Abstract
The precise DNA arrangement at chromosomal ends and the proteins involved in its maintenance are of crucial importance for genome stability. For the yeast Saccharomyces cerevisiae, this constitutive DNA configuration has remained unknown. We demonstrate here that G-tails of 12–14 bases are present outside of S phase on normal yeast telomeres. Furthermore, the Mre11p protein is essential for the proper establishment of this constitutive end-structure. However, the timing of extended G-tails occurring during S phase is not affected in strains lacking Mre11p. Thus, G-tails are present on yeast chromosomes throughout the cell cycle and the MRX complex is required for their normal establishment.
Keywords: Telomeres, G-tails, MRX complex, yeast
The physical ends of eukaryotic chromosomes, the telomeres, have a very conserved structure and are essential for genome stability (for review, see Blackburn 2001; Chakhparonian and Wellinger 2003). Short, direct DNA repeats constitute the underlying telomeric DNA and the strand running 5′ to 3′ toward the end of the chromosomes is usually rich in guanines (the G-rich strand). Lagging-strand synthesis always occurs on this G-rich strand and will leave a short gap at the 5′ end of the newly synthesized C-rich strand. This gap cannot be filled in by repair, and a 3′ G-rich overhang, called G-tail, remains. On the other end, leading-strand synthesis is thought to produce a blunt extremity. However, studies of the terminal DNA arrangement in a variety of organisms suggest that a G-tail is a conserved motif for all telomeres (Chakhparonian and Wellinger 2003). Thus, the question arises as to how the blunt-ended DNA ends generated by leading-strand synthesis are converted into ends with a G-tail.
Studies in the yeast Saccharomyces cerevisiae have shown that its telomeres acquire detectable G-tails late in S phase, after conventional replication (Wellinger et al. 1993a,b). Moreover, at least on the ends of a linear plasmid, G-tails occur on both, leading- and lagging-strand ends (Wellinger et al. 1996). Surprisingly, these S-phase-specific G-tails can also be detected in cells lacking telomerase, the main activity responsible for replicating telomeric G-strands (Dionne and Wellinger 1996). Collectively, these results suggest that the blunt end left after completion of leading-strand synthesis is processed into an end with a G-tail, presumably by nuclease/helicase activities (Wellinger et al. 1996). Analyses of the requirements to establish a normal telomeric DNA end-structure are hampered by the fact that for wild-type yeast cells, the precise DNA arrangement outside of S phase is unknown.
Recent studies on the Mre11p/Rad50p/Xrs2p (MRX) proteins, an evolutionarily conserved complex involved in a number of processes in mitosis and meiosis, revealed that this complex may play a key role in telomere length maintenance in humans, plants, and yeasts (for review, see Haber 1998; D'Amours and Jackson 2002). Yeast cells harboring a deletion of any one of these genes are viable, but display shortened telomeric repeat tracts (Kironmai and Muniyappa 1997; Boulton and Jackson 1998). The Mre11p protein alone, or in association with other proteins, displays various nucleolytic activities (D'Amours and Jackson 2002), and it has been demonstrated that in a de novo telomere formation assay, the MRX complex is needed for the generation of telomeric G-strand DNA and the loading of G-tail-binding proteins (Diede and Gottschling 2001). In addition, the MRX complex in Schizosaccharomyces pombe might process telomeric DNA ends in the absence of a DNA double-stranded-binding protein Taz1p (Tomita et al. 2003).
To gain insights into the requirements to establish a normal DNA end at telomeres, we developed a stringently controlled hybridization assay as well as a primer-extension assay. The results demonstrate that most of the normal yeast telomeres end in a G-tail of ∼12–14 bases. Moreover, we show that in mre11Δ strains, this constitutive DNA end-structure is compromised; specifically, G-tails in such mutant strains are shorter. In addition, whereas the dynamics of the cell cycle-dependent increase of G-tail signals at the end of S phase occurs in both wild-type and mutant strains in an indistinguishable fashion, signals for G-tails at the end of S phase were also weaker in the mre11Δ strain. The data thus show that short 10–15-base G-tails are the proper ends of yeast chromosomes, and in mre11Δ cells, there is a deficiency in forming those G-tails, even though G-tail presence is not completely abolished. Therefore, the MRX complex does play a direct role in telomeric end-structure processing in normal yeast cells and could play a similar role at mammalian telomeres.
Results and Discussion
Telomeric end-structure is affected in the absence of MRX complex
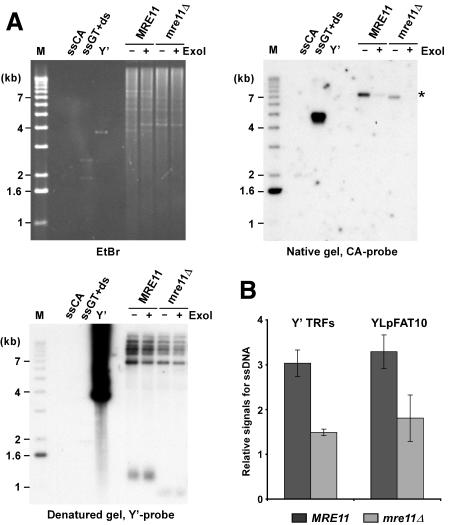
There is accumulating evidence that the three interacting proteins Mre11p, Rad50p, and Xrs2p (NBS1), forming the MRX or MRE11 complex, play a role in telomere maintenance in yeasts, plants, and mammals. If the MRX complex were involved in the processing of the blunt ends left after completion of leading-strand synthesis at the telomeres, one would expect to detect shorter G-tails (either transiently or constitutively) on yeast telomeres in strains lacking any of the components of the complex. The presence of G-tails can be assessed using a nondenaturing in-gel hybridization technique that we developed previously (Dionne and Wellinger 1996). Such analyses on DNA derived from asynchronously grown wild-type cell cultures yielded very faint signals for chromosomal terminal restriction fragments (TRFs). It remained unclear whether these signals were due to constitutive short G-tails or whether they were due to a fraction of cells being in late S phase, when G-tails are easily detectable on yeast telomeres (Wellinger et al. 1993b; Dionne and Wellinger 1996). However, such signals on telomeres of a high-copy linear plasmid of 7.5 kb, called YLpFAT10, are concentrated in a small area of the gel and become readily visible, even on DNA derived from G1-arrested cultures (Fig. 1A [top right] B). FACS analysis and counting of unbudded versus budded cells of these cultures established that the vast majority of cells (≥95%) were indeed in G1 (data not shown; see Supplementary Fig. 1). More importantly, when genomic DNA isolated from mre11Δ strains was analyzed in the same fashion, a significant reduction of the signals for the G-tails is observed (Fig. 1A [top right, cf. lanes marked MRE11 and those marked mre11Δ], B). Treatment of the DNA with exonuclease I prior to the end-structure analyses (ExoI+ lanes in Fig. 1A) indicates that the detected signal corresponds to terminal single-stranded G-strands. Furthermore, the same gel shown in Figure 1A was also hybridized to a G-rich probe and no telomeric C-rich signals were detected (data not shown). Quantification of the signals for G-tails confirmed that they are ∼1.3- to 2-fold lower on DNA derived from mre11Δ cells as compared with wild-type cells (Fig. 1B). Similar results were obtained for a rad50Δ strain, with strains of different genetic backgrounds and when chromosomal telomeres were analyzed (Fig. 1B; Supplementary Fig. 2; data not shown). Collectively, these results demonstrate that in G1, G-tails can be detected on telomeres of yeast cells, and that the signal for G-tails in mre11Δ cells is reduced when compared with wild-type cells.
Figure 1.
G-tail signals are reduced in mre11Δ strains. (A) Genomic DNA derived from α-factor-arrested cells was digested with XhoI. G-tails on YLpFAT10 (indicated by asterisk) were analyzed by native in-gel hybridization for wild-type and mre11Δ strains (MLY50 and MLY51). DNAs were either mock treated (labeled ExoI-), or treated with Escherichia coli Exonuclease I (labeled ExoI+), before in-gel analysis. (Top, left) Ethidium-bromide-stained gel. (Top, right) The gel after nondenaturing hybridization to the CA probe. (Bottom, left) The DNA was denatured in the gel, transferred to a nylon membrane, and the membrane hybridized to a Y′ probe. Single-stranded phagemid DNA containing yeast telomeric repeats of the G-rich strand (ssGT) and of the C-rich strand (ssCA) served as positive and negative controls, respectively. The ssGT control was mixed with PvuI-digested pMW55, the latter serving as double-stranded control (ds, a fragment of 1.9 kb). Linearized pVZY′K plasmid (3.9 kb) served as a positive control for the hybridization with the Y′ probe (labeled Y′). (M) End-labeled 1-kb ladder DNA serving as a size standard. (B) Relative amounts for G-tails (relative signals for ssDNA, in arbitrary units) were obtained by calculating the ratio of the respective signals on the native gels over the total amount of Y′ TRF signals or YLpFAT10 signals. Three to six individual nonsynchronous cultures were tested for each strain, and the standard deviation is indicated.
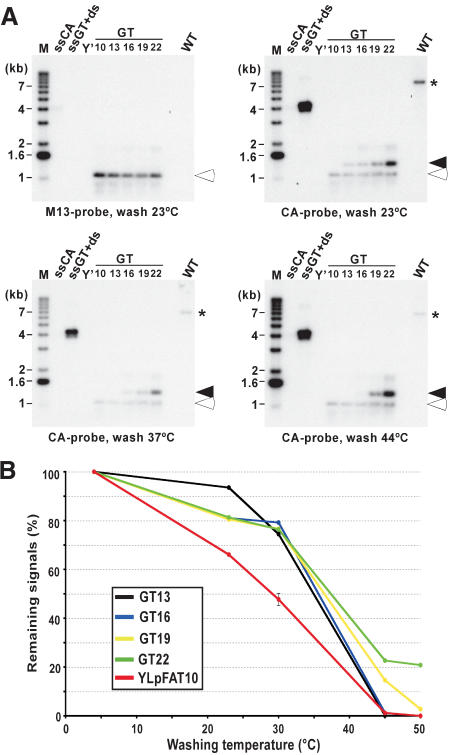
We next investigated the length of the telomeric G-tails for cells in G1. As a means of comparison, constructs containing a stretch of nucleotides corresponding to telomeric repeats were used. The plasmid with the longest tract (GT22) contains 22 nucleotides that are perfectly complementary to the CA probe used for the in-gel hybridization technique. In addition, derivatives of GT22 were obtained (GT19 to GT10). These constructs harbor stepwise-shortened tracts of telomeric sequences, each being 3 nucleotides shorter than the previous one. The different plasmids generated were double digested and heat denatured prior to loading onto a gel (Fig. 2A, lanes GT10–GT22). Note that a fragment migrating to an apparent length of 1.1 kb of all plasmids carries a sequence complementary to T7 and M13 probes, and that the G-rich telomeric repeat tract of indicated size resides on a slightly larger fragment. On the same gel, genomic DNA derived from G1-arrested wild-type cells was analyzed for G-tails (Fig. 2A, lanes marked WT). The gel was first hybridized to an M13 probe to show about equal loading of the GTxx plasmids (Fig. 2A, top left, open arrowhead). Next, the gel was hybridized to the 22-mer CA probe, revealing the fragment containing the G-rich strand of the different GT-plasmids (Fig. 2A, closed arrowheads), as well as G-tails on YLpFAT10 (Fig. 2A, *). An exposure was obtained after each washing step and the relative signals for all bands were quantified. By comparing the loss of signals occurring at each individual washing temperature for the known lengths of GT-tracts on the plasmid DNA to the loss of signals on YLpFAT10 telomeres, an estimation of the lengths of G-tails on YLpFAT10 could be derived (Fig. 2B). Clearly, at lower washing temperatures (20°C–30°C), the signals for the telomeres on YLpFAT10 are reduced to a greater extent than the signals for even the shortest model target plasmid, GT13 (Fig. 2B, red vs. black curve). If the overhangs were completely complementary to the 22-mer CA probe, these results suggest that they are shorter than 13 nucleotides. However, given that telomeric overhangs are of a variable sequence composition, it is likely that a considerable fraction of telomeres contains a number of mismatches with respect to the probe. We compared all theoretical possibilities of 16-nucleotide overhangs to the sequence of the probe, and found that all can yield at least 13-nucleotide matches to it (data not shown). Thus, this experiment suggests that G-tails on telomeres of wild-type cells are shorter than 16 bases, and only a minority could bear G-tails longer than that.
Figure 2.
Most G-tails on telomeres in G1-arrested cells are shorter than 16 bases. (A) The individual pGTxx plasmids, each containing telomeric repeat tracts ranging from 10 to 22 bp, were double digested and heat denatured prior to loading on an agarose gel. A common DNA fragment serves as a loading control after hybridization to an M13 probe (open arrowheads), whereas a fragment migrating at ∼1.3 kb of each plasmid contains the TG1–3 repeats of varying lengths serving to compare the hybridization of the CA probe with different telomeric repeat lengths (closed arrowheads). Wild-type cells (MLY30 strain) were arrested in G1 with α-factor, and XhoI-digested DNAs derived from those cells were loaded on the gel (lane marked WT). After hybridization to the M13 probe (top, left), the same gel was then hybridized at 4°C to the CA probe and washed at different temperatures as indicated at the bottom of gels shown. The signals obtained for G-tails on YLpFAT10 (indicated by asterisk) were compared with those detected on the 1.3-kb DNA fragments derived from the pGTxx plasmids after different washing temperatures. Controls and molecular-size standards are as described in Figure 1. (B) Remaining signals on pGTxx fragments, as well as signals for G-tails on YLpFAT10, were determined by using native gels washed at different temperatures. Signals are plotted as percent (%) with respect to the same gel washed at 4°C (designated at 100%).
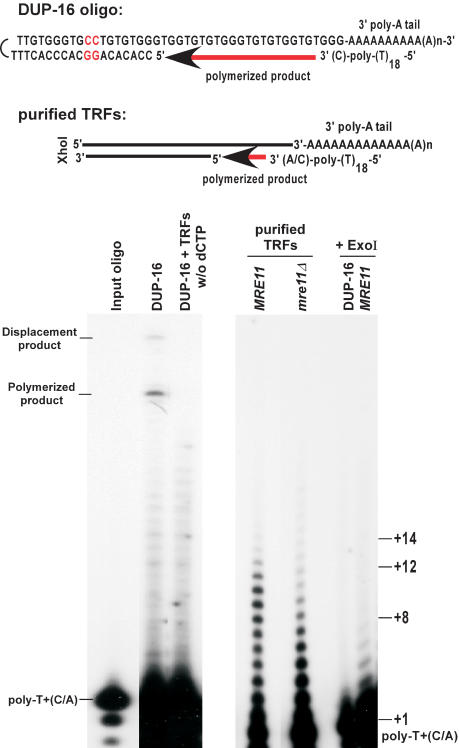
The above provided a rough estimate for the lengths of G-tails on a population of telomeres in wild-type cells, but could not yield a precise determination of G-tail lengths on individual telomeres. We therefore developed a modified primer-extension method, in which poly(A)-tailed purified TRFs were annealed to an end-labeled poly-T oligonucleotides ending with a C or an A at their 3′-end (Fig. 3, top). The annealing products were then extended using T4 DNA polymerase in conditions that minimized strand displacement at the double-stranded to single-stranded transition point. To verify the reliability of the method, conditions were established using a model oligonucleotide substrate with a sequence composition similar to the one expected on the TRFs (DUP-16, Fig. 3, left). TRFs purified from the various strains were first analyzed for telomeric DNA end-structure using nondenaturing in-gel hybridization to make sure that no alterations occurred during DNA manipulations (data not shown). Optimized conditions were then applied to such purified TRFs derived from wild-type and mre11Δ cells (Fig. 3). For wild-type cells, extension products up to 12–14 nucleotides are readily detectable. Remarkably, products of 9–12 nucleotides were more abundant than shorter products. In contrast, short products (+2 to +8) were much more abundant than the longer products when DNA derived from mre11Δ cells was used (Fig. 3). However, we note that even with DNA derived from these mutant cells, weak signals for G-tails of the same length as those seen with DNA derived from wild-type cells could be detected. Quantification of the signals revealed that with DNA derived from wild-type cells, about three- to fourfold more signal is in the range of +8 and longer as compared with the mre11Δ cells (data not shown; see Fig. 3). Therefore, whereas all strains can have G-tails of 12–14 bases, the fraction of telomeres ending with a G-tail of 8 nucleotides and longer is significantly reduced in mre11Δ cells. At least qualitatively, these results are consistent with the results obtained using hybridization techniques (Figs. 1, 2) and confirm that a large fraction of the constitutive G-tails observed on telomeres in G1 is shorter in mre11Δ cells, when compared with wild-type cells. Taken together, the data show that the constitutive chromosomal DNA end-structure for wild-type yeast cells encompasses overhangs of the G-rich strand that vary in length, with most of them being shorter than 14 nucleotides. Moreover, the constitutive G-tails on telomeres of mre11Δ cells clearly are shorter, most of them being shorter than 8 bases.
Figure 3.
Constitutive G-tails are shorter in mre11Δ cells. (Top) DNA structure of the annealed model substrate DUP-16 and expected structure for TRFs. In DUP-16, the two nucleotides not corresponding to telomeric sequence, in which strand displacement synthesis stops, are indicated in red. (Bottom) Analysis of extension products by PAGE. DUP-16 and/or TRFs were poly(A) tailed, annealed to end-labeled poly-T+(C/A) oligo, and subjected to an extension reaction using T4 DNA polymerase. (Left gel) Control reactions using DUP-16 alone; w/o dCTP; dCTP was omitted from the extension reaction. Poly-T+(C/A) indicates the position of the end-labeled oligo alone, and corresponds to position +1 in terms of overhang size. (Polymerized product) Product of the extension reaction on DUP-16 covering the overhang only (+24); (displacement product) product of the extension reaction reaching the two inversed G:C base pairs (+31). (Right gel) Purified TRFs from the indicated strains were subjected to the procedure. In the last two lanes, either DUP-16 or purified TRFs from a wild-type strain were treated with ExoI before the tailing reaction, and then processed as the other samples. Size indications on right denote overhang size.
The majority of yeast telomeres carry constitutive overhangs
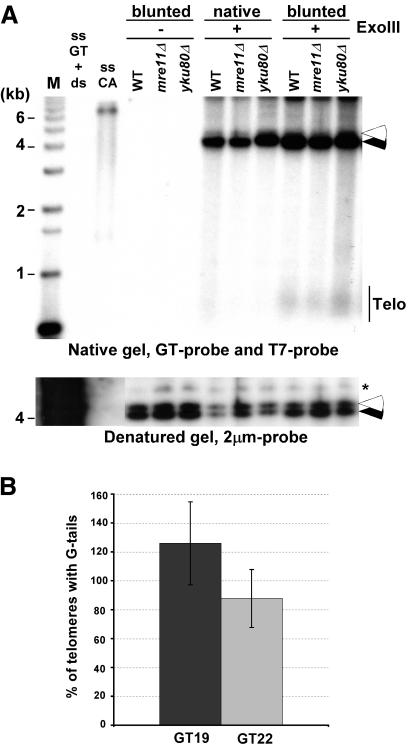
Next, we wished to examine what fraction of telomeres does carry such 3′-overhangs and whether some telomeres carried a blunt-end or a short 5′-overhang. Genomic DNA with a native telomeric end-structure was treated with Exonuclease III. This strand-specific exonuclease degrades the strands with the 3′-end on dsDNA that is blunt or possesses a 5′-overhang. As positive controls, we used genomic DNA on which telomeres were blunt-ended by ExoI treatment as in Figure 1, prior to ExoIII treatment. On this DNA, the generation of 5′-overhangs is detectable by a GT probe after nondenaturing in-gel hybridization (Fig. 4A). However, native ends are, by and large, resistant to ExoIII treatment (Fig. 4A). Quantification of the gel revealed that the signals for blunted ends were 5- to 10-fold increased when compared with those obtained with native telomeres. In a second and independent approach, we used YLpFAT10 telomeres to determine the amount of ends harboring G-tails. Because YLpFAT10 as well as the pGTxx plasmid series each contained one sequence element complementary to the T7 probe, the estimated number of telomeres ending with G-tails could be derived by comparing signals obtained on pGTxx plasmids with those on YLpFAT10. DNA in gels such as shown in Figure 2A was denatured, hybridized to a T7 probe, and the signals for the GT22, GT19 plasmids and YLpFAT10 were quantified. This yielded an approximate ratio of YLpFAT10 molecules with respect to the control pGTxx DNAs on the gels. In parallel, the same DNA was analyzed by nondenaturing in-gel hybridization using the CA-telomeric probe, washed at 4°C, and signal intensities for plasmids GT22, GT19 and YLpFAT10 were again quantified. These latter signal intensities were corrected for the fact that each YLpFAT10 molecule contains two telomeres, whereas the pGTxx plasmids only contained one target sequence for the CA probe. The obtained values were then adjusted to the molecular ratio of the molecules. Assuming equal chances of hybridization to each of the targets, the obtained values indicate the fraction of YLpFAT10 telomeres hybridizing to the probe (Fig. 4B). Although there is some variability, the data corroborate the results obtained with the ExoIII experiments, in that at least 80% of the telomeres on YLpFAT10 hybridized to the probe when the gels were washed at 4°C. Taken together, these data indicate that the vast majority, and most likely, all native telomeres have a 3′-overhang.
Figure 4.
Most yeast telomeres end with a G-tail. (A, top) Genomic DNAs isolated from the indicated strains were first treated with Exonuclease I (labeled blunted) or mock treated (labeled native). DNAs were then mock treated (labeled ExoIII-), or treated with Exonuclease III (labeled ExoIII+) before native gel analysis. (Closed arrowhead) Internal positive control for the ExoIII treatment (Acc65I-digested pRS304); (open arrowhead) internal negative control (KpnI-digested pRS314), revealed by the T7 probe. KpnI-digested TRFs are seen with the GT probe only in lanes in which genomic DNA was blunted with ExoI prior to ExoIII reaction. (Bottom) Same gel as above after denaturation and rehybridization to a 2-μm probe to show equal loading of DNA (indicated by asterisk). Controls and molecular size standards are as in Figure 1. (B) Percentage of telomeres ending with a G-tail was estimated by quantifying the amount of G-tail signals found on YLpFAT10 on native gels washed at 4°C (see Fig. 2A). The values obtained were compared with signals detected for pGT19 and pGT22 fragments and were adjusted for the molecular ratio of hybridization targets on pGT19/pGT22 vs. the YLpFAT10 plasmid (see Supplemental Material). DNAs derived from three independent cultures of wild-type cells were used for quantification, and the standard deviation is indicated.
The dynamics of the cell cycle-dependent changes in G-tail detection is not affected in mre11Δ cells
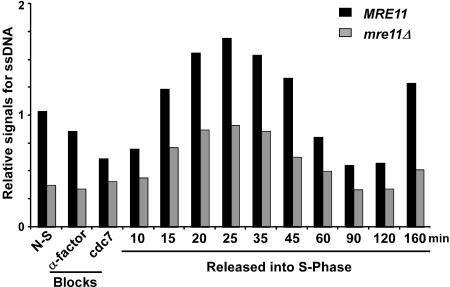
Because we observed that mre11Δ cells display shorter G-tails than wild-type cells, we were interested in determining whether the S-phase-dependent increase in G-tail signals observed in wild-type cells was decreased or abolished in the mutants. MRE11 and mre11Δ cells were synchronized, and DNA analyzed at different time points by in-gel hybridization (Supplementary Figs. 1, 2; data not shown). A quantification of the relative signals shows a significant reduction for telomeric G-tail signals in α-factor and cdc7-arrested mre11 cells when compared with wild-type cells (Fig. 5). For the cells that were released, the relative signals for G-tails increase for both strains in S phase (Fig. 5). From these data, it is clear that mre11Δ cells display a cell cycle-dependent change in the G-tail signals as wild-type cells do. Throughout the experiment, a significant difference between G-tail signal intensity for wild-type and mre11Δ strains was observed, suggesting that not only G1-specific G-tails, but also the S-phase-specific G-tails may be shorter in the mutant strain. However, due to the techniques used, such comparisons between cultures are challenging, and it remains uncertain whether this constant difference is real. Nevertheless, these results are consistent with previous data, which suggested that nucleolytic processing of a DNA double-strand break generated at the MAT locus during mating-type switching is inefficient in strains harboring deletions of the RAD50 or XRS2 genes (Ivanov et al. 1994). It is therefore tempting to speculate that the lower levels of the cell cycle-dependent G-tails observed in our synchrony experiments with mre11Δ cells are due to inefficient processing. Importantly though, the data do show that in the mutant strain, S-phase-dependent long G-tails are generated, suggesting that it is a processing event that probably occurs after DNA replication, which should establish a normal end-structure for the rest of the cell cycle, that is affected in these cells.
Figure 5.
S-phase-dependent G-tail formation in mre11Δ cells. A cell synchronization experiment using MRE11 and mre11Δ cells (AR120 and MLY40 strains) was performed as described in Supplementary Figure 1. DNA isolated from each cell aliquot was analyzed for telomeric DNA end-structures by nondenaturing in-gel hybridization (Supplementary Fig. 2; data not shown). The relative amounts for telomeric ssDNA on YLpFAT10 (in arbitrary units) were obtained by calculating the ratio of the G-tail signals retrieved on the native gel over the total amount of YLpFAT10 detected.
There is previous evidence that the MRX complex could be involved in a chromosome-end processing event. For example, with respect to G-strand replication, the MRX complex is in the same epistasis group as telomerase components (Nugent et al. 1998). Furthermore, in a telomere formation assay that is based on HO-cutting and exposing a short telomeric repeat tract, the MRX complex was shown to be needed for Cdc13p binding, and hence, G-tail formation. In mre11Δ cells, telomere formation was delayed, but not completely abolished (Diede and Gottschling 2001). Our results reported here are consistent with the idea that mre11Δ cells have an altered telomeric end-structure, consisting of shorter G-tails (Figs. 1, 3). However, Cdc13p binding to telomeres appears not to be reduced in mre11Δ cells (Tsukamoto et al. 2001), which suggests that in vivo, the length requirements for binding of this protein to G-tails may be <11 bases, the minimal length required for binding in vitro (Hughes et al. 2000). Alternatively, there may be a different mode of Cdc13p binding on yeast telomeres with very short overhangs. Taken together, the data suggest that in the absence of the MRX complex, alternative activities establish an altered and shortened G-tail on yeast telomeres. This also suggests that the establishment of such an end-structure and the binding of Cdc13p are essential features for telomere capping.
These suggested roles for the MRX complex in processing yeast telomeres may be conserved in mammals. For example, the functional homolog of the yeast Xrs2p in humans, NBS1, associates transiently with a telomeric complex of hRAD50/hMRE11/TRF2 (Zhu et al. 2000). This association occurs in S phase of the cell cycle, and it has been suggested that the association of NBS1 with hRAD50 and hMRE11 at telomeres either is necessary to prepare the telomeres for replication, or for a post-replicative processing event, such as the generation of appropriate G-tails and/or the formation of t-loops (Zhu et al. 2000). Furthermore, there is evidence for an S-phase-specific regulatory role for the NBS1 protein in maintaining telomeres in the absence of telomerase in human cell lines (Wu et al. 2000). The MRX complex is also involved in intra-S checkpoint signaling (D'Amours and Jackson 2001). On telomeres, the DNA strand replicated by leading-strand synthesis is thought to become a transiently blunt-ended molecule. Such ends may be sensed by this checkpoint, and processing to expose a single-stranded 3′ end could be initiated. Therefore, our results raise the possibility that such leading-strand ends may have particular requirements for recognition and processing, which would involve the MRE11 complex in yeast and humans.
Materials and methods
Yeast cell growth, DNA manipulations, and analyses followed standard and published protocols. For details, see the Materials and Methods section in the Supplemental Material.
Primer extension analysis of G-tails
A total of 0.1 ng of the oligo DUP-16 were boiled for 10 min in PE1 buffer (20 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol) and left to anneal at 23°C for more than 1 h. Chromosomal terminal restriction fragments (TRFs) were gel purified using a QIAquick gel extraction kit (Qiagen). Standard poly(A)-tailing reactions were performed with annealed DUP-16 oligo or gel-purified TRFs (details to be published elsewhere). Radiolabeled poly-T+(C/A) oligo [5′-CGGAATTCC(T)18M-3′; M = C or A] was annealed to these 3′-terminally tailed products in PE1 buffer at 16°C for 16 h. Primer-extension reactions were performed on annealed substrates in PE1 buffer, which also contained 0.325 mM dATP, 0.325 mM dCTP, and 20 μg BSA at 12°C for 20 min. In these conditions, strand displacement occurs on <10% of products (data not shown; Fig. 3). Reactions were stopped and loaded on a 15% polyacrylamide gel.
Acknowledgments
We thank P. Szankasi for yeast strains and S. Martin for help with the pGTxx plasmids. We also thank D. Gottschling for a critical reading of the manuscript, anonymous reviewers for comments and suggestions, and all members of the Wellinger laboratory for many valuable discussions. This work was supported by a research grant (MOP 12616), and a core group grant (GRP86284) from the Canadian Institutes for Health Research. M.L. was supported by an MRC studentship, C.L. by a studentship of NSERC, and R.J.W. is a Chercheur National of the FRSQ.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1199404.
References
- Blackburn E.H. 2001. Switching and signaling at the telomere. Cell 106: 661-673. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakhparonian M. and Wellinger, R.J. 2003. Telomere maintenance and DNA replication: How closely are these two connected? Trends Genet. 19: 439-446. [DOI] [PubMed] [Google Scholar]
- D'Amours D. and Jackson, S.P. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes & Dev. 15: 2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. The Mre11 complex: At the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3: 317-327. [DOI] [PubMed] [Google Scholar]
- Diede S.J. and Gottschling, D.E. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11: 1336-1340. [DOI] [PubMed] [Google Scholar]
- Dionne I. and Wellinger, R.J. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. 93: 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 1998. The many interfaces of Mre11. Cell 95: 583-586. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Weilbaecher, R.G., Walterscheid, M., and Lundblad, V. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. 97: 6457-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E.L., Sugawara, N., White, C.I., Fabre, F., and Haber, J.E. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai K.M. and Muniyappa, K. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2: 443-455. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E., and Lundblad, V. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8: 657-660. [DOI] [PubMed] [Google Scholar]
- Tomita K., Matsuura, A., Caspari, T., Carr, A.M., Akamatsu, Y., Iwasaki, H., Mizuno, K., Ohta, K., Uritani, M., Ushimaru, T., et al. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 23: 5186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Taggart, A.K., and Zakian, V.A. 2001. The role of the Mre11–Rad50–Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328-1335. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Wolf, A.J., and Zakian, V.A. 1993a. Origin activation and formation of single-strand TG1–3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13: 4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1993b. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72: 51-60. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Ethier, K., Labrecque, P., and Zakian, V.A. 1996. Evidence for a new step in telomere maintenance. Cell 85: 423-433. [DOI] [PubMed] [Google Scholar]
- Wu G., Lee, W.H., and Chen, P.L. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. J. Biol. Chem. 275: 30618-30622. [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster, B., Mann, M., Petrini, J.H., and Lange, T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25: 347-352. [DOI] [PubMed] [Google Scholar]