Abstract
Tumor development is dependent upon the inactivation of two key tumor-suppressor networks, p16Ink4a–cycD/cdk4–pRB–E2F and p19Arf–mdm2–p53, that regulate cellular proliferation and the tumor surveillance response. These networks are known to intersect with one another, but the mechanisms are poorly understood. Here, we show that E2F directly participates in the transcriptional control of Arf in both normal and transformed cells. This occurs in a manner that is significantly different from the regulation of classic E2F-responsive targets. In wild-type mouse embryonic fibroblasts (MEFs), the Arf promoter is occupied by E2F3 and not other E2F family members. In quiescent cells, this role is largely fulfilled by E2F3b, an E2F3 isoform whose function was previously undetermined. E2f3 loss is sufficient to derepress Arf, triggering activation of p53 and expression of p21Cip1. Thus, E2F3 is a key repressor of the p19Arf–p53 pathway in normal cells. Consistent with this notion, Arf mutation suppresses the activation of p53 and p21Cip1 in E2f3-deficient MEFs. Arf loss also rescues the known cell cycle re-entry defect of E2f3-/- cells, and this correlates with restoration of appropriate activation of classic E2F-responsive genes. Our data also demonstrate a direct role for E2F in the oncogenic activation of Arf. Specifically, we observe recruitment of the endogenous activating E2Fs, E2F1, and E2F3a, to the Arf promoter. Thus, distinct E2F complexes directly contribute to the normal repression and oncogenic activation of Arf. We propose that monitoring of E2F levels and/or activity is a key component of Arf's ability to respond to inappropriate, but not normal cellular proliferation.
Keywords: Tumor surveillance, p19Arf–p53, oncogenic activation, proliferation, E2F3
The development of mammalian tumors is dependent upon the disruption of two key biological activities, the control of cellular proliferation and the apoptotic response (Hanahan and Weinberg 2000). Remarkably, the Ink4a/Arf locus encodes two distinct tumor-suppressor proteins, p16Ink4a and p19Arf (p14Arf in humans), that influence one or both of these processes (Chin et al. 1998; Sherr 2001). p16Ink4a is a core component of the cell cycle control machinery (Sherr and Roberts 1999). It controls the activity of the G1 kinase, cyclinD · cdk4/6, and consequently, the phosphorylation status of the pocket protein family. This family includes the retinoblastoma protein (pRB) tumor suppressor and its relatives, p107 and p130. In the unphosphorylated state, the pocket proteins bind to the E2F family of transcription factors and prevent the expression of genes that are essential for entry into, and passage through the cell cycle (Trimarchi and Lees 2002). This inhibition occurs through two distinct mechanisms. pRB binds to the activating E2Fs, E2F1, E2F2, and E2F3a, and blocks their transcriptional activity. At the same time, the repressive E2Fs, E2F4, and E2F5 recruit p107 or p130 and their associated histone deactylases to E2F-responsive promoters. Under these conditions, the cell is blocked in G0/G1. Mitogenic signaling activates cell cycle re-entry by allowing cyclinD · cdk4/6 to overcome the repression by p16Ink4a. The consequent phosphorylation of the pocket proteins causes them to dissociate from E2F, enabling activation of E2F-responsive genes. In normal cells, the p16Ink4a–cyclinD · cdk4/6–pRB–E2F pathway responds to both positive and negative growth regulatory signals to determine whether or not a cell will divide (Sherr and Roberts 1999). This pathway is disrupted in most, if not all, mammalian tumors through loss of p16Ink4a, up-regulation of cyclinD · cdk4/6 or loss of pRB (Sherr 1996). The resulting deregulated proliferation is due, at least in part, to the inappropriate activation of E2F (Pan et al. 1998; Tsai et al. 1998; Yamasaki et al. 1998; McCaffrey et al. 1999; Ziebold et al. 2001, 2003).
The second product of the Ink4a/Arf locus, p19Arf,isa key component of the p53 tumor-surveillance network (Sherr 2001). p19Arf exists at low or undetectable levels in most normal cell and tissue types (Zindy et al. 2003). However, its expression is specifically activated by abnormal proliferative signals. These include the continued in vitro culturing of mouse embryonic fibroblasts (MEFs; Kamijo et al. 1997) and the inappropriate expression of proliferative oncogenes including activated ras, c-myc, E2F, E1A, and v-Abl (Serrano et al. 1997; de Stanchina et al. 1998; Palmero et al. 1998; Radfar et al. 1998; Zindy et al. 1998; Dimri et al. 2000). Once it is expressed, p19Arf inhibits the p53 ubiquitin ligase, mdm2, allowing activation of the p53 tumor suppressor (Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998; Honda and Yasuda 1999; Weber et al. 1999; Llanos et al. 2001). Depending on the cellular context, p53 triggers either cell cycle arrest (via induction of the cdk inhibitor, p21Cip1) or apoptosis (through activation of various apoptosis inducers). In either case, this counteracts the effect of the abnormal proliferative signals. Essentially, p19Arf acts as a defense to oncogenic signals. The recent analysis of a mouse strain that expresses GFP in place of p19Arf confirms that Arf is induced by the oncogenic signals present in incipient tumors (Zindy et al. 2003). This explains why inactivation of the p19Arf–p53 network is essential for the survival and proliferation of tumor cells in vivo (Sherr 2001).
The ability of Arf to specifically respond to inappropriate, but not normal proliferative signals must require a careful balance of transcriptional signals. Understanding how this is achieved remains a major challenge. Numerous studies have implicated E2F in this process (Phillips and Vousden 2001). The Arf promoter contains consensus E2F-binding sites and the overexpression of E2F1 is sufficient to trigger its transcriptional activation (DeGregori et al. 1997; Bates et al. 1998). However, it is unclear whether this regulation is direct because the identified E2F sites are not required for E2F-dependent activation (Parisi et al. 2002; Berkovich et al. 2003). There is also considerable debate as to which E2F family members might activate Arf (Trimarchi and Lees 2002). Some groups conclude that this is an E2F1-specific activity, whereas others propose that this is a shared property of the activating E2Fs. Certainly, E2F1 is not required for Arf induction in numerous settings (Palmero et al. 2002; Baudino et al. 2003) and p19Arf itself is dispensable for E2F-dependent apoptosis (Russell et al. 2002; Tolbert et al. 2002; Tsai et al. 2002). These findings could reflect redundancy; perhaps multiple E2Fs can activate a large panel of apoptotic inducers that includes p19Arf. Alternatively, E2F may not contribute to Arf activation in vivo. Others have suggested that Arf is regulated by repressive E2F · pocket protein complexes (Rowland et al. 2002). However, unlike classic E2F-responsive genes, Arf is not appreciably induced during cell cycle entry. Thus, if Arf is a genuine E2F target, it must be regulated in a distinct manner from classic E2F-responsive genes. In this study, we use E2f3-deficient MEFs to probe the role of E2F in Arf regulation. This analysis shows that a single member of the E2F family, E2f3, is required to maintain the transcriptional repression of Arf under normal proliferative conditions.
Results
E2F3 loss causes induction of p19Arf and activation of p53 in primary cells
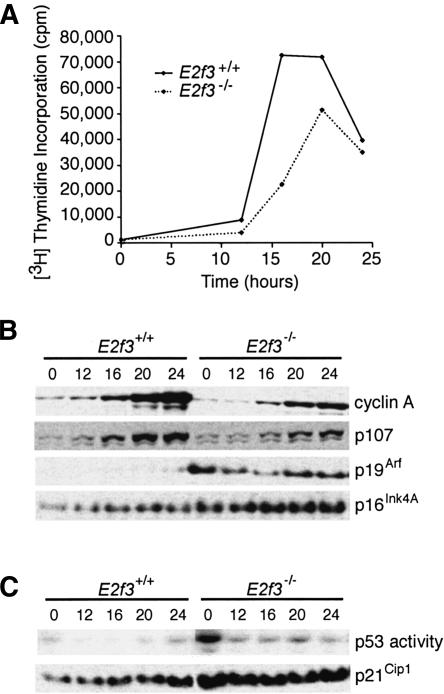
Two E2F family members, E2F3a and E2F3b, are encoded by a single locus through the use of different promoters and 5′-coding exons (Leone et al. 2000). These proteins share domains required for DNA binding, heterodimerization, and pocket protein binding, but have distinct N termini comprising either 122 (E2F3a) or 6 (E2F3b) amino acids. We have previously generated an E2f3 mutant mouse strain that inactivates both of these proteins. For simplicity, we refer to these mice as E2f3-/- or E2F3 deficient. MEFs derived from E2f3-/- animals typically have a reduced rate of proliferation (Humbert et al. 2000). They also display a major defect in mitogen-induced cell cycle re-entry and a corresponding impairment in the activation of all known E2F-responsive genes examined (Humbert et al. 2000; Fig. 1A,B). Given these observations, we hypothesized that p19Arf expression might be altered in E2F3-deficient MEFs if it is a bona fide E2F-target gene. To test this notion, we compared the levels of p19Arf in early passage wild-type versus E2f3-/- MEFs during cell cycle re-entry (Fig. 1). Consistent with previous studies (Sherr and DePinho 2000), p19Arf was barely detectable in the wild-type cells, and its expression did not vary significantly during the cell cycle (Fig. 1B). Strikingly, whereas the expression of the classic E2F-responsive targets, cyclin A and p107, was lower in the E2f3-/- MEFs than the wild-type controls, that of p19Arf was greatly increased (Fig. 1B). Thus, E2F3 loss affects p19Arf expression, but in an entirely distinct manner from other E2F-responsive genes.
Figure 1.
E2f3-/- MEFs have increased levels of p19Arf, leading to activation of p53. (A) Wild-type MEFs (solid line) and E2f3-/- MEFs (dotted line) were synchronized by serum starvation and cell cycle re-entry was monitored by [3H]thymidine incorporation. (B,C) Total protein extracts were prepared at the indicated times after stimulation with 10% serum and subjected to Western blotting for cyclin A, p107, p19Arf, p16Ink4a,orp21Cip1 (B,C), or electrophoretic mobility shift assay for active p53 (C).
The predominant function of p19Arf is to activate p53 by inhibiting its negative regulator, the ubiquitin ligase mdm2. We therefore used electrophoretic mobility shift assays (EMSA) to determine whether the induction of p19Arf in the E2f3-/- MEFs affected p53 activity (Figs. 1C, 2B). In wild-type MEFs, p53 DNA-binding activity remained low throughout the cell cycle. p53 activity was elevated in E2F3-deficient MEFs, especially during quiescence. The p53 protein also showed characteristic hall-marks of activation; there was an increase in the levels of p53 that was phosphorylated on Ser 15 and a subtle increase in the total p53 protein levels (data not shown). Thus, deregulation of p19Arf in E2f3-/- MEFs is accompanied by the activation of p53. A known downstream target of p53, the cdk inhibitor p21Cip1, was also expressed at higher levels in the E2F3-deficient MEFs (Fig. 1C), as was p16Ink4a, the other protein expressed from the Ink4a/Arf locus (Fig. 1B). It seemed likely that the elevated levels of p19Arf, p53, and p21Cip1 might contribute to the defective cell cycle entry in the E2F3-deficient MEFs.
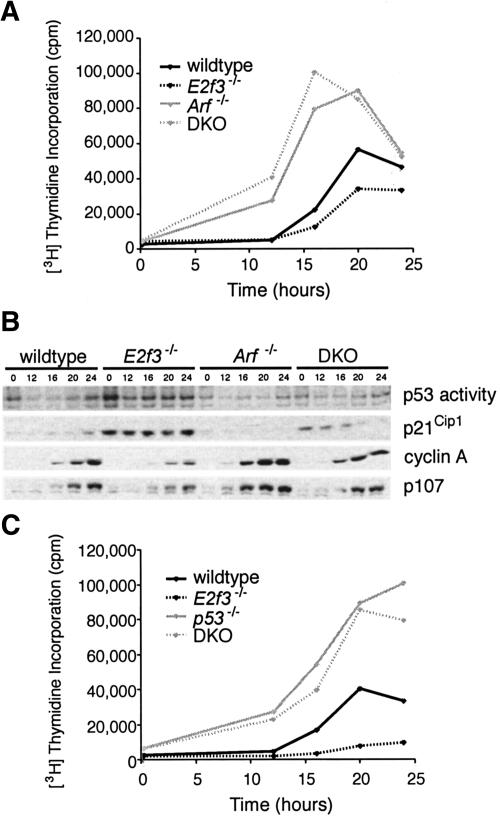
Figure 2.
Elimination of p19Arf rescues cell cycle re-entry and p53 activation defects of E2f3-deficient MEFs. (A) Wild-type MEFs (solid black line), E2f3-/- MEFs (dotted black line), Arf-/- MEFs (solid gray line), and E2f3-/-;Arf-/- DKO MEFs (dotted gray line) were synchronized by serum starvation and cell cycle re-entry was monitored by [3H]thymidine incorporation. (B) Total protein extracts were prepared at the indicated times after serum stimulation and subjected to Western blotting for cyclin A, p107, and p21Cip1, or electrophoretic mobility shift assay for active p53. (C) Wild-type MEFs (solid black line), E2f3-/- MEFs (dotted black line), p53-/- MEFs (solid gray line), and E2f3-/-;p53-/- DKO MEFs (dashed gray line) were analyzed exactly as described in A.
The up-regulation of p19Arf accounts for the cell cycle re-entry defect of the E2f3-/- MEFs
To address the biological consequences of p19Arf upregulation, we intercrossed E2f3 and Arf mutant mice to generate E2f3-/-;Arf-/- double-mutant (DKO) MEFs. We then examined the ability of these cells to re-enter the cell cycle relative to wild-type, E2f3-/-, and Arf-/- controls (Fig. 2A). Consistent with previous reports (Kamijo et al. 1997), the Arf-/- MEFs initiated DNA synthesis more rapidly than the wild-type counterparts. Significantly, the cell cycle kinetics of the E2f3-/-;Arf-/- DKO MEFs were indistinguishable from those of the Arf-/- MEFs. Consistent with this rescue, Arf mutation also suppressed the defective mitogen-induced activation of classic E2F-responsive targets, such as cyclin A and p107, which exists in the E2f3-/- cells (Fig. 2B). Thus, p19Arf loss is sufficient to over-ride the cell cycle reentry defect of the E2f3-/- MEFs. EMSA and Western blotting experiments showed that Arf mutation also suppressed the activation of p53 and the induction of p21Cip1 in the E2f3-/- MEFs (Fig. 2B), confirming that these events are dependent upon the up-regulation of p19Arf. Importantly, p53 mutation suppressed the cell cycle re-entry defect of the E2f3-/- MEFs in an analogous manner to Arf mutation (Fig. 2C). This correlated with suppression of the p21Cip1 induction and restoration of appropriate E2F-responsive gene activation (data not shown). Taken together, our findings indicate that E2f3 is required to inhibit Arf expression, and this plays an important role in regulating normal cell cycle re-entry by preventing inappropriate activation of p53.
E2F3 directly contributes to the transcriptional regulation of Arf in normal cells
Although numerous studies implicate E2F in Arf control, it has not been shown that any E2F family member is a direct transcriptional regulator of this gene. To address this question, we first asked whether E2F3 loss affects Arf at the transcriptional level. Using quantitative RT–PCR, we examined the mRNA levels of both Arf and a well-characterized E2F-responsive gene, p107, in wild-type versus E2F3-deficient MEFs (Fig. 3A). As in our previous studies (Humbert et al. 2000), the mitogen-induced activation of the p107 transcript was greatly impaired in the E2f3-/- MEFs. In contrast, Arf mRNA levels were significantly higher in E2f3-/- MEFs than the wild-type controls (Fig. 3A). This up-regulation was most striking in the G0 time point (Fig. 3A), where we observed the greatest increase in the levels of the p19Arf protein (Fig. 1B). Thus, E2F3 loss is acting, at least in part, through changes in the levels of Arf mRNA. To determine whether E2F3 is directly involved in Arf regulation, we used chromatin immunopreciptiation (ChIP) to examine binding of E2F family members to the Arf and p107 promoters (Fig. 3B). In asynchronously growing wild-type MEFs, we detected no significant enrichment of any E2F at a control sequence lacking E2F sites, 1 kb upstream of the E2F1 promoter. In contrast, several E2F family members could be detected at the p107 promoter including E2F1, E2F2, E2F3, and E2F4 (Fig. 3B; data not shown). This is entirely consistent with the previous characterization of archetypal mammalian E2F-responsive genes (Takahashi et al. 2000). Remarkably, the spectrum of E2F proteins at the Arf promoter differed completely from this norm; we observed only a single E2F, E2F3 (Fig. 3B). This specificity was not restricted to murine cells, as E2F3 was also the sole E2F bound to the p14Arf promoter in both primary (WI-38) and transformed (T98G, BJ-T) human cell lines (data not shown). In E2f3-/- MEFs, there was no longer any enrichment of the Arf or p107 promoter sequences by the anti-E2F3 antibodies, confirming their specificity (Fig. 3B). Strikingly, E2F3 deficiency caused a slight increase in the level of E2F1 associated with the p107 promoter, but there was no evidence of E2F1, or any other E2F family member binding to the Arf promoter in these cells (Fig. 3B). Taken together, these data show that E2F3 is directly involved in the regulation of Arf, and that loss of E2F3, without apparent recruitment of E2F1, is sufficient to allow Arf activation. On the basis of these findings, we conclude that E2F3 represses Arf expression in normal cells.
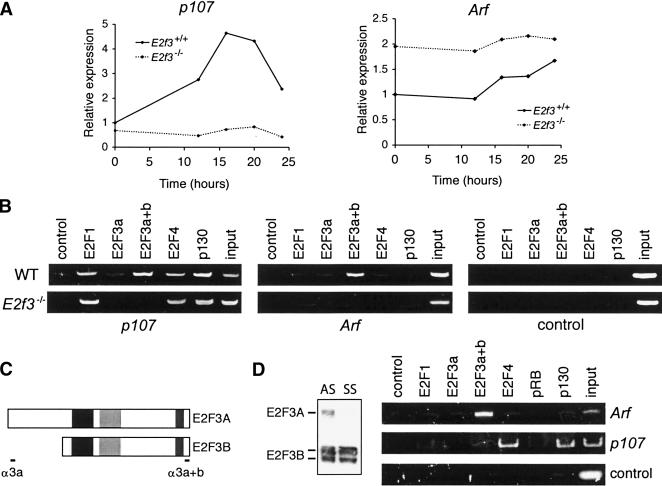
Figure 3.
E2F3 is a direct repressor of Arf. (A) Wild-type (solid lines) or E2f3-/- (dotted lines) MEFs were synchronized by serum starvation, and RNA was extracted at the indicated times after stimulation. Quantitative real-time RT–PCR analysis of p107 or Arf mRNA is shown. (B) ChIP analysis of asynchronous wild-type (WT) or E2f3-/- MEFs. Sonicated, cross-linked chromatin was immunoprecipitated with the indicated antibodies, and the purified DNA was analyzed by PCR with primers specific for the p107 or Arf promoters, or a control sequence lacking E2F sites (1 kb upstream of the E2f1 promoter). Input, 0.5% of chromatin in IP reactions was analyzed by PCR. (C) Schematic of the E2F3a and E2F3b proteins, and the antibodies used in ChIP analysis. (Black box) DNA-binding domain; (light gray box) dimerization domain; (dark gray box) transactivation domain. (D) Cell lysates of asynchronous (AS) wild-type MEFs or wild-type MEFs incubated in 0.1% serum for 3 d (SS) were subjected to Western blotting for the two E2F3 isoforms. ChIP assays for the indicated E2F and pocket protein family members were performed on the serum-starved wild-type MEFs.
It is well established that the E2F proteins can contribute to the repression of E2F-responsive genes through recruitment of the pocket proteins, pRB, p107, and p130, and their associated histone deacetylases (Trimarchi and Lees 2002). The E2F3 proteins have been reported to bind to pRB, but not p107 or p130, in vivo (Moberg et al. 1996; He et al. 2000; Leone et al. 2000). Given this finding, we used ChIP assays to determine whether one or more pocket proteins were recruited to the Arf promoter along with E2F3 (Fig. 3B). Consistent with previous studies, p130, but not pRB, was readily detected at the p107 promoter. In contrast, none of the pocket proteins were detected at the Arf promoter (Fig. 3B,D; data not shown). It is entirely possible that pRB is present, but cannot be detected due to limitations in the assay, the antibodies or the structure of the pRB/E2F3 repressive complex. However, we consistently detect pRB at differentiation-associated promoters using the same ChIP conditions (T. Yuan and J.A. Lees, unpubl.). This raises the possibility that E2F3 mediates Arf repression in a pocket protein-independent manner.
The E2F3 locus encodes two proteins, E2F3a and E2F3b, which differ only in their N-terminal sequences (Leone et al. 2000; Fig. 3C). E2F3a has been linked to the transcriptional activation of E2F-responsive genes, but this does not rule out a role in repression. The transcriptional properties of E2F3b have not been established. To discern which species of E2F3 is responsible for regulation of Arf, we made use of two antibodies raised against the E2F3 proteins. The sc-879x antibody recognizes an epitope on the unique N terminus of E2F3a, and therefore, does not cross-react with E2F3b. In contrast, a C-terminal antibody (sc-878x) recognizes both E2F3a and E2F3b. For clarity, we will refer to these antibodies as E2F3a specific or anti-E2F3a+b. By detecting ectopically expressed E2F3a and E2F3b proteins, we have confirmed the specificity of these two reagents (data not shown). These experiments also showed that the E2F3a-specific antibody has a considerably lower activity in ChIP assays than anti-E2F3a+b (data not shown). Despite its low avidity, the E2F3a-specific antibody yielded a detectable ChIP signal at the p107 promoter (Fig. 3B). Under the same conditions, we did not see any evidence of E2F3a binding to Arf (Fig. 3B); however, because of the poor avidity of the E2F3a-specific antibody, we cannot rule out that E2F3a contributes to the repression of Arf, yet falls beneath the detection limit of this reagent. To address this in an alternative way, we took advantage of the differential expression patterns of the two E2F3 species. E2F3a expression is cell cycle regulated, peaking during the G1/S transition, whereas E2F3b expression is constant throughout the cell cycle. Consequently, E2F3b is the only E2F3 isoform expressed during quiescence (Fig. 3D). Therefore, we performed ChIP from MEFs that had been arrested in G0/G1 by serum deprivation (Fig. 3D). In this setting, the p107 promoter was specifically occupied by E2F4 and p130, the key components of the archetypal repressive E2F/pocket protein complex. At the same time, we still specifically detected E2F3 at the Arf promoter using the anti-E2F3a+b antibody. This analysis strongly implicates E2F3b in the repression of Arf in quiescent cells, yet does not exclude contribution of E2F3a, along with E2F3b, in asynchronous cells.
The activating E2Fs are directly involved in activation of Arf in response to oncogenic stress
It is well documented that Arf is a key regulator of the mammalian tumor-surveillance-response network. Through an as yet undetermined mechanism, the inappropriate expression of numerous oncogenes induces the expression of p19Arf. The resulting p53 activation triggers either cell cycle arrest (via p21Cip1 induction) or apoptosis (through activation of pro-apoptotic genes), thereby circumventing the oncogenes' ability to drive inappropriate proliferation. Having established a direct role for E2F3 in repression of Arf in unstressed cells, we sought to determine what role E2F might play during the activation of Arf by oncogenic challenge.
E2F1 is a potent oncogene. Its over-expression activates both cellular proliferation and also the induction of p19Arf and high levels of apoptosis (DeGregori et al. 1997; Bates et al. 1998). This latter response greatly impedes investigation of the underlying molecular mechanisms. To overcome this problem, we have used MEFs in which exons 2 and 3 of the Ink4a/Arf locus are deleted (Ink4a/Arf-/- MEFs) and therefore p16Ink4a and p19Arf expression is disrupted (Serrano et al. 1996; Fig. 4A). This mutation does not affect the upstream regulatory region of Arf, which is ∼13 kb upstream of the deletion, allowing us to examine E2F binding at the Arf promoter in the absence of apoptosis or other secondary events that might result from p19Arf induction. We infected these Ink4a/Arf-/- MEFs with either control or E2F1-expressing retroviruses and used ChIP to assess E2F binding to either the Arf or p107 promoters (Fig. 4B). When Ink4a/Arf-/- MEFs were infected with a control retrovirus, the spectrum of E2F complexes detected at the Arf and p107 promoters was similar to that observed in wild-type, uninfected cells. Overexpression of E2F1 increased the level of E2F1 associated with the p107 promoter, and caused a coordinate decrease in levels of bound E2F3 and p130 (Fig. 4B). This is consistent with the E2F1's known ability to promote cell cycle progression and activate expression of classic E2F-responsive genes (Trimarchi and Lees 2002). Strikingly, we also detected significant levels of E2F1 at the Arf promoter, showing that E2F1 can directly contribute to the transcriptional activation of Arf when it is inappropriately expressed (Fig. 4B).
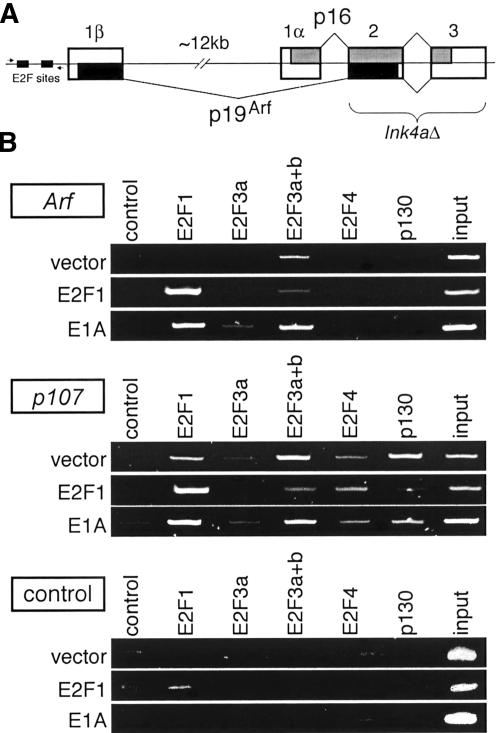
Figure 4.
The activating E2Fs bind to the Arf promoter during oncogenic challenge. (A) Schematic of the mouse Ink4a/Arf locus, indicating the exon structure, and the region deleted in the Ink4a/Arf-/- MEFs (bracket). Shaded regions in the exons indicate regions coding for the p16Ink4a (light gray) and p19Arf (black) proteins. The small black boxes indicate two consensus E2F-binding sites in the Arf promoter, and the two small arrows indicate the location of the primers used in ChIP analysis. (B) Ink4a/Arf-/- MEFs were infected with retrovirus overexpressing E2F1, E1A, or an empty virus (vector) and subjected to ChIP assays with the indicated antisera.
We wished to determine whether the endogenous E2F1 participates in the induction of Arf by other oncogenes. To address this question, we examined the effect of overexpressing the adenoviral oncoprotein, E1A (Fig. 4B). This promotes proliferation and tumorigenesis by sequestering the pocket proteins and relieving the transcriptional inhibition of their associated E2Fs (Ben-Israel and Kleinberger 2002). In Ink4a/Arf-/- MEFs expressing E1A, we observed a decrease in p130 binding to the p107 promoter, and an increase in E2F1 binding, as expected (Fig. 4B). Notably, we also detected E2F1 binding to the Arf promoter, showing that the endogenous E2F1 protein contributes to the activation of Arf arising from E1A expression. In addition, we detected a weak signal with the E2F3a-specific antibody, indicating that E2F3a was cooperating with E2F1 in the transcriptional regulation of Arf. Given these findings, we conclude that the endogenous activating E2Fs play a direct role in the oncogene-induced activation of Arf and the tumor-surveillance network.
Discussion
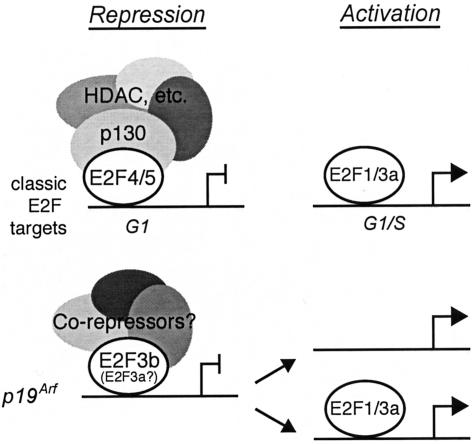
Taken together, our data show that the E2F proteins play a direct role in the transcriptional regulation of Arf. This firmly establishes Arf as a bona fide E2F-responsive gene. However, the nature of this regulation diverges considerably from that of archetypal E2F-responsive targets; for example, genes encoding key components of the cell cycle control and DNA replication machinery (Fig. 5). These classic E2F-responsive genes are expressed in a cell cycle-dependent manner that is orchestrated by the specific binding of the repressive E2F/pocket protein complexes (predominantly E2F4/p130) during G0/G1 or the activating E2Fs (E2F1, E2F2, and E2F3a) during late G1 and G1/S phase. In contrast, our data show that Arf is regulated by a more restricted subset of E2F complexes, whose activity is somehow determined by the stress status, rather than the cell cycle staging of the cell.
Figure 5.
Arf is regulated by E2F in a distinct manner from classic E2F-responsive genes.
In normal cells, Arf is constitutively repressed, and our data show that this correlates with the promoter binding of E2F3, and not other E2F family members. Because the E2F3a-specific antibody works poorly in ChIP assay, our inability to detect a signal with this reagent does not exclude the possibility that E2F3a contributes to Arf regulation. In contrast, the presence of a robust anti-E2F3a+b ChIP signal in quiescent cells that express E2F3b and not E2F3a, clearly shows that E2F3b is involved. At least in MEFs, the absence of E2F3 is sufficient to trigger the expression of Arf, without obligatory recruitment of the activating E2Fs. Thus, E2F3b (possibly in parallel with E2F3a) is required for the constitutive repression of Arf in normal cells. On the basis of its high expression in G0/G1 cells, E2F3b had been proposed to function as a transcriptional repressor (Leone et al. 2000). Our data provide the first direct evidence for this hypothesis.
Remarkably, our analysis of the double-mutant MEFs shows that Arf loss completely suppresses the cell cycle entry defect of the E2f3-/- MEFs. This correlates with the loss of activation of the p53 pathway and also restoration of the normal cell cycle-dependent activation of classic E2F-responsive genes. This genetic rescue experiment is entirely consistent with the notion that E2F3 is acting to repress Arf, and therefore p53, in a linear pathway. However, it is also possible that the loss of Arf (or p53) confers a proliferative advantage on the cells that outweighs or overrides the proliferative disadvantage that results from E2f3 loss. In this scenario, Arf and E2f3 could be acting in parallel pathways or, alternatively, E2f3 could exert its effects through both Arf/p53-dependent and Arf/p53-independent mechanisms. We currently favor the second alternative. Taken together, the presence of E2F3 at the Arf promoter and the derepression of Arf in E2f3-/- MEFs strongly support the existence of an Arf-dependent mechanism. However, additional experiments indicate that the p19Arf–p53 pathway does not fully account for the phenotypic consequences of E2F3 loss. First, whereas loss of Arf or p53 completely rescues the cell cycle reentry defect of the E2f3-/- MEFs, it only partially suppresses their asynchronous proliferation defect (A. Aslanian and J.A. Lees, unpubl.). Second, loss of Arf or p53 has no detectable effect on the developmental phenotypes and the resulting embryonic/neonatal lethality of the E2f3-/- mice (A. Aslanian and J.A. Lees, unpubl.). Thus, E2f3 has at least one essential function that is Arf/p53 independent. This second function remains to be established. We had previously concluded that the cell cycle re-entry defect of the E2f3-/- MEFs reflected a direct role for E2f3 in the activation of classic E2F-responsive genes. However, this current study shows that the defect in gene activation in the E2f3-/- MEFs is fully reversed by the loss of Arf or p53. On the basis of the same logic outlined above, these results can be explained in two ways. First, the defect in activation of classic E2F-responsive genes in the E2f3-/- cells could be an indirect consequence of activation of the p19Arf–p53 pathway. Alternatively, E2f3 may be required for the appropriate activation of classic E2F-responsive genes, but the consequent loss of this activation in E2F3-deficient cells is outweighed or overridden by the increased induction of these targets that results from the loss of Arf or p53. Given the extensive cross-talk between the pRB and p53 pathways, both models are highly plausible. More subtle experiments will be required to distinguish between these two possibilities.
Our analysis of the E2f3 mutant cells reinforces a growing body of evidence that the p19Arf–p53 network is the key determinant of the proliferation status of cultured primary fibroblasts (Sherr 2001). Several other transcriptional regulators, such as Bmi1, TBX2, TBX3, and Twist, have been linked to the repression of Arf and the inhibition of p53 signaling (Jacobs et al. 1999, 2000; Maestro et al. 1999; Brummelkamp et al. 2002; Lingbeek et al. 2002). There is limited insight into the mechanism(s) of action of these repressors. In vitro promoter mapping studies show that TBX2 and TBX3 can bind to a variant T-site located within the Initiator sequence of the human Arf promoter (Lingbeek et al. 2002). However, promoter binding has not been demonstrated in vivo. Similarly, antibodies against Bmi1 have not worked in ChIP assays. This is thought to be an issue of antibody accessibility, as Bmi1 is a component of the multiprotein polycomb complex (van Lohuizen 1998). However, it is unclear how the Bmi1–polycomb complex is recruited to Arf, with respect to either the target DNA sequence/chromatin structure or the identity of the component(s) that mediate the DNA/chromatin binding. Our identification of E2F3 as an additional repressor of Arf raises additional questions. First, what is the precise nature of the repressive E2F3 complex? Is it a unique function of E2F3b or does E2F3a contribute? Does it involve a higher order, antibody-inaccessible pRB complex, or does it function in a pocket protein-independent manner? Second, what directs E2F3, but not other E2F complexes, to bind specifically to the Arf promoter in normal cells? Finally, what is the relationship between E2F3 and the other known Arf repressors? Do they function independently of one another or work cooperatively to ensure the repression of Arf? Careful analysis of both the repressor complexes and the Arf promoter will be required to unravel this complexity.
Our data show that E2F is also involved in the activation of Arf in response to oncogenic signals. Although there is extensive literature suggesting a link between E2F and Arf (Trimarchi and Lees 2002), this is the first study to demonstrate that the action of these proteins is direct. During oncogenic activation, we see recruitment of the endogenous E2F1 and, to a lesser extent, the other activating E2Fs, E2F2 (P. Iaquinta and J.A. Lees, unpubl.), and E2F3a. It is important to note that we do not completely lose the anti-E2F3a+b signal at the Arf promoter under conditions of oncogenic stress (Fig. 4B). We believe that this partially reflects differences in the level of oncogenic activation within the population of infected cells; some have sufficient E2F activation to disrupt the E2F3b repression, whereas others do not. The continued presence of p130 at the p107 promoter in E1A-infected cells, strongly suggests that some cells express insufficient E1A to fully dissociate the pocket protein/E2F complexes. A second possibility is that the sustained anti-E2F3a+b signal at the induced Arf promoter reflects recruitment of transcriptionally active E2F3a. We suspect that this is the case, at least in the E1A-expressing cells, because we see a weak signal with the E2F3a-specific antibody and an enhanced signal with the anti-E2F3a+b antibody. Finally, as there are at least two E2F consensus-binding sites in the Arf promoter, we cannot rule out the possibility that E2F3b remains bound to the Arf promoter during the stress response, despite recruitment of E2F1 or other activating E2Fs. In this scenario, the activating E2Fs must somehow override or negate the repressive function of E2F3b to ensure Arf induction.
Importantly, this study provides considerable insight into the cross-talk between the two key tumor-suppressor networks, pRB–E2F and p19Arf–p53. It is already well established that p53's growth-suppressive properties are at least partially dependent upon the ability of its downstream target, p21Cip1, to inhibit phosphorylation of pRB and thereby promote its ability to repress E2F (Sherr 2001). In this study, we now show that E2F also plays a direct role in regulation of the p19Arf–p53 pathway. Consistent with previous hypotheses, the activating E2Fs directly contribute to the induction of Arf that occurs in response to oncogenic stress, frequently referred to as the tumor-surveillance response. In addition, our data show that E2F3 plays a key role in maintaining the p19Arf–p53 network in the repressed state, when there is no oncogenic stress signal. There has been considerable debate as to how a cell could know whether it initiates inappropriate, as opposed to appropriate, proliferation, and regulate Arf accordingly (Lowe and Sherr 2003). It is well known that the state of E2F complexes is a key determinant of whether normal cells will divide. Our current finding that various E2F complexes contribute to both the repressed and activated states of Arf regulation strongly suggests that monitoring of E2F levels and/or activity is likely to lie at the heart of this tumor-surveillance mechanism.
Materials and methods
Generation of mouse strains and MEF preparation
E2f3+/-;Arf+/- and E2f3+/-;p53+/- mice were generated by inter-crossing E2f3+/- mice (Humbert et al. 2000) with either Arf-/- mice (Kamijo et al. 1997) or p53+/- (Williams et al. 1994), respectively. Genotyping was performed as previously described. Double heterozygous mutant mice were intercrossed, and MEFs were prepared from e13.5 embryos as previously described (Humbert et al. 2000). Ink4a/Arf-/- mice (Serrano et al. 1996) were used to generate Ink4a/Arf-/- MEFs.
Serum starvation and release experiments
Passage 4 MEFs were plated in triplicate onto 3.5-cm dishes at 2 × 105 cells/dish and cell cycle re-entry was performed as previously described (Humbert et al. 2000). After 48 h, cells were washed twice with DME and then incubated in low serum (0.1% FCS) for 72 h. Cells were subsequently fed with medium containing 10% FCS. For each timepoint, cells were labeled with 5 μCi of [3H]thymidine for 1 h. Cells were harvested and [3H]thymidine incorporation was measured as previously described (Moberg et al. 1996).
RNA preparation and Quantitative Real-time PCR analysis
Passage 4 MEFs were plated onto 15-cm dishes at 3 × 106 cells/dish and cell cycle re-entry was performed as described above. For each time point, cells were harvested and total RNA was isolated using RNeasy (Qiagen), and an on-column DNAse step was performed according to the manufacturer's instructions. cDNAs were generated with Superscript First Strand Synthesis System (Invitrogen). Quantitative realtime PCR reactions using SYBR Green dye (Applied Biosystems) were run in triplicate on an ABI Prism 7000 Real-time PCR machine (Applied Biosystems). Control 18S rRNA reactions (Applied Biosystems) were also run to normalize ΔCt values. Fold change was calculated as 2^(ΔΔCt). Primer sequences for Arf were 5′-CACCGGAATCCTGGACCAG-3′ and 5′-GCAGTTCGAATCTGCACCGT-3′, and for p107 were 5′-GATGCTCATCTGACCGGAGT-3′ and 5′-ATAAGTCACGTAGGCGCACA-3′.
Protein preparation, Western blotting, and gel-retardation assays
Passage 4 MEFs were plated onto 15-cm dishes at 3 × 106 cells/dish and cell cycle re-entry was performed as described above. For each time point, cells were harvested and total protein was isolated as described previously (Moberg et al. 1996). Western blotting was performed with 100 μg of whole-cell extract using anti-cyclin A (Santa Cruz sc-596), anti-p107 (Santa Cruz sc318), anti-p19Arf (Novus NB200-106), anti-p16Ink4a (Santa Cruz sc-1207), and anti-p21Cip1 (Santa Cruz sc-6246). In Figure 3D, extracts were prepared as above from asynchronous MEFs, or MEFs incubated in 0.1% serum for 3 d. Western blotting was performed on 30 μg of extract using the anti-E2F3a+b antibody (sc-878, Santa Cruz). p53 gel-retardation assays were performed in the presence of supershifting antibody using NuShift (Geneka) according to the manufacturer's instructions.
Retroviral infection
Infections were performed exactly as described (Serrano et al. 1997), except that 15-cm plates were used at all steps, and the procedure was scaled up accordingly. Target Ink4a/Arf-/- MEFs wereselectedfor2din2mg/mLpuromycin, grown for a further 2 d, and then subjected to ChIP analysis as described below. pBabe-Puro, pBabe-Puro-E2F1 (Humbert et al. 2000), and LPC-12S-E1A have been described (de Stanchina et al. 1998).
ChIP
ChIP was performed essentially as described (Takahashi et al. 2000). Early-passage (passage 3–5) MEFs were used for all experiments. Sonicated, cross-linked chromatin corresponding to ∼3 × 106 cells was immunoprecipitated with the following antibodies: E2F1, sc-193; E2F3a, sc-879x; E2F3a+b, sc-878x; E2F4, sc-1082x; p130, sc-317x (all from Santa Cruz); pRB, MS-594 and MS-595 (Neomarkers); control (anti-Luciferase) 05-603 (Upstate Biotechnology). A total of 3%–4% of the precipitated DNA, or 0.5% input DNA, was amplified by 30 cycles of PCR using the following primer sequences: p107 (5′-TTAGAGTCCGAGGTCCATCTTCT-3′ and 5′-GGGCTCGTCCTCGAACATATCC-3′), Arf (5′-GCTGGCTGTCACCGCGAT-3′ and 5′-GCGTTGAGGCACCTCGAGA-3′), and E2f1-upstream (control, 5′-TGGAGGTCAAGTAGTGGCCCAAA-3′ and 5′-ACAATGTCTGGTTTGCTCCGCCC-3′). PCR products were resolved on 8% polyacrylamide gels and stained with ethidium bromide.
Acknowledgments
We thank Chuck Sherr, Tyler Jacks, and Ron DePinho for providing Arf, p53, and Ink4a/Arf mutant mice, and also for stimulating discussions of this project. We are also grateful to Phil Sharp, Steve Bell, and members of the Lees lab for helpful discussions during this study and the preparation of this manuscript. J.A.L. is a Daniel K. Ludwig Scholar. This work was supported by a grant from the NIH (PO1-CA42063) to J.A.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1196704.
References
- Bates S., Phillips, A.C., Clark, P.A., Stott, F., Peters, G., Ludwig, R.L., and Vousden, K.H. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395: 124-125. [DOI] [PubMed] [Google Scholar]
- Baudino T.A., Maclean, K.H., Brennan, J., Parganas, E., Yang, C., Aslanian, A., Lees, J.A., Sherr, C.J., Roussel, M.F., and Cleveland, J.L. 2003. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol. Cell 11: 905-914. [DOI] [PubMed] [Google Scholar]
- Ben-Israel H. and Kleinberger, T. 2002. Adenovirus and cell cycle control. Front. Biosci. 7: d1369-d1395. [DOI] [PubMed] [Google Scholar]
- Berkovich E., Lamed, Y., and Ginsberg, D. 2003. E2F and Ras synergize in transcriptionally activating p14ARF expression. Cell Cycle 2: 127-133. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Kortlever, R.M., Lingbeek, M., Trettel, F., MacDonald, M.E., van Lohuizen, M., and Bernards, R. 2002. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J. Biol. Chem. 277: 6567-6572. [DOI] [PubMed] [Google Scholar]
- Chin L., Pomerantz, J., and DePinho, R.A. 1998. The INK4a/ARF tumor suppressor: One gene—two products—two pathways. Trends Biochem. Sci. 23: 291-296. [DOI] [PubMed] [Google Scholar]
- de Stanchina E., McCurrach, M.E., Zindy, F., Shieh, S.Y., Ferbeyre, G., Samuelson, A.V., Prives, C., Roussel, M.F., Sherr, C.J., and Lowe, S.W. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes & Dev. 12: 2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Miron, A., Jakoi, L., and Nevins, J.R. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. 94: 7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Itahana, K., Acosta, M., and Campisi, J. 2000. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol. Cell. Biol. 20: 273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg, R.A. 2000. The hallmarks of cancer. Cell 100: 57-70. [DOI] [PubMed] [Google Scholar]
- He Y., Armanious, M.K., Thomas, M.J., and Cress, W.D. 2000. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene 19: 3422-3433. [DOI] [PubMed] [Google Scholar]
- Honda R. and Yasuda, H. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18: 22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert P.O., Verona, R., Trimarchi, J.M., Rogers, C., Dandapani, S., and Lees, J.A. 2000. E2f3 is critical for normal cellular proliferation. Genes & Dev. 14: 690-703. [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom, K., Marino, S., DePinho, R.A., and van Lohuizen, M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164-168. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Keblusek, P., Robanus-Maandag, E., Kristel, P., Lingbeek, M., Nederlof, P.M., van Welsem, T., van de Vijver, M.J., Koh, E.Y., Daley, G.Q., et al. 2000. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat. Genet. 26: 291-299. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy, F., Roussel, M.F., Quelle, D.E., Downing, J.R., Ashmun, R.A., Grosveld, G., and Sherr, C.J. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649-659. [DOI] [PubMed] [Google Scholar]
- Leone G., Nuckolls, F., Ishida, S., Adams, M., Sears, R., Jakoi, L., Miron, A., and Nevins, J.R. 2000. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 20: 3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingbeek M.E., Jacobs, J.J., and van Lohuizen, M. 2002. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J. Biol. Chem. 277: 26120-26127. [DOI] [PubMed] [Google Scholar]
- Llanos S., Clark, P.A., Rowe, J., and Peters, G. 2001. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 3: 445-452. [DOI] [PubMed] [Google Scholar]
- Lowe S.W. and Sherr, C.J. 2003. Tumor suppression by Ink4a–Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 13: 77-83. [DOI] [PubMed] [Google Scholar]
- Maestro R., Dei Tos, A.P., Hamamori, Y., Krasnokutsky, S., Sartorelli, V., Kedes, L., Doglioni, C., Beach, D.H., and Hannon, G.J. 1999. Twist is a potential oncogene that inhibits apoptosis. Genes & Dev. 13: 2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey J., Yamasaki, L., Dyson, N.J., Harlow, E., and Griep, A.E. 1999. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell. Biol. 19: 6458-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg K., Starz, M.A., and Lees, J.A. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16: 1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I., Pantoja, C., and Serrano, M. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395: 125-126. [DOI] [PubMed] [Google Scholar]
- Palmero I., Murga, M., Zubiaga, A., and Serrano, M. 2002. Activation of ARF by oncogenic stress in mouse fibroblasts is independent of E2F1 and E2F2. Oncogene 21: 2939-2947. [DOI] [PubMed] [Google Scholar]
- Pan H., Yin, C., Dyson, N.J., Harlow, E., Yamasaki, L., and Van Dyke, T. 1998. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol. Cell 2: 283-292. [DOI] [PubMed] [Google Scholar]
- Parisi T., Pollice, A., Di Cristofano, A., Calabro, V., and La Mantia, G. 2002. Transcriptional regulation of the human tumor suppressor p14(ARF) by E2F1, E2F2, E2F3, and Sp1like factors. Biochem. Biophys. Res. Commun. 291: 1138-1145. [DOI] [PubMed] [Google Scholar]
- Phillips A.C. and Vousden, K.H. 2001. E2F-1 induced apoptosis. Apoptosis 6: 173-182. [DOI] [PubMed] [Google Scholar]
- Pomerantz J., Schreiber-Agus, N., Liegeois, N.J., Silverman, A., Alland, L., Chin, L., Potes, J., Chen, K., Orlow, I., Lee, H.W., et al. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713-723. [DOI] [PubMed] [Google Scholar]
- Radfar A., Unnikrishnan, I., Lee, H.W., DePinho, R.A., and Rosenberg, N. 1998. p19(Arf) induces p53-dependent apoptosis during abelson virus-mediated pre-B cell transformation. Proc. Natl. Acad. Sci. 95: 13194-13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland B.D., Denissov, S.G., Douma, S., Stunnenberg, H.G., Bernards, R., and Peeper, D.S. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2: 55-65. [DOI] [PubMed] [Google Scholar]
- Russell J.L., Powers, J.T., Rounbehler, R.J., Rogers, P.M., Conti, C.J., and Johnson, D.G. 2002. ARF differentially modulates apoptosis induced by E2F1 and Myc. Mol. Cell. Biol. 22: 1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D., and DePinho, R.A. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27-37. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin, A.W., McCurrach, M.E., Beach, D., and Lowe, S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593-602. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. 1996. Cancer cell cycles. Science 274: 1672-1677. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell. Biol. 2: 731-737. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and DePinho, R.A. 2000. Cellular senescence: Mitotic clock or culture shock? Cell 102: 407-410. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts, J.M. 1999. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 13: 1501-1512. [DOI] [PubMed] [Google Scholar]
- Stott F.J., Bates, S., James, M.C., McConnell, B.B., Starborg, M., Brookes, S., Palmero, I., Ryan, K., Hara, E., Vousden, K.H., et al. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17: 5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B., and Dynlacht, B.D. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 14: 804-816. [PMC free article] [PubMed] [Google Scholar]
- Tolbert D., Lu, X., Yin, C., Tantama, M., and Van Dyke, T. 2002. p19(ARF) is dispensable for oncogenic stress-induced p53-mediated apoptosis and tumor suppression in vivo. Mol. Cell. Biol. 22: 370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees, J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3: 11-20. [DOI] [PubMed] [Google Scholar]
- Tsai K.Y., Hu, Y., Macleod, K.F., Crowley, D., Yamasaki, L., and Jacks, T. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2: 293-304. [DOI] [PubMed] [Google Scholar]
- Tsai K.Y., MacPherson, D., Rubinson, D.A., Crowley, D., and Jacks, T. 2002. ARF is not required for apoptosis in Rb mutant mouse embryos. Curr. Biol. 12: 159-163. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M. 1998. Functional analysis of mouse Polycomb group genes. Cell. Mol. Life Sci. 54: 71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.D., Taylor, L.J., Roussel, M.F., Sherr, C.J., and Bar-Sagi, D. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1: 20-26. [DOI] [PubMed] [Google Scholar]
- Williams B.O., Remington, L., Albert, D.M., Mukai, S., Bronson, R.T., and Jacks, T. 1994. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat. Genet. 7: 480-484. [DOI] [PubMed] [Google Scholar]
- Yamasaki L., Bronson, R., Williams, B.O., Dyson, N.J., Harlow, E., and Jacks, T. 1998. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/-) mice. Nat. Genet. 18: 360-364. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiong, Y., and Yarbrough, W.G. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725-734. [DOI] [PubMed] [Google Scholar]
- Ziebold U., Reza, T., Caron, A., and Lees, J.A. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes & Dev. 15: 386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebold U., Lee, E.Y., Bronson, R.T., and Lees, J.A. 2003. E2F3 loss has opposing effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol. Cell. Biol. 23: 6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Eischen, C.M., Randle, D.H., Kamijo, T., Cleveland, J.L., Sherr, C.J., and Roussel, M.F. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 12: 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Williams, R.T., Baudino, T.A., Rehg, J.E., Skapek, S.X., Cleveland, J.L., Roussel, M.F., and Sherr, C.J. 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc. Natl. Acad. Sci. 100: 15930-15935. [DOI] [PMC free article] [PubMed] [Google Scholar]