (65) between PvLEA6 monomers.
Even though some of the data indicated that polar-to-polar interactions have a predominant role in dimer stabilization, calorimetric analysis showed that monomer self-association was entropically driven, accompanied by a small favorable enthalpy change. This thermodynamic signature is typically considered to be an indication of hydrophobic interactions, although enthalpically driven profiles are more commonly observed in predominantly electrostatic interactions. Nevertheless, it is worth recalling that the overall binding enthalpy between polar groups depends largely on the stereochemical details of the interaction (66). For instance, at room temperature the purely electrostatic recognition between magnesium and ATP ions is entropically driven (TΔS = 10.6 kcal/mol), opposed by a substantial endothermic enthalpy (ΔH = 4.4 kcal/mol) (67). In contrast, if magnesium interacts with ATP previously bound to a protein's binding site, where the octahedral coordination of the cation is fully achieved, the enthalpic component becomes favorable (67, 68). In this regard, it is interesting to compare PvLEA6 behavior with that of serine/threonine mammalian sterile 20-like kinase, MST1, an intrinsically unstructured protein, whose homodimerization energetics have also been characterized by IDC (69). MST1 monomers show affinity in the low micromolar range (Kd = 1 μm), forming a well structured antiparallel coiled-coil motif. By contrast with PvLEA6, MST1 evolves a large self-association enthalpy (ΔHdim = −16 kcal/mol). Accordingly, it can be concluded that PvLEA6 dimerization is driven by the formation of polar interactions at a not-fully structured interfacial environment. This observation is in agreement with the finding that PvLEA6 oligomers maintain a level of structural disorder similar to that detected for the monomer.
The BiFC system revealed that PvLEA6 dimers could be formed in living plant cells and showed that these dimeric complexes are localized to the cytosol and nuclei, a similar distribution to that heretofore described by immunolocalization (17). Dimer fluorescence also showed that this protein complex might be excluded from the nucleolus, in contrast to other LEA proteins also targeted to this organelle (5, 70–72). Given that the dimer dissociation constant indicated that this is a weak transient interaction, the rather general subcellular distribution of PvLEA6 dimers suggests that monomers are continually interacting with each other implying a functional relevance for this structural organization.
Although the PvLEA6 function remains unknown, the determination of PvLEA6 protein competence to form homo-oligomers, not only in vitro but also in vivo, uncovers alternative functional mechanisms to accomplish and or modulate its role during the plant response to water deficit. Furthermore, given the high conservation among the proteins in this group, it allows proposing oligomerization as a common property for this protein family. Although the formation of oligomers has been described for very few LEA proteins, all of them from group 2 or dehydrins (73–75), our finding calls the attention to explore this property in LEA proteins from the different groups, which may impact their in vivo function(s) and offer alternative models for their mechanisms of action.
Acknowledgments
We are grateful to César Cuevas for critical reading of the manuscript, Federico del Río for support on NMR techniques at the beginning of this work, and the following facilities of the Instituto de Biotecnologia-Universidad Nacional Autónoma de México for their services at different stages of this work: Laboratorio Universitario de Proteómica, Laboratorio Nacional de Microscopía Avanzada, Unidad de Síntesis y Secuenciación de ADN, and Unidad de Cómputo. We also thank the Laboratorio Nacional de Estructura de Macromoléculas for NMR analyses (Universidad Nacional Autónoma de México-Universidad del Estado de Morelos).
This work was supported in part by Consejo Nacional de Ciencia y Tecnología-Mexico Grant 132258 and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México) Grant IN-208212.
- LEA
- late embryogenesis abundant protein
- BiFC
- bimolecular fluorescence complementation
- IDC
- isothermal dilution calorimetry
- TFE
- trifluoroethanol
- PICUP
- photo-induced cross-linking of unmodified protein
- PPII
- polyproline II
- PEG
- polyethylene glycol
- rPvLEA6
- recombinant PvLEA6
- Ru(II)bpy32+
- Tris-bipyridylruthenium (II) di-cation
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- IDP
- intrinsically disordered protein.
REFERENCES
- 1. Tweddle J. C., Dickie J. B., Baskin C. C., Baskin J. M. (2003) Ecological aspects of seed desiccation sensitivity. J. Ecol. 91, 294–304 [Google Scholar]
- 2. Ingram J., Bartels D. (1996) The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403 [DOI] [PubMed] [Google Scholar]
- 3. Bewley J. D., Bradford K., Hilhorst H., Nonogaki H. (2013) Seeds: Physiology of Development, Germination and Dormancy, 3rd Ed., pp. 124–140, Springer, New York [Google Scholar]
- 4. Shinozaki K., Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227 [DOI] [PubMed] [Google Scholar]
- 5. Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., Covarrubias A. A. (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275, 5668–5674 [DOI] [PubMed] [Google Scholar]
- 7. Oldfield C. J., Dunker A. K. (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584 [DOI] [PubMed] [Google Scholar]
- 8. Hincha D. K., Thalhammer A. (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 40, 1000–1003 [DOI] [PubMed] [Google Scholar]
- 9. Tompa P. (2012) Intrinsically disordered proteins: a 10-year recap. Trends Biochem. Sci. 37, 509–516 [DOI] [PubMed] [Google Scholar]
- 10. Xue B., Dunker A. K., Uversky V. N. (2012) Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30, 137–149 [DOI] [PubMed] [Google Scholar]
- 11. Sun X., Rikkerink E. H., Jones W. T., Uversky V. N. (2013) Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25, 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tompa P., Szász C., Buday L. (2005) Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 30, 484–489 [DOI] [PubMed] [Google Scholar]
- 13. Tompa P. (2005) The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 14. Campos F., Cuevas-Velazquez C., Fares M. A., Reyes J. L., Covarrubias A. A. (2013) Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archaeae and bacteria domains. Mol. Genet. Genomics 288, 503–517 [DOI] [PubMed] [Google Scholar]
- 15. Battaglia M., Covarrubias A. A. (2013) Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 4, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Colmenero-Flores J. M., Campos F., Garciarrubio A., Covarrubias A. A. (1997) Characterization of Phaseolus vulgaris cDNA clones responsive to water deficit: identification of a novel late embryogenesis abundant-like protein. Plant Mol. Biol. 35, 393–405 [DOI] [PubMed] [Google Scholar]
- 17. Colmenero-Flores J. M., Moreno L. P., Smith C. E., Covarrubias A. A. (1999) Pvlea-18, a member of a new late-embryogenesis-abundant protein family that accumulates during water stress and in the growing regions of well irrigated bean seedlings. Plant Physiol. 120, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreno-Fonseca L. P., Covarrubias A. A. (2001) Downstream DNA sequences are required to modulate Pvlea-18 gene expression in response to dehydration. Plant Mol. Biol. 45, 501–515 [DOI] [PubMed] [Google Scholar]
- 19. Hundertmark M., Popova A. V., Rausch S., Seckler R., Hincha D. K. (2012) Influence of drying on the secondary structure of intrinsically disordered and globular proteins. Biochem. Biophys. Res. Commun. 417, 122–128 [DOI] [PubMed] [Google Scholar]
- 20. Carrari F., Fernie A. R., Iusem N. D. (2004) Heard it through the grapevine? ABA and sugar cross-talk: the ASR story. Trends Plant Sci. 9, 57–59 [DOI] [PubMed] [Google Scholar]
- 21. Hanin M., Brini F., Ebel C., Toda Y., Takeda S., Masmoudi K. (2011) Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal. Behav. 6, 1503–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fancy D. A., Kodadek T. (1999) Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. U.S.A. 96, 6020–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 24. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 25. Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, pp. 5–12, CANTINA Verlag, Germany [Google Scholar]
- 26. Velazquez-Campoy A., Leavitt S. A., Freire E. (2004) Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 261, 35–54 [DOI] [PubMed] [Google Scholar]
- 27. Belda-Palazón B., Ruiz L., Martí E., Tárraga S., Tiburcio A. F., Culiáñez F., Farràs R., Carrasco P., Ferrando A. (2012) Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7, e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J. D. (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56, 169–179 [DOI] [PubMed] [Google Scholar]
- 29. Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 [DOI] [PubMed] [Google Scholar]
- 30. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vacic V., Uversky V. N., Dunker A. K., Lonardi S. (2007) Composition profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 8, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mészáros B., Simon I., Dosztányi Z. (2009) Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 5, e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Disfani F. M., Hsu W. L., Mizianty M. J., Oldfield C. J., Xue B., Dunker A. K., Uversky V. N., Kurgan L. (2012) MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 28, i75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Receveur-Bréchot V., Bourhis J. M., Uversky V. N., Canard B., Longhi S. (2006) Assessing protein disorder and induced folding. Proteins 62, 24–45 [DOI] [PubMed] [Google Scholar]
- 35. Rucker A. L., Creamer T. P. (2002) Polyproline II helical structure in protein unfolded states: lysine peptides revisited. Protein Sci. 11, 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Z., Olson C. A., Rose G. D., Baldwin R. L., Kallenbach N. R. (2002) Polyproline II structure in a sequence of seven alanine residues. Proc. Natl. Acad. Sci. U.S.A. 99, 9190–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi Z., Chen K., Liu Z., Ng A., Bracken W. C., Kallenbach N. R. (2005) Polyproline II propensities from GGXGG peptides reveal an anticorrelation with β-sheet scales. Proc. Natl. Acad. Sci. U.S.A. 102, 17964–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tiffany M. L., Krimm S. (1969) Circular dichroism of the “random” polypeptide chain. Biopolymers 8, 347–359 [Google Scholar]
- 39. Chen K., Liu Z., Kallenbach N. R. (2004) The polyproline II conformation in short alanine peptides is noncooperative. Proc. Natl. Acad. Sci. U.S.A. 101, 15352–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kjaergaard M., Nørholm A. B., Hendus-Altenburger R., Pedersen S. F., Poulsen F. M., Kragelund B. B. (2010) Temperature-dependent structural changes in intrinsically disordered proteins: Formation of α-helices or loss of polyproline II? Protein Sci. 19, 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park S. H., Shalongo W., Stellwagen E. (1997) The role of PII conformations in the calculation of peptide fractional helix content. Protein Sci. 6, 1694–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tiffany M. L., Krimm S. (1972) Effect of temperature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers 11, 2309–2316 [DOI] [PubMed] [Google Scholar]
- 43. Uversky V. N., Li J., Fink A. L. (2001) Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem. 276, 10737–10744 [DOI] [PubMed] [Google Scholar]
- 44. Uversky V. N. (2009) Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 28, 305–325 [DOI] [PubMed] [Google Scholar]
- 45. Soulages J. L., Kim K., Walters C., Cushman J. C. (2002) Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol. 128, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soulages J. L., Kim K., Arrese E. L., Walters C., Cushman J. C. (2003) Conformation of a group 2 late embryogenesis abundant protein from soybean. Evidence of poly(l-proline)-type II structure. Plant Physiol. 131, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeong J. K., Shin H. J., Kim J. W., Lee C. H., Kim H. D., Lim W. K. (2003) Fluorescence and folding properties of Tyr mutant tryptophan synthase β-subunits from Escherichia coli. Biochem. Biophys. Res. Commun. 300, 29–35 [DOI] [PubMed] [Google Scholar]
- 48. Gally J. A., Edelman G. M. (1962) The effect of temperature on the fluorescence of some aromatic amino acids and proteins. Biochim. Biophys. Acta 60, 499–509 [DOI] [PubMed] [Google Scholar]
- 49. Liyanage M. R., Bakshi K., Volkin D. B., Middaugh C. R. (2014) Fluorescence spectroscopy of peptides. Methods Mol. Biol. 1088, 237–246 [DOI] [PubMed] [Google Scholar]
- 50. Johnson D. E., Xue B., Sickmeier M. D., Meng J., Cortese M. S., Oldfield C. J., Le Gall T., Dunker A. K., Uversky V. N. (2012) High-throughput characterization of intrinsic disorder in proteins from the protein structure initiative. J. Struct. Biol. 180, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Georgakopoulou S., Möller D., Sachs N., Herrmann H., Aebi U. (2009) Near-UV circular dichroism reveals structural transitions of vimentin subunits during intermediate filament assembly. J. Mol. Biol. 386, 544–553 [DOI] [PubMed] [Google Scholar]
- 52. Luo P., Baldwin R. L. (1997) Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36, 8413–8421 [DOI] [PubMed] [Google Scholar]
- 53. Buck M. (1998) Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355 [DOI] [PubMed] [Google Scholar]
- 54. Hackl E. V. (2014) Limited proteolysis of natively unfolded protein 4E-BP1 in the presence of trifluoroethanol. Biopolymers 101, 591–602 [DOI] [PubMed] [Google Scholar]
- 55. Gentile F., Amodeo P., Febbraio F., Picaro F., Motta A., Formisano S., Nucci R. (2002) SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric β-glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS. Stabilizing role of the A-C intermonomeric interface. J. Biol. Chem. 277, 44050–44060 [DOI] [PubMed] [Google Scholar]
- 56. Pazos F., Pietrosemoli N., García-Martín J. A., Solano R. (2013) Protein intrinsic disorder in plants. Front. Plant Sci. 4, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tiffany M. L., Krimm S. (1968) Circular dichroism of poly-l-proline in an unordered conformation. Biopolymers 6, 1767–1770 [DOI] [PubMed] [Google Scholar]
- 58. Hoekstra F. A., Golovina E. A., Buitink J. (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci. 6, 431–438 [DOI] [PubMed] [Google Scholar]
- 59. Hand S. C., Menze M. A., Toner M., Boswell L., Moore D. (2011) LEA proteins during water stress: not just for plants anymore. Annu. Rev. Physiol. 73, 115–134 [DOI] [PubMed] [Google Scholar]
- 60. Tunnacliffe A., Hincha D., Leprince O., Macherel D. (2010) in Dormancy and Resistance in Harsh Environments (Lubzens E., Cerda J., Clark M., eds) pp. 91–108, Springer-Verlag, Berlin [Google Scholar]
- 61. Demchenko A. P. (2001) Recognition between flexible protein molecules: induced and assisted folding. J. Mol. Recognit. 14, 42–61 [DOI] [PubMed] [Google Scholar]
- 62. Marsh J. A., Teichmann S. A., Forman-Kay J. D. (2012) Probing the diverse landscape of protein flexibility and binding. Curr. Opin. Struct. Biol. 22, 643–650 [DOI] [PubMed] [Google Scholar]
- 63. Mittag T., Kay L. E., Forman-Kay J. D. (2010) Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 23, 105–116 [DOI] [PubMed] [Google Scholar]
- 64. Gunasekaran K., Tsai C. J., Nussinov R. (2004) Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J. Mol. Biol. 341, 1327–1341 [DOI] [PubMed] [Google Scholar]
- 65. Perkins J. R., Diboun I., Dessailly B. H., Lees J. G., Orengo C. (2010) Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 66. Wilcox D. E. (2008) Isothermal titration calorimetry of metal ions binding to proteins: An overview of recent studies. Inorganica Chimica Acta 361, 857–867 [Google Scholar]
- 67. Pulido N. O., Salcedo G., Pérez-Hernández G., José-Núñez C., Velázquez-Campoy A., García-Hernández E. (2010) Energetic effects of magnesium in the recognition of adenosine nucleotides by the F1-ATPase β subunit. Biochemistry 49, 5258–5268 [DOI] [PubMed] [Google Scholar]
- 68. Salcedo G., Cano-Sánchez P., de Gómez-Puyou M. T., Velázquez-Campoy A., García-Hernández E. (2014) Isolated noncatalytic and catalytic subunits of F1-ATPase exhibit similar, albeit not identical, energetic strategies for recognizing adenosine nucleotides. Biochim. Biophys. Acta 1837, 44–50 [DOI] [PubMed] [Google Scholar]
- 69. Constantinescu Aruxandei D., Makbul C., Koturenkiene A., Lüdemann M. B., Herrmann C. (2011) Dimerization-induced folding of MST1 SARAH and the influence of the intrinsically unstructured inhibitory domain: low thermodynamic stability of monomer. Biochemistry 50, 10990–11000 [DOI] [PubMed] [Google Scholar]
- 70. Amara I., Odena A., Oliveira E., Moreno A., Masmoudi K., Pagès M., Goday A. (2012) Insights into maize LEA proteins: from proteomics to functional approaches. Plant Cell Physiol. 53, 312–329 [DOI] [PubMed] [Google Scholar]
- 71. Duan J., Cai W. (2012) OsLEA3–2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 7, e45117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sasaki K., Christov N. K., Tsuda S., Imai R. (2014) Identification of a novel LEA protein involved in freezing tolerance in wheat. Plant Cell Physiol. 55, 136–147 [DOI] [PubMed] [Google Scholar]
- 73. Ceccardi T. L., Meyer N. C., Close T. J. (1994) Purification of a maize dehydrin. Protein Expr. Purif. 5, 266–269 [DOI] [PubMed] [Google Scholar]
- 74. Kazuoka T., Oeda K. (1994) Purification and characterization of COR85-oligomeric complex from cold-acclimated spinach. Plant Cell Physiol. 35, 601–611 [Google Scholar]
- 75. Nakayama K., Okawa K., Kakizaki T., Honma T., Itoh H., Inaba T. (2007) Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 144, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]