Background: The role of the circadian clock component Period (PER) remains unclear.
Results: PER1 and PER2, but not PER3, protected CLOCK phosphorylation against Cryptochrome (CRY).
Conclusion: PER may counteract CRY function via the protection of CLOCK phosphorylation.
Significance: Detailed understanding of the control mechanism of CLOCK phosphorylation may reveal the true role of PER in the molecular clock machinery.
Keywords: Circadian Clock, Circadian Rhythm, Clock Gene, Gene Expression, Phosphorylation
Abstract
The circadian transcription factor CLOCK exhibits a circadian oscillation in its phosphorylation levels. Although it remains unclear whether this phosphorylation contributes to circadian rhythm generation, it has been suggested to be involved in transcriptional activity, intracellular localization, and degradative turnover of CLOCK. Here, we obtained direct evidence that CLOCK phosphorylation may be essential for autonomous circadian oscillation in clock gene expression. Importantly, we found that the circadian transcriptional repressors Cryptochrome (CRY) and Period (PER) showed an opposite effect on CLOCK phosphorylation; CRY impaired BMAL1-dependent CLOCK phosphorylation, whereas PER protected the phosphorylation against CRY. Interestingly, unlike PER1 and PER2, PER3 did not exert a protective action, which correlates with the phenotypic differences among mice lacking the Per genes. Further studies on the regulatory mechanism of CLOCK phosphorylation would thus lead to elucidation of the mechanism of CRY-mediated transcriptional repression and an understanding of the true role of PER in the negative feedback system.
Introduction
Almost all living organisms exhibit circadian rhythms in physiology and behavior. These rhythms are driven by the internal circadian clock, which enables adaptational synchrony with the earth's environmental period. The central circadian clockwork consists of the clock gene-driven negative feedback loop of transcription (1, 2). Expression of the circadian transcription repressor genes Period (Per) and Cryptochrome (Cry) is driven by the circadian transcription factors CLOCK and BMAL1. Subsequently, the PER proteins, together with CRY, serve to negatively regulate the CLOCK-BMAL1 complex.
It is well known that CLOCK is phosphorylated following dimerization with BMAL1 (3). A few reports have indicated the functional significance of CLOCK phosphorylation. First, BMAL1-dependent phosphorylation of CLOCK may be required for nuclear translocation of the BMAL1-CLOCK dimer (4). Second, phosphorylated CLOCK (pCLOCK) may be an active form as a transcription factor and an unstable form that leads to degradation (5). Third, in contrast to the report mentioned above, CLOCK may become transcriptionally inactive by phosphorylation (6). Based on electrophoretic patterns of CLOCK in the liver collected around the clock, CLOCK has been suggested to have multiple phosphorylation sites (7), each of which may play a different role in a phase-dependent manner. Investigation of CLOCK phosphorylation remains insufficient, however, and any definitive conclusion on the necessity of CLOCK phosphorylation in the circadian clockwork requires further study.
Here, we provide direct evidence that CLOCK phosphorylation may be essential for autonomous circadian oscillation in clock gene expression. In addition, our present results indicate that CRY may inhibit transcription by attenuating BMAL1-dependent CLOCK phosphorylation consistently with a previous report (8). Finally, we found the surprising phenomenon that PER protects CLOCK phosphorylation against CRY.
MATERIALS AND METHODS
Plasmid Construction
The promoter regions were isolated and cloned into the pGL3-Basic vector (Promega) (9). The mPer2, hPer3, hCry1, hCry2, and hBmal1 regions range from −2811 to +110, −779 to +170, −3195 to +417, −2925 to +41, and −3465 to +57, respectively (+1 is the putative transcriptional start site). mPer1-luc was a kind gift from Hajime Tei (10). The mPer1, mPer2, mPer3, mCry1, mCry2, mBmal1, and mClock coding sequences were subcloned into the pcDNA3 expression vector (Invitrogen). These clock proteins are expressed as a fusion protein with a c-Myc or hemagglutinin (HA) tag. The CRY1 mutant was constructed by PCR-based mutagenesis and verified by DNA sequencing. The conserved Leu-251 and Ser-252 residues of the flavin-binding sites were replaced by alanines.
Cell Culture and Transfection
In this study, NIH3T3 fibroblasts were used in all of the cultured cell-based assays. NIH3T3 cells are a well used cell line, known to contain the functional cell-autonomous circadian clock system (11). Cells were grown in DMEM (supplemented with antibiotics and 10% FBS) and cultured in 5% CO2. Transfection of plasmid DNA into cells was performed with Lipofectamine Plus (Invitrogen) as described previously (12). We used the Screen-WellTM kinase inhibitor library (Enzo Life Sciences, 2832(V2)A) for screening inhibitors of CLOCK phosphorylation. PF-670462, a selective casein kinase Iϵ (CKIϵ)3 and CKIδ inhibitor, was purchased from Sigma-Aldrich.
Western Blotting
NIH3T3 cells were cultured in 6-well plates and transfected with the indicated combination of vectors. The total amount of DNA was adjusted to 1000 ng with empty pcDNA3. The cells were lysed, and after removal of debris (15,000 rpm for 15 min), the lysates were treated with Laemmli sample buffer and subjected to Western blot analyses. Western blot signals were analyzed with the software Image Gauge (Fuji Film). Anti-Myc (mouse monoclonal, 9E10) and anti-HA (rabbit polyclonal) antibodies were purchased from Santa Cruz Biotechnology, Inc. and Clontech, respectively. Anti-PER2 antibody was purchased from Alpha Diagnostic International Inc. Anti-BMAL1 antibody was produced in rabbits in accordance with a previous report (13).
Luciferase Reporter Assay
NIH3T3 cells were cultured in 24-well plates and transfected with the indicated combination of vectors (a promoter-driven firefly luciferase vector and a Renilla luciferase vector as an internal control plus clock protein expression vectors). The total amount of DNA was adjusted to 400 ng/well with empty pcDNA3, and transfection was performed. About 20 h after transfection, the cells were immediately frozen in liquid nitrogen and stored at −80 °C. The cell lysates were used in the Dual Luciferase Assay System (Promega), as described previously (9). Luminescence was measured with a GloMax 20/20 luminometer (Promega). Data (Fluc/Rluc in relative light units (RLU)) show relative firefly luciferase activity (Fluc RLU), which was normalized by Renilla luciferase activity (Rluc RLU). Student's t test was used for comparisons between two groups. Differences were considered to be significant when the p value was less than 0.05.
mRNA Determination
Total RNA was reverse-transcribed using ReverTraAce (Toyobo), and real-time PCR was performed using SYBR Green (ABI) and a volume of the reverse transcription product. Data were obtained using PRISM7300 (ABI) and corrected by expression levels of 18 S rRNA. Primers were selected when there was no nonspecific amplification in dissociation curves and when amplification efficiency was relatively favorable. The primers were purchased from FASMAC Co., Ltd. (Japan). Their sequences are shown in our previous paper (14). Circadian peak time and amplitude were calculated with the single cosinor procedure program (Acro.exe, version 3.5, designed by Dr. Roberto Refinetti), and statistical significance between two groups was determined using Student's t test.
Animals
Per2-luciferase knock-in mice were a gift from Dr. Joseph Takahashi (15). Mice were maintained on a 12-h/12-h light dark cycle (light on at 9:00 a.m.) and were allowed ad libitum access to food and water. All experiments were in accordance with the rules of the Yamaguchi University Animal Usage Committee.
Explant Cultures and Bioluminescence Measurement
Coronal brain slices, including the suprachiasmatic nucleus (SCN) (300–400-μm thickness) were prepared from adult knock-in mice. Paired SCN were excised and explanted from coronal brain sections and placed on a culture membrane (Millicell-CM, PICM030-50; Millipore) in a covered and sealed Petri dish. Bioluminescence was measured with a photomultiplier tube (LM2400, Hamamatsu).
Real-time Monitoring of the Luciferase Activity in Living Cells
NIH3T3 cells were transfected with the indicated combinations of expression vectors and, after a serum shock, moved into a photomultiplier tube assembly (LM2400, Hamamatsu). Light emissions were measured using a photomultiplier tube in the presence of 0.1 mm luciferin and integrated for 1 min at 15-min intervals, as described previously (16).
RESULTS
Phosphorylation of CLOCK May Be Necessary for Circadian Gene Expression
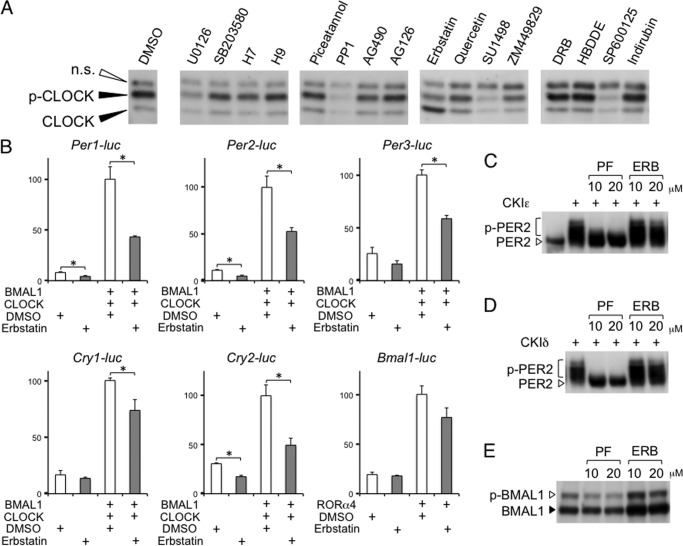
BMAL1-CLOCK dimerization leads to post-translational modifications of CLOCK, which correlate with their transcriptional activities (4, 8, 17). The CLOCK protein with an amino acid substitution in its phosphorylation site attenuates transcription (5). However, it remains unclear whether CLOCK phosphorylation is essential for autonomous circadian oscillations in gene expression. Therefore, we used a pharmacological approach to obtain evidence that CLOCK phosphorylation is indeed involved in circadian transcription. First, we tested 80 kinase inhibitors to identify a drug that could suppress CLOCK phosphorylation and found that erbstatin analog, a tyrosine kinase inhibitor, partially inhibited BMAL1-dependent CLOCK phosphorylation (Fig. 1A). Among the 80 inhibitors, including those targeting mitogen-activated protein kinase cascades, glycogen synthase kinase-3β, casein kinase I, casein kinase II, and protein kinase C, only erbstatin analog showed an obvious effect on BMAL1-induced CLOCK phosphorylation. It is unfortunately difficult to identify the kinase responsible for CLOCK phosphorylation because the specificity of erbstatin analog is low.
FIGURE 1.
Screening of kinase inhibitors affecting BMAL1-induced CLOCK phosphorylation. A, NIH3T3 cells were transfected with the BMAL1 and CLOCK expression vectors and treated with 80 kinase inhibitors at a concentration of 10 μm. Phosphorylation levels of CLOCK (p-CLOCK) were analyzed by Western blotting. Experiments were repeated twice with 80 kinase inhibitors. A part of the data set is shown in the figure. B, NIH3T3 cells were transfected with the indicated expression vectors and treated with DMSO or 10 μm erbstatin analog (ERB). Transcription levels of clock genes were evaluated using a luciferase assay. The data are represented as means ± S.E. (error bars) of three independent experiments. Maximum values were set to 100. Student's t test was used for comparisons between two groups. Differences were considered significant when the p value was less than 0.05. C–E, NIH3T3 cells were transfected with the casein kinase I and PER2 expression vectors (C and D) or the BMAL1 and CLOCK expression vectors (E) and treated with PF-670462, a selective CKIϵ and CKIδ inhibitor, or erbstatin analog at the indicated concentrations. Phosphorylation levels were analyzed by Western blotting. Experiments were repeated three times, and representative data are shown.
As expected, luciferase assays indicated that BMAL1- and CLOCK-mediated activation of Per1, Per2, Per3, and Cry2 were attenuated by erbstatin treatment (Fig. 1B). However, erbstatin analog did not completely block their expression. One reason may be that the erbstatin concentration was insufficient to effectively block overexpressed BMAL1 and CLOCK, as shown in the Western blotting data (Fig. 1A). Unexpectedly, we found that Cry1 expression was not obviously inhibited compared with other E-box-controlled genes. As expected, RORα-mediated activation of Bmal1 transcription was not significantly affected by erbstatin treatment.
To evaluate the specificity of the inhibitory effect of erbstatin analog on phosphorylation, we examined whether the inhibitor affected PER2 and BMAL1 phosphorylation (Fig. 1, C–E). Whereas PF-670462, a selective CKIϵ and CKIδ inhibitor, strongly inhibited CKI-induced PER2 phosphorylation, erbstatin analog showed no effect on it. Likewise, CLOCK-induced BMAL1 phosphorylation was not affected by erbstatin treatment. These results suggest that erbstatin analog specifically inhibits CLOCK phosphorylation.
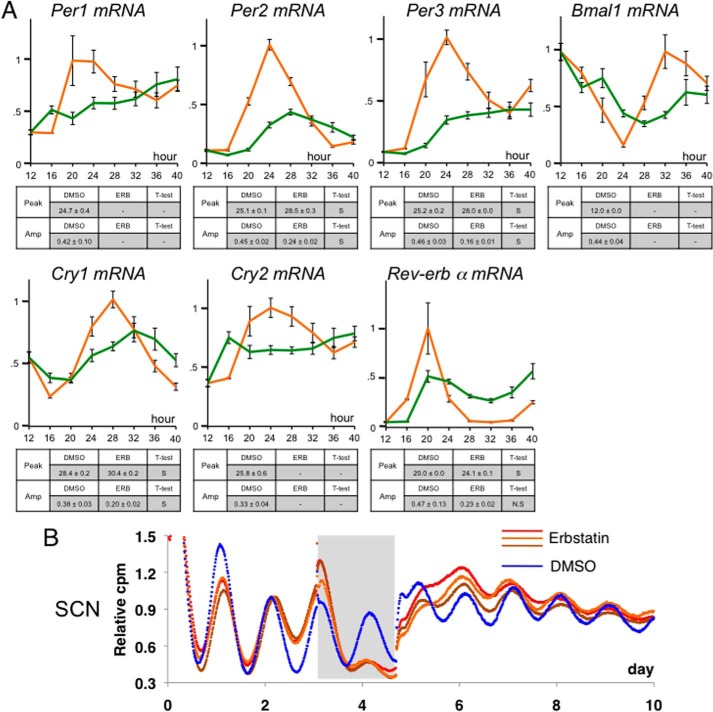
Additionally, to confirm the effect of erbstatin analog on endogenous clock gene expression, by using synchronized NIH3T3 cells, we investigated the circadian pattern of endogenous clock gene expression levels in the presence of erbstatin analog (Fig. 2A). Whereas circadian activation of the Per1, Per2, Per3, and Cry2 genes was strongly inhibited, that of the Rev-erb α, Cry1, and Bmal1 genes was not largely affected. These results are consistent with those shown in luciferase assays (Fig. 1B). Circadian expression of the Cry1 gene is regulated not only through E-box elements but also through ROR elements; it has been reported that two ROR elements are essential for circadian transcription of Bmal1 (18). This may be a reason why Cry1 expression was not substantially affected by erbstatin analog.
FIGURE 2.
Impairment of CLOCK phosphorylation results in reduction of circadian amplitude and phase delay. A, NIH3T3 cells were stimulated with 50 nm dexamethasone for circadian synchronization. 12 h after the shock, the cells were cultured in the presence of DMSO or 10 μm erbstatin analog and then stored in a deep freezer at 4-h intervals. After extracting total RNA, RT-PCR was performed to monitor transcriptional oscillation of clock genes. The data were corrected by expression levels of 18 S rRNA and represented as means ± S.E. (error bars) of three independent experiments. The orange and green lines show the data obtained with DMSO and erbstatin analog, respectively. Maximum values were set to 1. Circadian peak time and amplitude of the normalized data were determined using the single cosinor procedure program (Acro.exe, version 3.5, designed by Dr. Refinetti). When cosine fitting resulted in “not significant” more than once among three independent experiments, the temporal pattern of gene expression was determined to be arrhythmic (shown as a minus sign in the tables). Student's t test was used for comparisons between two groups (DMSO versus erbstatin (ERB)). Differences were considered significant when the p value was less than 0.05. S and N.S., significant and not significant, respectively. B, paired SCNs were excised from the coronal brain sections of Per2-luc knock-in mice and cultured in a culture membrane in a covered and sealed Petri dish. Bioluminescence was measured using a photomultiplier tube. The gray box indicates the treatment duration with 10 μm erbstatin analog or vehicle. Triplicate slices treated with 10 μm erbstatin analog are shown with a control sample.
Furthermore, erbstatin treatment dampened the circadian expression of Per2 in a reversible manner in an ex vivo culture of the SCN obtained from Per2-luc knock-in mice (Fig. 2B). These data provide additional evidence that CLOCK phosphorylation may be necessary for circadian oscillations in gene expression.
Phosphorylation Levels of CLOCK Strongly Correlate with Transcriptional Activity
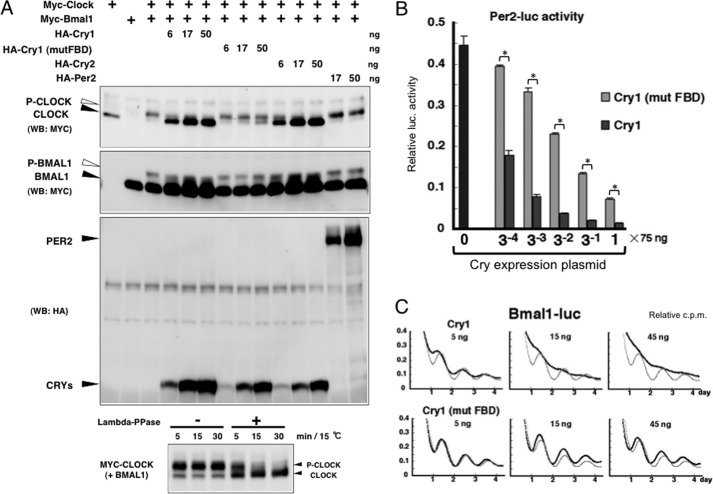
When BMAL1 and CLOCK were co-overexpressed, the post-translational modifications of both proteins produced an electrophoretic mobility shift (Fig. 3A). We confirmed that the shifted CLOCK band was eliminated by phosphatase treatment (Fig. 3A, bottom), indicating that CLOCK phosphorylation was closely associated with the transcriptional activities. We then examined the effects of CRY expression on CLOCK phosphorylation. As reported previously (8), CRY1 and CRY2 expression strongly reduced the phosphorylated forms of not only CLOCK but also BMAL1 in a dose-dependent manner, whereas PER2 showed no such effect.
FIGURE 3.
CRY-mediated transcriptional suppression is associated with impairment of CLOCK phosphorylation. A, top, NIH3T3 cells were transfected with the indicated expression vectors. Western blotting (WB) was performed with the indicated antibodies. Bottom, lysates were prepared from cells that expressed MYC-CLOCK and BMAL1, which were treated with λ-phosphatase (Lambda-PPase) for the indicated periods. The phosphorylation levels of CLOCK were analyzed by Western blotting. B, the effects of the overexpression of CRY1 or CRY1 (mut FBD) proteins on Per2 transcription were evaluated using a luciferase assay. The data are represented as means ± S.E. (error bars) of four independent experiments. Student's t test was used for comparisons between two groups. Differences were considered significant (asterisk) when the p value was less than 0.05. C, the transcriptional oscillation of Bmal1 was monitored using a cell culture-based luminescent reporter assay in the presence of overexpressed CRY1 or CRY1 (mut FBD). NIH3T3 cells were transfected with the hBmal1(−3465 to +57)-luc construct and stimulated with a high concentration of serum. To facilitate an accurate comparison, the light gray line shows the data obtained with an empty vector-transfected control. Experiments were repeated three times, and representative data are shown.
To further confirm that the suppression of CLOCK phosphorylation is an important process during CRY-mediated transcriptional repression, we constructed a CRY1 mutant with defects in its flavin-binding sites. Mutation of the CRY flavin-binding sites causes dysfunctional CRY-mediated transcriptional repression (19, 20). Similar results were obtained by investigating Per2 transcription using a CRY1 mutant, which carried point mutations different from those in previous studies (Fig. 3B). In NIH3T3 cells synchronized by serum shock, CRY1 overexpression severely disrupted the circadian transcription of Bmal1, whereas the CRY1 mutant only caused a phase shift (Fig. 3C). If the suppression of CLOCK phosphorylation is a pivotal event in CRY-induced transcription inhibition, a CRY1 mutant with a lower ability to inhibit transcription should have a reduced ability to suppress the phosphorylation of CLOCK. Consistent with this hypothesis, we found that the CRY1 mutant did not cause the strong suppression of CLOCK phosphorylation (Fig. 3A). These results demonstrate that there is a strong correlation between CRY-mediated transcriptional repression and the suppression of CLOCK phosphorylation.
PER Protects Phospho-CLOCK from CRY
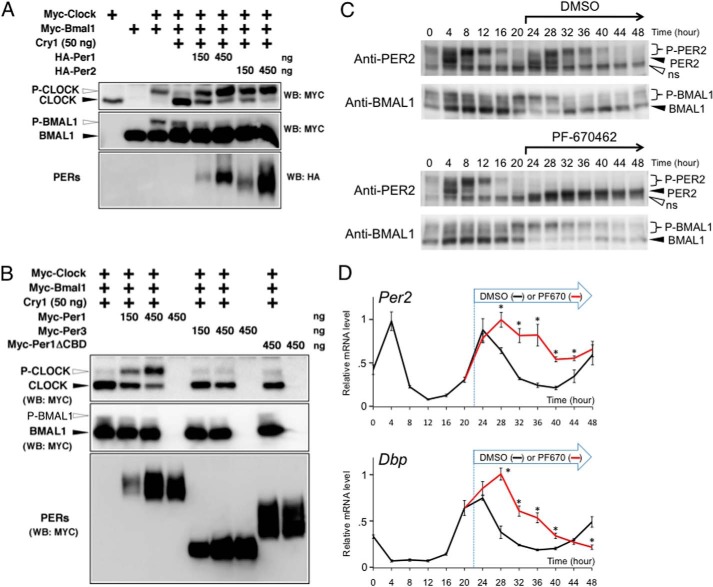
Fig. 3 indicates that the inhibition of CLOCK phosphorylation might be an important process in CRY-mediated transcriptional repression. Next, we examined whether PER expression has any effect on the CRY-induced reduction in the phosphorylation of CLOCK. Surprisingly, in NIH3T3 cells, PER1 or PER2 expression led to the strong protection of pCLOCK from CRY (Fig. 4A). However, PER alone, without CRY, had no effect on the phosphorylation levels of CLOCK (Fig. 3A). PER1 or PER2 expression did not restore the phosphorylation levels of BMAL1. Interestingly, as shown in Fig. 4B, PER3 expression did not protect pCLOCK from CRY. Consistent with the finding that the CRY-binding domain on PER was essential for the physical interaction of PER with CRY, the domain was also indispensable for PER-mediated protection of pCLOCK from CRY (Fig. 4B). Overall, these results demonstrate the possibility that PER1 and PER2, but not PER3, at least partially maintain the activity of the BMAL1-CLOCK complex by protecting pCLOCK from CRY, thereby protecting Per2 transcriptional activity against CRY.
FIGURE 4.
PER protects pCLOCK against CRY. A and B, NIH3T3 cells were transfected with the indicated expression vectors. Western blotting (WB) was performed with the indicated antibodies. We evaluated the effects of PER1 or PER2 expression (A) and PER3 or PER1ΔCBD expression (B) on the CRY-mediated suppression of pCLOCK. Experiments were repeated three times, and representative data are shown. C and D, NIH3T3 cells were stimulated with 50 nm dexamethasone for circadian synchronization (time 0). 22 h after the shock, the cells were cultured in the presence of DMSO or 10 μm PF-670462 and then stored in a deep freezer at 4-h intervals. C, phosphorylation levels of endogenous PER2 and BMAL1 were analyzed by Western blotting. Experiments were repeated four times, and representative data are shown. D, after extracting total RNA, RT-PCR was performed to monitor transcriptional oscillation of Per2 and Dbp. The data were corrected by expression levels of 18 S rRNA and represented as means ± S.E. (error bars) of four independent experiments. Maximum values were set to 1. Student's t test was used for comparisons between two groups (DMSO versus PF-670462). Differences were considered significant (asterisk) when the p value was less than 0.05.
In synchronized NIH3T3 cells, the circadian phase of transcriptional repression correlated with that of endogenous PER2 phosphorylation levels (Fig. 4, C and D). Experiments with PF-670462, a selective CKIϵ and CKIδ inhibitor, clearly showed that the PER2 phosphorylation is completely dependent on CKIϵ and CKIδ activity (Fig. 4C). Based on the temporal correlation between this transcriptional repression and PER2 phosphorylation levels, we hypothesized that CKIϵ and CKIδ are involved in the switching process from an inactive form of PER-CRY to an active inhibitor against BMAL1 and CLOCK. Unfortunately, we were unable to detect endogenous CLOCK phosphorylation with Western blot analysis and therefore evaluated BMAL1 and CLOCK complex activity by assessing endogenous BMAL1 phosphorylation levels, on the basis that CRY inhibited both BMAL1 and CLOCK phosphorylation, as shown in Fig. 3A. Whereas BMAL1 phosphorylation levels showed a circadian oscillation in negative controls, PF-670462 treatment caused a constitutive hyperphosphorylation of BMAL1 (Fig. 4C). These data indicate that the inhibition of PER phosphorylation resulted in activation of the BMAL1 and CLOCK complex, which is consistent with the finding that E-box-mediated transcription of clock genes remains highly activated in the presence of PF-670462 (Fig. 4D). Together, CKI-mediated phosphorylation of PER may play a role in the switching process from an inactive form of PER-CRY to an active inhibitor.
Significance of Transient Silencing of the Nuclear PER-CRY Complex
PER thus counteracts CRY in the regulation of BMAL1-dependent CLOCK phosphorylation. Because pCLOCK may be a transcriptionally active form, PER may inhibit CRY function. Although our present data may be insufficient for a conclusive evaluation, a very recent study indicates that PER indeed inhibits CRY-mediated repression of BMAL1 and CLOCK transcriptional activity (21). Furthermore, in terms of crystal structure, PER2 winds around the helical CRY1 domain covering the binding sites of CLOCK-BMAL1 (22). Our present results are consistent with these two previous studies.
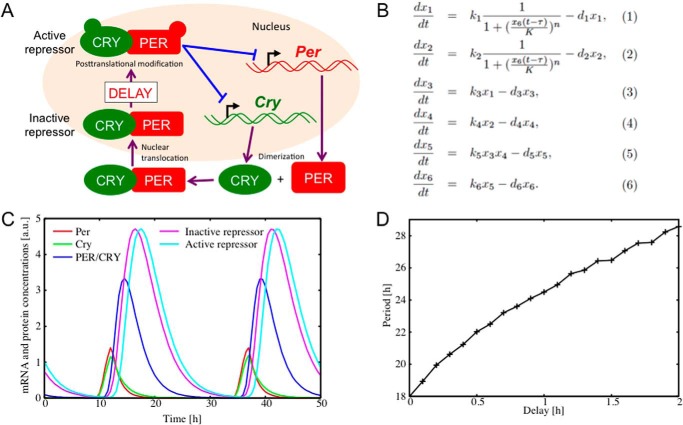
In the negative feedback process, if the nuclear PER-CRY complex immediately suppresses transcription, generation of a circadian period would be difficult. PER-mediated time delay might be essential for generating stable and long duration circadian gene expression. To study the significance of transient silencing of the nuclear PER-CRY complex in circadian rhythmicity, we constructed a mathematical model that takes into account the inactive phase of the repressor complex. The Goodwin model is often used to mathematically describe the circadian transcriptional feedback loop (23), and we therefore modified this model based on our findings. Fig. 5A shows a schematic view of the model. To focus on the essence of the transcription-translation feedback loop, a negative feedback loop of the Per and Cry genes was modeled by a set of differential equations, as shown in Fig. 5B. The variables x1 and x2 represent mRNA levels of the Per and Cry genes, respectively, whereas x3 and x4 correspond to cytoplasmic concentrations of the PER and CRY proteins, respectively. x5 and x6 stand for cytoplasmic and nuclear concentrations of the PER-CRY complex, respectively. ki (where i = 1, 2, … 6) are the production constants, whereas di (where i = 1, 2, … 6) determine the degradation rates. The term 1/(1 + (x6(t − τ)/K)n) is known as the Hill function, which models switchlike behavior of the transcription factor CLOCK-BMAL1 complex (23). Of particular note is the nuclear PER-CRY concentration x6(t), whose repressor function is inactivated for a time interval of τ. Transcription is therefore repressed by the time-delayed term of x6(t − τ).
FIGURE 5.
Mathematical model of the circadian oscillator. A, schematic view of the translation-transcription feedback loop. The CLOCK-BMAL1 complex activates transcription of the Per and Cry genes, resulting in an increase in Per and Cry mRNAs. Increased levels of PER and CRY proteins enhance formation of their complex, which is transported to the nucleus. The nuclear PER-CRY complex is initially inactive. After some time delay, however, it starts to inhibit the transcription of the Per and Cry genes as an active repressor. B, the variables x1 and x2 represent mRNA levels of the Per and Cry genes, respectively, whereas x3 and x4 correspond to cytoplasmic concentrations of the PER and CRY proteins, respectively. x5 and x6 stand for cytoplasmic and nuclear concentrations of the PER-CRY complex, respectively. ki (where i = 1, 2, … 6) are the production constants, whereas di (where i = 1, 2, … 6) represent the degradation rates. The repressor function is attenuated by a time interval of τ. C, time series of the Per and Cry mRNAs and the proteins generated from the model. Per mRNA (x1(t), red), Cry mRNA (x2(t), green), cytoplasmic PER-CRY protein (x5(t), blue), and nuclear PER-CRY protein in the inactive state (x6(t), purple) and active state (x6(t − τ), sky blue) oscillate with a period of 24.5 h. The parameter values were set as k1 = 1.4, k2 = 1, k3 = 1.4, k4 = 1.4, k5 = 0.84, k6 = 0.84, d1 = 0.7, d2 = 0.5, d3 = 0.7, d4 = 0.7, d5 = 0.42, d6 = 0.42, K = 0.03, n = 10, and τ = 1. a.u., arbitrary units. D, dependence of oscillation period on the time delay τ.
Fig. 5C shows a circadian rhythmicity generated from the model. The Per and Cry gene mRNAs and their proteins oscillate with a period of 24.5 h. Fig. 5D shows the dependence of the oscillation period on the time delay τ. It is shown that the system has a relatively short periodicity of 18 h in the case of no time delay (τ = 0). As the time delay is increased, the oscillation period is monotonically increased. Due to the nonlinear property of the transcription function, the amount of period lengthening is not merely equal to the value of the time delay. With a time delay of around τ = 1 h, the system achieves a circadian periodicity.
To examine the generality of the present result, we have constructed more comprehensive models for the transcription-translation feedback loop, including a positive feedback loop of Bmal1 transcription (24). The results thereby obtained were essentially the same. This implies that the effect of the time delay is independent of the modeling details and that the time delay caused by the inactive phase of the repressor provides an important component for controlling the circadian periodicity.
The present results indicate that PER-mediated transient inhibition of CRY leads to a time delay in the negative feedback process. Thus, PER may play a role in generating stable and long duration transcriptional feedback.
DISCUSSION
There is little doubt that CRY functions as an inhibitor of BMAL1 and CLOCK because there is little contradiction between data obtained in in vivo phenotypic analysis and in vitro cell-based assays. However, although it is known that the inhibition occurs via physical interaction between CRY, PER, and BMAL1 (25), few studies have investigated the molecular mechanism by which CRY inhibits BMAL1-CLOCK activity. Our present results suggest the possibility that CRY suppresses transcription via a decrease in the phosphorylation levels of CLOCK. This finding may provide new insights in our understanding of the mechanism. However, further studies are required to address many issues, including identification of the phosphorylation site that is involved in CRY-mediated transcriptional repression. In addition, it is completely inconclusive whether CRY inhibits the kinase reaction or accelerates the phosphatase reaction.
On the other hand, the following discrepancies mean that the true role of PER in the transcriptional negative feedback loop remains unclear. Although Per knock-out mice exhibit severe circadian defects (26–29), the repression effect of PER on transcription in reporter assays is much lower than that of CRY. In contrast to Cry-deficient mice, Per and Cry expression levels are reduced in Per knock-out mice, which contradicts the current molecular model, in which PER functions as a transcriptional repressor of the Per and Cry genes (26, 28, 29).
Our data indicate that PER counteracts CRY in the regulation of BMAL1-dependent CLOCK phosphorylation. This suggests that PER may inhibit CRY function via the protection of CLOCK phosphorylation, given our data showing that pCLOCK may be a transcriptionally active form. In the negative feedback process, if the nuclear PER-CRY complex immediately suppresses transcription, generation of a circadian period in clock gene expression would be difficult. Given the present results, CRY, a potent transcriptional repressor, might be temporally inactivated by PER, thereby leading to a time delay in the negative feedback process. Subsequently, the PER-CRY complex would become active as a transcriptional repressor via an undefined mechanism. Although we have no experimental evidence on the mechanism by which the inactive PER-CRY complex is switched to an active one, we speculate that it may involve post-translational modification. Together, PER may play a role in generating stable and long duration transcriptional feedback.
However, there may be multiple possible mechanisms for maintaining the circadian period, and our model might therefore represent only one of them. For example, the stable and long period transcriptional feedback may be controlled by the balance between protein kinases and protein phosphatases (30, 31).
In our present data, PER3 did not protect BMAL1-dependent CLOCK phosphorylation against CRY1. According to gene knock-out studies (28, 32), unlike PER1 and PER2, PER3 is considered to play only a minor role in the core clock. However, the mechanism that underlies the functional differences among these three PER molecules remains undefined. Our present finding that PER3 does not have the ability to counteract CRY in the regulation of BMAL1-dependent CLOCK phosphorylation indicates that the role of PER3 in the core clock is distinct from that of PER1/PER2.
Taken together, our findings suggest that a detailed understanding of the mechanism of control of CLOCK phosphorylation may indicate how CRY inhibits transcription and reveal the true role of PER in the molecular clock machinery.
Acknowledgments
We thank Rie Tashiro and Junko Sumino for expert technical assistance. The Per2-luciferase knock-in mice were kindly provided by Joseph Takahashi.
This work was supported by fellowships from the Nakajima Foundation, the Naito Foundation, the Takeda Science Foundation, and the Japan Society for the Promotion of Science.
- CKIϵ and CKIδ
- RLU, relative luciferase units
- SCN
- suprachiasmatic nucleus
- pCLOCK
- phosphorylated CLOCK.
REFERENCES
- 1. Reppert S. M., Weaver D. R. (2001) Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676 [DOI] [PubMed] [Google Scholar]
- 2. Schibler U., Sassone-Corsi P. (2002) A web of circadian pacemakers. Cell 111, 919–922 [DOI] [PubMed] [Google Scholar]
- 3. Baker C. L., Dunlap J. C. (2010) Circadian rhythms: phosphorylating the CLOCK. Cell Cycle 9, 231–232 [PMC free article] [PubMed] [Google Scholar]
- 4. Kondratov R. V., Chernov M. V., Kondratova A. A., Gorbacheva V. Y., Gudkov A. V., Antoch M. P. (2003) BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 17, 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spengler M. L., Kuropatwinski K. K., Schumer M., Antoch M. P. (2009) A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8, 4138–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshitane H., Takao T., Satomi Y., Du N. H., Okano T., Fukada Y. (2009) Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol. Cell. Biol. 29, 3675–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 8. Dardente H., Fortier E. E., Martineau V., Cermakian N. (2007) Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem. J. 402, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akashi M., Ichise T., Mamine T., Takumi T. (2006) Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol. Biol. Cell 17, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hida A., Koike N., Hirose M., Hattori M., Sakaki Y., Tei H. (2000) The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics 65, 224–233 [DOI] [PubMed] [Google Scholar]
- 11. Akashi M., Nishida E. (2000) Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645–649 [PMC free article] [PubMed] [Google Scholar]
- 12. Akashi M., Tsuchiya Y., Yoshino T., Nishida E. (2002) Control of intracellular dynamics of mammalian period proteins by casein kinase I ϵ (CKIϵ) and CKIδ in cultured cells. Mol. Cell. Biol. 22, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J. J., Sassone-Corsi P. (2005) Circadian clock control by SUMOylation of BMAL1. Science 309, 1390–1394 [DOI] [PubMed] [Google Scholar]
- 14. Matsumura R., Okamoto A., Node K., Akashi M. (2014) Compensation for intracellular environment in expression levels of mammalian circadian clock genes. Sci. Rep. 4, 4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akashi M., Takumi T. (2005) The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12, 441–448 [DOI] [PubMed] [Google Scholar]
- 17. Tamaru T., Isojima Y., van der Horst G. T., Takei K., Nagai K., Takamatsu K. (2003) Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells 8, 973–983 [DOI] [PubMed] [Google Scholar]
- 18. Etchegaray J. P., Lee C., Wade P. A., Reppert S. M. (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182 [DOI] [PubMed] [Google Scholar]
- 19. Froy O., Chang D. C., Reppert S. M. (2002) Redox potential: differential roles in dCRY and mCRY1 functions. Curr. Biol. 12, 147–152 [DOI] [PubMed] [Google Scholar]
- 20. Zhu H., Green C. B. (2001) A putative flavin electron transport pathway is differentially utilized in Xenopus CRY1 and CRY2. Curr. Biol. 11, 1945–1949 [DOI] [PubMed] [Google Scholar]
- 21. Akashi M., Okamoto A., Tsuchiya Y., Todo T., Nishida E., Node K. (2014) A positive role for PERIOD in mammalian circadian gene expression. Cell Rep. 7, 1056–1064 [DOI] [PubMed] [Google Scholar]
- 22. Schmalen I., Reischl S., Wallach T., Klemz R., Grudziecki A., Prabu J. R., Benda C., Kramer A., Wolf E. (2014) Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157, 1203–1215 [DOI] [PubMed] [Google Scholar]
- 23. Goodwin B. C. (1965) Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 3, 425–438 [DOI] [PubMed] [Google Scholar]
- 24. Becker-Weimann S., Wolf J., Herzel H., Kramer A. (2004) Modeling feedback loops of the mammalian circadian oscillator. Biophys. J. 87, 3023–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ko C. H., Takahashi J. S. (2006) Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277 [DOI] [PubMed] [Google Scholar]
- 26. Zheng B., Larkin D. W., Albrecht U., Sun Z. S., Sage M., Eichele G., Lee C. C., Bradley A. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173 [DOI] [PubMed] [Google Scholar]
- 27. Cermakian N., Monaco L., Pando M. P., Dierich A., Sassone-Corsi P. (2001) Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 20, 3967–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536 [DOI] [PubMed] [Google Scholar]
- 29. Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z. S., Eichele G., Bradley A., Lee C. C. (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 [DOI] [PubMed] [Google Scholar]
- 30. Lee H. M., Chen R., Kim H., Etchegaray J. P., Weaver D. R., Lee C. (2011) The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. U.S.A. 108, 16451–16456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Partch C. L., Shields K. F., Thompson C. L., Selby C. P., Sancar A. (2006) Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc. Natl. Acad. Sci. U.S.A. 103, 10467–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shearman L. P., Jin X., Lee C., Reppert S. M., Weaver D. R. (2000) Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol. 20, 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]