Abstract
Although high-density lipoprotein (HDL) is known to inhibit endothelial adhesion molecule expression, the mechanism for this anti-inflammatory effect remains obscure. Surprisingly, we observed that HDL no longer decreased adhesion of U937 monocytoid cells to tumor necrosis factor (TNF)α-stimulated human endothelial cells (EC) in the presence of the general lipase inhibitor tetrahydrolipstatin. In considering endothelial mechanisms responsible for this effect, we found that endothelial lipase (EL) overexpression in both EC and non-EL–expressing NIH/3T3 mouse embryonic fibroblasts cells significantly decreased TNFα-induced VCAM1 expression and promoter activity in a manner dependent on HDL concentration and intact EL activity. Given recent evidence for lipolytic activation of peroxisome proliferator-activated receptors (PPARs)—nuclear receptors implicated in metabolism, atherosclerosis, and inflammation—we hypothesized HDL hydrolysis by EL is an endogenous endothelial mechanism for PPAR activation. In both EL-transfected NIH cells and bovine EC, HDL significantly increased PPAR ligand binding domain activation in the order PPAR-α≫-γ>-δ. Moreover, HDL stimulation induced expression of the canonical PPARα-target gene acyl-CoA-oxidase (ACO) in a PPARα-dependent manner in ECs. Conditioned media from EL-adenovirus transfected cells but not control media exposed to HDL also activated PPARα. PPARα activation by EL was most potent with HDL as a substrate, with lesser effects on LDL and VLDL. Finally, HDL inhibited leukocyte adhesion to TNFα-stimulated ECs isolated from wild-type but not PPARα-deficient mice. This data establishes HDL hydrolysis by EL as a novel, distinct natural pathway for PPARα activation and identifies a potential mechanism for HDL-mediated repression of VCAM1 expression, with significant implications for both EL and PPARs in inflammation and vascular biology.
Keywords: adhesion molecules, endothelial cells, HDL cholesterol, high-density lipoproteins, lipase, PPARs, transcriptional regulation
Epidemiologic studies establish a strong inverse relationship between plasma high-density lipoprotein (HDL) levels and cardiovascular events.1,2 These atheroprotective benefits of HDL are often ascribed to the role of HDL in reverse cholesterol transport and the efflux of cholesterol from foam cells within arterial plaque. Recent studies have also identified anti-inflammatory properties of HDL that may also contribute to this relationship between HDL and atherosclerosis. One important anti-inflammatory HDL effect is its well-established inhibition of adhesion molecule expression and monocyte adhesion to endothelial cells (EC), key early steps in atherosclerosis.3–5 Levels of soluble adhesion molecules are predictive of cardiovascular risk while physiologic concentrations of HDL can inhibit cytokine-induced expression of endothelial adhesion molecules like VCAM1, ICAM1, and E-selectin.4,5
The mechanisms underlying HDL’s anti-inflammatory effects have remained largely obscure. HDL may limit the effects of pro-inflammatory mediators by either physical binding or active catalysis, as suggested by lipid hydroperoxide destruction by paraoxonases present in HDL particles.6,7 HDL reportedly inhibits endothelial cell sphingosine kinase, resulting in decreased NF-κB activation.8 HDL may also increase nitric oxide levels.9 Much less is known about HDL-dependent pathways in the endothelium that may account for the ability of HDL to regulate transcription and specifically repress adhesion molecule expression.
In exploring this HDL/endothelial interaction, we noted that inhibition of endothelial leukocyte adhesion by HDL was blocked in the presence of the general lipase inhibitor tetrahydrolipstatin. Recently we reported evidence for specific mechanisms through which lipoprotein metabolism can regulate distinct transcriptional responses through peroxi-some proliferator-activated receptors (PPARs),10,11 ligand-activated nuclear receptors involved in the transcriptional regulation of metabolism, inflammation, and atherosclerosis.12,13 For example, very-low-density lipoprotein (VLDL) hydrolysis by lipoprotein lipase (LPL) can generate peroxisome proliferator-activated receptor alpha (PPARα) ligands.10 In macrophages, LPL-treated VLDL was found to activate PPARδ.14 These findings suggest an underappreciated role for lipoproteins as transcriptional regulators, with effects dictated by the nature of the lipoprotein particle and its uptake, including the biology of specific lipases. Although synthetic PPAR agonists, identified largely through serendipitous drug screening, have been variably described as decreasing adhesion molecule expression,15–17 endogenous mechanisms for PPAR activation remain poorly understood despite their potential molecular and clinical implications.
We hypothesized that endothelial lipase (EL), a unique member of the triacylglycerol lipase family expressed in the endothelium, limits VCAM1 expression by activating PPARs via HDL hydrolysis. We present evidence here establishing and characterizing HDL hydrolysis by EL as a distinct, specific lipolytic pathway in the endothelium that preferentially activates PPARα, regulates VCAM1 expression, and can limit leukocyte adhesion.
Materials and Methods
An expanded Materials and Methods appears in the online data supplement (http://circres.ahajournals.com).
Cell Culture
Bovine aortic endothelial cells (BAEC) and NIH cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 1% glutamine, penicillin, streptomycin, and fungizone as per standard procedure.10 Human umbilical vein endothelial cells (HUVEC) (“human ECs” throughout) were grown in medium M199 with similar components except for 20% FBS and endothelial cell growth factor (ECGF). The human hematopoietic U937 and mouse monocyte-macrophage-like J774A.1 cell lines were cultured in RPMI medium under standard conditions.15
Generation of EL-Conditioned Media
For EL-conditioned medium, COS cells were infected (serum-free DMEM, 48 hours) with full-length human EL.20,21 After heparinization (10 U/mL, 30 minutes) to release surface EL and subsequent centrifugation, EL-conditioned media was tested for HDL hydrolysis, quantified as 500 nmol of free fatty acid released/mL from HDL (100 µg/mL, 4 hours, 37°C).
Isolation of Primary ECs From Mice
Murine hearts were obtained from 1-month-old PPARα+/+ (129S3/SvImJ; Jackson Laboratories, Bar Harbor, Me) and PPARα−/− (129S4/SvJae, generous gift, F. Gonzalez, National Institutes of Health, Bethesda, Md) mice. Murine ECs from these hearts were isolated using double selection with intercellular adhesion molecule 2 (ICAM2) and platelet endothelial cell adhesion molecule 1 (PECAM1) antibodies (BD PharMingen) bound to Dynabeads (Dynal) as before.22
Lipoprotein Isolation
Lipoprotein isolation by gradient ultracentrifugation of plasma pooled from multiple healthy donors was performed using density ranges of VLDL 1.006 to 1.019 g/mL, LDL 1.019 to 1.063 g/mL, and HDL 1.063 to 1.210 g/mL.19,23 All lipoprotein concentrations (BSA assay; Pierce) are expressed as µg protein/mL unless otherwise noted. Specific triglyceride and phospholipid content in lipoproteins was determined using L-Type TG H and Phospholipids B kit (Wako Pure Chemical Industries).
Cell Transfection and PPAR-LBD Assays
Transient transfection was performed using Fugene (Roche Molecular Biochemicals) in 1% delipidated plasma (DLP)/DMEM as per manufacturer’s instructions. A human EL cDNA (1.5 kb construct subcloned into the EcoRV site of the expression vector pcDNA3.1) was transfected as noted and responses compared with transfection of pcDNA3.1 vector alone.18 Catalytically inactive EL was generated by mutating serine 149 to alanine using reverse-transcription polymerase chain reaction (RT-PCR).19 PPAR LBD-GAL4 assays were performed using standard procedures as before.10 A human VCAM1 promoter construct was transfected using 1% Nutridoma SP (Roche Molecular Biochemicals),10 allowed to recover (14 hours) before additional stimulation.
Northern Blot Analysis
Standard Northern blot analysis was performed using VCAM1, acyl-CoA-oxidase (ACO), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (both ATCC) cDNA probes before densitometry quantification (ImageJ 1.33u; NIH).
Leukocyte Adhesion Assay
Near confluent HUVECs, isolated murine PPARα+/+ or PPARα−/− ECs monolayers (all 24-well plates) were washed (DPBS, 3 times) after stimulation and before adding either human monocytoid U937 cells or mouse monocyte–macrophage-like J774A.1 previously labeled with 2′,7′-bis(2-carboxy)-fluorescein acetoxymethylester (Molecular Probes) in serum free RPMI medium (4×105 labeled cells/well, 37°C, 30 minutes, DPBS).24 Nonadherent cells were removed by rinsing (DPBS, 3 times) before measuring fluorescence intensity (Cytofluor II, PerSeptive Biosystems).
Results
HDL-Mediated Inhibition of TNFα-Stimulated Leukocyte Adherence to Human ECs Requires Intact Lipase Activity
TNFα stimulation significantly increased the adhesion of fluorescently labeled U937 monocytoid cells to a monolayer of HUVECs (Figure 1A, upper left panel). As expected, pre-treating EC (3 hours) with the PPARα agonist WY14643 (10 µmol/L) or HDL (90 µg/mL) significantly decreased leukocyte adhesion by 37% and 31%, respectively (P<0.01 for each, Figure 1B). Surprisingly, the repression of leukocyte adhesion by HDL was abolished if ECs were pretreated with the general lipase inhibitor tetrahydrolipstatin (10 µmol/L, Figure 1A, lower right panel). These results suggest that the HDL-mediated decrease in leukocyte adhesion derives at least in part from lipase-mediated catalysis.
Figure 1.
HDL inhibits adhesion of U937 monocytoid cells to TNFα-stimulated ECs but not in the presence of the lipase inhibitor tetrahydrolipstatin. A, Fluoresce-in-labeled U937 cells were added to TNFα-stimulated (50 ng/mL) human EC monolayers after pretreatment with the PPARα agonist WY14643 (10 µmol/L), HDL (90 µg/mL), or HDL (90 µg/mL) in the presence of the lipase inhibitor tetrahydrolipstatin (10 µmol/L). Fluorescent microscopy shows adherent U937 cells (green) on a near-confluent EC layer. B, Quantification of U937 adherence on EC monolayers as determined by fluorescence assay. Results are expressed as percentage of leukocytes bound to TNFα-stimulated cells. Bars represent mean±SEM (n=3); #P<0.01 TNFα-stimulated ECs vs control; *P<0.01 WY and HDL vs TNFα-stimulated ECs.
HDL Suppresses TNFα-Induced VCAM1 Expression and Promoter Responses in a Concentration-Dependent Manner
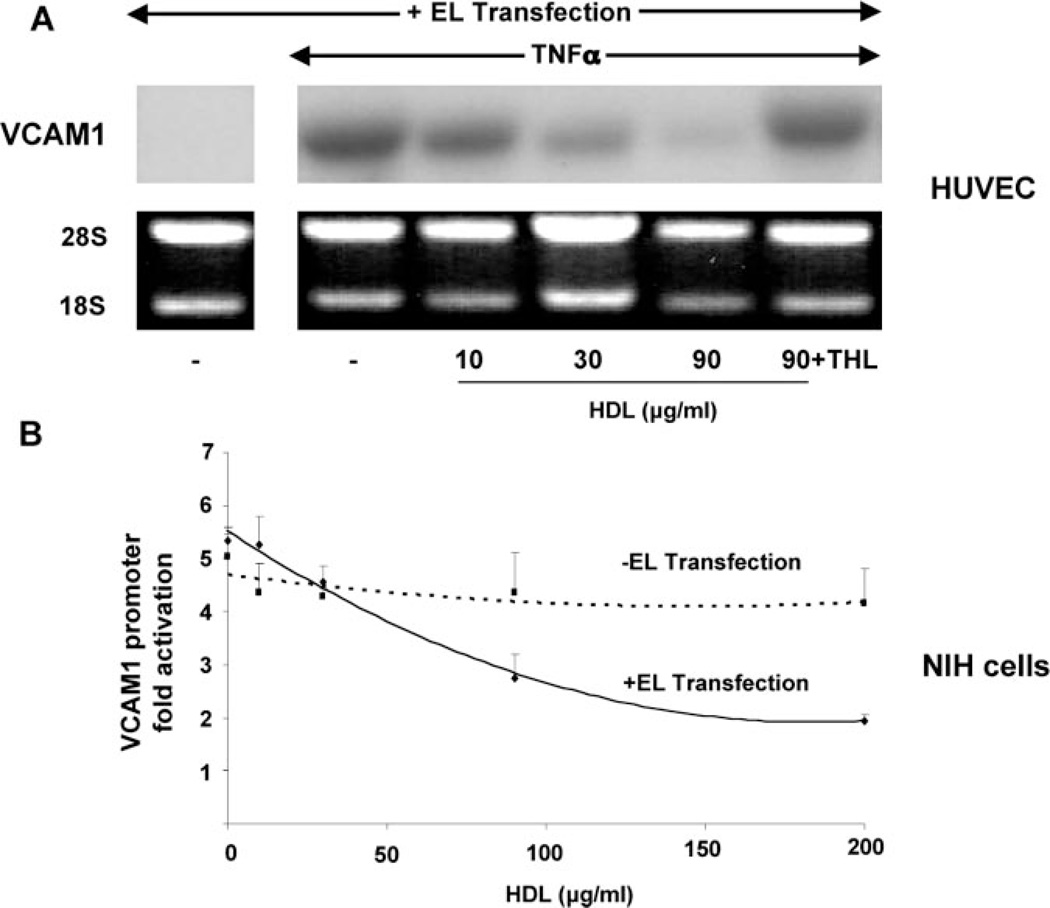
Because EL is considered the major lipase in ECs,28,32 we began by considering the endothelial mechanism(s) underlying the observations in Figure 1 by testing the effects of HDL on VCAM1 expression and promoter activity in cells over-expressing EL. Human ECs were transfected with EL, allowed to recover, then stimulated with TNFα and/or HDL at the concentrations shown before VCAM1 Northern blot analysis. HDL stimulation decreased TNFα-induced VCAM1 mRNA expression in a concentration-dependent manner (Figure 2A) but not in the presence of the lipase inhibitor tetrahydrolipstatin, even at the highest HDL concentration tested (90 µg/mL, Figure 2A). One representative Northern blot of three is shown; Northern blot analysis confirmed EL overexpression in transfected cells (supplemental Figure I).
Figure 2.
EL/HDL inhibits TNFα-mediated VCAM1 mRNA expression and promoter activity. A, Northern analysis of VCAM1 mRNA expression was performed on human EC (HUVEC) transfected with EL and then pretreated with increasing HDL concentrations (µg/mL, 3 hours) before TNFα (20 ng/mL, 16 hours) stimulation. In these EL-transfected cells, HDL decreased TNFα-induced VCAM1 expression but not in the presence of the lipase inhibitor tetrahydrolipstatin (10 µmol/L). B, TNFα- mediated VCAM1 promoter activation was tested in NIH cells over a concentration range of HDL in the presence (solid line) or absence (dashed line) of EL transfection. Cells were pretreated with HDL (3 hours) and then stimulated with TNFα (30 ng/mL, 12 hours) before performing promoterluciferase assays. In the presence of EL transfection, HDL significantly repressed VCAM1 promoter activation by 63% compared with 17% in EL nontransfected cells (*P<0.05 at 200 µg/mL of HDL; n=3, in triplicate for all). For comparison, the synthetic PPARα agonist WY14643 (250 µmol/L) suppressed TNFα-induced VCAM1 promoter activity from 5.3-fold to 1.7-fold; EL/HDL had no effect on the VCAM1 promoter in the presence of the lipase inhibitor tetrahydrolipstatin (10 µmol/L, data not shown).
We next tested if this EL-treated HDL effect was evident at the level of the VCAM1 promoter, supporting a transcriptional mechanism at work. Previous studies indicate NIH cells lack endogenous EL,25 a finding we confirmed using RT-PCR and Northern blotting (data not shown). NIH cells were cotransfected with a human VCAM1 promoterluciferase and either the EL-pcDNA3.1 expression construct or pcDNA3.1 alone (here and throughout) before exposure to increasing amounts of HDL. In EL-transfected cells, HDL treatment significantly reduced VCAM1 promoter activity in a concentration-dependent manner (−63% in EL-transfected cells versus −17% in pcDNA3.1-transfected cells, both at 200 µg/mL of HDL, P<0.05, Figure 2B). These results suggest that HDL repression of cytokine-induced VCAM1 involves transcriptional regulation through HDL hydrolysis by EL.
EL Preferentially Acts on HDL to Activate PPARα
Given previous reports that synthetic PPARα agonists repress VCAM1 expression, we performed standard PPARα ligand binding domain (LBD) assays to test PPARα activation through EL interaction with different lipoprotein substrates. Because EL has predominant phospholipase activity, the phospholipid phosphotidylcholine is well-established as a model EL substrate. In EL-transfected human EC, phosphatidylcholine treatment (100 µmol/L) increased PPARα–LBD activation 16.9-fold (Figure 3A).
Figure 3.
EL preferentially hydrolyzes HDL to activate PPARα in a manner dependent on EL catalysis. Standard PPARα-LBD-GAL4 assays as described in Methods were performed in BAECs either with or without cotransfection of human EL. Results are expressed as fold activation over control after normalization. A, EL preferentially acts on phosphotidylcholine (PC) and HDL to activate the PPARα– LBD. EC transfected with either empty vector or the EL construct were exposed to PC, LDL, VLDL, and HDL at the concentrations shown and PPARα–LBD activity determined. In EL-transfected cells, the most potent activation was seen with HDL (*P<0.05); more modest activation was also evident with LDL and VLDL (all lipoproteins at 60 µg protein/mL). For comparison, the PPARα-LBD response to WY14643 (10 µmol/L) is shown. B, In untransfected BAEC, HDL increases PPARα-LBD activation in a concentration-dependent manner but not in the presence of tetrahydrolipstatin (10 µmol/L). C, Tetrahydrolipstatin preincubation (30 minutes, room temperature) decreases PPARα-LBD activation in a concentration-dependent manner as evident in control transfected (not shown) and EL-transfected BAEC exposed to HDL (60 µg/mL). D, PPARα activation by EL/HDL requires catalytically-active EL. PPARα–LBD-GAL4 assays were performed in BAEC after transfection of either wild-type or a catalytically-inactive EL point mutant in the presence of increasing concentrations of HDL. In cells expressing the catalytically-inactive EL mutant, no significant increase in PPARα–LBD activation is seen above basal levels across the HDL concentration tested. For comparison, concomitant PPARα-LBD activation in cells transfected with wild-type EL is shown; tetrahydrolipstatin (10 µmol/L) abrogated the EL/HDL/PPARα response.
To begin characterizing this EL pathway, we next examined EL-mediated PPARα activation in response to common circulating human lipoproteins, namely VLDL, LDL, and HDL isolated from normal subjects. Treatment of ELtransfected ECs with either VLDL (60 µg/mL) or LDL (0 to 60 µg/mL) only modestly increased PPARα activation (Figure 3A, supplemental Figure II). The most potent PPARα activation occurred after HDL exposure to EL-transfected EC, with a concentration-dependent response that reached ≈30-fold at the maximal HDL concentrations tested (60 µg/mL), as compared with 18-fold in untransfected EC (*P<0.05, Figure 3A). Of note, the PPARα activation seen with this relatively modest HDL concentration, being even less than circulating HDL levels in vivo, equaled the responses seen with the synthetic PPARα agonist WY14643 at 10 µmol/L (Figure 3A). To better define this response to HDL and in EC under basal conditions without EL over-expression, PPARα-LBD experiments were repeated in untransfected BAEC. HDL increased PPARα-LBD activation in BAEC in a concentration-dependent manner (Figure 3B). Tetrahydrolipstatin completely blocked this response (Figure 3B). EL-mediated PPARα activation by HDL, VLDL, and LDL was also analyzed as a function of the triglyceride and phospholipid content of all these lipoprotein particles. These data support HDL as the preferential substrate for PPARα activation through EL (supplemental Figure II).
Given our initial observation that HDL-mediated repression of leukocyte adhesion required intact lipase activity as well as the noncatalytic effects of lipases including EL,19,26 we next asked if EL-mediated PPARα activation required EL catalysis. Tetrahydrolipstatin inhibited the HDL effect on PPARα-LBD activation in a dose-dependent fashion in EL-transfected cells (Figure 3C). To further test this and eliminate any possible nonspecific effect of tetrahydrolipstatin, we next compared HDL-induced PPARα–LBD activation in BAEC transfected with either wild-type or a catalytically inactive EL point mutant. Although HDL treatment activated PPARα in a concentration-dependent manner in wild-type EL-transfected cells, HDL had no effect in cells expressing a catalytically inactive EL point mutant (Figure 3D). This apparent dominant negative effect of catalytically inactive EL on endogenous EL function has been previously reported and may stem from EL dimerization.27 As before, tetrahydrolipstatin completely inhibited PPARα activation in the presence of EL transfection, even at the highest HDL concentrations (Figure 3D). These observations strongly support hydrolysis of HDL as being required for HDL-mediated PPARα activation. These results also suggest that the PPARα activation seen with HDL in untransfected EC was likely attributable to the known endogenous expression of EL in these cells. To definitively address this issue, similar EL transfection experiments were next performed in NIH cells that lack endogenous EL.25
The Presence of EL Is Necessary and Sufficient to Confer HDL-Induced PPARα Activation
Standard PPARα–LBD assays were performed in NIH cells after exposure to increasing HDL concentrations (maximum 200 µg/mL) in either the absence or presence of EL transfection as before. In the absence of EL transfection, HDL stimulation had no significant effect on PPARα activation (maximum 1.7-fold activation at 200 µg/mL of HDL); in contrast, in the presence of EL transfection, HDL stimulation increased PPARα activation 7.3-fold at 200 µg/mL of HDL (P<0.005, Figure 4A).
Figure 4.
Heterologous EL expression Confers PPARα Activation In Response to HDL. Standard PPARα–LBD-GAL4 assays were performed as before but in NIH cells known to lack endogenous EL expression. A, NIH cells transfected with either EL or the empty vector were exposed to increasing concentrations of HDL before PPARα–LBD activation was determined. HDL activates the PPARα–LBD in a concentration-dependent manner but only in EL transfected cells. B, PPARα–LBD assays were performed in NIH cells exposed to HDL (100 µg/mL) and increasing amounts of either EL protein conditioned medium (prepared and quantified from EL adenovirus-expressing cells as described in Methods) or control-transfected medium (50 µL/well to 500 µL/well of medium, 24-well plates). The extent of PPARα-LBD activation seen was linearly related to the amount of EL-conditioned medium added; control medium had no effect.
To further establish that EL itself can activate PPARα in the presence of HDL, PPARα–LBD experiments were repeated in NIH cells exposed to increasing volumes of conditioned medium from EL-overexpressing cells (from 50 µL/well to 500 µL/well of conditioned medium in 24-well plates). At a constant concentration of HDL (100 µg/mL), PPARα–LBD activation increased linearly as a function of the amount of EL-conditioned medium added (3.87-fold PPARα activation at 500 µL/well of EL-conditioned medium versus 1.25-fold with control medium, P<0.005, Figure 4B). The HDL hydrolytic activity for this conditioned medium was 500 nmol FFA formed per mL after 4 hours at 37°C.
PPAR Isotype-Specific Effects of EL/HDL
Because EC express all PPAR isotypes to varying degrees,36 we next characterized the relative PPAR isotype selectivity through HDL hydrolysis by EL. LBD assays were repeated in BAEC overexpressing EL as before but now after transfection of PPARα, PPARγ, or PPARδ–LBD constructs, comparing responses to the maximal activation seen with well-characterized agonists to each PPAR isotype. In the presence of EL transfection, HDL (90 µg/mL) induced activation that was 86% of the PPARα agonist (WY14643, 10 µmol/L), 49.5% of the PPARγ agonist (BRL, 1 mmol/L), and 39% of the PPARδ agonist (bezafibrate, 10 µmol/L, Figure 5).
Figure 5.
Hydrolysis of HDL by EL activates PPAR isotypes in the order -α>-γ >δ. The activation of each PPAR-LBD isotype (α, γ, or δ) in response to EL expression in the presence of increasing HDL concentrations was tested using LBD assays in NIH cells as before. EL/HDL most potently activated PPARα, followed by PPARγ, and then PPARδ at the maximal HDL concentration tested (90 µg/mL). The EL/HDL response was expressed as a percentage of the PPAR–LBD activation seen with known PPAR isotype-specific ligands as follows: PPARα-WY14643 (10 µmol/L); PPARγ-BRL49653 (1 µmol/L); PPARδ- bezafibrate (10 µmol/L).
EL/HDL Induces PPARα Target Gene Expression
If HDL hydrolysis by EL activates PPARα, then it should induce expression of positively regulated, canonical PPARα target genes that contain a known PPAR response element, doing so in a PPARα-dependent manner. Acyl-CoA-oxidase (ACO) is a well-established PPARα-regulated target gene that fulfills such requirements.10 Microvascular ECs from PPARα+/+ and PPARα−/− mice were transfected with EL or control plasmid and stimulated with either HDL (at 90 or 200 µg/mL) or the synthetic PPARα agonist WY14643 (10 µmol/L) before performing ACO Northern blot analysis. In the genetic presence of PPARα, HDL induced ACO expression to a similar level as the PPARα agonist WY in EL-transfected cells; this effect was absent in PPARα-deficient ECs (Figure 6). Densitometry and quantification reveal that ACO expression (normalized to GAPDH) in these microvascular ECs increases 1.92-fold and 2.47-fold in response to WY or HDL, respectively, as compared with the control.
Figure 6.
EL/HDL induces mRNA expression of the PPARα target gene acyl CoA oxidase (ACO) but only in the genetic presence of PPARα. ECs from PPARα wild-type (+/+) and deficient (−/−) mice were transfected with EL or a control plasmid and stimulated with either WY14643 (10 µmol/L) or HDL (90 or 200 µg/mL) before performing Northern blot analysis for ACO. Densitometry and quantification analysis as compared with GAPDH expression indicates WY and HDL increased ACO 1.92-fold and 2.47-fold, respectively, in EL-transfected cells.
Hydrolysis of HDL Is a Distinct Lipolytic Mechanism for PPARα Activation
The unique biologic role of EL among lipases establishes the importance of these observations for EL/HDL-mediated PPARα activation. Nevertheless, because LPL can generate PPARα ligands,10 we next investigated if hydrolysis of HDL by EL is distinct from LPL in terms of PPAR activation. To address this, we asked if the presence of EL modulated the maximal PPARα activation seen with LPL after exposure to either VLDL or HDL. As previously reported,10 in the presence of VLDL (5 µg/mL), LPL increased PPARα–LBD activation in a concentration-dependent manner (maximum 9.9-fold, Figure 7A). This concentration of VLDL used here produces robust PPARα activation in the presence of LPL, whereas the use of even higher VLDL concentrations induces cell death.10 EL overexpression had no impact on the LPL/VLDL responses seen (maximum 9.6-fold, Figure 7A). In contrast, when HDL (90 µg/mL) was the substrate, the presence of EL transfection increased PPARα activation significantly (*P<0.05, Figure 7B); in contrast LPL, even at maximal concentrations, had no effect on the EL/HDL/ PPARα response (Figure 7B). These results support preferential hydrolysis of HDL by EL as a unique pathway for PPARα distinct from LPL/VLDL-mediated responses.
Figure 7.
EL and LPL are distinct mechanisms for lipolytic PPARα activation. The additional PPARα–LBD activation seen with EL vs LPL in response to VLDL or HDL was compared in BAEC. A, Cells were exposed to increasing concentrations of LPL as shown in the presence of a fixed concentration of VLDL (5 µg/mL). As previously reported,10 LPL increased PPARα activation in a concentration-dependent manner. The presence or absence of EL-transfection had no impact on the PPAR activation seen by LPL/VLDL. B, Similar experiments as in (A) were repeated with increasing concentrations of LPL but in the presence of a fixed concentration of HDL (90 µg/mL). With HDL as the substrate, the presence of EL overexpression significantly increases the PPARα activation seen at any given LPL concentration (P<0.05 for each LPL concentration).
HDL Inhibits Leukocyte Adhesion to TNFα-Stimulated Mouse ECs in a PPARα-Dependent Manner
Given this evidence for HDL hydrolysis by EL as a mechanism for PPARα activation, we returned to our initial observation that a lipase inhibitor blocks HDL-mediated repression of leukocyte adhesion to ask if this response required the presence of PPARα. Similar adhesion assays as in Figure 1 were repeated but measuring adhesion of fluorescently-labeled murine monocyte-macrophage J774A.1 cells to microvascular ECs isolated from either wild-type (PPARα+/+) or PPARα-deficient (PPARα−/−) mouse hearts. As with HUVEC, both the PPARα agonist WY14643 (10 µmol/L) or HDL (90 µg/mL) significantly decreased leukocyte adhesion by 43% and 35%, respectively, but not when tetrahydrolipstatin was added with HDL (compared with TNFα stimulation alone, Figure 8A, upper panel). In the genetic absence of PPARα, HDL and WY had no effect (Figure 8A, lower panel). Quantification of these responses demonstrated a significant reduction in leukocyte adhesion by WY14643 and HDL (P<0.05) in a PPARα-dependent manner (Figure 8B). Interestingly, in the absence of PPARα, leukocyte adhesion is increased under both basal and TNFα-stimulated conditions (P<0.05).
Figure 8.
HDL inhibits leukocyte adhesion to TNFα-stimulated mouse ECs in a PPARα-dependent manner. A, Fluorescein-labeled murine J774A.1 mouse monocyte-macrophage-like cells were added to TNFα-stimulated (30 ng/mL) mouse PPARα+/+ and PPARα−/− ECs after pretreatment with either the PPARα agonist WY14643 (10 µmol/L), HDL (90 µg/mL), or HDL (90 µg/mL) in the presence of the lipase inhibitor tetrahydrolipstatin (10 µmol/L). Fluorescence microscopy shows adherent J774A.1 cells (green) on a confluent mouse EC layer. B, Quantification of J774A.1 adherence on mouse EC monolayers as determined by fluorescence assay. Results are expressed as percentage of leukocytes bound to TNFα-stimulated cells. Bars represent mean±SD (n=3); #P<0.05 (TNFα-stimulated alone vs TNFα-unstimulated control EC); **P<0.01 (TNFα-stimulated +/+ vs −/− ECs); *P<0.05 (TNFα and WY or HDL vs TNFα-stimulated alone +/+ ECs); †P<0.02 (unstimulated +/+ vs −/− ECs).
Discussion
We have observed that HDL-mediated repression of leukocyte adhesion to endothelial cells is markedly decreased after inhibition of lipase activity. Subsequent studies presented here identify hydrolysis of HDL by endothelial lipase (EL) as a pathway for PPARα activation and subsequent VCAM 1 repression, as evident in LBD assays, PPARα target gene induction, and a genetic requirement for PPARα for these EL/HDL effects to be seen. EL expression was both necessary and sufficient to confer PPARα responsiveness to HDL as shown in EL-nonexpressing cells after EL transfection or exposure to EL-conditioned media. Characterization of this EL/PPAR pathway reveals selectivity as to lipoprotein substrate (HDL≫LDL>VLDL) and the targeted PPAR isotype (PPAR-α≫-γ>-δ). These findings reveal HDL hydrolysis by EL as a mechanism for PPAR activation and identify a plausible mechanism potentially contributing to HDL effects on limiting leukocyte adhesion to the endothelium and vascular inflammation.
LPL hydrolysis of VLDL has been shown to activate PPARs.10,14 We reported that LPL acts on triglyceride-rich lipoproteins like VLDL to most potently activate PPARα, with lesser responses to LDL and even less with HDL as LPL substrates.10 In cell-free radioligand displacement assays, LPL-treated VLDL displaced high affinity synthetic PPAR agonists from expressed PPAR proteins following the order PPARα≫PPARδ>PPARγ. Chawla et al concurrently reported that LPL-treated VLDL activated PPARδ in mouse macrophages, cells that are thought to have low if not undetectable PPARα levels, in contrast to human macrophages.14 Comparing these findings to the data presented here, EL-mediated PPAR activation appears distinct from the LPL/PPAR pathway given differences in substrate preferences and the targeted PPAR isotypes. This conclusion is further supported by the demonstration that EL-treated HDL activation of PPARα is not altered by LPL regardless of its concentration (Figure 7B). The divergence we find between LPL and EL mechanisms of PPAR activation is consistent with previous studies defining specific differences between EL, LPL, and hepatic lipase as triacylglycerol hydrolysis family members.28,29 In addition to demonstrating and characterizing this novel EL/HDL/PPAR pathway, our results also identify PPAR activation as a previously unrecognized factor to be considered in interpreting studies on endothelial responses to HDL. More broadly, these findings support our working model that different lipases act on specific lipoproteins to direct distinct transcriptional responses. The specific limbs of such a network would be dictated by the nature of the lipoprotein substrate, the unique biologic role of specific lipases, as well as the distal consequences of activating a given PPAR isotype. These variables also suggest that establishing the relative potency between EL and LPL in terms of PPARα activation may be less relevant to PPAR activation in vivo via pathways of lipid metabolism. The distinctive, nonoverlapping roles for EL and LPL in PPAR activation are particularly intriguing given very recent evidence for EL and LPL effects on fatty acid uptake in adipocytes.20
EL is unique among triglyceride lipase family members given its predominant endothelial expression.18,25 The role of EL in vascular biology is not yet clearly defined. EL is known to participate in HDL metabolism,30,31 although EL-mediated nuclear receptor activation has not been previously reported. Like LPL, EL has nonenzymatic effects, including increased noncatalytic lipoprotein uptake.26 Such EL bridging effects are unlikely to contribute to PPARα activation given the absence of PPAR responses with either a catalytically inactive EL mutant or in the presence of a general lipase inhibitor (Figure 3B). Given EL’s high degree of phospholipase activity, one might predict the specific PPAR ligand released from HDL by EL to be a fatty acid derived from the SN-1 position of a phospholipid.18,25 This would also be consistent with HDL as a phospholipid-rich lipoprotein being the preferred substrate for EL-mediated PPAR activation.
EL expression is induced by inflammatory cytokines as well as physical forces such as shear stress.32,33 Repression of adhesion molecule expression as well as other reports of anti-inflammatory effects through PPARα activation raise the intriguing possibility that EL induction by these proinflammatory forces might foster counter-regulatory PPARα responses. Interestingly, endogenous PPARα activation has been previously implicated in terminating inflammatory responses, for example, through leukotrienes.37 Alternatively, because previous work suggests EL can increase lipid uptake and promote atherosclerosis,34 EL-mediated PPAR activation might contribute to these pro-atherogenic processes as well. Regardless of its net effect, the expression of EL in ECs and atherosclerotic plaque35 makes its activation of PPARα of obvious physiologic and pathologic relevance.
In summary, these data establish hydrolysis of HDL by EL as a mechanism for PPARα activation and PPARα regulation of VCAM1 expression. This EL-mediated PPAR activation identifies a specific mechanism through which HDL can exert transcriptional effects in the endothelium, thus representing an appealing mechanism contributing to the anti-inflammatory effects of HDL.
Supplementary Material
Acknowledgments
This work was supported by R01 HL79117 (J.P.), P0149245 (J.P.), the Donald W. Reynolds Foundation (W.A., J.P.), AHA SDG0530101N (O.Z.), and Boston Obesity Nutrition Research Center Award 5P30DK046200 (O.Z.). The authors thank Ruzena Tupy for excellent editorial assistance.
References
- 1.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 2.Castelli WP, Garrison RJ, Wilson PW, Abbott RD, Kalousdian S, Kannel WB. Incidence of coronary heart disease and lipoprotein cholesterol levels. The Framingham Study. JAMA. 1986;256:2835–2838. [PubMed] [Google Scholar]
- 3.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cel. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 4.Cockerill GW, Rye KA, Gamble JR, Vadas MA, Barter PJ. High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler Thromb Vasc Biol. 1995;15:1987–1994. doi: 10.1161/01.atv.15.11.1987. [DOI] [PubMed] [Google Scholar]
- 5.Calabresi L, Franceschini G, Sirtori CR, De Palma A, Saresella M, Ferrante P, Taramelli D. Inhibition of VCAM-1 expression in endothelial cells by reconstituted high density lipoproteins. Biochem Biophys Res Commun. 1997;238:61–65. doi: 10.1006/bbrc.1997.7236. [DOI] [PubMed] [Google Scholar]
- 6.Shih DM, Xia YR, Wang XP, Miller E, Castellani LW, Subbanagounder G, Cheroutre H, Faull KF, Berliner JA, Witztum JL, Lusis AJ. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J Biol Chem. 2000;275:17527–17535. doi: 10.1074/jbc.M910376199. [DOI] [PubMed] [Google Scholar]
- 7.Tward A, Xia YR, Wang XP, Shi YS, Park C, Castellani LW, Lusis AJ, Shih DM. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation. 2002;106:484–490. doi: 10.1161/01.cir.0000023623.87083.4f. [DOI] [PubMed] [Google Scholar]
- 8.Xia P, Vadas MA, Rye KA, Barter PJ, Gamble JR. High density lipoproteins (HDL) interrupt the sphingosine kinase signaling pathway. A possible mechanism for protection against atherosclerosis by HDL. J Biol Chem. 1999;274:33143–33147. doi: 10.1074/jbc.274.46.33143. [DOI] [PubMed] [Google Scholar]
- 9.Shaul PW, Mineo C. HDL action on the vascular wall: is the answer NO? J Clin Invest. 2004;113:509–513. doi: 10.1172/JCI21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziouzenkova O, Perrey S, Asatryan L, Hwang J, MacNaul KL, Moller DE, Rader DJ, Sevanian A, Zechner R, Hoefler G, Plutzky J. Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: evidence for an antiinflammatory role for lipoprotein lipase. Proc Natl Acad Sci U S A. 2003;100:2730–2735. doi: 10.1073/pnas.0538015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziouzenkova O, Asatryan L, Sahady D, Orasanu G, Perrey S, Cutak B, Hassell T, Akiyama TE, Berger JP, Sevanian A, Plutzky J. Dual roles for lipolysis and oxidation in peroxisome proliferation-activator receptor responses to electronegative low density lipoprotein. J Biol Chem. 2003;278:39874–39881. doi: 10.1074/jbc.M306786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 13.Plutzky J. Peroxisome proliferator-activated receptors in vascular biology and atherosclerosis: emerging insights for evolving paradigms. Curr Atheroscler Rep. 2000;2:327–335. doi: 10.1007/s11883-000-0067-3. [DOI] [PubMed] [Google Scholar]
- 14.Chawla A, Lee CH, Barak Y, He W, Rosenfeld J, Liao D, Han J, Kang H, Evans RM. PPA. Rdelta is a very low-density lipoprotein sensor in macrophages. Proc Natl Acad Sci U S A. 2003;100:1268–1273. doi: 10.1073/pnas.0337331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marx N, Sukhova GK, Collins T, Libby P, Plutzky J. PPARalpha activators inhibit cytokine-induced vascular cell adhesion molecule-1 expression in human endothelial cells. Circulation. 1999;99:3125–3131. doi: 10.1161/01.cir.99.24.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson SM, Parhami F, Xi XP, Berliner JA, Hsueh WA, Law RE, Demer LL. Peroxisome proliferator-activated receptor activators target human endothelial cells to inhibit leukocyte-endothelial cell interaction. Arterioscler Thromb Vasc Biol. 1999;19:2094–2104. doi: 10.1161/01.atv.19.9.2094. [DOI] [PubMed] [Google Scholar]
- 17.Pasceri V, Wu HD, Willerson JT, Yeh ET. Modulation of vascular inflammation in vitro and in vivo by peroxisome proliferator-activated receptor-gamma activators. Circulation. 2000;101:235–238. doi: 10.1161/01.cir.101.3.235. [DOI] [PubMed] [Google Scholar]
- 18.Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- 19.Broedl UC, Maugeais C, Marchadier D, Glick JM, Rader DJ. Effects of nonlipolytic ligand function of endothelial lipase on high density lipoprotein metabolism in vivo. J Biol Chem. 2003;278:40688–40693. doi: 10.1074/jbc.M304367200. [DOI] [PubMed] [Google Scholar]
- 20.Kratky D, Zimmermann R, Wagner EM, Strauss JG, Jin W, Kostner GM, Haemmerle G, Rader DJ, Zechner R. Endothelial lipase provides an alternative pathway for FFA uptake in lipoprotein lipase-deficient mouse adipose tissue. J Clin Invest. 2005;115:161–167. doi: 10.1172/JCI15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin W, Millar JS, Broedl U, Glick JM, Rader DJ. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J Clin Invest. 2003;111:357–362. doi: 10.1172/JCI16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marelli-Berg FM, Peek E, Lidington EA, Stauss HJ, Lechler RI. Isolation of endothelial cells from murine tissue. J Immunol Methods. 2000;244:205–215. doi: 10.1016/s0022-1759(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 23.Hodis HN, Kramsch DM, Avogaro P, Bittolo-Bon G, Cazzolato G, Hwang J, Peterson H, Sevanian A. Biochemical and cytotoxic characteristics of an in vivo circulating oxidized low density lipoprotein (LDL-) J Lipid Res. 1994;35:669–677. [PubMed] [Google Scholar]
- 24.Cepek KL, Parker CM, Madara JL, Brenner MB. Integrin alpha E beta 7 mediates adhesion of T lymphocytes to epithelial cells. J Immunol. 1993;150:3459–3470. [PubMed] [Google Scholar]
- 25.Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- 26.Fuki IV, Blanchard N, Jin W, Marchadier DH, Millar JS, Glick JM, Rader DJ. Endogenously produced endothelial lipase enhances binding and cellular processing of plasma lipoproteins via heparan sulfate proteoglycan-mediated pathway. J Biol Chem. 2003;278:34331–34338. doi: 10.1074/jbc.M302181200. [DOI] [PubMed] [Google Scholar]
- 27.Broedl UC, Maugeais C, Millar JS, Jin W, Moore RE, Fuki IV, Marchadier D, Glick JM, Rader DJ. Endothelial lipase promotes the catabolism of ApoB-containing lipoproteins. Circ Res. 2004;94:1554–1561. doi: 10.1161/01.RES.0000130657.00222.39. [DOI] [PubMed] [Google Scholar]
- 28.McCoy MG, Sun GS, Marchadier D, Maugeais C, Glick JM, Rader DJ. Characterization of the lipolytic activity of endothelial lipase. J Lipid Res. 2002;43:921–929. [PubMed] [Google Scholar]
- 29.Duong M, Psaltis M, Rader DJ, Marchadier D, Barter PJ, Rye KA. Evidence that hepatic lipase and endothelial lipase have different substrate specificities for high-density lipoprotein phospholipids. Biochemistry. 2003;42:13778–13785. doi: 10.1021/bi034990n. [DOI] [PubMed] [Google Scholar]
- 30.Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T. Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003;111:347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maugeais C, Tietge UJ, Broedl UC, Marchadier D, Cain W, McCoy MG, Lund-Katz S, Glick JM, Rader DJ. Dose-dependent acceleration of high-density lipoprotein catabolism by endothelial lipase. Circulation. 2003;108:2121–2126. doi: 10.1161/01.CIR.0000092889.24713.DC. [DOI] [PubMed] [Google Scholar]
- 32.Hirata K, Ishida T, Matsushita H, Tsao PS, Quertermous T. Regulated expression of endothelial cell-derived lipase. Biochem Biophys Res Commun. 2000;272:90–93. doi: 10.1006/bbrc.2000.2747. [DOI] [PubMed] [Google Scholar]
- 33.Jin W, Sun GS, Marchadier D, Octtaviani E, Glick JM, Rader DJ. Endothelial cells secrete triglyceride lipase and phospholipase activities in response to cytokines as a result of endothelial lipase. Circ Res. 2003;92:644–650. doi: 10.1161/01.RES.0000064502.47539.6D. [DOI] [PubMed] [Google Scholar]
- 34.Choi SY, Hirata K, Ishida T, Quertermous T, Cooper AD. Endothelial lipase: a new lipase on the block. J Lipid Res. 2002;43:1763–1769. doi: 10.1194/jlr.r200011-jlr200. [DOI] [PubMed] [Google Scholar]
- 35.Azumi H, Hirata K, Ishida T, Kojima Y, Rikitake Y, Takeuchi S, Inoue N, Kawashima S, Hayashi Y, Itoh H, Quertermous T, Yokoyama M. Immunohistochemical localization of endothelial cell-derived lipase in atherosclerotic human coronary arteries. Cardiovasc Res. 2003;58:647–654. doi: 10.1016/s0008-6363(03)00287-6. [DOI] [PubMed] [Google Scholar]
- 36.Bishop-Bailey D. Peroxisome proliferator-activated receptors in the cardiovascular system. Br J Pharmacol. 2000 Mar;129(5):823–834. doi: 10.1038/sj.bjp.0703149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.