The crystal structure of the RNA chaperone Hfq in complex with RNA is presented at 0.97 Å resolution.
Keywords: Hfq, RNA chaperones, Escherichia coli
Abstract
In bacteria, small RNAs (sRNAs) silence or activate target genes through base pairing with the mRNA, thereby modulating its translation. A central player in this process is the RNA chaperone Hfq, which facilitates the annealing of sRNAs with their target mRNAs. Hfq has two RNA-binding surfaces that recognize A-rich and U-rich sequences, and is believed to bind an sRNA–mRNA pair simultaneously. However, how Hfq promotes annealing remains unclear. Here, the crystal structure of Escherichia coli Hfq is presented in complex with U6-RNA bound to its proximal binding site at 0.97 Å resolution, revealing the Hfq–RNA interaction in exceptional detail.
1. Introduction
The Hfq protein was first discovered as a host factor required for the replication of bacteriophage Qβ (Franze de Fernandez et al., 1972 ▶). Since then, Hfq has been shown to play essential roles in the bacterial life cycle and in virulence in general. It functions as an RNA chaperone and is a central player in post-transcriptional regulation of gene expression (Vogel & Luisi, 2011 ▶; Møller et al., 2002 ▶). It binds to sRNAs (small RNAs) and mRNAs and facilitates their unwinding and annealing. Moreover, it has also been implicated in interacting with many proteins involved in transcription, translation, protein folding and degradation as well as in RNA modification and degradation (Wilusz & Wilusz, 2005 ▶).
Hfq is highly conserved among bacteria and forms ring-shaped homohexamers. It belongs to the SM/LSM superfamily and has three distinct RNA-binding sites, referred to as ‘distal’, ‘lateral’ and ‘proximal’ (Aiba, 2007 ▶; Møller et al., 2002 ▶; Link et al., 2009 ▶; Sauter et al., 2003 ▶; Schumacher et al., 2002 ▶; Vogel & Luisi, 2011 ▶). The ‘distal’ face of the ring has a high affinity for A-rich sequences, the ‘lateral’ binding site is an accessory RNA-interaction site on the side of the ring and has little sequence specificity (Gottesman, 2004 ▶; Vogel & Luisi, 2011 ▶; Sauer & Weichenrieder, 2011 ▶; Link et al., 2009 ▶; Schumacher et al., 2002 ▶, Zhang et al., 2002 ▶, Sauer et al., 2012 ▶) and the ‘proximal’ face preferentially binds to U-rich RNA sequences. Since A-rich sequences are common in mRNAs while U-rich sequences are present at the 3′-end of most sRNAs, it is believed that Hfq binds simultaneously to an sRNA and an mRNA and thereby facilitates their pairing. Crystal structures of Hfq from several Gram-positive and Gram-negative bacteria, including Escherichia coli, have already been presented in the literature (Schumacher et al., 2002 ▶; Sauter et al., 2003 ▶; Nikulin et al., 2005 ▶; Nielsen et al., 2007 ▶; Bøggild et al., 2009 ▶; Link et al., 2009 ▶; Baba et al., 2010 ▶; Moskaleva et al., 2010 ▶; Beich-Frandsen, Večerek, Konarev et al., 2011 ▶; Beich-Frandsen, Večerek, Sjöblom et al., 2011 ▶; Sauer & Weichenrieder, 2011 ▶; Kadowaki et al., 2012 ▶; Someya et al., 2012 ▶; Wang et al., 2013 ▶; Yonekura et al., 2013 ▶; Robinson et al., 2014 ▶). Recent structural studies on Salmonella typhimurium Hfq also revealed RNA binding to the proximal binding site of Hfq and demonstrated that the free 3′-hydroxyl group at the end of the RNA is critical for RNA binding and 3′-end recognition (Sauer & Weichenrieder, 2011 ▶). However, despite these investigations the precise molecular mechanism of Hfq-mediated RNA annealing still remains ambiguous (Vogel & Luisi, 2011 ▶). Here, the crystal structure of the E. coli Hfq:U6-RNA complex is presented at a resolution exceeding 1 Å, which provides detailed insights into RNA recognition in E. coli Hfq and indicates that proximal-site RNA recognition is conserved between S. typhimurium and E. coli Hfq.
2. Material and methods
2.1. Protein purification
The protein construct used in this study contained amino acids 1–72 (Hfq) of the 102-residue E. coli Hfq protein. The hfq DNA was cloned C-terminal to a His6-SUMO tag in a pETM28-SUMO vector (PEP-Core, EMBL). Protein expression was conducted in E. coli BL21(DE3) in Terrific broth (TB) medium at 16°C for 20 h. The His6-SUMO-fusion protein was purified via affinity chromatography on a HisTrap (Ni Sepharose, GE Healthcare) column following the manufacturer’s instructions using a buffer consisting of 0.1 M HEPES–NaOH pH 8.0, 0.5 M NaCl, 0.005 M TCEP. To reduce the contamination with nucleic acids, the column was washed with 1 M LiCl before eluting the protein with imidazole. The eluate was then incubated with SenP2 protease [1:100(w/w), PEP-Core, EMBL] for 18 h at 4°C. To remove the cleaved SUMO tag a second Ni-purification step was performed. Finally, Hfq was further purified by size-exclusion chromatography on a Superdex 200 16/60 column. The resulting sample was 99% pure as judged by SDS–PAGE electrophoresis and was concentrated to 10 mg ml−1 and stored in 0.05 M HEPES–NaOH pH 8.0, 0.5 M NaCl at −80°C until further use.
2.2. RNA purification
The DNA template for RNA-OUT (5′-UUC GCA CAU CUU GUU GUC UGA UUA UUG AUU UUU CGC GAA ACC AUU UGA UCA UAU GAC AAG AUG UGU AUC CA-3′) was cloned into the pRAV23 vector and was amplified in E. coli XL1-Blue cells. The plasmid was extracted and purified via anion-exchange chromatography using Q-Sepharose beads. The purified DNA template was concentrated by ethanol precipitation and subjected to HindIII digestion in order to yield a run-off transcript template. To remove potential RNAse contamination, Proteinase K was added and subsequently denatured by heating to 95°C. For in vitro transcription the purified DNA template (0.1 mg ml−1) was incubated with T7 RNA polymerase (0.2 mg ml−1) and 20 units of pyrophosphatase (NEB) in TXN buffer [0.02 M MgCl2, 0.04 M of each rNTP, 0.3 M HEPES–NaOH pH 8.0, 0.005 M DTT, 0.001 M spermidine, 0.01%(v/v) Triton X-100] at 37°C for 18 h. After ethanol precipitation of the transcript, cleavage of the 3′ ribozyme GlmS from the RNA transcript was induced by addition of 0.05 M GlcN6P. Finally, RNA-OUT was purified via ion-exchange chromatography on a Resource Q column. The purity of the RNA was assessed on a denaturing urea gel.
2.3. Complex formation
For crystallization, complexes were formed by combining Hfq and RNA-OUT in a 1:1.2 molar ratio in HS buffer [2 M NaCl, 0.02 M HEPES–NaOH pH 8.0, 0.005 M MgCl2, 5%(v/v) glycerol] and dialyzing the solution against CX buffer [0.25 M NaCl, 0.02 M HEPES–NaOH pH 8.0, 0.05 M MgCl2, 10%(v/v) glycerol] at 20°C for 18 h. For crystallization trials the complex solution was concentrated to 5 mg ml−1 (protein concentration). Crystallization experiments were set up using sitting-drop vapour-diffusion plates at 20°C.
2.4. Data collection and processing
X-ray data collection was performed at −173°C; diffraction images were collected on beamline P13 at DESY/EMBL, Hamburg. Diffraction images were indexed and integrated using XDS (Kabsch, 2010 ▶) and scaled using AIMLESS (Winn et al., 2011 ▶; Evans & Murshudov, 2013 ▶). Data integration was performed in several consecutive XDS runs, during which refined unit-cell parameters, crystal orientation, mosaicity and beam divergence were used as starting values for the next run.
2.5. Structure solution and refinement
The structure was solved by molecular replacement (Phaser; McCoy et al., 2007 ▶) using PDB entry 1hk9 as a search model (Sauter et al., 2003 ▶). Coordinates were initially refined with good stereochemistry up to a resolution of 1.3 Å using REFMAC (Murshudov et al., 2011 ▶). Subsequently, water and H atoms were added, and the resolution was extended stepwise to the full resolution range and refined using PHENIX and REFMAC (Adams et al., 2010 ▶; Murshudov et al., 2011 ▶; Winn et al., 2011 ▶; Table 1 ▶). Additional and disordered residues were manually built into the structure using Coot (Emsley et al., 2010 ▶). Alternating steps of anisotropic refinement and minor structure adjustments were performed until the R values converged. Molecular images were generated in PyMOL (DeLano, 2002 ▶).
Table 1. Data-collection and refinement statistics for the Hfq:U6-RNA complex structure.
Values in parentheses are for the highest resolution shell.
| PDB code | 4pno |
| Data collection | |
| Space group | P6 |
| Unit-cell parameters (, ) | a = b = 61.11, c = 27.86, = = 90.0, = 120.0 |
| Resolution () | 52.920.97 (1.0050.97) |
| Wavelength () | 0.885611 |
| R merge | 0.033 (1.659) |
| R meas | 0.037 (1.896) |
| CC1/2 | 0.999 (0.585) |
| I/(I) | 26.0 (1.2) |
| Completeness (%) | 100.0 (100.0) |
| Observed reflections | 359537 (14597) |
| Unique reflections | 35377 (1762) |
| Multiplicity | 10.2 (8.3) |
| Refinement | |
| Resolution () | 30.55 0.97 |
| No. of reflections | 35355 |
| No. of reflections for R free | 1942 [5.49%] |
| R work | 0.142 (0.298) |
| R free | 0.164 (0.330) |
| B factors (2) | |
| Protein | 19.5 |
| RNA | 23.7 |
| Solvent | 32.5 |
| R.m.s. deviations | |
| Bond lengths () | 0.014 |
| Bond angles () | 1.71 |
3. Results and discussion
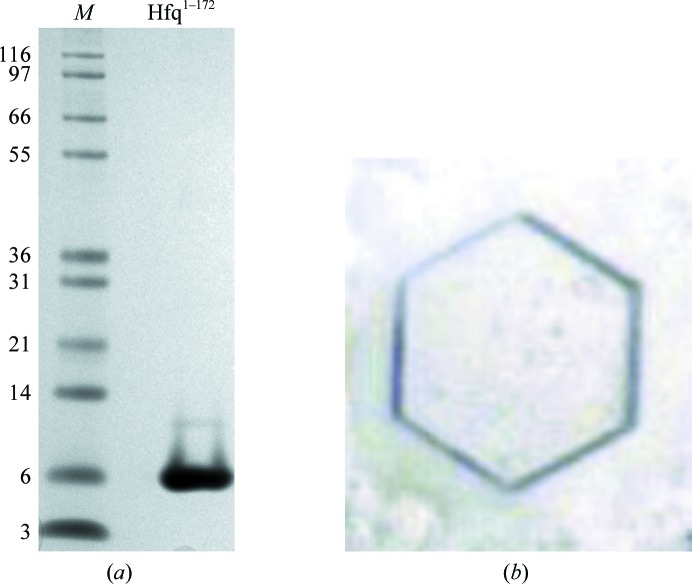
Crystals of Hfq:U6-RNA were grown at 20°C in sitting-drop vapour-diffusion plates by combining equal volumes of precipitant [12%(w/v) PEG 3350, 0.1 M HEPES–NaOH pH 8.0, 0.25 M KSCN] and the protein:RNA complex solution. Initial crystals were observed after 2–3 d, but the best diffracting crystals were obtained after 24 months (Fig. 1 ▶). Crystals were cryoprotected by transfer to precipitant solution containing 12%(v/v) 2,3-butanediol and were subsequently flash-cooled in liquid nitrogen prior to data collection. The crystals obtained in this study belonged to space group P6 (unit-cell parameters a = b = 61.11, c = 27.86 Å, α = β = 90, γ = 120°) with one monomer in the asymmetric unit. However, the sixfold crystal symmetry operation generates the biological homohexameric assembly (Fig. 2 ▶). The crystals diffracted to a resolution of 0.97 Å, and the structure was refined to R work and R free values of 14.2 and 16.4%, respectively (Table 1 ▶); the coordinates were deposited in the PDB as entry 4pno.
Figure 1.
(a) SDS–PAGE illustrating purified Hfq. Lane M contains molecular-mass markers (labelled in kDa). (b) A typical hexagonal crystal of the Hfq:U6-RNA complex. Crystals were observed after 2–3 d, but the best diffracting crystals were collected after 24 months.
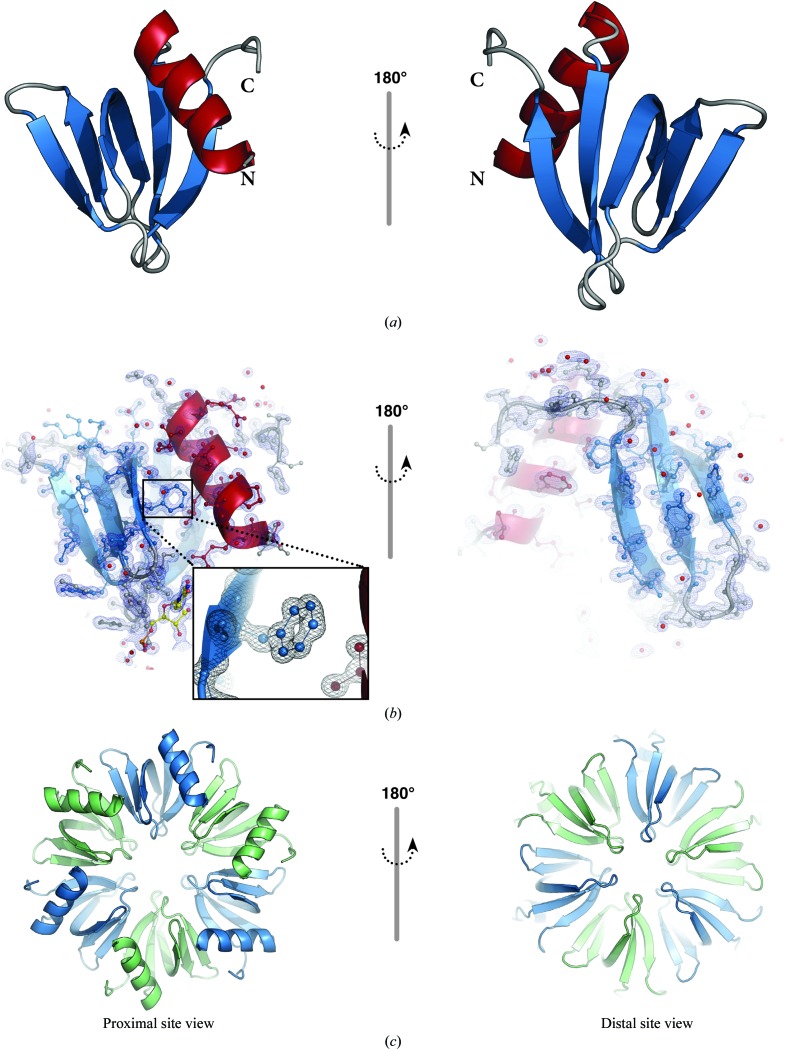
Figure 2.
(a) Cartoon representation of the Hfq monomer present in the asymmetric unit. The single α-helix is coloured red, β-strands are shown in blue and loop regions are shown in grey; the N- and C-termini are indicated. (b) Electron-density map (2F o − F c) contoured at the 1.5σ level. Protein residues and the uridine nucleotide are shown as ball-and-stick representations coloured as in (a) and uridine C atoms are shown in yellow. (c) The sixfold symmetry operation yields the biological assembly, a homohexamer; the individual chains are depicted as cartoons in alternating colours (blue and green).
The data yielded an electron-density map of overall excellent quality (Fig. 2 ▶ b). Residues 6–70 of the protein construct are clearly visible in the electron-density maps, with the exception of Asn48 (see below). A total of ten residues display alternate conformations, which are mainly located on the surface of Hfq. Nevertheless, the generally close crystal packing and the relatively low solvent content (39.8%) give rise to the highest resolution structure that has been reported for an Hfq:RNA complex to date.
The side chain of Asn48 was usually modelled in previous structures, although with highly elevated B factors. For example, in the S. typhimurium Hfq:U6-RNA complex (PDB entry 2ylc; Sauer & Weichenrieder, 2011 ▶) the side chain of Asn48 is stabilized by a hydrogen bond to the main-chain O atom of Gln33, but still has high B factors. In other structures (e.g. 2ylb; Sauer & Weichenrieder, 2011 ▶), Asn48 assumes multiple conformations depending on the crystal-packing environment. In the present structure Asn48 displays an even higher degree of disorder in its main-chain and side-chain density and no side-chain conformation could be refined with confidence. This may be due to its position in an exposed loop that has no direct crystal-packing contacts. The dynamic nature of this part of the structure is also reflected in elevated B factors and multiple alternative conformations of the neighbouring residues.
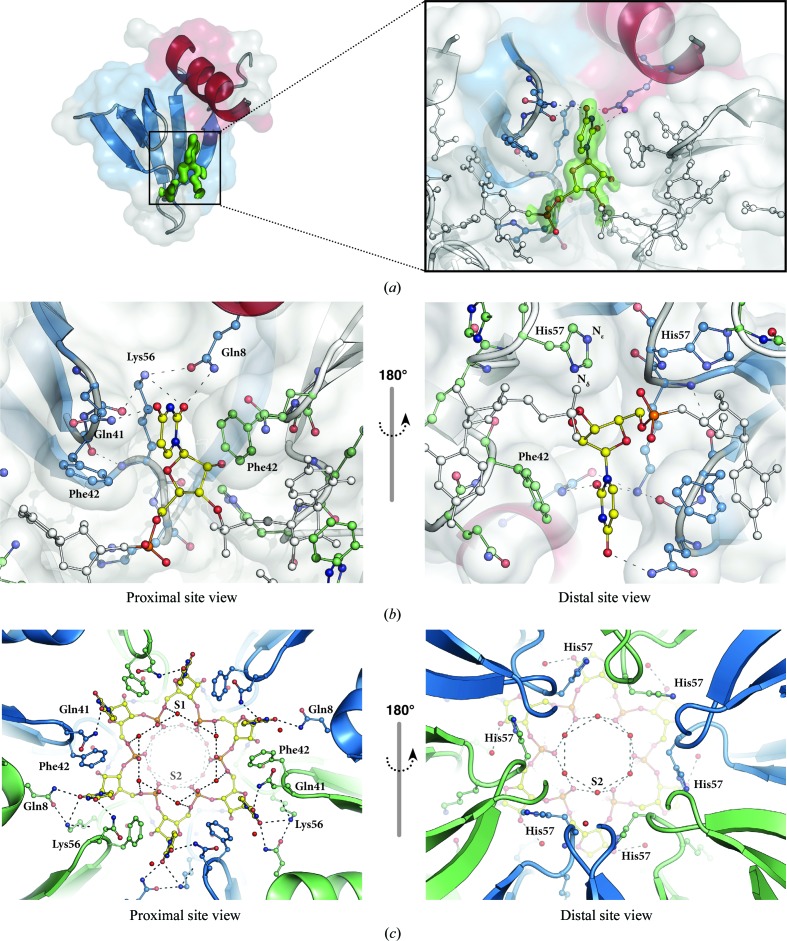
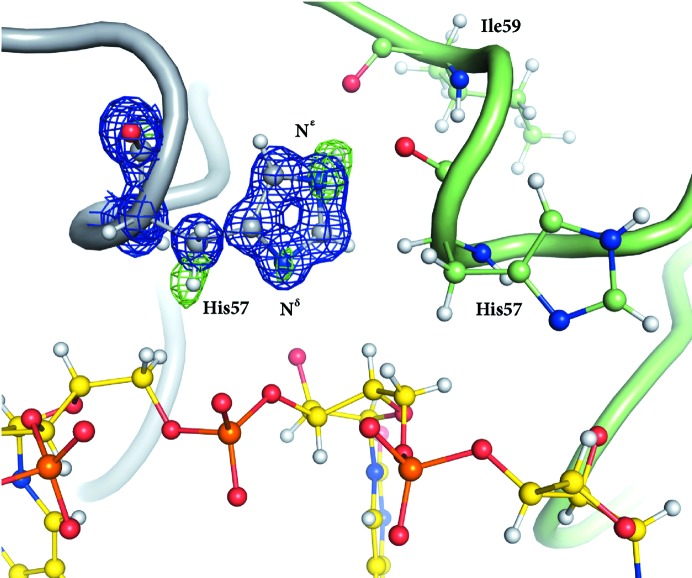
After initial refinement and modelling of alternate amino-acid conformations, a uridine monophosphate with the ribose in the C3′-endo conformation was built into the difference-map density at the proximal face of Hfq (Fig. 3 ▶). The uracil nucleotide makes hydrogen bonds to Gln8, Gln41 and Lys56 and is stabilized by a π-stacking interaction with Phe42 from the neighbouring Hfq chain (Fig. 3 ▶ b). The distal-site view also shows His57 of the neighbouring Hfq chain (Fig. 3 ▶ b), which has been shown to be important for RNA 3′-end recognition. In the present structure, His57 does not contact the nucleotide. This is consistent with previous observations that His57 does not interact with internal RNA nucleotides; rather, its N∊ atom forms a hydrogen bond to Ile59, leaving its Nδ atom unprotonated and preparing it to act as a hydrogen-bond acceptor for the terminal 3′-hydroxyl groups (Sauer & Weichenrieder, 2011 ▶). This protonation state of His57 can now clearly be identified in our structure from a hydrogen OMIT difference density map, unambiguously showing difference density for its N∊ H atom (Fig. 4 ▶). The above-described hydrogen-bonding network, together with water molecules (S1 and S2 in Fig. 3 ▶ c) that bridge neighbouring phosphates, keeps the RNA in a well defined stable conformation.
Figure 3.
(a) A half-transparent surface representation of an Hfq monomer (colours as in Fig. 2 ▶ a) is shown with F o − F c OMIT map (green, contoured at 3σ) for the uridine nucleotide at the proximal face. The enlarged view shows how the uridine nucleotide fits into the difference density and how it sits between two Hfq monomers. (b) Proximal- and distal-site views of the uridine-binding site on Hfq. The interactions of Gln8, Gln41, Phe42 and Lys56 with the uridine nucleotide are shown. (c) The hydrogen-bond network between the solvent atoms (red spheres, S1 and S2) and the RNA chain is illustrated.
Figure 4.
A hydrogen OMIT map illustrating the protonation state of His57: N∊ is clearly protonated pointing towards Ile59, leaving the Nδ atom unprotonated and allowing it to act as a hydrogen-bond acceptor of terminal hydroxyl groups. The electron-density map (2F o − F c) contoured at the 2.5σ level is coloured blue and the difference density map (F o − F c) is coloured green. Protein residues and the uridine nucleotide are shown in ball-and-stick representation.
Consistent with the sixfold symmetry of the Hfq hexamer, the electron density for the RNA nucleotides also forms a continuous circle of six uridine nucleotides. To create a continuous RNA chain, the uridine monophosphate was linked with a covalent bond to its symmetry-related counterpart. Further refinement resulted in electron density for the RNA that is of excellent quality. However, residual difference density required lowering the occupancy for the phosphate group. Owing to the symmetry, the 5′ and 3′ termini of the RNA chain could not be unambiguously identified. Even reprocessing the data in space group P1 and subsequent refinement did not lead to any differences in the position of the 5′ or 3′ RNA termini either. This perfect sixfold symmetry of the complex, including the RNA, further supports the view that 5′- and 3′-terminal nucleotides adopt a highly similar position as internal nucleotides (Sauer & Weichenrieder, 2011 ▶).
Although the Hfq:RNA-OUT complex was used for crystallization, only the protein component bound to a stretch of uridine nucleotides can be found in the crystals. One possibility is that a cellular U6-RNA fragment has co-purified and co-crystallized with Hfq. However, in light of the rather long incubation time and the low stability of RNA molecules, it is also possible that RNA-OUT was degraded during crystal growth. Such degradation may have resulted in a statistically bound U5 fragment that could well explain the density observed in our structure. This is supported by the fact that the uridine monophosphate can be refined best at an occupancy of 5/6, leading to slightly improved R values.
In comparison to the RNA complex structure reported for S. typhimurium (PDB entry 2ylc) only minor differences in the backbone with an r.m.s.d. of 0.29 Å (calculated for the Cα atoms) can be observed. Furthermore, the position and conformation of the U6-RNA and the side chains stabilizing its conformation are practically unaltered between the two structures, demonstrating that the RNA binding at the proximal site is conserved between S. typhimurium and E. coli Hfq.
Interestingly, another E. coli Hfq structure that was reported in complex with an AU6A-RNA bound to its proximal face (PDB entry 3rer; Wang et al., 2013 ▶) reveals a markedly different RNA conformation. While this difference could in principle reflect species-specific differences in the proximal RNA-binding mode (similar differences were reported previously for distal-face binding in Bacillus subtilis; Someya et al., 2012 ▶), our structure reveals that here it is more likely explained by the organization of the molecules in the crystalline environment. Namely, in the structure 3rer the first nucleotide of the AU6A–RNA complex (A1) is bound to the distal face of a symmetry-related Hfq hexamer, which alters the conformation of U2, U3 and U6, orienting them away from the binding pocket. However, the internal nucleotides U4, U5 and U7 are less affected and adopt conformations where the uridine nucleotide is bound by Gln8, Gln41 and Phe42 as seen in other proximal-site complexes (e.g. PDB entries 2ylc and 1kq2; Schumacher et al., 2002 ▶). On the other hand, in our E. coli Hfq:U6 structure the binding modes of all uridines are identical to each other and to those observed for S. typhimurium and Staphylococcus aureus (PDB entries 2ylc snd 1kq2) Hfq. This indicates a conserved RNA-binding mode irrespective of the source organism.
In summary, our 0.97 Å resolution structure provides the highest resolution structure of Hfq to date. While it is highly similar to previously reported Hfq structures, it identifies hydrogen positions relevant to RNA binding and will provide a valuable resource for further detailed mechanistic studies.
Supplementary Material
PDB reference: Hfq:RNA complex, 4pno
Acknowledgments
This work was supported by the EMBL, and the European Commission under the Marie Curie Intra-European Fellowship Programme to ECS (PIEF-GA-2011-301489). The authors would like to thank the beamline scientists of the EMBL Hamburg for support during data collection. We are grateful to Matthias Wilmanns for many helpful discussions and his continuous support.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Aiba, H. (2007). Curr. Opin. Microbiol. 10, 134–139. [DOI] [PubMed]
- Baba, S., Someya, T., Kawai, G., Nakamura, K. & Kumasaka, T. (2010). Acta Cryst. F66, 563–566. [DOI] [PMC free article] [PubMed]
- Beich-Frandsen, M., Večerek, B., Konarev, P. V., Sjöblom, B., Kloiber, K., Hämmerle, H., Rajkowitsch, L., Miles, A. J., Kontaxis, G., Wallace, B. A., Svergun, D. I., Konrat, R., Bläsi, U. & Djinović-Carugo, K. (2011). Nucleic Acids Res. 39, 4900–4915. [DOI] [PMC free article] [PubMed]
- Beich-Frandsen, M., Večerek, B., Sjöblom, B., Bläsi, U. & Djinović-Carugo, K. (2011). Acta Cryst. F67, 536–540. [DOI] [PMC free article] [PubMed]
- Bøggild, A., Overgaard, M., Valentin-Hansen, P. & Brodersen, D. E. (2009). FEBS J. 276, 3904–3915. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Franze de Fernandez, M. T., Hayward, W. S. & August, J. T. (1972). J. Biol. Chem. 247, 824–831. [PubMed]
- Gottesman, S. (2004). Annu. Rev. Microbiol. 58, 303–328. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kadowaki, M. A. S., Iulek, J., Barbosa, J. A. R. G., Pedrosa, F. de O., de Souza, E. M., Chubatsu, L. S., Monteiro, R. A., de Oliveira, M. A. S. & Steffens, M. B. R. (2012). Biochim. Biophys. Acta, 1824, 359–365. [DOI] [PubMed]
- Link, T. M., Valentin-Hansen, P. & Brennan, R. G. (2009). Proc. Natl Acad. Sci. USA, 106, 19292–19297. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Møller, T., Franch, T., Højrup, P., Keene, D. R., Bächinger, H. P., Brennan, R. G. & Valentin-Hansen, P. (2002). Mol. Cell, 9, 23–30. [DOI] [PubMed]
- Moskaleva, O., Melnik, B., Gabdulkhakov, A., Garber, M., Nikonov, S., Stolboushkina, E. & Nikulin, A. (2010). Acta Cryst. F66, 760–764. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nielsen, J. S., Bøggild, A., Andersen, C. B. F., Nielsen, G., Boysen, A., Brodersen, D. E. & Valentin-Hansen, P. (2007). RNA, 13, 2213–2223. [DOI] [PMC free article] [PubMed]
- Nikulin, A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S., Vassylyev, D., Garber, M. & Nikonov, S. (2005). Acta Cryst. D61, 141–146. [DOI] [PubMed]
- Robinson, K. E., Orans, J., Kovach, A. R., Link, T. M. & Brennan, R. G. (2014). Nucleic Acids Res. 42, 2736–2749. [DOI] [PMC free article] [PubMed]
- Sauer, E., Schmidt, S. & Weichenrieder, O. (2012). Proc. Natl Acad. Sci. USA, 109, 9396–9401. [DOI] [PMC free article] [PubMed]
- Sauer, E. & Weichenrieder, O. (2011). Proc. Natl Acad. Sci. USA, 108, 13065–13070. [DOI] [PMC free article] [PubMed]
- Sauter, C., Basquin, J. & Suck, D. (2003). Nucleic Acids Res. 31, 4091–4098. [DOI] [PMC free article] [PubMed]
- Schumacher, M. A., Pearson, R. F., Møller, T., Valentin-Hansen, P. & Brennan, R. G. (2002). EMBO J. 21, 3546–3556. [DOI] [PMC free article] [PubMed]
- Someya, T., Baba, S., Fujimoto, M., Kawai, G., Kumasaka, T. & Nakamura, K. (2012). Nucleic Acids Res. 40, 1856–1867. [DOI] [PMC free article] [PubMed]
- Vogel, J. & Luisi, B. F. (2011). Nature Rev. Microbiol. 9, 578–589. [DOI] [PMC free article] [PubMed]
- Wang, W., Wang, L., Wu, J., Gong, Q. & Shi, Y. (2013). Nucleic Acids Res. 41, 5938–5948. [DOI] [PMC free article] [PubMed]
- Wilusz, C. J. & Wilusz, J. (2005). Nature Struct. Mol. Biol. 12, 1031–1036. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yonekura, K., Watanabe, M., Kageyama, Y., Hirata, K., Yamamoto, M. & Maki-Yonekura, S. (2013). PLoS One, 8, e78216. [DOI] [PMC free article] [PubMed]
- Zhang, A., Wassarman, K. M., Ortega, J., Steven, A. C. & Storz, G. (2002). Mol. Cell, 9, 11–22. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Hfq:RNA complex, 4pno