Abstract
Aims
The C677T variant in the methylenetetrahydrofolate reductase (MTHFR; EC 1.5.1.20) enzyme, a key player in the folate metabolic pathway, has been associated with increased risk of migraine with aura. Other genes encoding molecular components of this pathway include Methionine synthase (MTR; EC 2.1.1.13), and Methionine synthase reductase (MTRR; EC 2.1.1.135) among others. We performed a haplotype analysis of migraine risk and MTHFR, MTR, and MTRR.
Methods
Study participants are from a random sub-sample participating in the population-based AGES-Reykjavik Study, including subjects with non-migraine headache (n=367), migraine without aura (n=85), migraine with aura (n=167), and no headache (n=1347). Haplotypes spanning each gene were constructed using Haploview. Association testing was performed on single SNPs and haplotypes using logistic regression, controlling for demographic and cardiovascular risk factors and correcting for multiple testing
Results
Haplotype analysis suggested an association between MTRR haplotypes and reduced risk of migraine with aura. All other associations were not significant after correcting for multiple testing.
Conclusions
These results suggest that MTRR variants may protect against migraine with aura in an older population.
Keywords: Migraine, folate, haplotype, genetics
Introduction
Migraine with aura (MA) and migraine without aura (MO) affect approximately 11% of adults worldwide.(1) MA has been associated with genetic variation in the gene encoding 5′, 10′-methylenetetrahydrofolate reductase (MTHFR), a key enzyme in the metabolism of folate. A recent meta-analysis found that the TT genotype at C677T (rs1801133) in MTHFR was associated with increased odds of migraine with aura,(2) and that an increasing number of the least common allele (T) was associated with MA as well (OR = 1.40; 95% CI 1.1–1.8; p < 0.007).(3) However, a complete analysis of the variation in MTHFR and other genes involved in the folate metabolic pathway has not yet been performed. A recent report analyzed 6 polymorphisms in folate-related genes and found that only one variation in the MTHFR gene (C677T) was associated with MA.(4) To our knowledge, no study has had sufficient genotyping coverage to analyze the haplotype structure across the entire MTHFR gene, or the haplotype structure of MTR and MTRR, two other genes involved in folate metabolism. We predicted that variation across the MTHFR gene as well as the MTR and MTRR genes, using a combined SNP – haplotype approach to maximize the potential to identify putative regions of causal variation, would be associated with migraine with aura.
Methods
Study Design
Study participants are from the Age, Gene/Environment Susceptibility (AGES)-Reykjavik Study, which has been described previously.(5) Individuals who participated in the Reykjavik Study from 1967–1996 were invited to participate in the AGES-Reykjavik Study from 2002–2006. These dates of participation comprise the mid-life and late-life visits, respectively. Of the 5,764 individuals who returned to participate in the AGES-Reykjavik Study, we analyzed data from 2300 individuals randomly selected for a candidate gene sub-study. Individuals returning for the late-life visit had a slightly better cardiovascular risk profile (lower midlife cholesterol, systolic blood pressure, and fewer smokers) compared to individuals who participated at mid-life and not late-life.(5) Midlife assessments included questions about headache, measurement of cardiovascular risk factors, and demographic characteristics as previously described.(6) Late-life assessments included DNA collection, measurement of cardiovascular risk factors, and history of cardiovascular disease. All AGES-Reykjavik Study procedures were approved by the Icelandic National Bioethics Committee (VSN-00-063) and the Institutional Review Board for the U.S. National Institute on Aging, National Institutes of Health.
Mid-life Visit
Headache Classification: Participants reporting one or more headaches per month were asked follow-up questions about five symptoms of migraine: nausea or vomiting, unilateral location, photophobia, together classified as non-aura symptoms; and visual disturbance before or during headache, and unilateral numbness before headache, together classified as aura symptoms. Participants were classified into four mutually exclusive headache categories: No headache once or more per month (reference category; NO), nonmigraine headache (NMH), migraine without aura (MO), and migraine with aura (MA). The migraine without aura category included individuals with headache plus at least two of the three non-aura symptoms and no aura symptoms. Individuals in the migraine with aura (MA) group had headache plus one or both of the aura symptoms; they may have had non-aura symptoms. Individuals in the non-migraine headache group (NMH) reported headache with none or one non-aura symptom. Aura symptoms took precedence over other symptoms. The classification scheme represents an approximation of the International Classification of Headache Disorders (ICHD, 1988) diagnostic criteria for migraine with or without aura, which were formalized after the midlife data were collected.(7) ICHD features for MO that are missing from these criteria include pulsatility, exacerbation with activity, and phonophobia. ICHD criteria for MA that are missing from these criteria include duration of aura (aura symptoms must last between 5 and 60 minutes) and speed of onset (aura symptoms must develop gradually over more than 5 minutes).
Genotyping
DNA samples were extracted from blood using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). Genotyping employed the proprietary GoldenGate assay by Illumina Genotyping Services (Illumina Inc., San Diego, CA). Initial quality control was performed by Illumnia to identify failed samples and SNPs; 156 duplicates were also included, 92 of which Illumina was blinded to. Using the duplicates, genotyping error rates were calculated to be <0.01%, and the percentage of failed samples was 0.07%. The highest percentage of missing data per sample was 2.1%, and the highest percentage of missing data for included SNPs was 4.98%.
Markers Selected to Construct Haplotypes
SNPs chosen to construct haplotypes in the study are reported in the Single Nucleotide Polymorphism database (dbSNP), a public-domain archive for a broad collection of simple genetic polymorphisms.(8) Markers chosen for the present project are listed by RefSNP ID (NCBI) in Table 1, including the distance between markers and heterozygosity data. Distance between markers is 4484 bp on average (minimum = 62 bp, maximum = 14874 bp), indicating a relatively dense map of markers.
Table 1.
Single Nucleotide Polymorphisms Used as Markers
| Gene | RefSNP ID | 5′ to 3′ Location | Chromosome Position | bp to next SNP | MAFb | hap-mkr | Pairwise LD with next markerc |
|---|---|---|---|---|---|---|---|
| MTRR | rs1801394 | Ile22Met, exon 2a | 7870973 | 7206 | .493 | not in block | .343 |
| 5p15.3 | rs1532268 | Ser661Leu, exon 5 | 7878179 | 1804 | .478 | A1 | 1.00 |
| rs6555501 | intron 5 | 7879983 | 5976 | .323 | A2 | .936 | |
| rs162036 | Lys350Arg, exon 7 | 7885959 | 14874 | .225 | B1 | 1.00 | |
| rs8659 | 3′UTR | 7900833 | - | .446 | B2 | n/a | |
| MTR | rs10925235 | intron 1 | 236962775 | 14368 | .323 | A1 | .995 |
| 1q43 | rs12138911 | intron 6 | 236977143 | 11926 | .187 | A2 | 1.00 |
| rs12060570 | intron 10 | 236989069 | 3684 | .461 | A3 | 1.00 | |
| rs1806505 | intron 13 | 236996575 | 4751 | .457 | A4 | .993 | |
| rs6668344 | intron 14 | 237001326 | 8531 | .463 | A5 | 1.00 | |
| rs3754255 | intron 15 | 237009857 | 12273 | .490 | A6 | 1.00 | |
| rs10925250 | intron 18 | 237022130 | 232 | .187 | A7 | 1.00 | |
| rs10925252 | intron 18 | 237022362 | 6202 | .466 | A8 | 1.00 | |
| rs10495386 | intron 22 | 237032808 | 3246 | .007 | A9 | 1.00 | |
| rs4659479 | intron 22 | 237036054 | 1203 | .319 | A10 | 1.00 | |
| rs10495387 | intron 23 | 237037257 | 62 | .029 | HWE | 1.00 | |
| rs10925257 | intron 25 | 237046160 | 2340 | .187 | A11 | 1.00 | |
| rs1805087 | Asp919Gly, exon 26, A3179G | 237048500 | 176 | .187 | A12 | 1.00 | |
| rs2275565 | intron 26 | 237048676 | 6629 | .215 | A13 | 1.00 | |
| rs10802569 | intron 29 | 237055305 | 5128 | .326 | A14 | .993 | |
| rs3820571 | intron 42 | 237060433 | 1872 | .325 | A15 | 1.00 | |
| rs1050993 | 3′UTR, exon 33 | 237062305 | - | .325 | A16 | 1.00 | |
| MTHFR | rs4846048 | 3′UTR, exon 12 | 11846252 | 1816 | .290 | A1 | .922 |
| 1p36.3 | rs2274976 | Arg594Gln, exon 12 A1793G | 11850927 | 1373 | .068 | HWE | 1.00 |
| rs1476413 | intron 10 | 11852300 | 2176 | .298 | A2 | .964 | |
| rs1801131 | Glu429Ala, exon 8 A1298C | 11854476 | 1902 | .342 | A3 | 1.00 | |
| rs1801133 | Ala222Val, exon 5 C677T | 11856378 | 4305 | .329 | A4 | .988 | |
| rs7533315 | intron 3 | 11860683 | 1649 | .261 | HWE | n/a | |
| rs7525338 | intron 2 | 11862332 | 639 | .000 | Removed | ||
| rs2066472 | Arg68Gln, exon 2 | 11862971 | 1003 | .000 | Removed | ||
| rs1931226 | intron 1 | 11863974 | - | .000 | Removed |
Exon 2 is determined using isoform 2 sequence for MTRR.
Minor allele frequency (MAF)
The measure of linkage disequilibrium (LD) reported here is D′, as calculated in Haploview.
HWE indicates that markers deviated significantly from HWE and were removed from haplotype analyses. Other SNPs were removed due to low Heterozygosity (Het).
UTR – un-translated region
bp to next SNP is the number of base pairs between the current marker and the next marker selected for the haplotype analysis.
Validation; please see text above for a description of the levels of validation with regard to the type of evidence submitted for validation of each marker.
Potential Confounders
Because certain cardiovascular and demographic factors have been associated with migraine,(9) the following mid-life variables were included as potential confounders in our analysis: sex, age, educational level (primary, secondary, college, university), self-reported current use of medication for hypertension, smoking history (never, former, current smoker), and history of diabetes, body mass index, systolic blood pressure, total cholesterol, and fasting blood glucose.
Data Analysis
We analyzed data from 1976 individuals (86% total) who completed the migraine assessment at the mid-life visit and had genotype data. Prior to reconstructing haplotypes from the marker genotypes, each marker was tested for deviation from Hardy-Weinberg Equilibrium (HWE) (Table 1) and those significantly deviating from HWE were removed. In addition, we excluded three SNPs in the MTHFR region that were monomorphic in our sample (rs7525338, rs2066472, and rs1931226). These SNPs have low rates of heterozygosity as reported in public databases (<6% predicted),(10) so our findings are unlikely to reflect errors in genotyping. Haplotype blocks were constructed using genotype information from the remaining markers in each gene separately. Phase estimation and pairwise linkage disequilibrium (LD) calculations, based on the genotyping data,(11) were used to determine if one or more blocks were present in a given gene in our sample.
Prior to tests of genetic association, groups were compared on demographic variables, such as age, using analysis of variance (ANOVA) for continuous variables and logistic regression for dichotomous variables (Table 2).
Table 2.
Demographic Characteristics of Study Participants; n(%) or M(SD).
| Variable | NOd | NMH | MO | MA | F |
|---|---|---|---|---|---|
| N(%) | 1357(68.7) | 367(18.6) | 85(4.3) | 167(8.5) | – |
| Mid-life Agee | 50.39(6.11) | 49.69(6.28) | 48.62(5.68) | 48.95(5.98) | 5.18** |
| Late-life Agef | 76.29(5.46) | 75.26(5.67) | 74.15(5.18) | 74.52(5.62) | 10.24*** |
| Years to follow-upg | 25.90(3.08) | 25.58(3.35) | 25.53(3.16) | 25.57(3.32) | 1.57 |
| Women | 689(50.8%) | 248(67.6%) | 73(85.9%) | 133(79.6%) | 102.01*** |
| Systolic BP | 132.06(16.33) | 132.05(16.56) | 129.72(13.03) | 126.49(16.19) | 6.26*** |
| Blood sugar mg/dL | 78.3(9.7) | 76.7(9.1) | 77.9(7.7) | 76.4(9.5) | 3.94** |
| Cholesterol | 6.38(1.07) | 6.17(1.08) | 6.15(1.21) | 6.43(1.15) | 5.80** |
| Hypertension Treatment | 70(5.2%) | 34(9.3%) | 5(5.9%) | 14(8.4%) | 9.80* |
| Smoking (Y/N) | 285(21.0%) | 61(16.6%) | 14(16.5%) | 36(21.6%) | 18.48** |
| Type 2 Diabetes | 18(1.3%) | 5(1.4%) | 1(1.2%) | 4(2.4%) | 1.27 |
|
| |||||
| NOd | NMH | MO | MA | X2 | |
|
| |||||
| Education | 21.73* | ||||
| Primary | 446(32.9%) | 133(36.2%) | 31(36.5%) | 66(39.5%) | – |
| Secondary | 648(47.8%) | 183(49.9%) | 31(36.5%) | 77(46.1%) | – |
| College | 163(12.0%) | 34(9.3%) | 19(22.4%) | 18(10.8%) | – |
| University | 100(7.4%) | 17(4.6%) | 4(4.7%) | 6(3.6%) | – |
p < 0.05.
p < 0.01.
p < 0.001.
Notes: Groups are No Symptoms (NO), non-migraine headache (NMH), migraine without aura (MO), and migraine with aura (MA). Mid-life Age is age at mid-life when the headache assessment was administered. Late-life Age is at the follow-up assessment at which DNA was collected. Years to follow-up is Late-life Age minus Mid-life Age.
Haplotype inference was performed using PHASE 2.1.1, removing haplotypes of < 2% frequency.(12) Bayesian methods were used to estimate haplotypes from population genotype frequencies as performed in the PHASE program. After haplotype reconstruction, specific haplotypes within separate blocks were tested for association with migraine using multinomial regression with four diagnostic categories (i.e., no symptoms, NO (reference category); non-migraine headache, NMH; migraine without aura, MO; and migraine with aura, MA). The independent variable was the presence or absence of one or two copies of a given haplotype, therefore this presumes an additive or dosage model for analysis. Haplotype analyses were used rather than diplotype analyses because there are no hypothesized gene interaction models. Results from both types of analysis are presented in Tables 3, 4, and 5 with the copies of each haplotype on a separate row to facilitate comparisons between genders and two levels of adjustment (Model 1 and Model 2). Model 1 controls for age and time to follow-up. Time to follow-up was included because the migraine assessment was performed during a mid-life visit, but DNA collection occurred about 26 years later and was not the same for all study participants. Model 2 was additionally adjusted for the demographic and medical history variables described above. All analyses were performed separately for men and women.
Table 3.
MTHFR Haplotype analysis of migraine status, OR(95% CI).
| minimally adjusted | fully adjusted | ||||||
|---|---|---|---|---|---|---|---|
| NMH | MO | MA | NMH | MO | MA | ||
| copies | Men | ||||||
| hap1 | 1 | 1.05(0.69–1.61) | 0.48(0.10–2.25) | 1.56(0.75–3.26) | 0.99(0.64–1.54) | 0.53(0.10–2.70) | 1.69(0.79–3.62) |
| 2 | 0.96(0.32–2.86) | 2.17(0.26–18.06) | 2.10(0.46–9.70) | 0.96(0.32–2.89) | 2.49(0.27–22.92) | 1.97(0.41–9.54) | |
| hap2 | 1 | 1.32(0.56–3.08) | 4.24(0.88–20.34) | 2.08(0.60–7.28) | 1.13(0.48–2.70) | 3.16(0.60–16.72) | 2.00(0.55–7.31) |
| 2 | – | – | – | – | – | – | |
| hap3 | 1 | 1.26(0.72–2.22) | 1.50(0.32–6.99) | 0.76(0.22–2.55) | 1.19(0.66–2.14) | 1.79(0.36–8.76) | 0.70(0.20–2.41) |
| 2 | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | |
| hap4 | 1 | 1.66(1.05–2.63).030 | 1.52(0.41–5.73) | 0.64(0.22–1.86) | 1.70(1.07–2.71).025 | 1.75(0.44–6.93) | 0.72(0.24–2.12) |
| 2 | 1.21(0.26–5.64) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 1.30(0.27–6.20) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | |
| hap5 | 1 | 0.38(0.12–1.25) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.39(0.12–1.29) | 0.00(0.00–0.00) | 0.00(0.00–0.00) |
| 2 | – | – | – | – | – | – | |
| hap6 | 1 | 0.64(0.42–0.97).036 | 0.95(0.30–3.04) | 0.73(0.34–1.55) | 0.63(0.41–0.97).035 | 0.93(0.28–3.06) | 0.73(0.34–1.58) |
| 2 | 0.84(0.36–1.95) | 0.00(0.00–0.00) | 0.87(0.19–3.90) | 0.91(0.39–2.14) | 0.00(0.00–0.00) | 0.82(0.18–3.80) | |
| hap7 | 1 | 0.99(0.65–1.51) | 0.72(0.20–2.58) | 1.63(0.77–3.47) | 1.05(0.68–1.61) | 0.56(0.15–2.12) | 1.60(0.74–3.46) |
| 2 | 1.11(0.59–2.07) | 1.35(0.27–6.85) | 1.04(0.28–3.79) | 1.17(0.62–2.20) | 1.14(0.21–6.13) | 1.01(0.27–3.76) | |
| minimally adjusted | fully adjusted | ||||||
| NMH | MO | MA | NMH | MO | MA | ||
| copies | Women | ||||||
| hap1 | 1 | 1.15(0.83–1.59) | 1.39(0.83–2.34) | 0.87(0.57–1.34) | 1.14(0.82–1.59) | 1.37(0.81–2.32) | 0.87(0.56–1.34) |
| 2 | 1.94(0.92–4.10) | 1.76(0.50–6.24) | 1.10(0.36–3.32) | 1.71(0.79–3.69) | 1.62(0.45–5.91) | 1.23(0.40–3.79) | |
| hap2 | 1 | 0.78(0.38–1.62) | 0.51(0.12–2.17) | 1.02(0.44–2.38) | 0.83(0.40–1.73) | 0.46(0.11–2.01) | 1.02(0.43–2.40) |
| 2 | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | |
| hap3 | 1 | 0.98(0.63–1.53) | 1.20(0.61–2.39) | 1.16(0.68–1.99) | 0.97(0.62–1.51) | 1.14(0.57–2.29) | 1.16(0.67–2.01) |
| 2 | 0.67(0.14–3.20) | 1.14(0.14–9.33) | 1.92(0.50–7.41) | 0.66(0.14–3.18) | 1.07(0.12–9.21) | 2.07(0.53–8.14) | |
| hap4 | 1 | 0.62(0.41–0.96).030 | 0.51(0.24–1.10) | 0.99(0.61–1.59) | 0.62(0.40–0.96).031 | 0.52(0.24–1.12) | 0.97(0.60–1.58) |
| 2 | 1.86(0.74–4.64) | 0.76(0.10–6.02) | 0.93(0.20–4.23) | 1.77(0.70–4.49) | 0.85(0.11–6.76) | 0.98(0.21–4.51) | |
| hap5 | 1 | 1.23(0.61–2.48) | 1.03(0.30–3.51) | 1.36(0.57–3.20) | 1.29(0.63–2.63) | 0.99(0.29–3.45) | 1.28(0.54–3.07) |
| 2 | 0.00(0.00–0.00) | 1.00(1.00–1.00) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 1.08(1.08–1.08) | 0.00(0.00–0.00) | |
| hap6 | 1 | 1.18(0.87–1.61) | 1.13(0.69–1.87) | 1.08(0.73–1.62) | 1.16(0.85–1.59) | 1.22(0.73–2.04) | 1.09(0.72–1.63) |
| 2 | 1.27(0.67–2.41) | 0.77(0.23–2.65) | 2.01(1.00–4.02).049 | 1.26(0.66–2.42) | 0.81(0.23–2.83) | 2.16(1.07–4.38).032 | |
| hap7 | 1 | 0.90(0.66–1.23) | 1.13(0.67–1.90) | 0.89(0.60–1.32) | 0.94(0.68–1.29) | 1.18(0.70–2.00) | 0.86(0.57–1.28) |
| 2 | 0.68(0.66–1.12) | 0.85(0.37–1.92) | 0.45(0.22–0.95).035 | 0.71(0.43–1.18) | 0.80(0.35–1.84) | 0.43(0.20–0.90).026 | |
Notes for Table 3:
NMH: non-migraine headache
MO: migraine without aura
MA: migraine with aura
Comparisons are minimally and fully adjusted (see methods). P-values < 0.05 are indicated. No haplotypes met the adjusted level of significance for MTHFR (p=0.0071).
Table 4.
MTR haplotype analysis of migraine status.
| OR(95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| minimally adjusted | fully adjusted | ||||||
| NMH | MO | MA | NMH | MO | MA | ||
| Block A | copies | Men | |||||
| hap1 | 1 | 1.06(0.66–1.68) | 2.68(0.56–12.78) | 0.67(0.31–1.44) | 1.03(0.64–1.65) | 2.79(0.58–13.53) | 0.63(0.29–1.37) |
| 2 | 1.54(0.90–2.61) | 1.64(0.23–11.76) | 0.70(0.26–1.88) | 1.52(0.89–2.59) | 1.54(0.20–11.89) | 0.64(0.23–1.79) | |
| hap2 | 1 | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 1.47(0.42–5.10) | 0.90(0.54–1.48) | 0.00(0.00–0.00) | 1.67(0.47–5.98) |
| 2 | 0.75(0.29–1.97) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 0.90(0.55–1.47) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | |
| hap3 | 1 | 0.80(0.51–1.25) | 0.46(0.10–2.13) | 1.00(0.46–2.15) | 0.82(0.52–1.29) | 0.44(0.09–1.05) | 1.01(0.46–2.21) |
| 2 | 0.26(0.03–1.96) | 0.00(0.00–0.00) | 0.99(0.13–7.82) | 0.27(0.04–2.06) | 0.00(0.00–0.00) | 1.28(0.16–10.50) | |
| hap4 | 1 | 1.06(0.70–1.59) | 3.01(0.79–11.48) | 1.68(0.79–3.56) | 1.03(0.68–1.56) | 3.15(0.79–12.51) | 1.55(0.72–3.34) |
| 2 | 0.71(0.35–1.44) | 1.38(0.14–13.47) | 1.39(0.43–4.44) | 0.69(0.33–1.43) | 1.60(0.16–16.21) | 1.60(0.49–5.23) | |
| minimally adjusted | fully adjusted | ||||||
| NMH | MO | MA | NMH | MO | MA | ||
| Block A | copies | Women | |||||
| hap1 | 1 | 1.36(0.97–1.91) | 0.76(0.43–1.33) | 1.05(0.68–1.62) | 1.39(0.99–1.96) | 0.76(0.43–1.35) | 1.05(0.67–1.62) |
| 2 | 1.23(0.80–1.90) | 1.33(0.70–2.50) | 1.49(0.89–2.48) | 1.24(0.80–1.92) | 1.35(0.71–2.56) | 1.47(0.87–2.46) | |
| hap2 | 1 | 0.67(0.33–1.36) | 0.42(0.10–1.79) | 0.98(0.45–2.17) | 0.67(0.32–1.38) | 0.42(0.10–1.81) | 0.99(0.44–2.20) |
| 2 | 0.83(0.09–8.08) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 1.02(0.10–10.28) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | |
| hap3 | 1 | 0.91(0.66–1.25) | 0.67(0.38–1.79) | 1.01(0.67–1.51) | 0.91(0.66–1.26) | 0.64(0.36–1.13) | 1.02(0.68–1.54) |
| 2 | 1.26(0.58–2.75) | 0.75(0.17–3.33) | 1.19(0.44–3.28) | 1.25(0.57–2.74) | 0.72(0.16–3.21) | 1.07(0.38–2.98) | |
| hap4 | 1 | 1.16(0.86–1.57) | 0.96(0.57–1.62) | 0.63(0.42–0.94).024 | 1.15(0.85–1.57) | 0.97(0.57–1.64) | 0.64(0.43–0.97).034 |
| 2 | 0.52(0.28–0.96).036 | 1.27(0.60–2.70) | 0.60(0.31–1.19) | 0.55(0.29–1.01) | 1.33(0.62–2.85) | 0.59(0.30–1.17) |
Notes for Table 4:
NMH: non-migraine headache
MO: migraine without aura
MA: migraine with aura
Comparisons are minimally and fully adjusted (see methods). P-values < 0.05 are indicated. No haplotypes met the adjusted level of significance for MTR (p=0.0125)
Table 5.
MTRR Haplotype analysis and migraine status.
| OR(95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| minimally adjusted | fully adjusted | ||||||
| NMH | MO | MA | NMH | MO | MA | ||
| Block A | copies | Men | |||||
| hap1-A | 1 | 0.93(0.61–1.43) | 1.18(0.33–4.23) | 0.83(0.40–1.75) | 0.87(0.56–1.34) | 1.09(0.29–4.08) | 0.84(0.39–1.82) |
| 2 | 1.27(0.70–2.32) | 1.66(0.30–9.27) | 1.15(0.40–3.33) | 1.13(0.61–2.08) | 1.54(0.26–9.14) | 0.91(0.31–2.72) | |
| hap2-A | 1 | 1.10(0.72–1.66) | 0.21(0.03–1.63) | 0.64(0.27–1.53) | 1.10(0.72–1.68) | 0.25(0.03–2.03) | 0.61(0.25–1.47) |
| 2 | 1.04(0.35–3.10) | 2.09(0.25–17.30) | 3.49(1.09–11.14).035 | 0.96(0.32–2.89) | 1.83(0.21–16.16) | 2.78(0.81–9.60) | |
| hap3-A | 1 | 0.83(0.54–1.29) | 1.42(0.36–5.56) | 0.82(0.38–1.77) | 0.88(0.56–1.38) | 1.33(0.32–5.45) | 0.90(0.40–1.99) |
| 2 | 0.83(0.48–1.46) | 1.03(0.17–6.26) | 0.79(0.29–2.16) | 0.95(0.54–1.69) | 1.00(0.15–6.54) | 1.05(0.37–2.99) | |
| Women | |||||||
| hap1-A | 1 | 1.03(0.75–1.41) | 1.23(0.73–2.07) | 0.99(0.67–1.48) | 1.01(0.73–1.39) | 1.26(0.74–2.14) | 0.98(0.65–1.47) |
| 2 | 0.75(0.47–1.21) | 0.67(0.28–1.60) | 0.77(0.42–1.10) | 0.73(0.45–1.18) | 0.64(0.27–1.56) | 0.75(0.41–1.39) | |
| hap2-A | 1 | 1.29(0.95–1.77) | 0.00(0.00–0.00) | 2.26(0.96–5.32) | 1.36(0.99–1.87) | 0.00(0.00–0.00) | 1.24(0.83–1.86) |
| 2 | 1.32(0.59–2.95) | 1.20(0.72–2.00) | 2.62(0.96–5.32) | 1.49(0.65–3.41) | 1.26(0.75–2.11) | 2.12(0.89–5.07) | |
| hap3-A | 1 | 1.08(0.78–1.50) | 1.17(0.67–2.06) | 0.89(0.9–1.36) | 1.09(0.78–1.52) | 1.13(0.64–2.02) | 0.91(0.60–1.39) |
| 2 | 0.88(0.57–1.36) | 1.17(0.58–2.36) | 0.81(0.47–1.39) | 0.85(0.55–1.31) | 1.12(0.55–2.28) | 0.85(0.49–1.48) | |
| Block B | Men | ||||||
| hap1-B | 1 | 1.50(0.73–3.08) | 1.21(0.14–10.61) | 0.274(0.10–0.79).016 | 1.58(0.76–3.28) | 1.22(0.14–10.98) | 0.32(0.11–0.96).043 |
| 2 | 1.20(0.58–2.47) | 1.29(0.15–11.01) | 0.57(.23–1.42) | 1.23(0.59–2.55) | 1.14(0.13–10.22) | 0.64(0.25–1.66) | |
| hap2-B | 1 | 1.05(0.69–1.60) | 0.19(0.03–1.52) | 0.52(0.21–1.29) | 1.05(0.69–1.60) | 0.24(0.03–1.89) | 0.49(0.19–1.25) |
| 2 | 0.75(0.26–2.19) | 1.48(0.18–12.02) | 3.17(1.11–9.05).031 | 0.70(0.24–2.08) | 1.38(0.16–11.99) | 2.57(0.85–7.80) | |
| Hap3-B | 1 | 1.18(0.74–1.89) | 2.10(0.62–7.13) | 0.59(0.20–1.70) | 1.25(0.78–2.02) | 2.34(0.66–8.30) | 0.64(0.22–1.89) |
| 2 | 0.71(0.09–5.76) | 0.00(0.00–0.00) | 2.38(0.28–19.92) | 0.56(0.06–4.94) | 0.00(0.00–0.00) | 2.94(0.32–26.65) | |
| Women | |||||||
| hap1-B | 1 | 1.04(0.63–1.71) | 1.91(0.65–5.57) | 1.27(0.64–2.55) | 1.04(0.62–1.72) | 1.89(0.64–5.59) | 1.28(0.64–2.58) |
| 2 | 0.84(0.50–1.39) | 1.87(0.64–5.46) | 1.16(0.58–2.33) | 0.82(0.49–1.37) | 1.75(0.59–5.20) | 1.17(0.58–2.37) | |
| hap2-B | 1 | 1.22(0.90–1.67) | 1.01(0.60–1.69) | 1.23(0.83–1.84) | 1.28(0.93–1.75) | 1.04(0.62–1.77) | 1.22(0.81–1.83) |
| 2 | 1.09(0.49–2.39) | 0.39(0.05–2.93) | 2.15(0.96–4.81) | 1.23(0.55–2.75) | 0.44(0.06–3.37) | 1.96(0.86–4.46) | |
| Hap3-B | 1 | 1.09(0.77–1.53) | 0.88(0.49–1.61) | 0.57(0.34–0.96).033 | 1.04(0.74–1.48) | 0.89(0.48–1.64) | 0.58(0.34–0.99).044 |
| 2 | 0.94(0.30–2.96) | 0.76(0.10–5.99) | 0.38(0.05–2.99) | 0.85(0.27–2.73) | 0.82(0.10–6.69) | 0.43(0.06–3.42) |
Notes for Table 5:
NMH: non-migraine headache
MO: migraine without aura
MA: migraine with aura
Comparisons are minimally and fully adjusted (see methods). P-values < 0.05 are indicated. No haplotypes met the adjusted level of significance for MTRR-A and -B is p = 0.0167. The one test of association falling below this value is denoted with an asterisk.
The critical level of significance for the haplotype-specific tests within each haplotype set was adjusted by Bonferroni correction, i.e., the conventional p < 0.05 was divided by the number of major haplotypes (>2%) observed in each case. The reported p values are two-tailed and are listed below tables. Post-hoc power was calculated for our sample using the Case-control Genetic Power Calculator for discrete traits.(13) Assuming a putative risk allele frequency of 0.20 and a relative risk of 1.5, marker frequency of 0.20 and 0.80 LD (D′) with the risk allele, we had 80% power to reject the null hypothesis of no association at p < 0.05. Logistic regression analyses were performed using SPSS (version 16; SPSS Inc., Chicago, IL, USA).
Results
Individuals varied in age between diagnostic groups (F3, 1975 = 5.18, p = .001) with individuals with migraine (MA or MO) being younger than those with non-migraine headache or no headache (Table 2). Mean (±SD) time between the mid-life and late-life assessments was 25.8 (±3.2) years and participants were on average 50.1 (±6.1) years of age at the mid-life exam and 75.9 (±5.5) years of age at the late-age exam.
Haplotype Reconstruction
Odds ratios were calculated as the measure of effect size and are reported for each block within each of the three genes tested followed by the 95% confidence interval (Figure 1). The haplotype analysis indicated one haplotype block in MTHFR, one block in MTR, and two blocks in MTRR, and regression analyses were performed separately within each block. The conventional significance level of p < 0.05 was divided by the number of major haplotypes observed in each case giving the following p values for each haplotype block; MTHFR, p = 0.0071; MTR, p = 0.0125, and MTRR-A and -B, p = 0.0167. The marker locations and LD block structure for the haplotype blocks are shown in Figure 1. Tables 7, 8, and 9 report the allele present for each marker of each haplotype, and the frequency of that haplotype in the population.
Figure 1. Estimated linkage disequilibrium (LD) and location of haplotype blocks across MTHFR (a), MTR (b) and MTRR (c) genes.
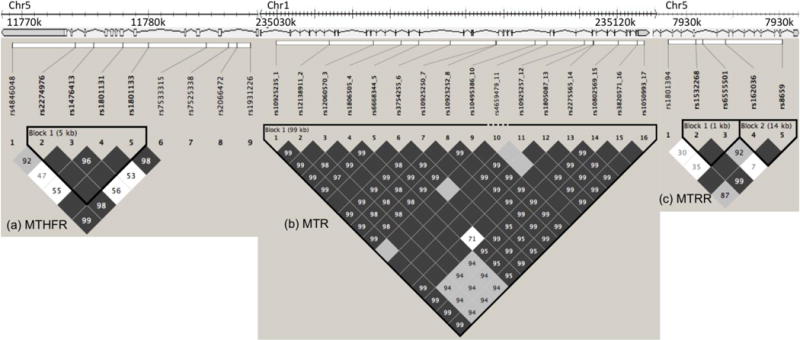
Chromosomal locations of these genes with their coding regions are shown above the LD plots, with 1k bases between marks (not to scale). The pair-wise LD (D′) numbers are reported within cells. Evidence for LD or recombination is shown as different shades of grey (white - low D′, shades of grey - high D′ but low LOD, black - high D′ and high LOD). The haploblocks were identified using a confidence interval algorithm in Haploview program (Barrett, Fry, & Daily, 2005). Haplotype blocks for the present study are contained within black triangles in the figure, and markers outside of the blocks are outside of these triangles. 95% confidence bounds on D′ were generated and each comparison was called “strong LD”, “inconclusive” or “strong recombination”. A block was created if 95% of informative comparisons were “strong LD”.
Table 7.
Risk Score Effect.
| OR(95% CI) | ||||||
|---|---|---|---|---|---|---|
| minimally adjusted | fully adjusted | |||||
| NMH | MO | MA | NMH | MO | MA | |
| Men | ||||||
| Risk-score | 0.98(0.86–1.12) | 0.85(0.57–1.24) | 0.95(0.76–1.20) | 0.97(0.85–1.10) | 0.86(0.58–1.26) | 0.98(0.77–1.24) |
| Women | ||||||
| Risk-score | 1.07(0.97–1.18) | 1.04(0.88–1.23) | 1.13(0.99–1.28) | 1.06(0.96–1.18) | 1.04(0.87–1.23) | 1.13(0.99–1.29) |
Notes for Table 7:
NMH: non-migraine headache
MO: migraine without aura
MA: migraine with aura
Comparisons are minimally and fully adjusted (see methods). No p-values were less than 0.05.
Table 8.
MTHFR Haplotypes and Frequencies.
| Haplotype | rs2274976 | rs1476413 | rs1801131 | rs1801133 | Frequency |
|---|---|---|---|---|---|
| 1 | G | G | A | T | .330 |
| 2 | G | G | A | C | .321 |
| 3 | G | A | C | C | .225 |
| 4 | A | A | C | C | .067 |
| 5 | G | G | C | C | .050 |
| 6 | A | G | C | T | .004 |
| 7 | A | G | A | T | .031 |
Notes: There was only one block in MTHFR.
Table 9.
MTR Haplotypes and Frequencies.
| Marker | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A14 | A15 | A16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Frequency | ||||||||||||||||
| 1 | T | T | C | C | C | C | A | C | C | A | A | A | A | C | T | A | 0.443 |
| 2 | C | T | G | T | T | T | A | T | A | G | A | A | A | G | G | G | 0.316 |
| 3 | T | C | G | T | T | T | G | T | C | A | G | G | C | C | T | A | 0.187 |
| 4 | T | T | G | T | T | C | A | T | C | A | A | A | C | C | T | A | 0.028 |
Notes: 1 refers to the major allele, and 2 denotes the minor allele. There was only one block in MTR.
The rs numbers for markers are listed in Table 1.
MTHFR
Two markers in MTHFR significantly deviated from HWE, rs2274976 (Arg594Gln/A1793G in exon 12; p = 0.0001) and rs7533315 (p = 0.0005), a noncoding SNP, and were removed from analyses leaving a four marker haplotype (A1-rs4846048, A2-rs1476413, A3-rs1801131 and A4-rs1801133; see Table 1). A single haplotype block spanning the MTHFR region was identified, and seven haplotypes over 2% frequency were identified in our sample (i.e., hap1 – hap7). The two coding variations, C677T and A1298C, had a D′ linkage disequilibrium value of 1. Other pairwise LD values for consecutive SNPs are shown in Table 1.
In men, hap4 (AACC) was associated with increased risk of non-migraine headache, although this association did not survive correction for multiple tests of association in both minimally and fully adjusted analyses (Table 3). In women we found the opposite: hap 4 was associated with decreased risk of non-migraine headache in minimally and fully adjusted analyses, although again this did not survive correction for multiple tests. This haplotype, AACC, carries the minor allele at the second and third SNPs (rs1476413 and rs1801131), as underlined. Rs1801131 is a coding variant, Glu429Ala (A1298C), and has previously been associated with MTHFR deficiency and depressed enzyme activity.(14,15) Hap6 (was associated with increased risk of non-migraine headache in men, but associated with increased risk of migraine with aura in women, although neither tests survived correction of the p value. Hap6, AGAC, consists of the major alleles on MTHFR. Hap7 was associated with migraine with aura in women only, but did not survive correction of the p value. Hap7, AGAT, carries the minor allele at rs1801133, which is the coding variant Ala222Val, commonly referred to as C677T. This SNP has previously been associated with increased risk of migraine with aura.(2) In tests of the individual SNPs in MTHFR (Table 6), only this one, rs1801133/C677T was associated with (decreased) risk of migraine with aura in women (OR = 0.45, 95%CI(0.21–0.94), p = 0.034 minimally adjusted analyses, OR = 0.58, 95%CI(0.34–0.99), p = 0.045 in fully adjusted analyses).
Table 6.
Individual Tests With Individual SNPs.
| OR(95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| minimally adjusted | fully adjusted | ||||||
| NMH | MO | MA | NMH | MO | MA | ||
| MTRR | copies | Men | |||||
| rs1801394 | 1 | 1.42(0.79–2.55) | 0.81(0.15–4.24) | 0.65(0.25–1.67) | 1.48(0.82–2.69) | 0.73(0.13–4.05) | 0.77(0.29–2.04) |
| Ile22Met | 2 | 1.02(0.54–1.92) | 1.02(0.19–5.40) | 0.74(0.29–194) | 1.04(0.55–1.97) | 0.94(0.17–5.23) | 0.84(0.31–2.25) |
| rs1532268 | 1 | 0.73(0.41–1.32) | 0.71(0.14–3.59) | 0.72(0.25–2.04) | 0.77(0.42–1.40) | 0.71(0.13–3.79) | 0.92(0.31–2.71) |
| Ser661Leu | 2 | 0.79(0.43–1.43) | 0.60(0.11–3.36) | 0.87(0.30–2.50) | 0.89(0.48–1.64) | 0.65(0.11–3.86) | 1.09(0.37–3.25) |
| rs6555501 | 1 | 0.83(0.53–1.28) | 1.41(0.36–5.54) | 0.81(0.37–1.76) | 0.88(0.56–1.37) | 1.33(0.32–5.43) | 0.89(0.40–1.98) |
| 2 | 0.83(0.47–1.46) | 1.02(0.17–6.21) | 0.78(0.28–2.14) | 0.95(0.53–1.68) | 1.00(0.15–6.51) | 1.03(0.36–2.96) | |
| rs162036 | 1 | 1.18(0.74–1.89) | 2.10(0.62–7.13) | 0.59(0.20–1.70) | 1.25(.78–2.02) | 2.34(0.66–8.30) | 0.64(0.22–1.89) |
| Lys350Arg | 2 | 0.71(0.09–5.76) | 0.00(0.00–0.00) | 2.38(0.28–19.92) | 0.56(.06–4.94) | 0.00(0.00–0.00) | 2.94(0.32–26.65) |
| rs8659 | 1 | 1.25(0.83–1.88) | 0.94(0.28–3.11) | 0.48(0.21–1.23) | 1.29(0.85–1.95) | 1.07(0.31–3.74) | 0.51(0.21–1.20) |
| 2 | 0.83(0.40–1.72) | 0.77(0.09–6.55) | 1.76(0.70–4.40) | 0.81(0.39–1.69) | 0.88(0.10–7.88) | 1.56(0.60–4.05) | |
| Women | |||||||
| rs1801394 | 1 | 1.13(0.74–1.73) | 1.05(0.52–2.136) | 1.06(0.62–1.82) | 1.17(0.76–1.79) | 1.05(0.50–2.13) | 1.05(0.61–1.80) |
| Ile22Met | 2 | 0.98(0.63–1.55) | 1.03(0.49–2.18) | 0.89(0.50–1.58) | 0.98(0.62–1.56) | 0.97(0.45–2.07) | 0.90(0.50–1.60) |
| rs1532268 | 1 | 1.37(0.86–2.18) | 1.83(0.792–4.24) | 1.30(0.72–2.35) | 1.38(0.86–2.23) | 1.96(0.83–4.61) | 1.31(0.71–2.39) |
| Ser661Leu | 2 | 1.33(0.83–2.13) | 1.49(0.63–3.54) | 1.31(0.71–2.35) | 1.38(0.85–2.24) | 1.55(0.64–3.76) | 1.33(0.72–2.47) |
| rs6555501 | 1 | 1.08(0.78–1.50) | 1.17(0.67–2.06) | 0.89(0.59–1.36) | 1.09(0.78–1.52) | 1.13(0.64–2.02) | 0.91(0.60–1.39) |
| 2 | 0.88(0.57–1.36) | 1.17(0.58–2.36) | 0.81(0.47–1.39) | 0.85(0.55–1.31) | 1.12(0.55–2.28) | 0.85(0.49–1.48) | |
| rs162036 | 1 | 1.09(0.77–1.54) | 0.89(0.49–1.61) | 0.57(0.34–0.96).034 | 1.05(0.74–1.49) | 0.89(0.48–1.64) | 0.58(0.34–0.99).045 |
| Lys350Arg | 2 | 0.94(0.30–2.97) | 0.76(0.10–5.99) | 0.38(0.05–2.30) | 0.855(0.27–2.73) | 0.82(0.10–6.69) | 0.43(0.06–3.43) |
| rs8659 | 1 | 1.25(0.92–1.71) | 0.99(0.60–1.65) | 1.16(0.78–1.72) | 1.28(0.94–1.76) | 1.04(0.62–1.74) | 1.17(0.78–1.74) |
| 2 | 1.20(0.72–2.00) | 0.53(0.18–1.56) | 0.91(0.45–1.83) | 1.23(0.74–2.07) | 0.56(0.19–1.67) | 0.90(0.44–1.83) | |
| MTHFR | copies | Men | |||||
| rs2274976 | 1 | 0.71(0.42–1.20) | 0.77(0.17–3.57) | 1.54(0.46–5.17) | 0.69(0.41–1.17) | 0.83(0.16–4.26) | 1.28(0.37–4.39) |
| Arg594Gln | 2 | – | – | – | – | – | – |
| rs1476413 | 1 | 0.76(0.40–1.43) | 0.07(0.01–0.72).025 | 0.77(0.24–2.42) | 0.71(0.37–1.37) | 0.05(0.01–0.50).012 | 0.75(0.23–2.46) |
| 2 | 0.67(0.35–1.26) | 0.50(0.13–1.94) | 0.74(0.24–2.30) | 0.66(0.34–1.26) | 0.31(0.07–1.35) | 0.67(0.21–2.13) | |
| rs1801131 | 1 | 1.07(0.70–1.64) | 0.47(0.12–1.90) | 1.07(0.51–2.23) | 1.03(0.67–1.58) | 0.46(0.11–1.93) | 1.19(0.56–2.54) |
| Glu429Ala | 2 | 1.43(0.79–2.57) | 1.82(0.44–7.46) | 1.03(0.33–3.22) | 1.39(0.77–2.54) | 2.28(0.51–10.14) | 1.15(0.36–3.68) |
| rs1801133 | 1 | 1.01(0.66–1.53) | 0.73(0.20–2.62) | 1.75(0.83–3.70) | 1.40(0.79–2.49) | 0.61(0.09–4.04) | 2.09(0.77–5.63) |
| Ala222Val | 2 | 1.13(0.61–2.12) | 1.39(0.28–7.05) | 1.07(0.29–3.89) | 1.31(0.52–3.33) | 3.05(0.42–22.24) | 1.69(0.37–7.67) |
| Women | |||||||
| rs2274976 | 1 | 0.99(0.64–1.52) | 2.06(0.81–5.27) | 0.76(0.46–1.27) | 1.03(0.66–1.59) | 2.18(0.84–5.64) | 0.77(0.46–1.29) |
| Arg594Gln | 2 | – | – | – | – | – | – |
| rs1476413 | 1 | 0.66(0.40–1.08) | 1.46(0.55–3.93) | 0.86(0.44–1.68) | 0.65(0.39–1.08) | 1.49(0.55–4.04) | 0.81(0.41–1.60) |
| 2 | 0.71(0.44–1.14) | 1.27(0.48–3.39) | 0.96(0.50–1.85) | 0.73(0.45–1.18) | 1.30(0.48–3.50) | 0.92(0.47–1.78) | |
| rs1801131 | 1 | 0.84(0.62–1.16) | 0.83(0.50–1.40) | 0.90(0.61–1.35) | 0.84(0.61–1.15) | 0.82(0.48–1.40) | 0.88(0.59–1.32) |
| Glu429Ala | 2 | 1.39(0.89–2.18) | 1.08(0.49–2.35) | 1.17(0.64–2.12) | 1.35(0.85–2.13) | 1.03(0.47–2.27) | 1.21(0.66–2.22) |
| rs1801133 | 1 | 0.87(0.64–1.18) | 1.14(0.68–1.92) | 0.87(0.59–1.29) | 1.05(0.71–1.56) | 1.22(0.65–2.30) | 1.15(0.70–1.88) |
| Ala222Val | 2 | 0.66(0.40–1.09) | 0.87(0.38–1.96) | 0.45(0.21–0.94).033 | 1.04(0.57–1.89) | 0.88(0.33–2.36) | 0.18(0.04–0.79).022 |
| MTR | Men | ||||||
| rs10925235 | 1 | 1.55(0.75–3.18) | 2.22(0.27–18.11) | 1.22(0.40–3.73) | 1.53(0.73–3.19) | 2.08(0.24–17.73) | 1.05(0.33–3.29) |
| 2 | 1.49(0.73–3.06) | 0.77(0.08–7.56) | 0.77(0.24–2.47) | 1.50(0.72–3.11) | 0.66(0.06–6.77) | 0.71(0.22–2.34) | |
| rs12138911 | 1 | 0.77(0.49–1.20) | 0.42(0.09–1.95) | 0.95(0.44–2.05) | 0.78(0.50–1.22) | 0.37(0.08–1.75) | 0.97(0.44–2.13) |
| 2 | 0.25(0.03–1.87) | 0.00(0.00–0.00) | 0.94(0.12–7.49) | 0.26(0.03–1.94) | 0.00(0.00–0.00) | 1.19(0.15–9.74) | |
| rs12060570 | 1 | 0.96(0.59–1.54) | 2.46(0.52–11.75) | 0.69(0.32–1.48) | 0.92(0.57–1.50) | 2.69(0.54–13.32) | 0.60(0.27–1.33) |
| 2 | 1.59(0.94–2.67) | 1.40(0.20–10.10) | 0.64(0.24–1.73) | 1.57(0.93–2.66) | 1.50(0.91–11.72) | 0.57(0.20–1.59) | |
| rs1806505 | 1 | 0.63(0.39–1.01) | 1.69(0.35–8.09) | 1.03(0.39–2.72) | 0.60(0.37–0.98).040 | 1.70(0.32–9.04) | 1.02(0.37–2.78) |
| 2 | 0.66(0.39–1.11) | 0.68(0.10–4.92) | 1.50(0.56–4.07) | 0.66(0.39–1.13) | 0.64(0.08–5.01) | 1.71(0.61–4.80) | |
| rs6668344 | 1 | 0.70(0.41–1.06) | 1.76(0.37–8.41) | 1.07(0.40–2.82) | 0.63(0.39–1.02) | 1.75(0.33–9.32) | 1.03(0.38–2.82) |
| 2 | 0.66(0.39–1.12) | 0.71(0.10–5.10) | 1.56(0.58–4.21) | 0.66(0.39–1.31) | 0.65(0.08–5.12) | 1.74(0.62–4.89) | |
| rs3754255 | 1 | 0.71(0.45–1.14) | 1.87(0.39–8.93) | 1.36(0.53–3.50) | 0.70(0.44–1.12) | 1.98(0.37–10.45) | 1.35(0.50–3.60) |
| 2 | 0.72(0.42–1.23) | 0.90(0.12–6.45) | 1.52(0.54–4.28) | 0.73(0.42–1.25) | 0.84(0.11–6.53) | 1.68(0.57–4.90) | |
| rs10925250 | 1 | 0.79(0.51–1.23) | 0.43(0.09–2.00) | 0.98(0.45–2.10) | 0.80(0.51–1.25) | 0.38(0.08–1.78) | 1.00(0.46–1.21) |
| 2 | 0.49(0.11–2.14) | 0.00(0.00–0.00) | 0.92(0.12–7.28) | 0.50(0.11–2.20) | 0.00(0.00–0.00) | 1.23(0.15–10.06) | |
| rs10925252 | 1 | 1.03(0.64–1.67) | 2.48(0.52–11.84) | 0.69(0.32–1.49) | 0.99(0.61–1.61) | 2.75(0.55–13.63) | 0.60(0.27–1.33) |
| 2 | 1.56(0.92–1.65) | 1.38(0.19–9.94) | 0.63(0.23–1.71) | 1.56(0.92–2.66) | 1.50(0.19–11.72) | 0.57(0.20–1.60) | |
| rs4659479 | 1 | 1.08(0.72–1.63) | 2.98(0.78–11.39) | 1.63(0.77–3.46) | 1.06(0.70–1.61) | 3.29(0.82–13.29) | 1.48(0.69–3.21) |
| 2 | 0.70(0.34–1.44) | 1.35(0.14–13.18) | 1.37(0.43–4.37) | 0.71(0.34–1.47) | 1.54(0.15–15.92) | 1.56(0.48–5.16) | |
| rs10925257 | 1 | 0.77(0.50–1.21) | 0.43(0.09–1.96) | 0.96(0.45–2.06) | 0.78(0.50–1.22) | 0.37(0.08–1.75) | 0.97(0.44–2.14) |
| 2 | 0.49(0.11–2.14) | 0.00(0.00–0.00) | 0.92(0.12–7.28) | 0.50(0.11–2.20) | 0.00(0.00–0.00) | 1.23(0.15–10.05) | |
| rs1805087 | 1 | 0.77(0.50–1.21) | 0.43(0.09–1.96) | 0.91(0.45–2.06) | 0.71(0.38–1.32) | 0.25(0.03–2.37) | 0.78(0.28–2.19) |
| Asp919Gly | 2 | 0.49(0.11–2.14) | 0.00(0.00–0.00) | 0.92(0.12–7.28) | 0.00(0.00–0.00) | 0.00(0.00–0.00) | 3.37(0.35–32.89) |
| rs2275565 | 1 | 1.69(0.49–5.85) | – | 0.90(0.19–4.32) | 1.64(0.47–5.69) | – | 0.75(0.15–3.71) |
| 2 | 2.17(0.65–7.27) | – | 0.89(0.20–4.03) | 2.10(0.62–7.10) | – | 0.72(0.15–3.37) | |
| rs10802569 | 1 | 1.16(0.77–1.75) | 2.91(0.76–11.10) | 1.50(0.70–3.21) | 1.14(0.75–1.73) | 3.21(0.80–13.00) | 1.40(0.64–3.06) |
| 2 | 0.77(0.38–1.54) | 1.28(0.13–12.53) | 1.62(0.55–4.76) | 0.76(0.38–1.55) | 1.44(0.14–14.88) | 1.72(0.57–5.17) | |
| rs3820571 | 1 | 1.16(0.77–1.75) | 2.91(0.76–11.10) | 1.50(0.70–3.21) | 1.14(0.75–1.73) | 3.21(0.80–13.00) | 1.40(0.64–3.06) |
| 2 | 0.77(0.38–1.54) | 1.28(0.13–12.53) | 1.62(0.55–4.76) | 0.76(0.38–1.55) | 1.44(0.14–14.88) | 1.72(0.57–5.17) | |
| rs1050993 | 1 | 1.51(0.75–3.01) | 2.26(0.28–18.48) | 0.92(0.33–2.60) | 1.50(0.74–3.04) | 2.23(0.26–18.89) | 0.82(0.28–2.36) |
| 2 | 1.30(0.65–2.61) | 0.78(0.08–7.61) | 0.62(0.21–1.81) | 1.31(0.64–2.66) | 0.69(0.07–7.14) | 0.58(0.19–1.76) | |
| Women | |||||||
| rs10925235 | 1 | 2.37(1.29–4.35).006 | 0.78(0.36–1.69) | 1.15(0.57–2.33) | 2.19(1.19–4.05).012 | 0.76(0.35–1.66) | 1.24(0.61–2.53) |
| 2 | 1.98(1.07–3.64).029 | 0.83(0.39–1.77) | 1.68(0.85–3.33) | 1.87(1.01–3.46).048 | 0.80(0.37–1.73) | 1.77(0.89–3.53) | |
| rs12138911 | 1 | 0.92(0.67–1.27) | 0.67(0.38–1.18) | 1.06(0.71–1.58) | 0.92(0.66–1.27) | 0.60(0.34–1.07) | 1.08(0.73–1.62) |
| 2 | 1.08(0.50–2.31) | 0.97(0.28–3.36) | 1.03(0.38–2.80) | 1.04(0.48–2.25) | 0.96(0.27–3.38) | 0.97(0.35–2.67) | |
| rs12060570 | 1 | 1.42(1.01–2.00).049 | 0.88(0.50–1.57) | 1.13(0.72–1.80) | 1.44(1.01–2.04).042 | 0.91(0.51–1.62) | 1.11(0.71–1.75) |
| 2 | 1.28(0.83–1.96) | 1.44(0.76–2.74) | 1.70(1.02–2.82).042 | 1.27(0.82–1.96) | 1.52(0.80–2.93) | 1.69(1.01–2.83).046 | |
| rs1806505 | 1 | 1.07(0.73–1.57) | 0.56(0.30–1.02) | 0.68(0.43–1.07) | 1.09(0.74–1.62) | 0.54(0.29–1.01) | 0.66(0.42–1.06) |
| 2 | 0.77(0.50–1.19) | 0.73(0.39–1.38) | 0.59(0.35–0.99).045 | 0.78(0.50–1.20) | 0.69(0.36–1.31) | 0.59(0.35–0.99).047 | |
| rs6668344 | 1 | 1.03(0.71–1.50) | 0.59(0.32–1.08) | 0.68(0.43–1.08) | 1.05(0.71–1.54) | 0.57(0.31–1.06) | 0.68(0.43–1.07) |
| 2 | 0.73(0.48–1.13) | 0.75(0.40–1.41) | 0.56(0.33–0.94).027 | 0.74(0.48–1.14) | 0.70(0.37–1.34) | 0.55(0.33–0.93).027 | |
| rs3754255 | 1 | 1.12(0.78–1.61) | 0.71(0.39–1.30) | 0.67(0.43–1.04) | 1.13(0.78–1.63) | 0.67(0.36–1.23) | 0.67(0.43–1.05) |
| 2 | 0.78(0.51–1.20) | 0.95(0.50–1.80) | 0.63(0.38–1.05) | 0.79(0.51–1.21) | 0.88(0.46–1.68) | 0.62(0.37–1.04) | |
| rs10925250 | 1 | 0.90(0.65–1.24) | 0.67(0.38–1.18) | 1.06(0.71–1.58) | 0.89(0.64–1.24) | 0.60(0.34–1.07) | 1.08(0.73–1.63) |
| 2 | 1.08(0.50–2.31) | 0.97(0.28–3.36) | 1.03(0.38–2.80) | 1.04(0.48–2.25) | 0.96(0.27–3.38) | 0.97(0.35–2.67) | |
| rs10925252 | 1 | 1.43(1.01–2.03).047 | 0.94(0.53–1.67) | 1.11(0.71–1.75) | 1.44(1.01–2.05).045 | 0.96(0.54–1.72) | 1.11(0.71–1.76) |
| 2 | 1.36(0.89–2.08) | 1.37(0.71–2.63) | 1.71(1.03–2.85).039 | 1.34(0.87–2.07) | 1.45(0.75–2.83) | 1.72(1.03–2.88).039 | |
| rs4659479 | 1 | 1.16(0.86–1.57) | 1.00(0.59–1.68) | 0.61(0.41–0.92).017 | 1.15(0.84–1.56) | 1.00(0.59–1.70) | 0.62(0.41–0.93).022 |
| 2 | 0.52(0.28–0.96).037 | 1.30(0.61–2.76) | 0.66(0.34–1.28) | 0.55(0.30–1.02) | 1.32(0.61–2.85) | 0.63(0.32–1.23) | |
| rs10925257 | 1 | 0.92(0.67–1.27) | 0.67(0.38–1.18) | 1.06(0.71–1.58) | 0.92(0.66–1.27) | 0.60(0.34–1.07) | 1.09(0.73–1.62) |
| 2 | 1.08(0.50–2.31) | 0.97(0.28–3.36) | 1.03(0.38–2.80) | 1.04(0.48–2.25) | 0.96(0.27–3.38) | 0.97(0.35–2.67) | |
| rs1805087 | 1 | 0.92(0.67–1.27) | 0.67(0.38–1.18) | 1.06(0.71–1.58) | 0.78(0.52–1.17) | 0.58(0.29–1.15) | 1.13(0.68–1.88) |
| 2 | 1.08(0.50–2.31) | 0.97(0.28–3.36) | 1.03(0.38–2.80) | 0.98(0.38–2.54) | 0.40(0.05–3.23) | 1.19(0.36–3.91) | |
| rs2275565 | 1 | 0.74(0.37–1.47) | 0.60(0.19–1.91) | 0.90(0.37–2.19) | 0.76(0.38–1.52) | 0.63(0.19–2.03) | 0.97(0.40–2.38) |
| 2 | 0.90(0.46–1.75) | 1.11(0.37–3.31) | 0.93(0.39–2.22) | 0.93(0.47–1.82) | 1.23(0.40–3.73) | 0.98(0.41–2.36) | |
| rs10802569 | 1 | 1.15(0.85–1.56) | 0.97(0.57–1.64) | 0.60(0.40–0.90).014 | 1.14(0.84–1.55) | 0.97(0.57–1.66) | 0.61(0.40–0.93).020 |
| 2 | 0.57(0.32–1.01) | 1.36(0.66–2.83) | 0.74(0.40–1.38) | 0.59(0.33–1.05) | 1.39(0.66–2.92) | 0.71(0.38–1.36) | |
| rs3820571 | 1 | 1.15(0.85–1.56) | 0.97(0.57–1.65) | 0.60(0.40–0.90).015 | 1.14(0.84–1.56) | 0.97(0.57–1.66) | 0.61(0.41–0.93).021 |
| 2 | 0.57(0.32–1.01) | 1.36(0.66–2.83) | 0.74(0.40–1.38) | 0.59(0.33–1.05) | 1.39(0.66–2.92) | 0.71(0.38–1.34) | |
| rs1050993 | 1 | 2.03(1.13–3.62).017 | 0.72(0.34–1.50) | 0.81(0.42–1.57) | 1.95(1.09–1.19).025 | 0.70(0.33–150) | 0.86(0.45–1.68) |
| 2 | 1.76(0.99–3.15) | 0.74(0.35–1.53) | 1.36(0.73–2.54) | 1.71(0.95–3.07) | 0.72(0.34–1.52) | 1.41(0.75–2.65) |
Notes for Table 6:
NMH: non-migraine headache
MO: migraine without aura
MA: migraine with aura
Comparisons are minimally and fully adjusted (see methods). P-values < 0.05 are indicated in superscript.
No SNPs met the adjusted level of significance.
MTR
The haplotype block across MTR had four haplotypes in greater than 2% frequency (Table 1). None of the haplotypes in MTR were significantly associated with headache in men or women. In women, one copy of hap4 decreased the risk of MA, but this association did not survive correction for multiple tests, OR = 0.63, 95%CI(0.42–0.94), p = 0.024 in the minimally adjusted analysis, and OR = 0.64, 95%CI(0.43–0.97), p = 0.034 in the fully adjusted analysis (Table 4). This haplotype, hap4, TTGCCCATGGAACGGA carries the most common allele (underlined in hap4) at rs1805087 (Asp919Gly, A3179G), suggesting that this might be a weakly protective allele, decreasing the risk of MA when present on hap4. This allele is also present on hap1 and hap2 in MTR.
MTRR
Two haplotype blocks were identified in MTRR. Our data combined with publicly available on-line data (NCBI) indicated that rs1801394 (Ile22Met, exon 2) was not in either haplotype block, so it is analyzed separately (Table 6). In women, hap3B (GT) reduced risk of MA (Table 5), but this association did not survive the corrected error rate for this set of analyses (p < 0.0167). In men, hap1B (AA) was associated with reduced risk for migraine with aura and this analysis did survive correction (OR = 0.27, 95%CI(0.10–0.79), p = 0.016 in minimally adjusted analyses. The finding did not survive correction in fully adjusted analyses, OR = 0.32, 95%CI(0.11–0.96), p = 0.043. This hap1B, AA, carries the most common alleles for the two SNPs comprising this haplotype, rs162036 and rs8659. The first SNP, rs162036 is a missense variation (Lys350Arg), and was tested separately as well, but was not associated with risk of migraine (Table 6). Although this haplotype, with the common alleles for both SNPs, reduces risk of MA in men, we would expect that copies of hap2B (AT) or 3B (GT) to increase risk for MA, and indeed the odds ratio’s for 2 copies of 2B and 3B were above 1, but these tests were not statistically significant.
Risk score
In addition to the haplotype analysis, a risk score was calculated as the sum of the number of minor alleles for each of the coding variations (i.e., rs1801394, rs1532268, rs162036, rs1805087, rs2274976, rs1801131, rs1801133, rs2066472). This risk score would equal 4 for an individual heterozygous at each of the 8 loci, as we assigned a score of 0 for the major genotype, 0.5 for the heterozygous genotype, and 1 for the homozygous minor genotype. Therefore, the risk score assumes an additive rather than interactive or non-additive relationship between these loci. The score was analyzed as a continuous variable, and compared across groups, with full and minimal adjustment analogous to all previous analyses. As shown in Table 7, this risk score was not associated with headache type.
Discussion
Summary
In summary, hap1B in MTRR was associated with reduced risk of MA in men, and this haplotype carries the most common alleles for both markers on the haplotype, including rs162036 (Lys350Arg). This was also found in women at p = .043, ns. In the case of rs162036 in MTRR, the most common allele was associated with decreased risk of MA in men. This SNP in MTRR has been associated with spina bifida and congenital heart defects,(16) and methionine synthase reductase (MSR) is necessary for folate metabolism.(17) However, it is not known if rs162036 leads to deficient levels of MSR or otherwise differential function.
One SNP in MTHFR, rs1801133 (C677T), was associated with reduced risk of MA in women, p = 0.034, ns. This is in contrast to a recent meta-analysis of 13 studies, which reported the TT genotype of the MTHFR 677C>T polymorphism was associated with an increased risk for only migraine with aura, but not for migraine without aura, and primarily in non-Caucasian populations.(2) However, we note that a different study also in an older sample (mean 54.1 years) also found a moderate protective effect of C677T on MA.(18) In addition, a more recent article found a protective effect of C677T on MA but only among individuals with fewer than 6 migraine attacks per year.(19) Participants in the present study were on average 76 years of age when they provided a DNA sample at the late-life visit. It has been reported that individuals with migraine with aura are at increased risk of overall mortality compared to others.(20,21) MA was also associated with increased mortality risk in the Reykjavik cohort.(22) If participants in our study reporting MA at the mid-life visit were more likely to die before the late-life visit (mean age 76) when DNA was collected, and if mortality risk varied within the MA group depending on genotype, our observed protective association between C677T in MTHFR (as well as hypothetically other protective associations reported herein) could have been influenced by selective mortality. The selective mortality effect on this region is tested and described further in a recent paper. (24) Specifically, the TT and CT genotypes at C677T in MTHFR may be linked with somewhat reduced longevity. The effect this may have on our observed association between MA and C677T is to attenuate a ‘real’ deleterious effect of the genotype, and essential reveal a protective effect of individuals with this genotype. In the simulation by Scher et al. (24), the authors demonstrate that even small effects on survival of this allele might obscure a genetic risk factor, and even suggest a protective factor in a population comprised of older individuals as is the case in this study.
Because analyses above compared all 4 groups of individuals, we performed post-hoc tests comparing just the MA (n = 167) and MO (n = 85) groups to determine if any of the genes were specifically associated with aura, rather than migraine status. With the caveat that our sample size for this analysis was low, and these analyses are likely to have been underpowered, we found no significant associations between any of the haplotypes or specific loci and the MO vs. MA group comparison.
Strengths
Our study analyzed data from a large, well-characterized sample giving us the statistical power to stratify analyses by gender. Participants were interviewed about migraine history in middle age. Participants in the study were followed up as part of a comprehensive epidemiologic study, allowing us to adjust for plausible confounding risk factors.
Limitations
Headache symptoms were only assessed in participants reporting at least monthly attacks, so individuals with less frequent headaches or those with aura but no headache would probably have been included in the no headache group. However, we note that our prevalence of migraine overall is consistent with prior reports.(1) Our prevalence of MA (8.5%) is higher than that reported previously, perhaps because subjects with non-specific visual symptoms such as blurred vision might have been included in the MA group. The effect of these misclassification errors would likely have been to attenuate towards the null any true association between haplotypes / SNPs and headache rather than overstating an association.
It should be noted a recent genome-wide association study (GWAS) employing multiple separate samples found only one variation, rs1835740 on chromosome 8q22.1, to be associated with migraine,(25) and this variation is on a different chromosome than the three genes in this study. The markers that were genotyped and available for this study provided sufficient coverage of the MTR and MTRR genes, but MTHFR was not covered between introns 1 and 3 because available markers in that region were monomorphic in our sample, and no other markers on the chip covered that region of the gene. Future genotyping efforts should include sufficient coverage of polymorphic markers in this region so that the haplotype analysis is more complete. However, many studies report two marker haplotypes for MTHFR utilizing even fewer than the three coding SNPs in MTHFR used in our study (i.e., Arg594Gln, Glu429Ala, Ala222Val).(23)
Others have found that MTRR A66G (rs1801394) is associated with coronary artery disease, and attributed this association to A66G’s capacity to interfere with reductive methylation of cobalmin in the folate pathway,(26) leading to hyperhomocysteinaemia or high levels of homocysteine.(27) Our results provide the first evidence of an association between MTRR and migraine with aura, adding to evidence that the folate-metabolic pathway may have etiological significance in migraine with aura. However, replication of this study will be needed to confirm our results.
Table 10.
MTRR Haplotypes and Frequencies.
| Block | Haplotype | Frequency | ||
|---|---|---|---|---|
| rs1532268 | rs6555501 | |||
| A | 1 | T | T | .434 |
| 2 | C | C | .372 | |
| 3 | T | C | .194 | |
| rs162036 | rs8659 | Frequency | ||
| B | 1 | A | A | .119 |
| 2 | A | T | .673 | |
| 3 | G | T | .207 |
Top 5 references, including contact information for the senior or corresponding author.
-
Schürks M, Rist PM and Kurth T. MTHFR 677C>T and ACE D/I polymorphisms in migraine: a systematic review and meta-analysis. Headache 2010: 588–599.
M. Schürks, Division of Preventive Medicine, Brigham and Women’s Hospital, 900 Commonwealth Avenue East, 3rd fl, Boston, MA 02215–1204, USA.
-
Oterino A, Toriello M, Valle N, et al. The relationship between homocysteine and genes of folate-related enzymes in migraine patients. Headache 2010; 50: 99–168.
Oterino, A., University Hospital Marques De Valdecilla, Avda Valdecilla Sn Santander Cantabria Spain 39008, Avda Valdecilla Sn, Santander, Cantabria, 39008, Spain.
-
Goncalves FM, Martins-Oliveira A, Speciali JG, et al. Vascular endothelial growth factor genetic polymorphisms and haplotypes in women with migraine. DNA Cell Biol 2010; 29: 357–362.
Jose E. Tanus-Santos, M.D., Ph.D., Department of Pharmacology, Faculty of Medicine of Ribeirao Preto, University of Sao Paulo, Av. Bandeirantes, 3900, Ribeirao Preto, Sao Paulo 14049–900, Brazil, E-mail: tanus@fmrp.usp.br tanussantos@yahoo.com
-
Corominas R, Sobrido MJ, Ribases M, et al. Association study of the serotoninergic system in migraine in the Spanish population. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 177–184.
B. Cormand, Ph.D., Associate Professor of Genetics, Departament de Genetica, Facultat de Biologia, Universitat de Barcelona, Av. Diagonal 645, edifici annex 3a planta, 08028 Barcelona, Spain; bcormand@ub.edu
-
Todt U, Netzer C, Toliat M, et al. New genetic evidence for involvement of the dopamine system in migraine with aura. Hum Genet 2009; 125: 265–279.
Christian Kubisch, Institute of Human Genetics, University of Cologne, Karpener Strasse 34, 50931, Cologne, Germany. E-mail: christian.kubisch@uk-koeln.de
Acknowledgments
We would like to thank the participants of the study and the Icelandic Heart Association clinic staff for their invaluable contribution.
Funding
This study was supported by a grant from the National Institutes of Health (N01-AG-1-2100), the National Institute on Aging Intramural Research Program, the Hjartavernd (the Icelandic Heart Association) and the Althingi (the Icelandic Parliament).
Footnotes
Conflict of Interest Statement
The Authors declare that there is no conflict of interest.
References
- 1.Stovner L, Hagen K, Jensen R, et al. The global burden of headache: a documentation of headache prevalence and disability worldwide. Cephalalgia. 2007;27:193–210. doi: 10.1111/j.1468-2982.2007.01288.x. [DOI] [PubMed] [Google Scholar]
- 2.Schürks M, Rist PM, Kurth T. MTHFR 677C>T and ACE D/I polymorphisms in migraine: a systematic review and meta-analysis. Headache. 2010;50:588–599. doi: 10.1111/j.1526-4610.2009.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher AI, Terwindt GM, Verschuren WM, et al. Migraine and MTHFR C677T genotype in a population-based sample. Ann Neurol. 2006;59:372–375. doi: 10.1002/ana.20755. [DOI] [PubMed] [Google Scholar]
- 4.Oterino A, Toriello M, Valle N, et al. The relationship between homocysteine and genes of folate-related enzymes in migraine patients. Headache. 2010;50:99–168. doi: 10.1111/j.1526-4610.2009.01484.x. [DOI] [PubMed] [Google Scholar]
- 5.Harris TB, Launer LJ, Eiriksdottir G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher AI, Gudmundsson LS, Sigurdsson S, et al. Migraine headache in middle age and late-life brain infarcts. JAMA. 2009;301:2563–2570. doi: 10.1001/jama.2009.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain: Headache Classification Committee of the International Headache Society. Cephalalgia. 1988;8(suppl 7):1–96. [PubMed] [Google Scholar]
- 8.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scher AI, Terwindt GM, Picavet HS, et al. Cardiovascular risk factors and migraine: the GEM population-based study. Neurology. 2005;64:614–620. doi: 10.1212/01.WNL.0000151857.43225.49. [DOI] [PubMed] [Google Scholar]
- 10.Database of Single Nucleotide Polymorphisms (dbSNP) Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine; dbSNP accession:{rs7525338, rs2066472, rs1931226}, (dbSNP Build ID: {116/132}). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 12.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 14.Tonetti C, Amiel J, Munnich A, et al. Impact of new mutations in the methylenetetrahydrofolate reductase gene assessed on biochemical phenotypes: a familial study. J Inherit Metab Dis. 2001;24:833–842. doi: 10.1023/a:1013988123902. [DOI] [PubMed] [Google Scholar]
- 15.Selzer RR, Rosenblatt DS, Laxova R, et al. Adverse effect of nitrous oxide in a child with 5,10-methylenetetrahydrofolate reductase deficiency. New Eng J Med. 2003;349:45–50. doi: 10.1056/NEJMoa021867. [DOI] [PubMed] [Google Scholar]
- 16.Shaw GM, Lu W, Zhu H, et al. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Med Genet. 2009;10:49. doi: 10.1186/1471-2350-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada K, Gravel RA, Toraya T, et al. Human methionine synthase reductase is a molecular chaperone for human methionine synthase. Proc Natl Acad Sci U S A. 2006;103:9476–9481. doi: 10.1073/pnas.0603694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schürks M, Zee RY, Buring JE, et al. Interrelationships among the MTHFR 677C>T polymorphism, migraine, and cardiovascular disease. Neurology. 2008;71:505–513. doi: 10.1212/01.wnl.0000316198.34558.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ligthart L, de Vries B, Smith AV, et al. Meta-analysis of genome-wide association for migraine in six population-based European cohorts. Eur J Hum Genet. 2011;19:901–907. doi: 10.1038/ejhg.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurth T, Gaziano MJ, Cook NR, et al. Migraine and risk of cardiovascular disease in women. Journal of the American Medical Association. 2006;296:283–291. doi: 10.1001/jama.296.3.283. [DOI] [PubMed] [Google Scholar]
- 21.Berger K, Evers S. Migraine with aura and the risk of increased mortality. BMJ. 2010;341:c4410. doi: 10.1136/bmj.c4410. [DOI] [PubMed] [Google Scholar]
- 22.Gudmundsson LS, Scher AI, Aspelund T, et al. Migraine with aura and risk of cardiovascular and all cause mortality in men and women: prospective cohort study. BMJ. 2010;341:c3966. doi: 10.1136/bmj.c3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakutani Y, Kowa H, Kusumi M, et al. A haplotype of the methylenetetrahydrofolate reductase gene is protective against late-onset Alzheimer’s disease. Neurobiol Aging. 2004;25:291–294. doi: 10.1016/S0197-4580(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 24.Scher A, Eiriksdottir G, Garcia M, et al. Lack of association between the MTHFR C677T variant and migraine with aura in an older population: Could selective survival play a role? Cephalalgia. 2012 doi: 10.1177/0333102412469739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anttila V, Stefansson H, Kallela M, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet. 2010;42:869–873. doi: 10.1038/ng.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijaya Lakshmi SV, Naushad SM, Rupasree Y, et al. Interactions of 5′-UTR thymidylate synthase polymorphism with 677C -> T methylene tetrahydrofolate reductase and 66A -> G methyltetrahydrofolate homocysteine methyl-transferase reductase polymorphisms determine susceptibility to coronary artery disease. J Atheroscler Thromb. 2011;18:56–64. doi: 10.5551/jat.5702. [DOI] [PubMed] [Google Scholar]
- 27.Laraqui A, Allami A, Carrie A, et al. Influence of methionine synthase (A2756G) and methionine synthase reductase (A66G) polymorphisms on plasma homocysteine levels and relation to risk of coronary artery disease. Acta Cardiol. 2006;61:51–61. doi: 10.2143/AC.61.1.2005140. [DOI] [PubMed] [Google Scholar]


