Abstract
DBP (albumin D-site-binding protein), HLF (hepatic leukemia factor), and TEF (thyrotroph embryonic factor) are the three members of the PAR bZip (proline and acidic amino acid-rich basic leucine zipper) transcription factor family. All three of these transcriptional regulatory proteins accumulate with robust circadian rhythms in tissues with high amplitudes of clock gene expression, such as the suprachiasmatic nucleus (SCN) and the liver. However, they are expressed at nearly invariable levels in most brain regions, in which clock gene expression only cycles with low amplitude. Here we show that mice deficient for all three PAR bZip proteins are highly susceptible to generalized spontaneous and audiogenic epilepsies that frequently are lethal. Transcriptome profiling revealed pyridoxal kinase (Pdxk) as a target gene of PAR bZip proteins in both liver and brain. Pyridoxal kinase converts vitamin B6 derivatives into pyridoxal phosphate (PLP), the coenzyme of many enzymes involved in amino acid and neurotransmitter metabolism. PAR bZip-deficient mice show decreased brain levels of PLP, serotonin, and dopamine, and such changes have previously been reported to cause epilepsies in other systems. Hence, the expression of some clock-controlled genes, such as Pdxk, may have to remain within narrow limits in the brain. This could explain why the circadian oscillator has evolved to generate only low-amplitude cycles in most brain regions.
Keywords: PAR bZip proteins, circadian transcription factors, epilepsy, pyridoxal kinase
DBP (albumin D-site-binding protein), HLF (hepatic leukemia factor), and TEF (thyrotroph embryonic factor) are the three members of the proline and acidic amino acid-rich basic leucine zipper (PAR bZip) transcription factor family. These proteins display high sequence similarity within their C-terminal basic DNA-binding domain, their leucine zipper dimerization domain, and a proline and acidic amino acid-rich domain immediately N-terminal to the bZip region with yet unknown function (Mueller et al. 1990; Drolet et al. 1991; Inaba et al. 1992). The extraordinarily high sequence conservation of PAR bZip transcription factors during mammalian evolution—92%–98% amino acid sequence identity between mouse and man—suggests that they fulfill important physiological functions.
The most striking feature of PAR bZip transcription factors is their circadian accumulation. In the liver and other peripheral organs, Dbp and Tef mRNA and protein accumulation displays robust circadian oscillation (Wuarin and Schibler 1990; Fonjallaz et al. 1996). In the rat liver Hlf specifies multiple mRNA species through the usage of multiple promoters, alternative splice sites, and multiple polyadenylation sites (Falvey et al. 1995). These mRNAs produce two proteins differing in N-terminal sequences, one fluctuating with a high and the other with a low circadian amplitude. While Dbp and Tef are expressed in most peripheral cell types, Hlf expression appears to be confined mainly to liver, kidney, and brain. All three PAR bZip genes are also expressed in a cyclic fashion in the suprachiasmatic nucleus (SCN), the master circadian pacemaker in mammals (Lopez-Molina et al. 1997; Mitsui et al. 2001).
The circadian expression of Dbp is driven directly by components of the molecular clock (Ripperger et al. 2000), which consists of two interconnected feedback loops of clock gene expression (Reppert and Weaver 2002). Thus, Dbp is activated by BMAL1 and CLOCK, the members of the positive limb of the circadian oscillator, and repressed by CRY and PER proteins, the members of the negative limb (Ripperger et al. 2000). Tef and Hlf transcription may be regulated by similar mechanisms, but these have not yet been studied in detail.
The high-amplitude cycles in PAR bZip and clock gene expression observed in peripheral tissues and in the SCN are in stark contrast to their nearly invariable expression patterns in most brain regions (Fonjallaz et al. 1996). The mechanisms by which the molecular oscillator generates high amplitude oscillations in the SCN and peripheral tissues, but only low amplitude fluctuations in most brain regions are not known. However, we consider it likely that this difference is physiologically relevant. To address the physiological importance of PAR bZip proteins, we initiated genetic loss-of-function experiments several years ago, starting with Dbp, the gene producing the most abundant PAR bZip protein (Lopez-Molina et al. 1997). Surprisingly, Dbp-/- mice manifested only relatively mild phenotypes, including a moderate reduction in circadian period length, a diminished circadian amplitude of slow wave sleep, an augmented sleep fragmentation, a reduction of spontaneous locomotor activity, and an amplitude reduction or phase difference in the expression patterns of putative PAR bZip target genes (Lopez-Molina et al. 1997; Franken et al. 2000). We thus suspected that some or all of the DBP functions are redundant and can be assumed by TEF and/or HLF in Dbp-null mice.
To examine the functions of all PAR bZip proteins, we generated Hlf- and Tef-deficient mice in addition to Dbp null mice. By using conventional breeding strategies, we then created mice containing two or three disrupted PAR bZip genes. Here we report that a large fraction of animals deficient for all three PAR bZip proteins succumb to lethal audiogenic and spontaneous epileptic seizures, and propose that the down-regulation of pyridoxal kinase (Pdxk) expression in PAR bZip-deficient mice may participate in the seizure phenotype.
Results
PAR bZip and clock gene expression in liver and brain
We used TaqMan RT–PCR assays to examine the temporal expression patterns of PAR bZip transcripts and mRNAs encoding central clock components in liver and brain. As shown in Figure 1A and B, Dbp mRNA levels oscillate with a daily amplitude of >50-fold in the liver but with an amplitude of only about twofold in the brain. Similarly, Tef transcripts, whose accumulation is strongly circadian in the liver (amplitude ninefold), are expressed at nearly constant levels in the brain. Hlf transcript levels display only a moderate circadian amplitude in the liver (1.6-fold) and are nearly flat in the brain. In the case of Hlf only one out of three promoters initiates transcription in a circadian fashion in mouse liver (F. Damiola and U. Schibler, unpubl.). This explains the low daily amplitude recorded for hepatic Hlf mRNAs. Nevertheless, DNA binding studies show that PAR bZip protein:DNA complexes follow a robustly circadian pattern (Lopez-Molina et al. 1997, and this paper). Hence, proteins translated from constitutively accumulating Hlf mRNAs do not contribute significantly to the DNA-binding activity of PAR bZip proteins at times of the day when TEF and DBP are expressed at nadir levels.
Figure 1.
Temporal expression of mRNAs encoding PAR bZip transcription factors and clock proteins in mouse liver and brain. mRNA levels were estimated by TaqMan real-time RT–PCR and normalized to the values obtained for Gapdh mRNA. The values for Dbp, Tef, and Hlf transcripts can be directly compared, since the values were corrected for PCR efficiencies (see Materials and Methods). Mean values ± S.E.M. obtained from four animals are given in all diagrams. The Zeitgeber times (ZT) at which the animals were sacrificed are indicated on the abscissa of the diagrams. (A) Circadian expression PAR bZip transcription factor transcripts in the mouse liver. (B) Temporal expression of Dbp, Tef, and Hlf mRNAs in the brain. (C–F) Temporal expression of mPer1 (C), mPer2 (D), Rev-erbα (E), and BMal1 (F) transcripts in mouse liver and brain.
As expected, a low amplitude of clock gene expression accompanies the low amplitude of PAR bZip mRNA accumulation in the brain. Thus, mPer1 transcript levels, which display a 12-fold circadian amplitude in the liver, are nearly invariable in the brain (Fig. 1C). Likewise, the mRNAs encoding mPer2, Rev-Erbα, and Bmal1 oscillate strongly in the liver but only weakly, if at all, in the brain (Fig. 1D–F).
Disruption of the Tef and Hlf genes
We have previously established a mouse strain with a Dbp null allele, in which the entire coding region had been replaced with a lacZ coding region and a PGK-Neomycin gene (Fig. 2A). To gain insight into the physiological roles of the two other PAR bZip transcription factors and to examine whether the three PAR bZip transcription factors might be functionally redundant, we decided to interrupt Tef and Hlf as well. In contrast to Dbp, which spans only ∼6 kb of DNA, Tef and Hlf are relatively large genes, encompassing 24 kb and 52 kb from the transcription initiation sites to the polyadenylation sites, respectively. Thus we could not delete the entire coding regions of Tef and Hlf, as we have previously done for Dbp, but instead replaced exon 2 of Tef and Hlf with a DNA segment containing a lacZ and a PGK-Neomycin gene (Fig. 2B,C; for details, see Supplemental Material). In both genes exon 2 encodes the transactivation domain, and mutant proteins lacking 40 amino acids of this domain have been demonstrated to be completely silent in cotransfection experiments (Hunger et al. 1996). Moreover, as most transcripts initiated at the Tef and Hlf promoters are expected to be polyadenylated at the strong SV40 polyadenylation site following the lacZ open reading frame, we did not anticipate that significant levels of Tef and Hlf transcripts would accumulate in the corresponding mutant mouse strains.
Figure 2.
Disruption of the Dbp, Tef, and Hlf alleles by homologous recombination. (A) Structure of the Dbp-null allele. The entire coding sequence was replaced by a LacZ–Neo cassette (for details, see Lopez-Molina et al. 1997). (B) Strategy used to delete the activation domain of the Hlf allele. The cartoon displays a map of the Hlf allele, with its three promoters (α, β, and γ) and its four polyadenylation sites (vertical arrows). The positions of the recognition sites for the following restriction endonucleases are given. (E) EcoRI; (B) BamHI; (X) XbaI; (No) NotI. ATG and TAA indicate the positions of the initiation and termination codons, respectively. The structure of the targeting vector, in which part of exon 2 containing the activation domain and part of intron 2 have been replaced by a LacZ–Neo cassette, is given below the Hlf locus. (C) Strategy used to delete the activation domain of Tef allele. The cartoon displays a map of the Tef allele, with its two promoters (α and β). The positions of the recognition sites for the following restriction endonucleases are given. (N) NcoI; (X) XbaI; (No) NotI. ATG and TAA indicate the positions of the initiation and termination codons, respectively. The structure of the targeting vector, in which part of exon 2 containing the activation domain and part of intron 2 have been replaced by a LacZ–Neo cassette, is given below the Tef locus. (D,E) Expression of the disrupted Hlf allele in mouse liver (D) and brain (E). The levels of liver Hlf and HlfΔAD transcripts were estimated by TaqMan real-time RT–PCR as in Figure 1A. The results represent means ± S.E.M. from four animals. (F,G) Expression of the mutated Tef allele in mouse liver (F) and brain (G). The levels of liver Tef and TefΔAD were estimated by TaqMan real-time RT–PCR as in Figure 1A. The results represent means ± S.E.M. from four animals.
We obtained morphologically normal and fertile animals homozygous for Hlf and Tef mutant alleles. Even animals devoid of two (Dbp/Hlf; Dbp/Tef; and Hlf/Tef) or all three PAR bZip proteins are anatomically normal and fertile, although the animals devoid of all PAR bZip alleles display a dramatically shortened life span (see below). To examine whether the Hlf and Tef mutant alleles really correspond to loss-of-function alleles, we used highly sensitive techniques to check for the possible presence of RNA and protein products specified by the disrupted Hlf and Tef alleles. As a first step, we applied TaqMan RT–PCR to probe liver and brain RNA from mice with disrupted Hlf and Tef alleles for the presence of Hlf and Tef mutant transcripts. As shown in Figure 2, these assays were unable to detect Hlf transcripts in mice with a disrupted Hlf allele (Fig. 2D,E). Hence, the Hlf mutant allele can be considered as a true null allele. TaqMan RT–PCR did however reveal a small amount of Tef transcripts in the liver and brain of Tef mutant mice (Fig. 2F,G), and we deemed it necessary to determine the exact nature of these transcripts. RACE experiments revealed a weakly expressed transcript that initiates at the Pα promoter of the Tef mutant allele. This transcript could potentially encode a DNA-binding protein containing 167 amino acids. However, in spite of using highly sensitive techniques, we were unable to detect such a polypeptide in Tef knockout mice (see Supplementary Fig. 1; Supplemental Material), and we thus concluded that the mutant Tef allele is indeed a loss-of-function allele.
Mice deficient of all three PAR bZip proteins are susceptible to lethal seizures
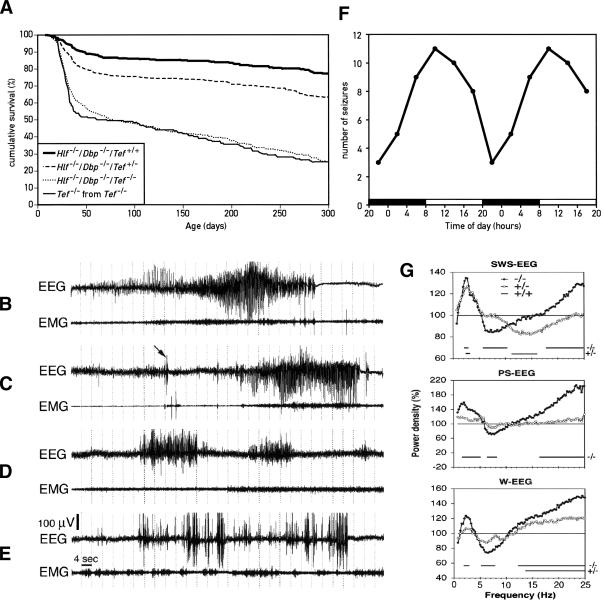
We noticed that a large fraction of young animals homozygous for the Tef mutant allele and at least one more PAR bZip null allele (Dbp-/-/Tef-/-; Hlf-/-/Tef-/-; and Hlf-/-/Dbp-/-/Tef-/-) succumbed to a sudden death. Not surprisingly, the death rate was highest in Hlf-/-/Dbp-/-/Tef-/- triple-knockout mice. In contrast, Hlf-/-/Dbp-/- double-knockout mice did not die prematurely when compared with wild-type mice. To examine the effect of Tef deficiency in greater detail, we recorded survival curves for mice with the three genotypes Hlf-/-/Dbp-/-/Tef+/+, Hlf-/-/Dbp-/-/Tef+/-, and Hlf-/-/Dbp-/-/Tef-/- (Fig. 3A). For the sake of simplicity, we will refer to these mice as double-knockout mice, Tef heterozygous, double-knockout mice, and triple-knockout mice, respectively, in the following sections. Nearly 50% of PAR bZip triple-knockout mice died within the first two months after birth. The mortality was significantly reduced in Tef heterozygous, double-knockout animals, and the life expectancy was nearly normal in double-knockout mice.
Figure 3.
Phenotypes of the Hlf/Dbp/Tef triple-knockout mice. (A) Increased mortality in young triple-knockout mice. About 50% of homozygous triple-knockout mice (Hlf-/-/Dbp-/-/Tef-/-) died between 20 and 40 d of age, irrespective of whether these animals were the offspring of Tef heterozygous, double-knockout mice (Hlf-/-/Dbp-/-/Tef+/-) or homozygous triple-knockout parents (Hlf-/-/Dbp-/-/Tef-/-, labeled as Tef-/- in the figure). After this age, triple-knockout animals continued to die progressively and had an expected life span inferior to that of double-knockout animals. All double-knockout mice (Hlf-/-/Dbp-/-) and Tef heterozygous, double-knockout mice (Hlf-/-/Dbp-/-/Tef+/-) were the offspring from Tef heterozygous, double-knockout mice (Hlf-/-/Dbp-/-/Tef+/-). (B,C) Spontaneous epileptic seizure in triple-knockout animals. All three genotypes were implanted with chronic EEG and EMG electrodes to characterize the seizure activity. Spontaneous tonic–clonic seizures only occurred in triple KO mice and primarily during slow-wave sleep (SWS). The arrow in the EEG from the individual presented in C points to an abrupt sharp wave (myoclonus) followed by a generalized epileptic seizure. (D) Absence seizure in a Tef heterozygous, double-knockout animal. Several absence-like seizures without any sign of EEG hypoactivity were also recorded in heterozygous mice. These occurred during SWS as well, particularly at the transition from waking to sleep. (E) Epileptic EEG activity in a Tef heterozygous, double-knockout animal. The most common form of epileptic activity in such mice was characterized by high-voltage sharp waves, similar to those observed in homozygous triple-knockout mice before generalized seizures initiated. We never recorded any generalized seizures in mice containing at least one Tef wild-type allele. (F) Temporal occurrence of seizures. The cartoon represents the occurrence of seizure in triple-knockout mice during 4-h intervals during the day. The lights were turned on and off at 8 a.m. and 8 p.m., respectively. For esthetic reasons the values were double-plotted. (G) EEG spectral analysis of homozygous and Tef heterozygous, double-knockout mice. The sleep EEG was recorded during a 12-h daytime period in nine triple, six heterozygous, and six double-knockout mice. Spectral analysis was performed on all 4-sec epochs without any seizure activity. Mean spectral profiles for each vigilance state were calculated over each genotype and expressed as a percentage of the mean values in double-knockout mice. Horizontal lines on the bottom of each panel connect those frequency bins in which power density is significantly different from that of double-knockout mice (indicated as 100% throughout the frequency range).
A serendipitous observation helped us to identify the cause of death of PAR bZip triple-knockout mice. Our daily records of deceased animals indicated that more animals died on Mondays and Thursdays, when the animal facility personnel used the vacuum cleaner, than on other weekdays. Indeed, ∼50% of young triple-knockout mice placed in close proximity to a vacuum cleaner exhibited “wild running” and tonic/clonic convulsions, symptoms that are typical for audiogenic seizures. Routine brain histopathological examination did not identify any noticeable difference between genotypes.
Although these sound-induced epileptic attacks explained the higher death rates observed on Mondays and Thursdays, they only accounted for ∼25% of all deceased animals. We suspected that the PAR bZip mutant mice also developed spontaneous seizures. We thus implanted mice of all three genotypes shown in Figure 3A with chronic bipolar electroencephalogram (EEG; frontal-somatosensory) and electromyogram (EMG; neck muscle) electrodes and continuously recorded EEG and EMG to characterize the seizure activity. EEGs recorded during 5 consecutive days revealed spontaneous seizures in all triple-knockout animals. However the number of seizures per animal was quite variable, amounting to one to nine seizures per individual during this period. Spontaneous tonic–clonic seizures occurred exclusively in triple mutant mice (Fig. 3B,C) and predominantly within slow-wave sleep (SWS) during the first 2 min after transition to sleep. Several seizure-like activities could also be observed in some Tef heterozygous, double-knockout mice, but in contrast to the attacks observed in homozygous triple-knockout mice these never developed into generalized epilepsies. The most common form was characterized by high voltage spikes (Fig. 3E), similar to those observed preceding generalized seizures. However, a few absence-like seizures without any sign of EEG hypoactivity were also recorded (Fig. 3D). Spontaneous, high-voltage spikes and absence-like seizures were also observed in the EEGs recorded from all six Tef-/- single knockout mice (see Supplementary Fig. 2C–G). However, these activities were less frequent than in triple-knockout mice and never resulted in generalized tonic–clonic epilepsies.
Analysis of the temporal occurrence of seizures in triple-knockout animals indicated a clear circadian trend similar to the circadian distribution of sleep. Thus, nearly four times more seizures were recorded during early light period hours (major sleep period) than during early dark period hours (major active period; Fig. 3F). Continuous video-EEG recordings were also performed in two triple-knockout mice to clinically characterize their seizure activities. Typical spontaneous seizures (example in Supplementary Video 1) started with abrupt myoclonic jerks with sharp EEG spikes (arrow in Fig. 3C), generalized tremor, high-amplitude rhythmic theta activity (5–8 Hz, Fig. 3B) followed by head and forelimb clonus, tail extension, and retropulsion. The attacks ended with tonic flexion and flattening of the EEG. Recovery was associated with jerks after 30–80 sec. We also noticed that most inter-ictal sharp EEG spikes were in fact accompanied by myoclonic jerks.
We analyzed the sleep EEG of a 12-h light period recording in nine homozygous triple-knockout mice, six Tef heterozygous, double-knockout mice, and six double-knockout mice. Spectral analysis was performed on all three consecutive 4-sec epochs without any artifact or seizure activity. Mean spectral profiles for each vigilance state were calculated for each genotype and expressed as a percentage of the mean values in double-knockout mice homozygous for the Tef wild-type allele (Fig. 3G). In all three vigilance states, a significant increase in slow-wave δ activity (0.75–4.5 Hz) and a significant decrease in theta activity (5–8 Hz) were found in homozygous triple-knockout mice. This trend was much less pronounced in Tef heterozygous, double-knockout mice. Also, activity in all frequency bins >15 Hz (the β frequency range) was significantly increased during all vigilance states of homozygous triple-knockout animals and in waking of Tef heterozygous, double-knockout mice.
PAR bZip proteins regulate the expression of pyridoxal kinase, an enzyme involved in the activation of vitamin B6
To elucidate the molecular link between PAR bZip transcription factors and epileptic seizures, we sought PAR bZip-regulated genes that could provide insight into the control of brain electrical activity. To this end we compared brain mRNA populations from double-knockout mice, which are not prone to epileptic attacks, and triple-knockout animals, which are susceptible to audiogenic and spontaneous seizures, by Affymetrix high-density oligonucleotide microarray hybridization. Three pools of brain RNA, composed of equivalent amounts of RNA from five males and five females each, were prepared for the two genotypes and compared by hybridization with Affymetrix oligonucleotide microarrays containing probe features for ∼12,000 genes (representing 30%–50% of all genes). This resulted in a total of nine comparisons.
The transcripts that are underexpressed and overexpressed in TEF-deficient mice by at least 50% are listed in Table 1. As expected, animals homozygous for the disrupted Tef allele contained considerably less Tef transcripts than animals homozygous for a Tef wild-type allele (see Fig. 2G). Interestingly, NFIL3/E4BP4, a putative antagonist of PAR bZip transcription factors (Mitsui et al. 2001), is also down-regulated in TEF-deficient animals, suggesting that TEF stimulates the production of its own competitive inhibitor. In the brain, TEF also appears to stimulate the expression of mPer2 and Dec1, two transcriptional regulators that had been implicated in the negative limb of the circadian oscillator (Kawamoto et al. 2004).
Table 1.
Affymetrix microarray analysis of brain transcripts in double- and triple-knockout mice
| Name | Product/homology | GenBank | Fold change | N/9 |
|---|---|---|---|---|
| A | ||||
| Tef | Thyrotroph embryonic factor | AI850638 | 7.42 ± 1.39 | 9 |
| Nfil3/E4BP4 | Nuclear factor, interleukin 3, regulated | U83148 | 2.59 ± 0.97 | 8 |
| EST (PDXK) | RIKEN cDNA 2310036D04 gene (similar to Pyridoxal Kinase) | AI841777 | 2.05 ± 0.25 | 9 |
| EST | RIKEN cDNA 0910001L24 gene (putative epoxide hydrolase) | AI836883 | 1.88 ± 0.25 | 9 |
| Per2 | Period homolog 2 | AF036893 | 1.73 ± 0.22 | 6 |
| EST | RIKEN cDNA 1200013P24 gene (GCN5-related N-acetyltransferase) | AI846961 | 1.60 ± 0.08 | 9 |
| Csflr | Colony stimulating factor 1 receptor | X06368 | 1.59 ± 0.18 | 7 |
| Bhlhb2/Dec1 | Basic helix–loop–helix domain containing, class B2 (Dec1) | Y07836 | 1.59 ± 0.15 | 8 |
| Baiap2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | AI844631 | 1.56 ± 0.15 | 8 |
| Igfbp6 | Insulin-like growth factor binding protein 6 | X81584 | 1.52 ± 0.15 | 7 |
| EST | RIKEN cDNA B930035K21 gene | AI846522 | 1.50 ± 0.15 | 8 |
| Cx3cl1 | Chemokine (C-X3-C motif) ligand 1 | U92565 | 1.50 ± 0.19 | 7 |
| B | ||||
| Anp32a | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | U73478 | 6.92 ± 5.19 | 7 |
| EST | Murine (DBA/2) mRNA fragment for gag related peptide | X05546 | 3.34 ± 2.51 | 6 |
| EST | Mus musculus prosaposin gene, promoter and exon 1 | AF037437 | 3.06 ± 2.80 | 6 |
| Kif1a | Kinesin heavy chain member 1A | D29951 | 2.54 ± 1.25 | 6 |
| EST | RIKEN cDNA C330012F17 gene | AA189811 | 2.47 ± 0.44 | 8 |
| Gna12 | Guanine nucleotide binding protein, alpha 12 | M63659 | 2.40 ± 1.70 | 6 |
| Sty1 | Synaptotagmin 1 | D37792 | 2.38 ± 1.06 | 6 |
| Timp2 | Tissue inhibitor of metalloproteinase 2 | X62622 | 2.31 ± 1.45 | 6 |
| Kif5a | Kinesin family member 5A | AF053473 | 2.25 ± 1.37 | 6 |
| Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit a isoform 1 | U13836 | 2.14 ± 1.42 | 6 |
| Ptpns1 | Protein tyrosine phosphatase, nonreceptor type substrate 1 | D85785 | 2.09 ± 1.20 | 6 |
| Zfp179 | Zinc finger protein 179 | AB013097 | 1.99 ± 1.37 | 6 |
| Ptpns1 | Protein tyrosine phosphatase, nonreceptor type substrate 1 | D85785 | 1.84 ± 1.07 | 6 |
| Ptn | Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | D90225 | 1.79 ± 0.52 | 6 |
| Hspc121 | Butyrate-induced transcript 1 (HSP20-like chaperone) | Z97207 | 1.71 ± 0.73 | 6 |
| Usp22 | Ubiquitin specific protease 22 | AW125800 | 1.54 ± 0.42 | 6 |
| Ppp1cb | Protein phosphatase 1, catalytic subunit, beta isoform | M27073 | 1.51 ± 0.24 | 8 |
| Vamp2 | Vesicle-associated membrane protein 2 (synaptobrevin 2) | U60150 | 1.50 ± 1.11 | 6 |
| Gria1 | Glutamate receptor, ionotropic, AMPA1 (α 1) | AW048549 | 1.50 ± 0.55 | 6 |
Transcripts that are less abundant or more abundant in triple knockout mice as compared to double knockout mice are listed in panel A and B, respectively. The “Fold change” is the average value ± S.D. of all nine comparisons (even if the values were not different in all nine comparisons). N/9 indicates the number of comparisons in which the hybridization signals were significantly different. Note that Pdxk mRNA yielded a lower hybridization signal with RNA from triple-knockout mice in nine out of nine comparisons, whereas not a single transcript listed in panel B (higher in triple- than in double-knockout mice) shows a difference in all nine comparisons.
Among the genes positively regulated by TEF, Pdxk caught our special attention. Pdxk mRNA levels scored about twofold less in Tef-deficient mice in nine out of nine comparisons (see above). PDXK performs the last step in the conversion of B6 vitamers into pyridoxal phosphate (PLP), a coenzyme of numerous decarboxylases and transaminases involved in amino acid and neurotransmitter metabolism (see Discussion). Even moderate reductions in PLP levels have previously been associated with a susceptibility to epileptic seizures and an increase in δ EEG activity (Sharma et al. 1994), and we thus examined the transcription of Pdxk in greater detail to evaluate whether this gene might be a direct target gene of TEF. As a first step, we determined the transcriptional start sites within the promoter of the murine Pdxk gene using RACE technology. The sequencing of 13 RACE products yielded two major and several minor cap sites, located between 65 bp and 91 bp upstream of the translation initiation codon (Fig. 4A). As shown by the alignment of the corresponding Pdxk sequences, a short conserved sequence block encompassing a PAR bZip recognition sequence (PARRE; Falvey et al. 1996) can be discerned in the first intron, 12–20 bp downstream of the exon–intron splice junction. We used a double-stranded oligonucleotide containing this putative binding site in electromobility shift assays (EMSA) with liver and brain nuclear extracts harvested at 4-h intervals around the clock to investigate whether this sequence element indeed binds PAR bZip proteins. As shown in Figure 4B, this element forms a prominent protein:DNA complex with liver nuclear proteins prepared from wild-type mice at times when DBP and TEF reach zenith levels (ZT10). A protein:DNA complex reflecting a liver nuclear protein with circadian accumulation is still discerned in double-knockout mice but not in triple-knockout mice, strongly suggesting that this complex contains TEF. Figure 4C shows that similar complexes can be observed in EMSA experiments with brain nuclear extracts prepared from animals of the three genotypes. As expected on the basis of the temporal mRNA accumulation profiles (Fig. 1B), the PAR bZip proteins revealed by these EMSA studies accumulate at high levels throughout the day in the brain of wild-type mice. Likewise, TEF levels do not appear to follow a circadian rhythm in the brain of double-knockout animals.
Figure 4.
The transcription of the pyridoxal kinase gene (Pdxk) is regulated by PAR bZip transcription factors. (A) Sequence comparison of the 5′ proximal region of the Pdxk genes from mouse, rat, and man. The transcription initiation sites, indicated by bent arrows, have been mapped on the mouse genomic sequence by RACE analysis. Three RACE products have been sequenced for each of the two start sites represented by more prominent arrows. Start sites represented by single RACE products are depicted by small arrows. A PAR bZIP transcription factor response element (PARRE) is located within the 5′-moiety of the first intron. The PARRE consensus sequence is given below the element (R = A or G, Y = C or T). (B) Binding of liver nuclear proteins to the intronic PARRE. Electomobility shift assays (EMSA) with a radio-labeled oligonucleotide encompassing the PARRE and liver nuclear proteins from triple-knockout mice, double-knockout mice and wild-type animals harvested at 4-h intervals around the clock. ZT stands for Zeitgeber time. The lights were turned on and off at ZT0 and ZT12, respectively. (C) Binding of brain nuclear proteins to the Pdxk PARRE. EMSA were carried out as in B, but with brain nuclear proteins. (D) Temporal expression of Pdxk RNA in the liver of double-knockout mice and triple-knockout mice. The relative accumulation of Pdxk mRNA was determined by TaqMan real-time RT–PCR with liver whole-cell RNAs prepared from animals sacrificed at the indicated Zeitgeber times (ZT). Mean values ± S.E.M. from four animals are shown. ANOVA revealed significant circadian rhythmicity only for the hepatic Pdxk mRNA expression of double-knockout animals (F[5,18] = 7.73, p < 0.0005). The difference between genotypes is highly significant (ANOVA F[1,46] = 32.03, p < 1.10-6). (E) Temporal expression of Pdxk mRNA in the brain of double-knockout and triple-knockout mice. The relative levels of Pdxk mRNA were determined as in D. The values are means ± S.E.M. from four animals. Although the transcript levels of double-knockout mice measured at ZT8 appear somewhat lower than those measured at other ZTs, ANOVA did not reveal a circadian Pdxk expression. The difference between genotype is highly significant (ANOVA F[1,46] = 41.61, p < 1.10-7). (F) Expression of Pdxk RNA in different regions of the brain. Coronal brain sections were prepared from double- or triple-knockout mice sacrificed at ZT10 and hybridized to a 35S-labeled antisense (top and middle panels) or a sense (bottom panel) Pdxk RNA probe. (CP) Caudate putamen (striatum); (LS) lateral septal nucleus; (MH) medial habenula; (HF) hippocampal formation; (BMA) baso-medial amygdaloid nucleus; (T) thalamus; (ZI) zona incerta; (DMH) dorsomedial hypothalamic nucleus; (IC) inferior colliculus; (PAG) peri-aqueductal gray; (GLC) granular layer of cerebellum; (CGP) central gray of the pons.
We used TaqMan RT–PCR technology to investigate the differences in Pdxk expression in the liver and brain of double- and triple-knockout mice. Figure 4D shows that in the liver of double-knockout mice, Pdxk mRNA accumulates in a circadian fashion, with a phase compatible with that of TEF accumulation (Fig. 4B). In triple-knockout mice, Pdxk transcript levels are nearly constant throughout the day and amount to about one-third of the zenith levels attained in double-knockout mice. As shown in Figure 4E, the Pdxk mRNA levels are also lower in the brain of triple-knockout mice (about two-fold on average). While the Pdxk mRNA concentrations determined in double-knockout mice are similar at most times of the day, the value obtained at ZT8 appears to be somewhat lower. However, in contrast to the clearly circadian expression pattern of liver Pdxk mRNA (ANOVA F[5,18] = 7.73, p < 0.0005), the temporal accumulation profile of brain Pdxk transcripts did not score as circadian by the ANOVA test (ANOVA F[5,18] = 0.78, p = 0.58). We also determined the daily accumulation of Pdxk mRNA in liver and brain of homozygous Tef single-knockout mice (Supplementary Fig. 2) and found that it was intermediate between double- and triple-knockout mice. This is in keeping with the low but significant seizure activities observed in the EEGs of Tef single-knockout animals.
As the TaqMan RT–PCR measurements of Pdxk mRNA were performed with total brain RNA, we considered it important to complement these studies by in situ hybridization experiments with sections from different brain regions. Figure 4F suggests that several brain regions show significantly lower signals in triple-knockout mice as compared with double-knockout mice. These include structures that have previously been implicated in the development of spontaneous and/or audiogenic epilepsies, such as cerebral cortex, amygadala, hippocampal formation, inferior colliculus, and periaqueductal gray (see Discussion).
Our DNA microarray hybridization experiments also unveiled some brain transcripts that accumulate to higher levels in triple-knockout mice (Table 1) than in double-knockout mice. Either the synthesis of these mRNA is negatively regulated by TEF, or it is induced by secondary mechanisms, for example, as a consequence of stress in TEF-deficient animals. At least in some instances, such as in the case of Synaptotagmin 1, Kinesin, or Gna 12, the increased expression may indeed be part of a stress response following epileptic seizures (Lason and Przewlocki 1994; Babity et al. 1997; Kirby et al. 2002).
The brains of PAR bZip triple-knockout mice contain reduced levels of PLP, serotonin, and dopamine, and increased levels of histamine
PLP is synthesized in the liver via the phosphorylation of B6 vitamers by PDXK, and a portion of the hepatic PLP is then exported into the bloodstream and transported to other sites, such as the brain. Before crossing the blood–brain barrier and the plasma membrane of target cells, PLP has to be dephosphorylated to pyridoxal (PL) by plasma phosphatases. In the target cells, PL is then rephosphorylated to PLP, which is the active coenzyme form of vitamin B6 (Bell and Haskell 1971; Bell et al. 1971;).
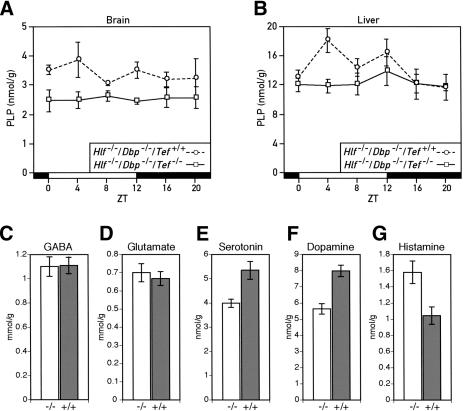
We examined whether the reduced Pdxk expression in the liver and brain of PAR bZip-deficient mice indeed affected the levels of PLP. Figure 5A and B show that both brain and liver levels of PLP are significantly reduced. Whereas PLP accumulation appears to follow a circadian rhythm in the liver of Tef wild-type mice, it is nearly invariable in the liver of PAR bZip triple-knockout mice and in the brain of double- and triple-knockout mice. As Pdxk expression is more strongly dependent on TEF in a subset of brain structures (see above), we suspect that in these structures the difference in PLP levels between TEF-proficient and TEF-deficient animals is considerably higher than the difference measured in extracts from total brains.
Figure 5.
Down-regulation of pyridoxal phosphate (PLP) levels and dysregulation of neurotransmitter metabolism in PAR bZip triple-knockout mice. (A) Temporal concentration of PLP in the brains of double- and triple-knockout mice. The PLP levels were measured by high-performance liquid chromatography (HPLC) of perchloric acid brain extracts prepared from animals sacrificed at the indicated Zeitgeber times (ZT). The values are means ± S.E.M. from three animals. The difference between the two genotypes is highly significant (ANOVA F[1,32] = 20.18, p < 1.10-4). (B) Temporal accumulation of PLP in the livers of double- and triple-knockout mice. PLP levels were determined as in A, and the values are means ± S.E.M. from at least six animals. ANOVA revealed significant circadian rhythmicity in PLP levels in double-knockout animals (F[5,32] = 2.60, p < 0.05). The difference between geno-types is also statistically significant (ANOVA F[1,73] = 5.31, p < 0.025). (C,D) GABA (C) and Glutamate (D) concentration in the brains of Hlf-/-/Dbp-/-/Tef-/- (-/-) and Hlf-/-/Dbp-/-/Tef+/+ (+/+) mice. No statistically significant difference could be found between the two genotypes. The values are means ± S.E.M. from 12 animals of each genotype. (E,F,G) Concentration of serotonin (E), dopamine (F) and histamine (G) in the brains of Hlf-/-/Dbp-/-/Tef-/- (-/-) and Hlf-/-/Dbp-/-/Tef+/+ (+/+) mice. The difference between genotype is statistically significant (Student's t test: p < 0.005 for serotonin, p < 0.0001 for dopamine and p < 0.01 for histamine). The values are means ± S.E.M. from 12 animals of each genotype.
In the brain, PLP is the coenzyme of several enzymes participating in the synthesis of neurotransmitters, such as γ-aminobutyric acid (GABA), serotonin, and dopamine, which had previously been associated with epileptic seizures (for review, see Tunnicliff and Ngo 1998). Moreover, changes in PLP levels can also influence the levels of glutamate (Glu) and histamines by determining the rate of conversion of these neurotransmitters to other compounds. We thus examined whether the reduced levels of PLP in the brains of PAR bZip triple-knockout mice affected the brain concentrations of GABA (Fig. 5C), Glu, (Fig. 5D), serotonin (Fig. 5E), dopamine (Fig. 5F), and histamine (Fig. 5G). The results revealed significantly reduced levels for serotonin and dopamine and increased levels of histamine in triple-knockout as compared with double-knockout animals.
PAR bZip proteins are clock output regulators rather than essential components of the circadian oscillator
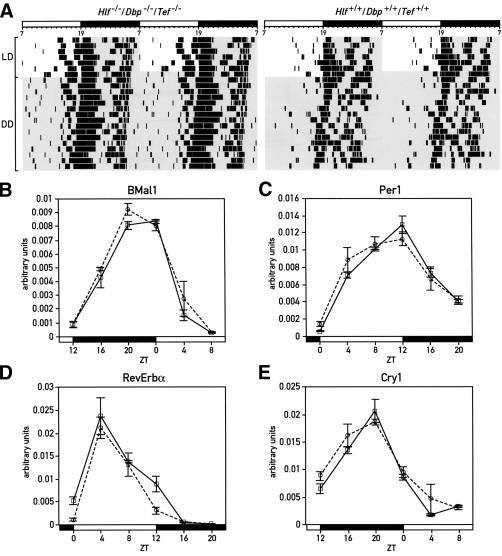
Since all three PAR bZip genes show circadian expression in the SCN and in peripheral tissues, we wished to examine whether these transcription factors are required for sustained rhythmicity in constant darkness (DD). The relevance of this issue is reinforced by the recent finding that PAR domain protein 1 (Pdp1), the only Drosophila PAR bZip domain protein, is an essential clock component in fruit flies (Cyran et al. 2003). We thus recorded the circadian locomotor activity of triple-knockout mice using wheel running assays. As seen in Figure 6A, PAR bZip-deficient mice are rhythmic in DD, suggesting that they still harbor a functional circadian pacemaker. Moreover, the period length of triple-knockout mice measured in DD is nearly identical to that measured for wild-type mice. Interestingly, Dbp single-knockout mice free-run with a period ∼30 min shorter than that of wild-type mice (Lopez-Molina et al. 1997), while Tef and Hlf single knockout mice free-run with periods ∼20–25 min longer than wild-type (F. Damiola, J. Ripperger, and U. Schibler, unpubl.). Apparently, these positive and negative period differences counteract each other in triple-knockout animals, yielding a period length that is close to that observed for wild-type mice. Although triple-knockout animals display a rhythmic behavior in constant darkness, we noticed that a large proportion of these animals show a bout of wheel running activity several hours before the onset of the dark phase in LD or the onset of the subjective night in DD (left panel in Fig. 6A). We did not yet investigate whether this anticipatory activity correlates with a phase advance of clock gene expression in the suprachiasmatic nucleus.
Figure 6.
Circadian behavior and gene expression in PAR bZip triple-knockout mice. (A) Circadian locomotor (wheel running) activity of Hlf/Dbp/Tef triple-knockout (left panel) and wild-type (right panel) mice. In each actogram, the first few days were recorded under LD conditions (lights on at 7 a.m.; lights off at 7 p.m.). Time spans during which the light was switched off are marked by gray shadowing. The free-running periods in constant darkness of these mice show no difference between geno-types (Hlf-/-/Dbp-/-/Tef-/-: 23.63 ± 0.33 h; wild-type: 23.68 ± 0.33 h). (B–E) Temporal expression of BMal1 (B), mPer1 (C), Rev-erbα (D), and Cry1 (E) transcripts in liver, as determined by real-time RT–PCR in Hlf/Dbp/Tef triple-knockout (solid line) and wild-type mice (dotted line). Mean values ± S.E.M. obtained from four animals are given in all diagrams. The Zeitgeber times (ZT) at which the animals were sacrificed are indicated on the abscissa of the diagrams.
We also compared the circadian expression of core clock components in the livers of triple-knockout and wild-type mice. As shown in Figure 6B–E, the rhythmic expression of the four clock genes Bmal1, Per1, RevErbα, and Cry1 is nearly identical in animals of these two genotypes. We thus conclude that PAR bZip proteins are dispensable for circadian rhythm generation, and hence that they are regulators of outputs of the mammalian circadian timing system rather than core components of the clock.
Discussion
We interrupted Dbp, Tef, and Hlf, the three genes of the murine PAR bZip transcription factor family, by homologous recombination in the mouse. In spite of the extraordinarily high sequence conservation of the three PAR bZip proteins in mammalian evolution, mice homozygous for loss-of-function alleles for one or multiple of these proteins appear to be anatomically normal, reach adulthood, and are fertile. Nevertheless, triple mutant mice display a dramatically reduced life span, in that <20% of these animals reach a life span >1 yr. Here we have identified generalized epileptic seizures as a major cause of premature death, but we consider it likely that other pathologies caused by the deficiency of PAR bZip proteins may also contribute to the shortened life span.
TEF regulates the expression of pyridoxal kinase in the liver and in seizure-relevant brain structures
The comparison of gene expression in seizure-resistant double-knockout mice and seizure-prone triple-knockout mice revealed a plausible TEF target gene, Pdxk, whose down-regulation in triple-knockout mice may explain at least part of the seizure phenotype. PDXK is required for the synthesis of PLP, and even moderate reductions in the concentration of this coenzyme have been shown to provoke epileptic seizures (Schlesinger and Lieff 1975; Sharma and Dakshinamurti 1992). Several observations suggest that TEF regulates Pdxk transcription by a direct mechanism in liver and brain. First, the Pdxk gene contains an evolutionarily conserved PAR bZip recognition sequence in the first intron that binds TEF and probably other PAR bZip proteins with high specificity and affinity. Second, in the liver of wild-type and double-knockout mice, Pdxk mRNA accumulation follows a circadian rhythm whose phase angle is consistent with that of cyclic TEF accumulation. While TEF appears to be the major PAR bZip regulator of Pdxk transcription in the liver and brain, the somewhat higher levels of Pdxk transcripts in Tef-/- single mutant mice (Supplementary Fig. 2A,B) as compared with triple mutant mice indicate that DBP and HLF also participate to some extent in the regulation of Pdxk transcription.
The spatial Pdxk mRNA expression pattern in the brains of double- and triple-knockout mice suggests that TEF regulates Pdxk transcription in many brain regions previously associated with epileptic seizures, including cerebral cortex, striatum, amygdala, hippocampus, inferior colliculus, and periaqueductal gray. For example, the inferior colliculus (IC) has previously been recognized as a key site in the amplification of audiogenic seizures (Kesner 1966; Willott and Lu 1980). The periaqueductal gray (PAG; N'Gouemo and Faingold 1999) and the striatum (Slaght et al. 2002) have also been implicated in epileptic attacks, and Pdxk expression in these regions is significantly diminished in triple-knockout mice.
Roles of PAR bZip proteins in neurotransmitter homeostasis and amino acid metabolism
By controlling Pdxk expression, PAR bZip transcription factors may play an important role in the finetuning of neurotransmitter homeostasis in the brain. PLP, the product of the PDXK reaction, serves as the coenzyme for a number of enzymes involved in the synthesis and degradation of neurotransmitters, and changes in PLP levels can result in severe neurotransmitter disequilibria and brain pathologies characterized by epilepsy (Tunnicliff and Ngo 1998). For example, the disruption of the gene encoding tissue-nonspecific alkaline phosphatase (TNAP) causes an approximately threefold reduction in brain PLP levels, and 100% of the Tnap mutant mice succumb to lethal epileptic seizures before 20 d of age (Waymire et al. 1995). γ-Aminobutyric acid (GABA), the major inhibitory neurotransmitter of the central nervous system, probably plays the most important role in preventing the uncontrolled activation of epileptic foci and the propagation/amplification of seizure waves through downstream regions. The function of PLP in determining the concentration of GABA is complex, as PLP is the coenzyme of both glutamate decarboxylase (GAD), which converts glutamate to GABA, and GABA transaminase (GABA-T), which degrades GABA to succinic semialehyde. Moreover, two GAD isoforms, GAD65 and GAD67, with possibly different functions are expressed in most glutamatergic neurons. Whereas GAD65 is believed to be mostly responsible for the synthesis of GABA used for neurotransmission in nerve endings, GAD67 may produce GABA that is used to a large extent for metabolic purposes involving the tricarboxylic acid cycle (Waagepetersen et al. 1999). GAD67 has a higher affinity for PLP than GAD65, and exists primarily in the holoenzyme form at physiological PLP levels. In contrast, a large fraction of GAD65 is found in the apoenzyme form, and the activity of this GAD isoform is thus more dramatically affected by changes in PLP concentration (Battaglioli et al. 2003). It appears that the bulk of brain GABA is synthesized by GAD67, as Gad65-/- mice (genetic backgound 129xC57BL/6) contain normal basal levels of brain GABA. As these mice are susceptible to spontaneous epileptic attacks, local reductions in GABA concentrations that are not revealed by measuring total brain GABA may be sufficient to provoke seizures (Kash et al. 1997). Hence, in spite of the virtually identical levels of total brain GABA measured in double- and triple-knockout mice, it remains possible that the susceptibility of PAR bZip triple-knockout animals to seizure is enhanced by a local reduction of GABA levels in key brain structures.
PLP also serves as the coenzyme of aromatic amino acid decarboxylase (AADC), an enzyme involved in the synthesis of the monoamines serotonin and dopamine (Rahman et al. 1982). Both of these neurotransmitters have been associated with epileptic attacks. For example, young adults of a mouse strain homozygous for a serotonin receptor 5-HT2C null allele develop audiogenic seizures (Tecott et al. 1995; Brennan et al. 1997) and mice homozygous for a dopamine receptor 2 (D2R) null allele develop epilepsies after injection of kainic acid at doses that do not provoke seizures in D2R wild-type littermates (Bozzi et al. 2000). Given these observations, we suspect that the significantly reduced serotonin and dopamine concentrations in the brains of PAR bZip triple-knockout mice contribute to the susceptibility of these mice to epileptic attacks.
Two PLP-dependent enzymes, histidine decarboxylase and histamine oxidase, are involved in histamine synthesis and catabolism, respectively (Ono and Hagen 1959; Blaschko and Buffoni 1965). Interestingly, high levels of PLP have been shown to result in a decrease of histamine levels, probably by promoting histamine degradation (Lee et al. 1988). Therefore, we speculate that the high level of histamine found in the brain of PAR bZip triple-knockout mice is caused by a decrease in histamine catabolism, due to diminished PLP levels. Whether or not histamine imbalances play a role in the generation of epilepsies is still somewhat controversial.
Mammalian PAR bZip proteins are output regulators of the circadian clock in the brain and peripheral tissues
All three PAR bZip genes are rhythmically expressed in the suprachiasmatic nucleus (Mitsui et al. 2001), and Pdp-1, the PAR bZip protein encoded by the Drosophila genome, plays an essential function in circadian rhythm generation (Cyran et al. 2003). These properties raised the question of whether PAR bZip proteins are essential core components of the mammalian circadian oscillator. This is clearly not the case, as PAR bZip triple-knockout mice still display rhythmic locomotor activity and clock gene expression (Fig. 6). Hence, PAR bZip proteins are output regulators, rather than core components of the mammalian circadian oscillator, and Pdxk is one of the many clock-controlled genes whose cyclic expression is governed by PAR bZip transcription factors in the liver (F. Gachon, P. Gos, and U. Schibler, unpubl.). Surprisingly, the phase of cyclic PLP accumulation differs by nearly 12 h from that of circadian Pdxk mRNA accumulation, perhaps implying that posttranscriptional mechanisms in addition to transcriptional mechanisms determine the rhythmic PDXK activity in the liver. Alternatively, the bulk of PLP synthesized during the dark phase, when Pdxk mRNA levels peak, is secreted into the blood stream and serves as a source of B6 vitamers for other tissues. Nevertheless, it is likely that the oscillation of hepatic PDXK expression contributes to the cyclic activity of enzymes used in amino acid metabolism, polyamine synthesis, and glycogen utilization (John 1995).
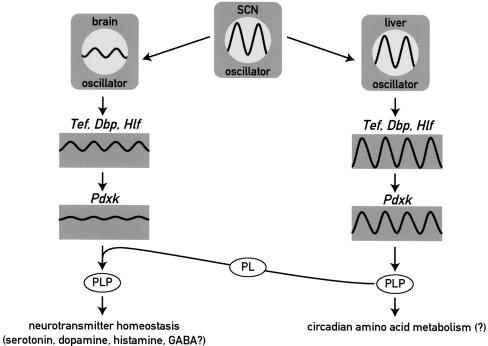
The cartoon in Figure 7 exhibits the different regulation and possible functions of PDXK in liver and brain. In both tissues, its expression is under the control of PAR bZip proteins, in particular TEF. The expression of PAR bZip proteins and that of its target gene Pdxk is strongly circadian in the liver. However, in most brain regions the circadian molecular clock generates only low amplitude-oscillations in clock gene expression, and as a consequence the levels of PAR bZip proteins remain nearly constant. Accordingly, the concentrations of Pdxk mRNA and PLP show little if any circadian fluctuations in this organ. On the basis of the results presented in this study, we speculate that high-amplitude circadian Pdxk expression in the brain might be dangerous, as daily reductions in PLP levels in the central nervous system could provoke life-threatening epileptic attacks. Clock-controlled genes such as Pdxk, whose minimal and/or maximal activities in the brain must remain within narrow limits, offer a physiological raison d'être for the low amplitude cycles of clock gene expression in most parts of the central nervous system.
Figure 7.
Model showing the regulation and function of Pdxk in brain and liver. The molecular circadian oscillator generates high amplitude cycles and low amplitude cycles of PAR bZip gene expression in liver and brain, respectively. Accordingly, target genes of PAR bZip transcription factors, such as Pdxk, are expressed in a strongly circadian manner in liver and at nearly constant levels in the brain. As PLP, the product of the PDXK reaction is a coenzyme for amino acid decarboxylases and aminotransferases, the rhythmic production of PLP in the liver may contribute to circadian amino acid metabolism. In the brain, nearly invariable Pdxk expression may be essential for regulating neurotransmitter homeostasis, as moderate decreases in PLP levels may result in neurotransmitter deficiencies and epileptic attacks (see text). (SCN) suprachiasmatic nucleus harboring the central circadian pacemaker; (PLP) pyridoxal phosphate; (PL) pyridoxal.
Spontaneous seizures in PAR bZip-deficient mice occur mainly during slow wave sleep
Continuous 5-d recordings revealed one to nine seizures in each individual, suggesting that all triple-knockout mice are seizure prone, but that epileptic attacks occur with variable frequency and severity in different individuals. Epileptic activities consisted in inter-ictal spikes, sharp waves, high-voltage spike-and-wave discharges typical for absence-like epilepsies. These activities were also observed in the EEG of Tef single-knockout and Tef heterozygous, double-knockout mice but never led to generalized tonic–clonic seizures as those seen in homozygous triple-knockout mice. The seizure occurrence followed a circadian trend closely paralleled by the distribution of sleep in mice (Franken et al. 1998). Seizures occur predominantly or exclusively during sleep in several epilepsy syndromes, including frontal lobe epilepsy, benign Rolandic epilepsy of childhood, some forms of temporal lobe epilepsy, and a variety of extratemporal lobe epilepsy syndromes (Mendez and Radtke 2001). Whereas waking and rapid-eye movement sleep are characterized by desynchronized cortical activity, slow-wave sleep represents a physiological neuronal synchronization characterized by coordinated synaptic activity similar to that necessary for seizure occurrence (Steriade et al. 1994). However, both in PAR bZip triple-knockout mice and humans (Minecan et al. 2002), the seizure frequency is highest during transitions from waking to sleep and not during sustained deep slow-wave sleep. This suggests a critical role of early neuronal recruitment at sleep onset in the initiation of seizures. We also showed that δ activity (slow, <4 Hz, EEG activity) is increased in the background EEG of all vigilance states of triple-knockout mice. Together these data suggest that a steady-state tendency for higher neuronal synchronization might constitute the basic susceptibility factor for both audiogenic and spontaneous seizures in our model.
Relevance to human epilepsies
Epilepsies are the most common neurological disorders, with a lifetime cumulative incidence of 3%. Up to 40% of childhood and 20% of adult epilepsies are presumed to be genetic and classified as idiopathic epilepsies. Mutations in >70 genes have been reported to cause epilepsies with the voltage- and ligand-gated channelopathies representing the large majority (Noebels 2003). Genetic animal models of epilepsies, although very numerous, rarely recapitulate all aspects of human counterparts. The data presented here indicate that homozygous triple-knockout mice develop spontaneous epilepsy within the first month after birth characterized by myoclonus, tonic–clonic, and possibly absence seizures, in addition to audiogenic seizure susceptibility. The pyridoxine dependent epilepsy is a very rare condition with similarities and dissimilarities to our homozygous triple-knockout mice. Dissimilarities include the presence of continuous rhythmic high voltage slow waves and a pattern of suppression-bursts in the EEG (Mikati et al. 1991). In addition, pyridoxine transport and metabolism appear normal and the unique gene localization (on chromosome 5q31) contains no obvious candidate gene (Cormier-Daire et al. 2000). Juvenile myoclonic epilepsy (JME) and Unverricht-Lundborg disease (ULD) also have several similarities to the condition described here in our mice. Whereas JME seems to be a complex multigeneic disorder, ULD is a recessive disorder caused by mutations in the cystatin B (Cstb) gene on chromosome 21 (Pennacchio et al. 1996). Interestingly, Cstb is tightly linked to Pdxk (the two genes are separated by <25 Kb) and because the most frequent mutation found in Cstb is a dodecamer expansion in its promoter region, probably resulting in Cstb down-regulation, changes in the expression of adjacent genes such as Pdxk may also contribute to the phenotype. Given the putative identification of Cstb as the relevant mutant gene in ULD and the close genetic linkage of Cstb and Pdxk in both men and mice, we also determined Cstb expression in our PAR bZip mutant mice. However, we found that Cstb mRNA levels are indistinguishable in Hlf/Dbp double- and Hlf/Dbp/Tef triple-knockout mice (see Supplementary Fig. 3), and hence consider it unlikely that Cstb is involved in the seizure phenotype of our triple-knockout mice.
In conclusion, we have shown that mice lacking all three PAR bZip proteins are highly seizure prone. Moreover, we have identified Pdxk as a target gene of these transcription factors whose down-regulation offers a plausible cause for the epilepsy phenotype in these animals. We are aware, however, that additional studies will be required to scrutinize the involvement of Pdxk in the epilepsy phenotype in an unequivocal fashion. In future experiments, we will thus examine whether additional Pdxk gene copies will alleviate the seizure proneness in transgenic PAR bZip triple-knockout mice.
Materials and methods
Animals
The mice were housed and their wheel-running activity monitored as described in Lopez-Molina et al. (1997).
Disruption of the Hlf and Tef alleles
The construction of Hlf and Tef targeting vectors displayed schematically in Figure 2B and C is described in detail in Supplemental Material. Electroporation of embryonic stem (ES) cells, selection of neomycin-resistant colonies, and injection of ES cells into blastocysts were accomplished as described previously (Preitner et al. 2002).
RNA isolation and analysis
For the quantification of RNA levels real-time TaqMan RT–PCR, 0.5 μg of whole cell RNA, isolated according to Schmidt and Schibler (1995), were reverse-transcribed using random hexamers and Superscript reverse transcriptase (Gibco). The cDNA equivalent to 20 ng of total RNA was PCR-amplified in an ABI PRISM 7700 detection system (PE-Applied Biosystems). The primers used for Gapdh, mPer2, Cry1, BMal1, and Rev-erbα have been previously described (Preitner et al. 2002). For the other transcripts the primers sequences are given in Supplemental Material. The relative level of each RNA was calculated on the basis of E-CT and normalized to the corresponding Gapdh RNA levels. To allow for a direct comparison of Dbp, Tef, and Hlf transcript levels, the values were corrected for the PCR efficiency according to Ramakers et al. (2003). These efficiencies were determined to be 0.822, 1.016, and 0.842 for Dbp mRNA, Tef mRNA, and Hlf mRNA, respectively. All TaqMan RT–PCR reactions were carried out in triplicate, and the average cycle threshold (CT) values were determined.
Affymetrix oligonucleotide microarray hybridization
Thirty double-knockout mice (15 males and 15 females) and 30 triple-knockout mice of approximately the same age (118 ± 12 and 114 ± 11 d, respectively) were killed at ZT15 and their brains were quickly removed, frozen in liquid nitrogen, and stored at -80°C. RNA pools for a given genotype were assembled by mixing equal amounts of RNA from 10 animals (5 males and 5 females). Fifteen micrograms of total RNA from each pool were used to synthesize biotinylated cRNA according to the Affymetrix protocol, and 15 μg of biotinylated cRNA were hybridized to mouse Affymetrix MG-U74Av2 chips, containing probe features for ∼12,000 transcripts. The chips were washed and scanned, and the fluorescence signals analyzed with Affymetrix software MAS 5.0 and DMT 3.0. The pairwise analyses of the DNA microarray data obtained with cRNA probes from three double-knockout RNA pools and three triple-knockout RNA pools resulted in nine comparison sets. Transcripts were considered as differentially expressed if their levels changed in the same direction in at least 6/9 comparisons, if the average median change was at least 1.5-fold, and if the median fold change p-value was ≤0.0005. Results represent the mean ± S.D. of the fold change in the nine comparisons.
In situ hybridization
In situ hybridizations with coronal cryosections of 12 μm were performed as described previously (Lopez-Molina et al. 1997). The Pdxk riboprobe, covering the entire coding sequence, was synthesized from the plasmid pKS + Bluescript–Pdxk. The cDNA insert of this plasmid was generated by RT–PCR from mouse brain RNA using the following primers: 5′-CGCGGATCCATGGAGGGCGAATGCCGGG-3′ and 5′-TCCCCCGGGTCACAGCACTGTGGCCTGC-3′.
Rapid amplification of cDNA ends (RACE)
RACE experiments were carried out as described (Preitner et al. 2002). For Tef (see Supplementary Fig. 1) and Pdxk RACE experiments, polyA+ RNA from triple-knockout and wild-type mouse liver were used respectively. The Tef specific primers were 5′-CTGCAAACCTGTGCTTCC-3′ and 5′-GGGTCGATGGGACTTGG-3′ for the first and second PCR step, respectively. The Pdxk specific primers were 5′-GTACAGCTCATGAAGTTCC-3′ and 5′-GAGTTCACGGCATCAACC-3′ for the first and second PCR step, respectively.
Electromobility shift assay
The radio-labeled double-stranded oligonucleotide probes were prepared by annealing two oligonucleotides (encompassing the intronic PARRE of Pdxk) and by filling in the 5′ overhang with α[32P]dCTP and Klenow DNA polymerase. The sequences of these oligonucleotides were 5′-CTCCACTGCTTATGTAAC-3′ and 5′-CGGCCCGGTTACATAAGCAGTGGAG-3′. Fifteen micrograms of protein extracted from highly purified liver or brain nuclei by the NaCl–Urea–NP40 (NUN) procedure were incubated with 0.1 pmol of double-stranded oligonucleotides in a 20 μL reaction containing 25 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 7.5% glycerol, 1 mM DTT, and 1.5 μg/μL salmon sperm DNA. After an incubation period of 10 min at room temperature, 2 μL of a 15% Ficoll solution were added, and the protein–DNA complexes were separated on a 5% polyacrylamide gel in 0.25× TBE.
EEG recordings
The methods used for EEG recording and analysis in mice have been described in detail elsewhere (Franken et al. 1998; Franken et al. 1999). All mice were adult males (10–12 wk old) individually housed in an experimental room under a 12-h light/dark cycle (lights on at 8:00). Food and water were available ad libitum. EEG and EMG electrodes were implanted under deep pentobarbital anesthesia. We allowed mice 10–14 d of recovery from surgery and habituation before EEG and EMG recording. EEG and EMG signals were monitored continuously for at least 24 h. Both signals were amplified, filtered and converted from analog to digital. EEG signals were subjected to fast-Fourier transform analysis, yielding power spectra between 0 and 25 Hz (0.25-Hz resolution) using a 4-sec window. We classified the behavior in each of these 4-sec epochs as slow-wave sleep (SWS), paradoxical sleep (PS), or wakefulness (W) by visual inspection of the EEG and EMG signals. Mean EEG spectra were obtained by averaging the spectra of all 4-sec epochs scored as either SWS, PS, or W during a 12-h daytime recording. The experimental protocols were approved by the local veterinary office (Office Vétérinaire Cantonal de Genève) and the ethics committee of the Geneva School of Medicine.
Quantification of PLP, monoamines, and amino acids
The concentrations of pyridoxal 5′-phosphate (PLP), serotonin (5-HT), dopamine (DA), γ-aminobutyric acid (GABA), glutamic acid (Glu), and histamine were measured in perchloric acid (PCA) extracts of total brain by the chromatographic procedures described in Supplemental Material.
Acknowledgments
We are grateful to Patrick Descombes, Mylène Docquier, and Olivier Schaad (Genomics facility of the NCCR program “Frontiers in Genetics”) for their invaluable help in performing the Affymetrix microarray hybridization experiments, and Drs. Naomi Sakai and Stephane Matile (Organic Chemistry Depart-ment) for their expert advice on high performance liquid chromatography experiments and for making their HPLC equipment available to us. We thank Nicolas Roggli for the artwork and Steven Brown for critical comments on the manuscript. This research was supported by the Swiss National Science Foundation (grants to U.S., D.D., and M.T.), the State of Geneva, the NCCR program “Frontiers in Genetics”, the Bonnizzi Theler Stiftung, and the Louis Jeantet Foundation of Medicine. F.G. received a postdoctoral fellowship from the Fondation pour la Recherche Médicale (France).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.301404.
References
- Babity J.M., Armstrong, J.N., Plumier, J.C., Currie, R.W., and Robertson, H.A. 1997. A novel seizure-induced synaptotagmin gene identified by differential display. Proc. Natl. Acad. Sci. 94: 2638-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli G., Liu, H., and Martin, D.L. 2003. Kinetic differences between the isoforms of glutamate decarboxylase: Implications for the regulation of GABA synthesis. J. Neurochem. 86: 879-887. [DOI] [PubMed] [Google Scholar]
- Bell R.R. and Haskell, B.E. 1971. Metabolism of vitamin B 6 in the I-strain mouse. I. Absorption, excretion, and conversion of vitamin to enzyme co-factor. Arch. Biochem. Biophys. 147: 588-601. [DOI] [PubMed] [Google Scholar]
- Bell R.R., Blanchard, C.A., and Haskell, B.E. 1971. Metabolism of vitamin B6 in the I-strain mouse. II. Oxidation of pyridoxal. Arch. Biochem. Biophys. 147: 602-611. [DOI] [PubMed] [Google Scholar]
- Blaschko H. and Buffoni, F. 1965. Pyridoxal phosphate as a constituent of the histaminase (benzylamine oxidase) of pig plasma. Proc. R. Soc. Lond. B. Biol. Sci. 163: 45-60. [DOI] [PubMed] [Google Scholar]
- Bozzi Y., Vallone, D., and Borrelli, E. 2000. Neuroprotective role of dopamine against hippocampal cell death. J. Neurosci. 20: 8643-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T.J., Seeley, W.W., Kilgard, M., Schreiner, C.E., and Tecott, L.H. 1997. Sound-induced seizures in serotonin 5-HT2c receptor mutant mice. Nat. Genet. 16: 387-390. [DOI] [PubMed] [Google Scholar]
- Cormier-Daire V., Dagoneau, N., Nabbout, R., Burglen, L., Penet, C., Soufflet, C., Desguerre, I., Munnich, A., and Dulac, O. 2000. A gene for pyridoxine-dependent epilepsy maps to chromosome 5q31. Am. J. Hum. Genet. 67: 991-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran S.A., Buchsbaum, A.M., Reddy, K.L., Lin, M.C., Glossop, N.R., Hardin, P.E., Young, M.W., Storti, R.V., and Blau, J. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329-341. [DOI] [PubMed] [Google Scholar]
- Drolet D.W., Scully, K.M., Simmons, D.M., Wegner, M., Chu, K.T., Swanson, L.W., and Rosenfeld, M.G. 1991. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes & Dev. 5: 1739-1753. [DOI] [PubMed] [Google Scholar]
- Falvey E., Fleury-Olela. F., and Schibler. U. 1995. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 14: 4307-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Marcacci, L., and Schibler, U. 1996. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: A single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem. 377: 797-809. [PubMed] [Google Scholar]
- Fonjallaz P., Ossipow, V., Wanner, G., and Schibler U. 1996. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 15: 351-362. [PMC free article] [PubMed] [Google Scholar]
- Franken P., Malafosse, A., and Tafti, M. 1998. Genetic variation in EEG activity during sleep in inbred mice. Am. J. Physiol. 275: R1127-R1137. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Genetic determinants of sleep regulation in inbred mice. Sleep 22: 155-169. [PubMed] [Google Scholar]
- Franken P., Lopez-Molina, L., Marcacci, L., Schibler, U., and Tafti, M. 2000. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J. Neurosci. 20: 617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger S.P., Li, S., Fall, M.Z., Naumovski, L., and Cleary, M.L. 1996. The proto-oncogene HLF and the related basic leucine zipper protein TEF display highly similar DNA-binding and transcriptional regulatory properties. Blood 87: 4607-4617. [PubMed] [Google Scholar]
- Inaba T., Roberts, W.M., Shapiro, L.H., Jolly, K.W., Raimondi, S.C., Smith, S.D., and Look, A.T. 1992. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science 257: 531-534. [DOI] [PubMed] [Google Scholar]
- John R.A. 1995. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta. 1248: 81-96. [DOI] [PubMed] [Google Scholar]
- Kash S.F., Johnson, R.S., Tecott, L.H., Noebels, J.L., Mayfield, R.D., Hanahan, D., and Baekkeskov, S. 1997. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. 94: 14060-14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Noshiro, M., Sato, F., Maemura, K., Takeda, N., Nagai, R., Iwata, T., Fujimoto, K., Furukawa, M., Miyazaki, K., et al. 2004. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem. Biophys. Res. Commun. 313: 117-124. [DOI] [PubMed] [Google Scholar]
- Kesner R.P. 1966. Subcortical mechanisms of audiogenic seizures. Exp. Neurol. 15: 192-205. [DOI] [PubMed] [Google Scholar]
- Kirby J., Menzies, F.M., Cookson, M.R., Bushby, K., and Shaw, P.J. 2002. Differential gene expression in a cell culture model of SOD1-related familial motor neurone disease. Hum. Mol. Genet. 11: 2061-2075. [DOI] [PubMed] [Google Scholar]
- Lason W. and Przewlocki R. 1994. Seizure-induced expression of G proteins in the rat hippocampus. Brain Res. Mol. Brain Res. 24: 65-69. [DOI] [PubMed] [Google Scholar]
- Lee N.S., Muhs, G., Wagner, G.C., Reynolds, R.D., and Fisher, H. 1988. Dietary pyridoxine interaction with tryptophan or histidine on brain serotonin and histamine metabolism. Pharmacol. Biochem. Behav. 29: 559-564. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Conquet, F., Dubois-Dauphin, M., and Schibler, U. 1997. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 16: 6762-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez M. and Radtke, R.A. 2001. Interactions between sleep and epilepsy. J. Clin. Neurophysiol. 18: 106-127. [DOI] [PubMed] [Google Scholar]
- Mikati M.A., Trevathan, E., Krishnamoorthy, K.S., and Lombroso, C.T. 1991. Pyridoxine-dependent epilepsy: EEG investigations and long-term follow-up. Electroencephalogr. Clin. Neurophysiol. 78: 215-221. [DOI] [PubMed] [Google Scholar]
- Minecan D., Natarajan, A., Marzec, M., and Malow, B. 2002. Relationship of epileptic seizures to sleep stage and sleep depth. Sleep 25: 899-904. [PubMed] [Google Scholar]
- Mitsui S., Yamaguchi, S., Matsuo, T., Ishida, Y., and Okamura, H. 2001. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes & Dev. 15: 995-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.R., Maire, P., and Schibler, U. 1990. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell 61: 279-291. [DOI] [PubMed] [Google Scholar]
- N'Gouemo P. and Faingold, C.L. 1999. The periaqueductal grey is a critical site in the neuronal network for audiogenic seizures: Modulation by GABA(A), NMDA and opioid receptors. Epilepsy Res. 35: 39-46. [DOI] [PubMed] [Google Scholar]
- Noebels J.L. 2003. The biology of epilepsy genes. Annu. Rev. Neurosci. 26: 599-625. [DOI] [PubMed] [Google Scholar]
- Ono S. and Hagen, P. 1959. Pyridoxal phosphate: A coenzyme for histidine decarboxylase. Nature (Suppl. 15) 184: 1143-1144. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Lehesjoki, A.E., Stone, N.E., Willour, V.L., Virtaneva, K., Miao, J., D'Amato, E., Ramirez, L., Faham, M., Koskiniemi, M., et al. 1996. Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1). Science 271: 1731-1734. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola, F., Luis Lopez, M., Zakany, J., Duboule, D., Albrecht, U., and Schibler, U. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251-260. [DOI] [PubMed] [Google Scholar]
- Rahman M.K., Nagatsu, T., Sakurai, T., Hori, S., Abe, M., and Matsuda, M. 1982. Effect of pyridoxal phosphate deficiency on aromatic L-amino acid decarboxylase activity with L-DOPA and L-5-hydroxytryptophan as substrates in rats. Jpn. J. Pharmacol. 32: 803-811. [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter, J.M., Deprez, R.H., and Moorman, A.F. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62-66. [DOI] [PubMed] [Google Scholar]
- Reppert S.M. and Weaver, D.R. 2002. Coordination of circadian timing in mammals. Nature 418: 935-941. [DOI] [PubMed] [Google Scholar]
- Ripperger J.A., Shearman, L.P., Reppert, S.M., and Schibler, U. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes & Dev. 14: 679-689. [PMC free article] [PubMed] [Google Scholar]
- Schlesinger K. and Lieff, B. 1975. Levels of pyridoxine and susceptibility to electroconvulsive and audiogenic seizures. Psychopharmacologia 42: 27-32. [DOI] [PubMed] [Google Scholar]
- Schmidt E.E. and Schibler, U. 1995. Cell size regulation, a mechanism that controls cellular RNA accumulation: Consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J. Cell Biol. 128: 467-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.K. and Dakshinamurti, K. 1992. Seizure activity in pyridoxine-deficient adult rats. Epilepsia 33: 235-247. [DOI] [PubMed] [Google Scholar]
- Sharma S.K., Bolster, B., and Dakshinamurti K. 1994. Picrotoxin and pentylene tetrazole induced seizure activity in pyridoxine-deficient rats. J. Neurol. Sci. 121: 1-9. [DOI] [PubMed] [Google Scholar]
- Slaght S.J., Paz, T., Mahon, S., Maurice, N., Charpier, S., and Deniau, J.M. 2002. Functional organization of the circuits connecting the cerebral cortex and the basal ganglia: Implications for the role of the basal ganglia in epilepsy. Epileptic Disord. (Suppl. 3) 4: S9-S22. [PubMed] [Google Scholar]
- Steriade M., Contreras, D., and Amzica, F. 1994. Synchronized sleep oscillations and their paroxysmal developments. Trends Neurosci. 17: 199-208. [DOI] [PubMed] [Google Scholar]
- Tecott L.H., Sun, L.M., Akana, S.F., Strack, A.M., Lowenstein, D.H., Dallman, M.F., and Julius, D. 1995. Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374: 542-546. [DOI] [PubMed] [Google Scholar]
- Tunnicliff G. and Ngo, T.T. 1998. Functional modification of proteins of the nervous system by pyridoxal 5′-phosphate. Cell Physiol. Biochem. 8: 117-129. [DOI] [PubMed] [Google Scholar]
- Waagepetersen H.S., Sonnewald, U., and Schousboe, A. 1999. The GABA paradox: Multiple roles as metabolite, neurotransmitter, and neurodifferentiative agent. J. Neurochem. 73: 1335-1342. [DOI] [PubMed] [Google Scholar]
- Waymire K.G., Mahuren, J.D., Jaje, J.M., Guilarte, T.R., Coburn, S.P., and MacGregor, G.R. 1995. Mice lacking tissue non-specific alkaline phosphatase die from seizures due to defective metabolism of vitamin B-6. Nat. Genet. 11: 45-51. [DOI] [PubMed] [Google Scholar]
- Willott J.F. and Lu, S.M. 1980. Midbrain pathways of audiogenic seizures in DBA/2 mice. Exp. Neurol. 70: 288-299. [DOI] [PubMed] [Google Scholar]
- Wuarin J. and Schibler, U. 1990. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 63: 1257-1266. [DOI] [PubMed] [Google Scholar]