Abstract
The spindle checkpoint arrests cells at the metaphase-to-anaphase transition until all chromosomes have properly attached to the mitotic spindle. Checkpoint proteins Mad2p and Mad3p/BubR1p bind and inhibit Cdc20p, an activator for the anaphase-promoting complex (APC). We find that upon spindle checkpoint activation by microtubule inhibitors benomyl or nocodazole, wild-type Saccharomyces cerevisiae contains less Cdc20p than spindle checkpoint mutants do, whereas their CDC20 mRNA levels are similar. The difference in Cdc20p levels correlates with their difference in the half-lives of Cdc20p, indicating that the spindle checkpoint destabilizes Cdc20p. This process requires the association between Cdc20p and Mad2p, and functional APC, but is independent of the known destruction boxes in Cdc20p and the other APC activator Cdh1p. Importantly, destabilization of Cdc20p is important for the spindle checkpoint, because a modest overexpression of Cdc20p causes benomyl sensitivity and premature Pds1p degradation in cells treated with nocodazole. Our study suggests that the spindle checkpoint reduces Cdc20p to below a certain threshold level to ensure a complete inhibition of Cdc20p before anaphase.
Keywords: Spindle checkpoint, Cdc20p, Mad2p, kinetochore, protein stability
Proper segregation of chromosomes during anaphase depends on bipolar attachment of all chromosomes to the mitotic spindle at metaphase. In the presence of a damaged spindle or even a single unattached kinetochore, the spindle checkpoint is triggered to delay the anaphase onset until all kinetochores have attached properly to spindle microtubules (Musacchio and Hardwick 2002; Yu 2002). In this way, the spindle checkpoint prevents missegregation of chromosomes from occurring.
Some of the spindle checkpoint proteins were first identified in the budding yeast Saccharomyces cerevisiae, including Mad1p, Mad2p, Mad3p (BubR1p in metazoans), Bub1p, Bub3p, and Mps1p (Hoyt et al. 1991; Li and Murray 1991; Weiss and Winey 1996). These six proteins are conserved during evolution, indicative of their important function (Musacchio and Hardwick 2002). When the mitotic spindle assembly is impaired by microtubule-depolymerizing agents, such as nocodazole or benomyl, spindle checkpoint mutants fail to delay their cell cycle progression and die quickly because of chromosome loss. In budding yeast, mutations in spindle checkpoint genes lead to an increased chromosome loss rate even under normal growing conditions (Hoyt et al. 1991; Li and Murray 1991; Warren et al. 2002). In metazoans, the spindle checkpoint is critical for life, as mad2 and bub3 homozygous knockout mice die during early embryogenesis (Dobles et al. 2000; Kalitsis et al. 2000). The heterozygous mad2 knockout mice develop tumors at a higher rate than mice with wild-type MAD2 (Michel et al. 2001). The spindle checkpoint proteins are localized to kinetochores during mitosis in all organisms that have been examined (Cleveland et al. 2003), consistent with the observation that the spindle checkpoint is triggered from kinetochores that lack microtubule attachment or are not under tension (Li and Nicklas 1995; Rieder et al. 1995; Nicklas et al. 2001; Stern and Murray 2001).
Anaphase occurs only after the spindle checkpoint is inactivated upon proper attachment of all chromosomes to the spindle. The metaphase-to-anaphase transition is triggered by degradation of the anaphase inhibitor securin (Pds1p in budding yeast), which liberates its binding partner separase (Esp1p in budding yeast) and allows separase to cleave the cohesin complex that holds sister chromatids together (Nasmyth 2002). Degradation of securin is dependent on its ubiquitination by an E3 ubiquitin ligase, the anaphase-promoting complex (APC). APC recognizes its substrates through its associated activators/specificity factors, a group of conserved proteins containing WD repeats. When coupled with Cdc20p, APC targets securin and initiates the degradation of mitotic cyclin, whereas complete degradation of the mitotic cyclin requires APCCdh1 (Harper et al. 2002). Recent evidence suggests that Cdc20p and Cdh1p serve as the substrate receptors for APC (Hilioti et al. 2001; Pfleger et al. 2001; Schwab et al. 2001). Cdc20p is essential for cell growth, and temperature-sensitive cdc20 mutants arrest before anaphase (Hartwell 1973).
The spindle checkpoint prevents Cdc20p from activating APC, thus blocking anaphase onset. Two-hybrid analysis identifies Mad2p as an interacting protein for Slp1, the Cdc20p homolog in fission yeast (Kim et al. 1998). The spindle checkpoint is impaired in cells containing specific mutations in Slp1 or Cdc20p that disrupt their interaction with Mad2p (Hwang et al. 1998; Kim et al. 1998). In addition, recombinant human Mad2p or BubR1p protein binds and inhibits APCCdc20 in vitro, with synergistic effect when both proteins are added together (Fang et al. 1998a; Tang et al. 2001; Fang 2002). Cdc20p is also found to coimmunoprecipitate with Mad2p, Bub3p, and Mad3p/BubR1p from various organisms (Hardwick et al. 2000; Fraschini et al. 2001; Chen 2002; Millband and Hardwick 2002). Purification from human HeLa cells yields a mitotic checkpoint complex (MCC) containing BubR1p, Bub3p, Cdc20p, and Mad2p, which inhibits APC in vitro (Sudakin et al. 2001). Unattached kinetochores are likely involved in the assembly of the complex between Cdc20p and the checkpoint proteins, because nocodazole treatment enhances the complex formation (Zhang and Lees 2001; Chen 2002; Chung and Chen 2002). In addition, blocking kinetochore association of the spindle checkpoint proteins prevents the complex formation (Chen 2002; Chung and Chen 2002). It is thought that unattached kinetochores may continuously stimulate the assembly and release of the checkpoint complex by bringing these proteins to close proximity and/or by their activation at kinetochores. Indeed, fluorescence recovery after photobleaching analysis (FRAP) shows a rapid turnover of Mad2p and Cdc20p at kinetochores (Howell et al. 2000; Kallio et al. 2002).
Cdc20p level fluctuates during the cell cycle, rising in S phase, peaking in mitosis, and declining upon exit from mitosis in both yeast and mammalian cells (Weinstein 1997; Prinz et al. 1998; Shirayama et al. 1998; Kramer et al. 2000). The change in the protein level is partly due to transcriptional control, as the mRNA of Cdc20p exhibits similar oscillation (Fang et al. 1998b; Prinz et al. 1998; Morris et al. 2003). Cdc20p is unstable throughout the cell cycle in budding yeast (Prinz et al. 1998; Goh et al. 2000). Transiently expressed Cdc20p is quickly degraded in cells arrested at G1, S, or in mitosis, with the shortest half-life observed in G1 cells (Prinz et al. 1998; Goh et al. 2000). There are conflicting reports with regard to whether APC plays a major role in Cdc20p degradation. Some reports show that Cdc20p is partially stabilized in APC mutants kept at the nonpermissive temperatures (Prinz et al. 1998; Shirayama et al. 1998), whereas one study reports that inactivating APC has no significant effect on the half-life of Cdc20p in G1 cells (Goh et al. 2000). Despite this discrepancy, Cdc20p remains unstable when APC is inactivated, suggesting that an APC-independent mechanism may also be involved in Cdc20p turnover (Prinz et al. 1998; Goh et al. 2000). Budding yeast Cdc20p contains two destruction-box sequences. Removing the first one slows down Cdc20p degradation, with a stabilizing effect more prominent in G1 than in S phase or mitosis (Prinz et al. 1998). Interestingly, there is no obvious destruction box in human Cdc20p. Instead, another destruction-motif KEN box is required for APCCdh1-dependent degradation of human Cdc20p (Pfleger and Kirschner 2000). APCCdh1 also mediates the degradation of Cdc20p in yeast (Huang et al. 2001).
In this study, we reveal that, in addition to binding and inhibiting Cdc20p, the spindle checkpoint proteins destabilize Cdc20p upon spindle damage.
Results
Elevated Cdc20p levels in spindle checkpoint mutants
To determine whether the spindle checkpoint regulates the Cdc20p level, we compared wild type with a spindle checkpoint mutant mad2-1 that behaves essentially like mad2Δ (Chen et al. 1999). To monitor Cdc20p, we expressed N-terminally 8myc-tagged protein from its genomic locus, which supported normal growth. In addition, cells containing 8mycCdc20p arrested at mitosis in the presence of nocodazole or benomyl (data not shown), indicating that the epitope tag does not interfere with the regulation of Cdc20p by the spindle checkpoint.
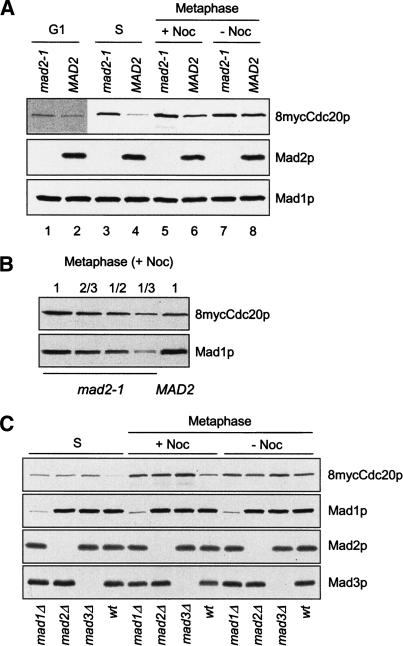
We examined Cdc20p levels at various stages of the cell cycle. We arrested mad2-1 and wild-type cells at metaphase through the expression of Pds1pΔdb, the non-degradable form of Pds1p that lacks its destruction box. Expression of this mutant from the GAL promoter arrests cells at metaphase for a prolonged period of time (Cohen-Fix and Koshland 1999; Tinker-Kulberg and Morgan 1999). When the mitotic spindle was disrupted by nocodazole during the metaphase arrest, the Cdc20p level in mad2-1 was about two- to threefold higher than that in wild type (Fig. 1A [lanes 5,6], B). Wild-type cells without nocodazole treatment contained more Cdc20p than those treated with nocodazole (Fig. 1A, lanes 6,8). There was little difference in Cdc20p levels between wild-type and mad2-1 cells without spindle checkpoint activation (Fig. 1A, lanes 7,8).
Figure 1.
Cdc20p levels are elevated in spindle checkpoint mutants. (A) mad2-1 and MAD2 cells containing 8mycCDC20 and a galactose-inducible nondegradable Pds1pΔdb (PGAL-pds1Δdb; RHC435 and RHC436) were arrested at G1 with α-factor (lanes 1,2), or at S phase with hydroxyurea (lanes 3,4) in YEPD at 30°C. For metaphase arrest, cells were first synchronized at G1 with α-factor in YEP containing raffinose (YEPR), followed by induction of Pds1pΔdb with 2% galactose for 1 h at 30°C during the G1 arrest. Cells were then washed and released into YEPD in the presence (lanes 5,6) or absence (lanes 7,8) of nocodazole. Pds1pΔdb ensures mitotic arrest of mad2-1, which lacks the spindle checkpoint and cannot be blocked at metaphase by nocodazole. Cell lysates were prepared at 100 min after the release for immunoblots of 8mycCdc20p, Mad2p, and Mad1p, using 9E10, anti-Mad2p, and anti-Mad1p antibodies, respectively. The Mad1p blot serves as a loading control. Because of the low level of Cdc20p at G1, a long exposure of the Cdc20p blot is presented for G1 samples (lanes 1,2). (B) The lysate prepared from mad2-1 cells arrested at metaphase by Pds1pΔdb in the presence of nocodazole was loaded in serial dilutions to compare its 8mycCdc20p level with that in the MAD2 cell lysate. (C) mad1Δ, mad2Δ, mad3Δ, or wild-type cells containing 8myc-CDC20 and PGAL-pds1Δdb (RHC472, RHC473, RHC474, and RHC476) were arrested at S phase with hydroxyurea, or at metaphase by Pds1pΔdb in the presence or absence of nocodazole, as described in A. Immunoblot analysis was performed as indicated. The weak signals present in mad1Δ lanes are from a cross-reacting protein recognized by anti-Mad1p antibody.
In cells arrested at G1 phase with α-factor, Cdc20p was present at a very low level. Nevertheless, we consistently found more Cdc20p in mad2-1 than in wild-type cells (Fig. 1A, lanes 1,2). Interestingly, hydroxyurea-arrested S-phase cells exhibited the most dramatic difference in Cdc20p levels between mad2-1 and wild-type cells (Fig. 1A, lanes 3,4). These results show that the absence of Mad2p leads to elevated Cdc20p levels in G1, S, and mitosis. At metaphase, spindle disruption further enhances the difference in Cdc20p levels between mad2-1 and wild-type cells.
To determine whether the level of Cdc20p is regulated by Mad2p specifically or by the spindle checkpoint in general, we examined Cdc20p levels in two other spindle checkpoint mutants, mad1Δ and mad3Δ. When arrested at S phase with hydroxyurea or at metaphase by Pds1pΔdb in the presence of nocodazole, mad1Δ, mad2Δ, and mad3Δ cells all contained higher levels of Cdc20p than wild-type cells did (Fig. 1C). Without spindle disruption, there was no significant difference in Cdc20p levels between the checkpoint mutants and wild-type cells. Therefore, all three spindle checkpoint mutants lose the ability to control Cdc20p levels, indicating that the spindle checkpoint in general, rather than Mad2p alone, is involved in the down-regulation of Cdc20p levels. Interestingly, there was consistently more Cdc20p in mad3Δ than in mad1Δ and mad2Δ under all conditions we examined.
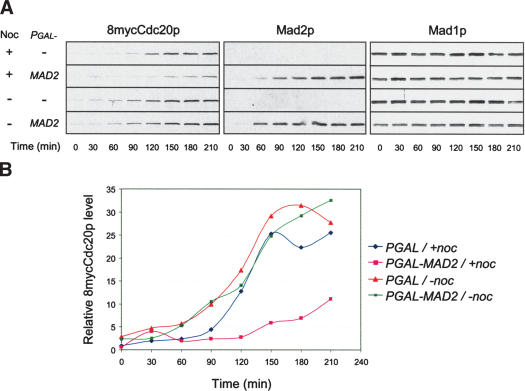
We then determined Cdc20p levels in cells overexpressing Mad2p. Synchronized G1 cells were released into the cell cycle in galactose-containing medium to induce both Mad2p and Pds1pΔdb. The latter blocked cells at metaphase by the end of the experiment. The levels of Cdc20p gradually increased as cells advanced from G1 to mitosis. In the presence of nocodazole, cells induced for Mad2p expression accumulated less Cdc20p than those without Mad2p, which was especially obvious after the 90-min time point (Fig. 2). On the other hand, Mad2p overexpression did not affect the Cdc20p level when nocodazole was absent. These results show that overexpression of Mad2p reduces the Cdc20p level when the spindle is disrupted.
Figure 2.
Overexpression of Mad2p reduces Cdc20p levels in the presence of nocodazole. (A) 8mycCDC20, PGAL-pds1Δdb cells containing PGAL vector alone or PGAL-MAD2 (RHC401 and RHC402) were first arrested at G1 with α-factor, then washed and released into YEPR in the presence or absence of nocodazole at 30°C. When ∼10% of the cells began to bud, 2% galactose was added to the medium. Samples were taken at 30-min intervals after galactose addition for immunoblot analysis. Mad1p blots serve as loading controls. (B) Signals of 8mycCdc20p in A were quantified and plotted against time.
Modest overexpression of Cdc20p impairs the spindle checkpoint
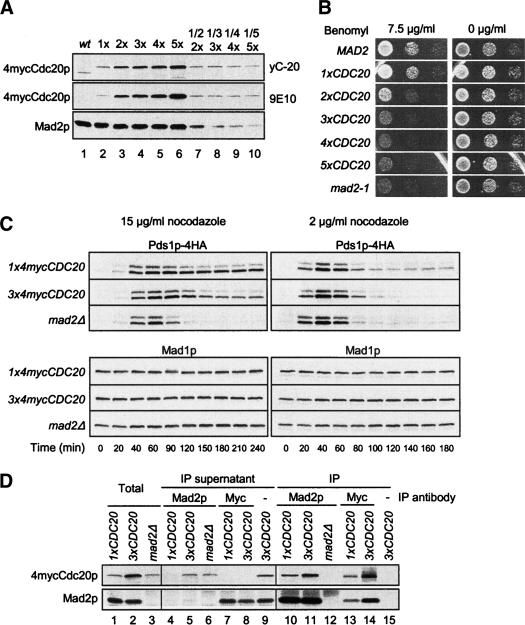
We asked if the control of the Cdc20p level is an essential part of the spindle checkpoint. It has been previously shown that overexpressing Cdc20p abolishes the mitotic arrest caused by microtubule-disrupting agents (Hwang et al. 1998; Schott and Hoyt 1998; Shirayama et al. 1998; Zhang and Lees 2001). Because these studies expressed Cdc20p to much higher levels than what we observed in the mad mutants, we tested the effect of a modest increase in Cdc20p levels on the spindle checkpoint. We replaced the endogenous CDC20 gene with one to five copies of 4mycCDC20 integrated into the genome. Cdc20p levels increase with increasing copies of CDC20 (Fig. 3A). In the benomyl sensitivity assay, cells containing a single copy of 4mycCDC20 (1×) grew similarly with wild-type cells (Fig. 3B), indicating that 4mycCdc20p responds to the spindle checkpoint inhibition just as the untagged protein does. As the Cdc20p level doubled through an additional copy of 4mycCDC20 (2×), growth was visibly reduced on the benomyl plate. Cells containing 4mycCdc20p at three to five times of the endogenous level grew as poorly as mad2-1 (Fig. 3B).
Figure 3.
Increased Cdc20p expression impairs the spindle checkpoint. (A) Cdc20p levels increase with additional copies of integrated 4mycCDC20. Cell lysates were prepared from wild-type and cdc20Δ cells containing one to five copies of 4mycCDC20 (1×, 2×, 3×, 4×, and 5×; RHC542–RHC546) temporarily arrested at metaphase with nocodazole. Immunoblots were performed with 40 μg of total protein (lanes 1–6), 20 μg of protein from the 2× strain (lane 7), 16.7 μg of protein from the 3× strain (lane 8), 10 μg of protein from the 4× strain (lane 9), and 8 μg of protein from the 5× strain (lane 10). 4mycCdc20p was detected with either anti-Cdc20p antibody yC-20 (top) or anti-myc antibody 9E10 (middle). The Mad2p blot serves as a loading control. (B) Increased Cdc20p expression leads to benomyl sensitivity. MAD2, mad2-1, and cdc20Δ cells containing one to five copies of integrated 4mycCDC20 (1×, 2×, 3×, 4×, and 5× CDC20) were spotted onto YEPD plates with or without 7.5 μg/mL benomyl. The cells were in 10-fold serial dilutions from left to right. (C) Cells overexpressing 4mycCDC20 by threefold are defective in maintaining Pds1p levels in the presence of nocodazole. PDS1-4HA cells containing one (RHC639) or three copies of 4mycCDC20 (RHC641), or one copy of 4mycCDC20 in mad2Δ (RHC644), were first arrested in G1 with α-factor. Cells were then released at time 0 into YEPD containing 15 μg/mL nocodazole at 25°C (left panel) or 2 μg/mL nocodazole at 30°C (right panel). α-Factor was added back at 1 h 20 min (left panel) or 1 h (right panel) to prevent re-entry into the next cell cycle. Samples were taken at the indicated time points for immunoblot analysis. Mad1p blots serve as loading controls. (D) A fraction of Cdc20p is not bound with Mad2p when Cdc20p is overexpressed. 1xCDC20 (RHC639), 3xCDC20 (RHC641), and mad2Δ (RHC644) cells were first arrested in G1 and then released into YEPD containing 15 μg/mL nocodazole at 25°C. Cell lysates were prepared 2 h after the release, followed by immunoprecipitation of Mad2p and 4mycCdc20p with anti-Mad2p and 9E10 antibodies, respectively. Mock immunoprecipitation was performed with the 3xCDC20 cell lysate as a control for 9E10 immunoprecipitation. The cell lysates (lanes 1–3), the supernatants from immunoprecipitations (lanes 4–9), and the immunoprecipitates (lanes 10–15) were immunoblotted for 4mycCdc20p and Mad2p as indicated.
To determine whether the increased benomyl sensitivity was due to spindle checkpoint defects, we monitored the levels of Pds1p in cells treated with nocodazole. When cells were first arrested at G1 and then released into medium containing nocodazole at 15 μg/mL, Pds1p levels in 1xCDC20 cells gradually increased and were stabilized for at least 4 h (Fig. 3C, left panel) and cells remained large-budded by the end of the time course (data not shown). This result indicates that anaphase is completely blocked by 15 μg/mL of nocodazole in cells containing a normal level of Cdc20p. As expected, Pds1p in mad2Δ cells accumulated normally, but started to degrade after 60 min, indicating that cells escaped cell cycle arrest. In cells containing 3xCDC20, Pds1p was partially degraded after 2 h (Fig. 3C, left panel), indicating that these cells also gradually escaped cell cycle arrest. When released into 2 μg/mL nocodazole, cells containing 1xCDC20 partially degraded Pds1p (Fig. 3C, right panel), and >50% of the cells completed mitosis and became unbudded by 180 min (data not shown). This result suggests that nocodazole at 2 μg/mL only imposes a delay in anaphase onset, and that cells were able to assemble a functional spindle after some lag, which mimics the benomyl plate assay (Fig. 3B). At this concentration of nocodazole, the rate of Pds1p degradation in 3xCDC20 cells was between that in 1xCDC20 and in mad2Δ cells (Fig. 3C, right panel). These results show that modest overexpression of Cdc20p causes premature Pds1p degradation in the presence of spindle damage.
As Cdc20p is normally inhibited in a complex with Mad2 upon spindle checkpoint activation, we examined the effect of an increased Cdc20p level on Mad2p–Cdc20p interaction. Our standard anti-Mad2p immunoprecipitation removed the vast majority of Mad2p from the cell lysates and left very little Mad2p in the supernatants (Fig. 3D, lanes 4,5). Most, if not all, of Cdc20p was coimmunoprecipitated with Mad2p from 1xCDC20 cells treated with nocodazole (Fig. 3D, lanes 4,10). In 3xCDC20 cells, the amount of Cdc20p associated with Mad2 increased (Fig. 3D, lanes 10,11), but a significant fraction of Cdc20p remained in the supernatant (Fig. 3D, lane 5). This result suggests that cells cannot incorporate all Cdc20p into a complex with Mad2p when Cdc20p is overexpressed by threefold. In addition, when Cdc20p was depleted from the cell lysates by anti-myc antibody (Fig. 3D, lanes 7,8), the level of Mad2p was only slightly reduced in the supernatants compared with that in the cell lysates (Fig. 3D, lanes 1,2,7,8), indicating that only a small fraction of Mad2p is able to complex with Cdc20p.
Spindle checkpoint reduces Cdc20p stability
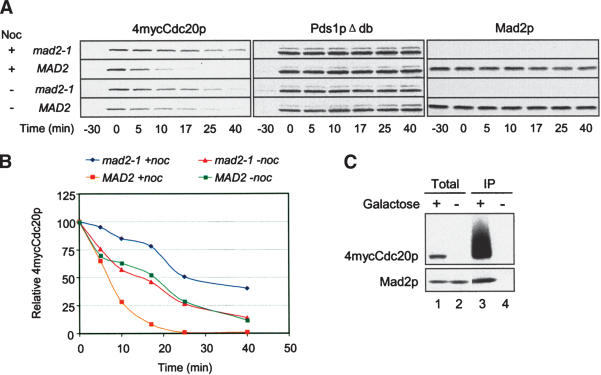
The spindle checkpoint may regulate the Cdc20p level by affecting transcription, protein synthesis, or stability. We specifically examined the half-life of Cdc20p in cells arrested at metaphase. During the metaphase arrest caused by Pds1pΔdb, we briefly induced 4mycCdc20p with galactose, and then suppressed the expression by adding glucose and the protein synthesis inhibitor cycloheximide (Fig. 4A). We determined the half-life of 4mycCdc20p by monitoring its disappearance over time after its expression was shut off. Galactose-induced 4mycCdc20p was quickly depleted upon glucose addition. In the representative experiment shown in Figure 4A and B, the half-life of Cdc20p was ∼26 min in mad2-1 cells and ∼7 min in MAD2 cells treated with nocodazole. In the absence of nocodazole, Cdc20p in mad2-1 and MAD2 cells degraded with similar kinetics. These results show that Cdc20p is less stable in wild-type than in mad2-1 cells when the spindle checkpoint is triggered. Without spindle checkpoint activation, Cdc20p stability is not affected by Mad2p. Coimmunoprecipitation study shows that Mad2p associated with 4mycCdc20p that was induced from the GAL promoter during nocodazole treatment, indicating that the ectopically produced Cdc20p behaved similarly to the endogenous protein (Fig. 4C).
Figure 4.
Spindle checkpoint activation reduces the stability of galactose-induced Cdc20p. (A) Galactose-induced Cdc20p has a longer half-life in mad2-1 than in MAD2 cells in the presence of nocodazole. mad2-1 and MAD2 cells containing PGAL-4mycCDC20 and PGAL-pds1Δdb (RHC421 and RHC422) were arrested at G1 with α-factor in YEPR, followed by a 40-min induction of Pds1pΔdb with 2% galactose at 30°C. Cells were then released into YEPR at 25°C in the presence or absence of nocodazole. The short induction of Pds1pΔdb was sufficient to arrest cells at metaphase. When >95% of the cells were arrested at metaphase (-30 min), expression of 4mycCdc20p was induced with 2% galactose for 30 min and then shut off with 2% glucose and 1 mg/mL cycloheximide (0 min). Samples were taken at the indicated time points for immunoblot analysis. Pds1pΔdb was detected with 16B12 as loading controls. (B) 4mycCdc20p levels were quantified and plotted against time. (C) 4mycCDC20 induced from the GAL promoter binds Mad2p. MAD2 cells containing PGAL-4mycCDC20 and PGAL-pds1Δdb (RHC422) were arrested at G1, briefly induced for Pds1pΔdb expression, and released into YEPR containing nocodazole as described in A. One-half of the culture was then induced for 4mycCdc20p expression with 2% galactose for 30 min (lanes 1,3), and the other half was kept uninduced (lanes 2,4). Cell lysates were prepared for anti-myc immunoprecipitation, followed by immunoblot analysis for 4mycCdc20p and Mad2p.
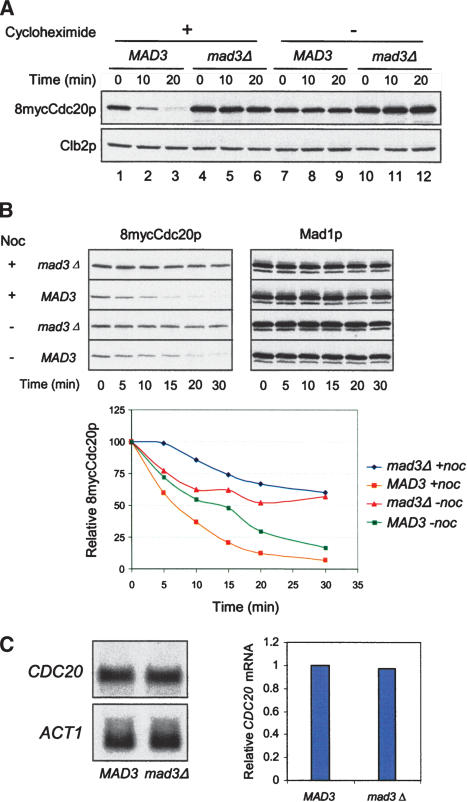
We also examined the stability of endogenous Cdc20p by treating mitotic cells with protein synthesis inhibitor cycloheximide. In the presence of nocodazole, most of the Cdc20p disappeared by 20 min after cycloheximide addition in wild-type cells (Fig. 5A, lanes 1–3), whereas the protein remained constant without cycloheximide treatment (Fig. 5A, lanes 7–9). In contrast, there was only a slight decrease in Cdc20p levels in mad3Δ cells treated with cycloheximide (Fig. 5A, lanes 4–6). These results indicate that Cdc20p is continuously synthesized and degraded in spindle checkpoint-active cells. On the other hand, the levels of mitotic cyclin Clb2p were constant in the presence of cycloheximide. Furthermore, the half-life of endogenous Cdc20p in wild type was shorter than that in mad3Δ in the presence of nocodazole (Fig. 5B). The difference in Cdc20p stability between wild-type and mad3Δ cells was reduced in the absence of nocodazole. These results are consistent with results obtained with galactose-induced Cdc20p (Fig. 4).
Figure 5.
Spindle checkpoint activation reduces the stability of endogenous Cdc20p. (A) Inhibition of protein synthesis rapidly depletes Cdc20p in spindle checkpoint-active cells. MAD3 (RHC476) and mad3Δ cells (RHC474) were arrested in mitosis by Pds1pΔdb in the presence of nocodazole at 30°C, as described for Figure 1A. Cycloheximide was then added at 1 mg/mL to half of the cultures, and samples were taken at the indicated time points for immunoblot analysis of 8mycCdc20p and Clb2p. The latter serves as controls for mitotic arrest and equal loading. (B) Endogenous Cdc20p has a longer half-life in mad3Δ cells than in MAD3 cells. MAD3 (RHC476) and mad3Δ cells (RHC474) were arrested in mitosis by Pds1pΔdb in the presence or absence of nocodazole at 25°C. Cycloheximide was then added at 1 mg/mL, and samples were taken at the indicated time points for immunoblot analysis. The Mad1p blots serve as loading controls. 8mycCdc20p levels were quantified and plotted against time. (C) Wild-type and mad3Δ cells contain similar levels of CDC20 mRNA in mitosis. Wild-type MAD3 (RHC292) and mad3Δ cells (RHC482) were arrested in mitosis by Pds1pΔdb in the presence of nocodazole at 30°C. Total RNA was prepared for Northern blot analysis of CDC20 and ACT1 mRNAs. The plot shows the relative CDC20 mRNA levels normalized with ACT1 signals.
To examine whether CDC20 transcription is affected by the spindle checkpoint, we compared CDC20 mRNA levels in wild type and mad3Δ cells arrested in mitosis in the presence of nocodazole. Northern blot analysis showed similar amounts of mRNA between these two strains (Fig. 5C), indicating that spindle checkpoint has no effect on the steady level of CDC20 mRNA.
Mutant Cdc20p that cannot bind Mad2p is present at elevated levels
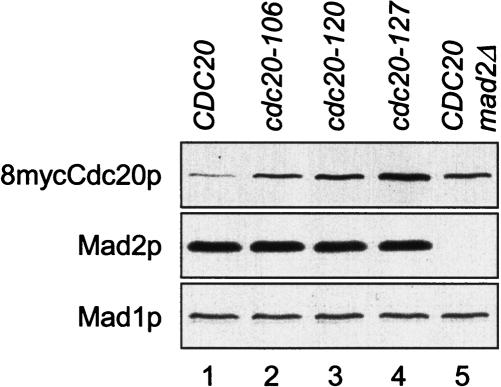
Spindle disruption enhances the interaction among Mad2p, Mad3p, and Cdc20p. It is possible that the formation of this complex contributes to Cdc20p degradation. We compared the levels of three cdc20 mutants, cdc20-106p, cdc20-120p, and cdc20-127p, that cannot bind to Mad2p and Mad3p (Hwang et al. 1998). When cells were arrested at metaphase by Pds1pΔdb in the presence of nocodazole, cdc20-106p, cdc20-120p, and cdc20-127p were all present at higher levels than was the wild-type protein (Fig. 6). This result suggests that the control of Cdc20p level requires the binding of Cdc20p to Mad2p.
Figure 6.
Mutant Cdc20p that cannot bind Mad2p is present at elevated levels. Cells containing 8myc-tagged CDC20, cdc20-106, cdc20-120, or cdc20-127 on centromeric plasmids (lanes 1–4, RHC554–RHC557), or mad2Δ cells containing 8mycCDC20 (lane 5, RHC558) were arrested at metaphase by Pds1pΔdb in the presence of nocodazole, as described for Figure 1A. Cell lysates were prepared for immunoblot analysis. The Mad1p blot serves as a loading control.
APC is required for down-regulation of Cdc20p by the spindle checkpoint
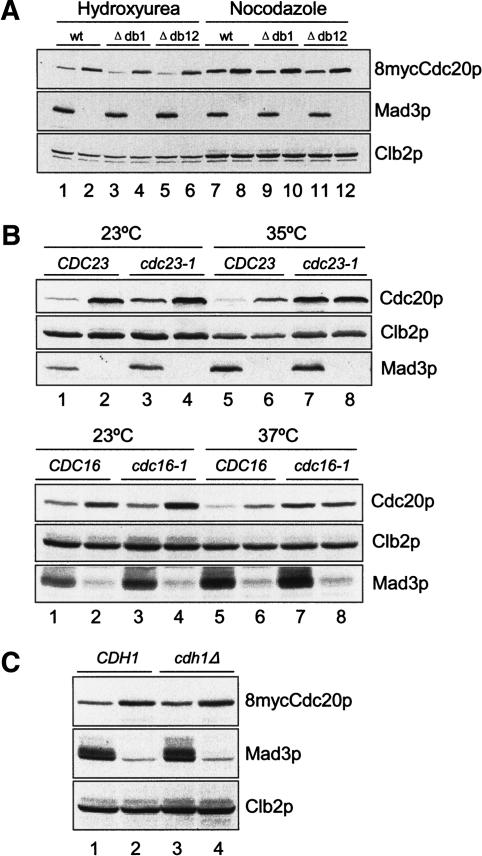
It is possible that the spindle checkpoint reduces the Cdc20p level through APC. To test this possibility, we first asked whether the spindle checkpoint can regulate the levels of Cdc20p mutants that lack either the first destruction box, Cdc20pΔdb1, or both destruction boxes, Cdc20pΔdb12. Under both hydroxyurea-induced S-phase arrest and Pds1pΔdb-induced mitotic arrest in the presence of nocodazole, Cdc20pΔdb1 and Cdc20pΔdb12 were present at a higher level in mad3Δ cells than in cells with a functional spindle checkpoint (Fig. 7A), indicating that the destruction boxes are not required for the down-regulation of Cdc20p levels by the spindle checkpoint.
Figure 7.
The spindle checkpoint regulates Cdc20p levels through APC. (A) The destruction-box sequences in Cdc20p are not important for spindle checkpoint-induced instability. MAD3 (odd lanes) or mad3Δ cells (even lanes) containing 8mycCDC20 (lanes 1,2,7,8), 8mycCDC20Δdb1 (lanes 3,4,9,10), or 8mycCDC20Δdb12 (lanes 5,6,11,12) on centromeric plasmids (RHC582–RHC587) were arrested at S phase with hydroxyurea (lanes 1–6) or at metaphase with Pds1pΔdb in the presence of nocodazole (lanes 7–12). Cell lysates were prepared for immunoblot analysis. (B) Regulation of Cdc20p levels by the spindle checkpoint is dependent on functional APC. (Upper panel) CDC23 (lanes 1,5, RHC292), CDC23 mad3Δ (lanes 2,6, RHC482), cdc23-1 (lanes 3,7, RHC576), and cdc23-1 mad3Δ (lanes 4,8, RHC581) cells were first arrested at G1 with α-factor in YEPR, followed by induction of Pds1pΔdb with 3% galactose for 1 h at 23°C. The cultures were then released into YEPR containing 3% galactose and split into two sets. The first set was kept at 23°C in the presence of 15 μg/mL nocodazole, and harvested 4.5 h later when >95% of the cells were arrested with large buds. The second set was shifted to 35°C with 30 μg/mL nocodazole and 10 μg/mL benomyl, and harvested 3 h later. (Lower panel) CDC16 (lanes 1,5, RHC292), CDC16 mad3Δ (lanes 2,6, RHC482), cdc16-1 (lanes 3,7, RHC647), and cdc16-1 mad3Δ (lanes 4,8, RHC648) cells were treated the same way as described for the upper panel, except that one set of the cells was released into 37°C instead of 35°C. Cell lysates were prepared for immunoblot analysis. (C) Regulation of Cdc20p levels by the spindle checkpoint is independent of Cdh1p. CDH1 (lane 1, RHC582), CDH1 mad3Δ (lane 2, RHC583), cdh1 (lane 3, RHC612), and cdh1 mad3Δ (RHC613) cells were arrested at metaphase by Pds1pΔdb in the presence of nocodazole at 30°C. Immunoblot analysis was performed for 8mycCdc20p, Clb2p, and Mad3p. The weak signals in mad3Δ lanes are cross-reacting proteins recognized by a certain batch of Mad3p antibody.
We next examined the effect of inactivating APC by using temperature-sensitive APC mutants cdc23-1 and cdc16-1. At 23°C, Cdc20p levels were higher in both cdc23-1 mad3Δ and cdc16-1 mad3Δ than in their isogenic MAD3 strains during mitotic arrest in the presence of nocodazole (Fig. 7B, lanes 3,4). At the nonpermissive temperature (35°C or 37°C), Cdc20p levels in cdc23-1 mad3Δ and cdc16-1 mad3Δ were similar to that in their isogenic MAD3 strains (Fig. 7B, lanes 7,8), whereas cells with functional APC contained less Cdc20p than isogenic mad3Δ cells did at both temperatures (Fig. 7B, lanes 1,2,5,6). These results show that inactivating APC abolishes the regulation of the Cdc20p level by the spindle checkpoint, indicating that APC is involved in the spindle checkpoint-induced Cdc20p degradation.
As Cdh1p has been shown to mediate Cdc20p degradation (Pfleger and Kirschner 2000; Sorensen et al. 2000; Huang et al. 2001), we asked whether Cdh1p was involved in destabilization of Cdc20p upon spindle checkpoint activation. The difference in Cdc20p levels between MAD3 and mad3Δ cells in the cdh1Δ background was similar to that in the CDH1 background (Fig. 7C), indicating that Cdh1p is not required for this process.
Discussion
Spindle checkpoint reduces Cdc20p stability
It is known that the spindle checkpoint prevents Cdc20p from activating APC through binding of the checkpoint proteins Mad2p and Mad3p/BubR1p to Cdc20p. We now show an additional mechanism by which the checkpoint inhibits APC function. We provide several lines of evidence to show that the spindle checkpoint destabilizes Cdc20p. First, wild-type cells contain a lower level of Cdc20p than the spindle checkpoint mutant mad1Δ, mad2Δ, and mad3Δ cells do. The difference is especially pronounced when the checkpoint is activated by spindle disruption (Fig. 1). Second, overexpressing Mad2p efficiently reduces the Cdc20p level when the spindle is disrupted (Fig. 2). Third, spindle checkpoint activation reduces Cdc20p stability in wild-type cells. Furthermore, the half-life of Cdc20p is shorter in cells with a functional spindle checkpoint than that in spindle checkpoint mutants upon spindle disruption (Figs. 4, 5). The two- to threefold increase of Cdc20p stability in checkpoint mutants quantitatively correlates with the two- to threefold increase of Cdc20p levels at metaphase. In addition, the difference in Cdc20p levels is not due to a change in transcription, as the steady levels of CDC20 mRNA are similar between wild type and mad3Δ in mitosis (Fig. 5).
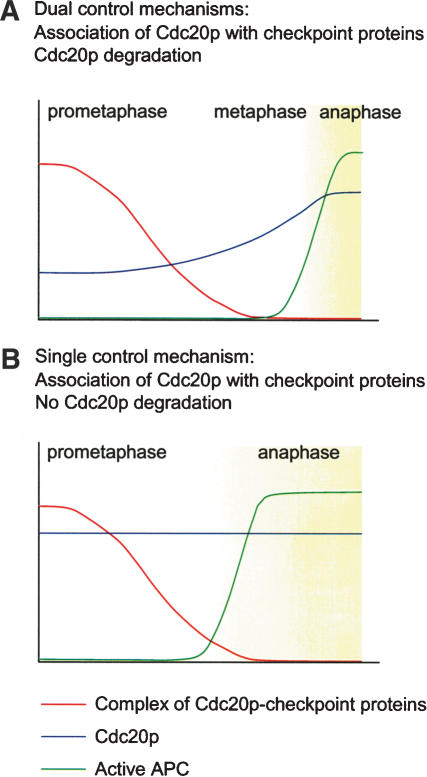
We further demonstrate that the tight control of the Cdc20p level is essential for the spindle checkpoint (Fig. 3). By expressing Cdc20p to various levels, we find a Cdc20p dosage-dependent sensitivity to benomyl. Even a twofold increase of Cdc20p renders the cells sensitive to benomyl. The benomyl sensitivity is indeed due to spindle checkpoint defect, because cells overexpressing Cdc20p by threefold (3xCDC20) cannot maintain Pds1p in the presence of nocodazole. In addition, coimmunoprecipitation study shows that a fraction of the overexpressed Cdc20p is not in a complex with Mad2, suggesting that such a modest increase in Cdc20p level can over-ride inhibition by the spindle checkpoint. We propose that the spindle checkpoint inhibits Cdc20p by at least two different mechanisms. First, the checkpoint proteins Mad2p and Mad3p/BubR1p bind and prevent Cdc20p from activating APC. Second, the checkpoint reduces the Cdc20p level to under a certain threshold to ensure a complete inhibition (Fig. 8A).
Figure 8.
The dual control mechanisms of the spindle checkpoint. (A) Activation of the spindle checkpoint early at prometaphase triggers Cdc20p degradation. The reduced level of Cdc20p ensures that the residual Cdc20p can be efficiently inhibited through binding with the spindle checkpoint proteins. Cdc20p remains inhibited even toward late prometaphase when only a few unattached kinetochores exist and the ability to assemble the Cdc20p–checkpoint protein complex has declined. After metaphase is achieved, both inhibitory mechanisms on Cdc20p are lifted. Cdc20p then accumulates and is able to activate APC. (B) Without degradation of Cdc20p in response to spindle checkpoint signal, the elevated level of Cdc20p overcomes the binding and inhibition by the spindle checkpoint proteins, especially toward late prometaphase. The inefficient inhibition of Cdc20p leads to premature anaphase before metaphase is achieved.
Even though the inhibition of Cdc20p by direct binding of the checkpoint proteins appears to be direct and efficient, this mechanism alone is not sufficient for the spindle checkpoint. This is probably because the formation of MCC is limited by the number of unattached kinetochores. We find that even though Mad2p is in excess of Cdc20, only a small fraction of Mad2p is able to bind Cdc20p upon spindle checkpoint activation (Fig. 3C). As discussed above, cells are not able to sequester all Cdc20p molecules in MCC when there is a modest increase of Cdc20p. In addition, the efficiency of forming MCC is expected to decrease when the cell progresses from prometaphase to metaphase and the number of unattached kinetochores gradually reduce. Furthermore, it is thought that MCC has a finite half-life and has to be regenerated at unattached kinetochores, as suggested by the dynamic behavior of Mad2p and Cdc20p at kinetochores. It is conceivable that inhibition of Cdc20p by direct binding of the checkpoint proteins may not be efficient in the presence of a single unattached kinetochore. This problem could be solved through Cdc20p degradation. The fully activated spindle checkpoint at early prometaphase triggers Cdc20p degradation and reduces the Cdc20p level, so that MCC can efficiently inhibit the residual proteins until the last lagging kinetochore captures microtubules. The degradation also reduces Cdc20p available for APC. Following attachment of all kinetochores to microtubules and termination of the spindle checkpoint, Cdc20p can then accumulate quickly to activate APC (Fig. 8A).
We observed the most pronounced difference in Cdc20p levels between spindle checkpoint mutants and wild-type cells upon hydroxyurea treatment, which activates the DNA replication checkpoint. It is possible that kinetochore–microtubule connections are not properly established in underreplicated centromeric regions in hydroxyurea-arrested cells, which may elicit the spindle checkpoint response that leads to Cdc20p reduction. This idea is consistent with the finding that the spindle checkpoint contributes to the pre-anaphase arrest under various conditions that activate the DNA replication checkpoint (Garber and Rine 2002). In addition, a recent report shows that hydroxyurea destabilizes Cdc20p in an Mec1p- and Rad53-dependent manner (Clarke et al. 2003). It remains a possibility that there might be interplay between the spindle checkpoint proteins and the DNA replication checkpoint proteins Mec1p and Rad53p in the control of Cdc20p level during S-phase arrest.
Interestingly, mad3Δ cells always contain slightly more Cdc20p than mad1Δ and mad2Δ cells do. In addition, Cdc20p in mad3Δ is more stable than that in MAD3 even in the absence of nocodazole (Fig. 5C), whereas the stability is similar in mad2Δ and MAD2 cells without spindle disruption (Fig. 4). These data suggest that Mad3p plays a more important or direct role in reducing the Cdc20p level. Alternatively, Mad3p may lie in a different pathway from Mad1p and Mad2p in the regulation of the Cdc20p level. In fact, it has been reported that cells lacking Mad2p or Mad3p/BubR1p exhibit differences in their chromosome loss rate, their response to microtubule toxins, and their ability to align chromosomes, supporting the notion that Mad3p/BubR1p may have functional bifurcations from Mad2p (Skoufias et al. 2001; Warren et al. 2002; Ditchfield et al. 2003).
Mechanisms of spindle checkpoint-induced Cdc20p degradation
Cdc20p is known to be inhibited when it is assembled into a complex with spindle checkpoint proteins. Does this complex also play a role in the control of Cdc20p stability? We show that several mutant Cdc20p proteins that cannot bind Mad2p are maintained at elevated levels, suggesting that the physical interaction between Cdc20p and Mad2p is required for the checkpoint-induced Cdc20p degradation.
The spindle checkpoint reduces Cdc20p level through APC, even though both of the known destruction-box sequences are not required for the spindle checkpoint-induced instability (Fig. 7A). It is possible that other degradation signals might be involved in the degradation of Cdc20p. Human Cdc20p is a substrate of APCCdh1 in vitro (Pfleger and Kirschner 2000), and overexpression of Cdh1p in mammalian cells prevents Cdc20p accumulation (Sorensen et al. 2000). In budding yeast, disruption of CDH1 increased the stability of ectopically expressed Cdc20p in G1 (Huang et al. 2001). However, Cdh1p is not active until late mitosis (Harper et al. 2002), after the spindle checkpoint has been inactivated. Indeed, cdh1Δ cells are still able to down-regulate Cdc20p upon spindle checkpoint activation (Fig. 7C). It is possible that a novel APC activator may specifically target Cdc20p before metaphase is achieved. Alternatively, APC may directly recognize Cdc20p as a substrate when Cdc20p is bound with the spindle checkpoint proteins, which is consistent with our observation that Cdc20p–Mad2p interaction is important for Cdc20p degradation. It remains to be determined whether the mechanism of Cdc20p degradation is related to that of other APC substrates that are degraded before metaphase, including cyclin A (Geley et al. 2001), the NIMA-related kinase Nek2 (Hames et al. 2001), and homeoprotein HOXC10 (Gabellini et al. 2003).
Our genetic and biochemical results suggest that the spindle checkpoint is composed of a dual control mechanism. First, unattached kinetochores stimulate the binding of Cdc20p with the spindle checkpoint proteins, so that Cdc20p cannot activate APC. Second, the checkpoint proteins trigger Cdc20p degradation and keep the available Cdc20p low until all kinetochores attach to spindle microtubules. Both mechanisms are integral parts of the spindle checkpoint (Fig. 8A).
Materials and methods
Growth of yeast
YEPD medium contained 1% yeast extract, 2% bacto-peptone, and 2% glucose. Glucose was substituted by 2% raffinose in YEPR medium. For induction from the GAL promoter, 2% or 3% galactose was added to YEPR medium. For G1 arrest, α-factor (Bachem) was added at 0.5 μg/mL for bar1 cells, or at 10 μg/mL for BAR1 cells from a 10 mg/mL stock in DMSO. Hydroxyurea (Sigma-Aldrich) was added directly to the medium at 200 mM for S-phase arrest. Nocodazole (Sigma-Aldrich) was added from a 10 mg/mL stock in DMSO to a final concentration of 15 μg/mL to disrupt the mitotic spindle at 30°C. To disrupt the spindle at 35°C, we used 30 μg/mL nocodazole and 10 μg/mL benomyl.
Construction of plasmids and yeast strains
Table 1 lists the strains used in this work. All strains are derivatives of W303. Standard genetic techniques were used to manipulate yeast strains (Ausubel et al. 2000). Western blots were used to confirm protein expression or gene deletions. All PCR-derived sequences were confirmed by sequencing.
Table 1.
Yeast strains
| Name | MAT | Relevant genotypea |
|---|---|---|
| RHC292 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1 |
| RHC401 | a | mad2-1, bar1Δ::KANr, 8mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11:PGAL:HIS3 |
| RHC402 | a | mad2-1, bar1Δ::KANr, 8mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11: PGAL-MAD2:HIS3 |
| RHC421 | a | mad2-1, bar1Δ::KANr, ura3-1:PGAL-4mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11:HIS3 |
| RHC422 | a | mad2-1, bar1Δ::KANr, ura3-1:PGAL-4mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11:MAD2:HIS3 |
| RHC435 | a | mad2-1, 8mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11:HIS3 |
| RHC436 | a | mad2-1, 8mycCDC20:URA3, trp1-1:PGAL-pds1Δdb:TRP1, his3-11:MAD2:HIS3 |
| RHC472 | a | bar1, mad1Δ::URA3, 8mycCDC20:HIS3, trp1-1:PGAL-pds1 Δdb:TRP1 |
| RHC473 | a | bar1, mad2Δ::URA3, 8mycCDC20:HIS3, trp1-1:PGAL-pds1 Δdb:TRP1 |
| RHC474 | a | bar1, mad3Δ::URA3, 8mycCDC20:HIS3, trp1-1:PGAL-pds1 Δdb:TRP1 |
| RHC476 | a | bar1, 8mycCDC20:HIS3, trp1-1:PGAL-pdsΔdb:TRP1 |
| RHC482 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3 |
| RHC513A | a/α | cdc20Δ::TRP/CDC20, ura3-1:4mycCDC20:URA3 (3x)/ura3-1, his3-11:4mycCDC20:HIS3 (1x)/his3-11 |
| RHC513B | a/α | cdc20Δ::TRP/CDC20, ura3-1:4mycCDC20:URA3 (3x)/ura3-1, his3-11:4mycCDC20:HIS3 (2x)/his3-11 |
| RHC542 | a | Segregant from RHC513A, cdc20Δ::TRP, his3-11:4mycCDC20:HIS3 (1x) |
| RHC543 | a | Segregant from RHC513B, cdc20Δ::TRP, his3-11:4mycCDC20:HIS3 (2x) |
| RHC544 | a | Segregant from RHC513A, cdc20Δ::TRP, ura3-1:4mycCDC20:URA3 (3x) |
| RHC545 | a | Segregant from RHC513A, cdc20Δ::TRP, ura3-1:4mycCDC20:URA3 (3x), his3-11:4mycCDC20:HIS3 (1x) |
| RHC546 | a | Segregant from RHC513B, cdc20Δ::TRP, ura3-1:4mycCDC20:URA3 (3x), his3-11:4mycCDC20:HIS3 (2x) |
| RHC554 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8mycCDC20 (CEN6, URA3) |
| RHC555 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8myccdc20-106 (CEN6, URA3 |
| RHC556 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8myccdc29-120 (CEN6, URA3) |
| RHC557 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8myccdc20-127 (CEN6, URA3) |
| RHC558 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad2 Δ::HIS3, 8mycCDC20 (CEN6, URA3) |
| RHC576 | a | bar1, cdc23-1, trp1-1:PGAL-pds1Δdb:TRP1 |
| RHC581 | a | bar1, cdc23-1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3 |
| RHC582 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8mycCDC20 (CEN6, HIS3) |
| RHC583 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3, 8mycCDC20 (CEN6, HIS3) |
| RHC584 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8mycCDC20 Δdb1 (CEN6, HIS3) |
| RHC585 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3, 8mycCDC20Δdb1 (CEN6, HIS3) |
| RHC586 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8mycCDC20 Δdb12 (CEN6, HIS3) |
| RHC587 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3, 8mycCDC20Δdb12 (CEN6, HIS3) |
| RHC612 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, 8mycCDC20 (CEN6, HIS3), cdh1 Δ::LEU2 |
| RHC613 | a | bar1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3, 8mycCDC20 (CEN6, HIS3), cdh1 Δ::LEU2 |
| RHC639 | a | bar1Δ::KANr, cdc20 Δ::TRP, PDS1-4HA:LEU2, his3-11:4mycCDC20:HIS3 (1x) |
| RHC641 | a | bar1Δ::KANr, cdc20 Δ::TRP, PDS1-4HA:LEU2, ura3-1:4mycCDC20:URA3 (3x) |
| RHC644 | a | bar1Δ::KANr, cdc20 Δ::TRP, PDS1-4HA:LEU2, his3-11:4mycCDC20:HIS3 (1x), mad2 Δ::URA3 |
| RHC647 | a | bar1, cdc16-1, trp1-1:PGAL-pds1Δdb:TRP1 |
| RHC648 | a | bar1, cdc16-1, trp1-1:PGAL-pds1Δdb:TRP1, mad3 Δ::URA3 |
bAll strains were prepared for this study.
All strains are derivative of W303(ade2-1, can1-100, his3-11, leu2-3,112, trp1-1, ura3-1)
RHC422 was made with pRS403-MAD2, which contains 0.5 kb of the 5′-UTR of MAD2, the open reading frame (ORF) of MAD2, and 1.6 kb of the 3′-UTR of MAD2. pRS403-MAD2 was cut with NheI for integration into the yeast genome.
CDC20 was tagged at its N terminus with 8myc at its genomic locus using pRS306-8mycCDC20N or pRS403-8mycCDC20N. Both plasmids contain 0.5 kb of the 5′-UTR of CDC20 followed by the first 700 bp of CDC20 ORF with eight copies of myc tag inserted right after the start codon ATG. The plasmids were cut with MluI within the CDC20 ORF for integration into the genome.
pRS403PGAL and pRS306PGAL were constructed by inserting the GAL1 promoter–MCS-CYC1 terminator cassette, which was amplified by PCR from pYES2 (Stratagene), at NotI and SacI sites in pRS403 and pRS306, respectively. MAD2 coding sequences were inserted at EcoRI site in pRS403PGAL, and the resulting plasmid was cut with NheI for integration. For pRS306PGAL-4mycCDC20, the complete CDC20 ORF with four copies of myc at the N terminus was placed between XhoI and KpnI sites in pRS306PGAL. The plasmid was then cut with StuI for integration.
For tagging Pds1p, four copies of HA sequence were inserted before the stop codon of PDS1 in the pRS405 vector, which contains the entire PDS1 ORF and 1 kb of the 3′-UTR. The plasmid was cut with BglII within ORF for integration at the genomic locus.
RHC513A and RHC513B were made with pRS306-4myc-CDC20 and pRS403-4mycCDC20. These plasmids contain 0.5 kb of the 5′-UTR of CDC20 followed by the entire CDC20 ORF with four copies of myc tag inserted right after the start codon ATG. These two constructs were cut with StuI and NheI, respectively, for integration. Transformants with multiple copies of integrated plasmids were selected based on their expression levels of 4mycCdc20p. 8myc-tagged CDC20 or its mutant forms cdc20-106, cdc20-120, and cdc20-127 were expressed from pRS316-based constructs that contain 0.5 kb of the 5′-UTR of CDC20 followed by the 8myc-tagged coding sequences.
Strains containing PGAL-pds1Δdb were made with pRTK-C2 (Tinker-Kulberg and Morgan 1999). Destruction box 1 (amino acids 17–25) and destruction box 2 (amino acids 60–68) of CDC20 were removed using the QuikChange site-directed mutagenesis kit (Stratagene).
BAR1 was deleted with pBlueScript-based bar1Δ construct pRC610, which contains the KANr gene flanked by 1.9 kb of the 5′-UTR and 1.7 kb of the 3′-UTR of BAR1. bar1Δ cells were identified as G418-resistant yeast transformants that were sensitive to 0.5 μg/mL α-factor.
CDH1 was deleted using pWS176 (Schwab et al. 1997), and the deletion was confirmed by PCR analysis.
Yeast cell lysates, Western blotting, and immunoprecipitation
Yeast cell lysates were prepared by bead-beating. For Western blotting, 5-mL cultures at O.D.600 = 0.5–1.5 were used to prepare lysates, and 20-mL cultures were used for immunoprecipitation. Cultures were cooled on ice before cells were harvested and washed once with cold TE (10 mM Tris at pH 7.5, 1 mM EDTA). Cell pellets were either quickly frozen and temporarily stored in liquid nitrogen, or beat immediately in 100–200 μL of lysis buffer (50 mM HEPES at pH 7.6, 75 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM β-glycerophosphate, 1 mM sodium vanadate, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF, and 10 μg/mL each of leupeptin, pepstatin, and chymostatin) together with Zirconia beads of the same volume in a minibead-beater (BioSpec Products) for 1 min at 4°C. Samples were then centrifuged at 14,000 rpm for 5 min at 4°C, and the supernatants were taken as yeast cell lysates. Protein concentration was determined with DC protein assay kit (Bio-Rad), and then normalized with lysis buffer.
For immunoprecipitation, anti-Mad2p antibody or anti-myc antibody 9E10 was incubated with lysates containing 1 mg of protein for 1 h on ice. Samples were then transferred to tubes containing protein A beads preblocked with 1 mg/mL BSA in lysis buffer, and incubated for 30 min at 4°C. The beads were then washed with lysis buffer four times, followed by denaturation in SDS sample buffer.
For Western blotting, 40 μg of the cell lysates or one-third of the immunoprecipitates were resolved by SDS-PAGE. Affinity-purified rabbit anti-Mad2p antibody was used at a dilution of 1:1000 (Chen et al. 1999), rabbit anti-Mad1p antibody at 1:4000 (Hardwick and Murray 1995), rabbit anti-Mad3p antibody at 1: 500 (Hardwick et al. 2000), mouse anti-myc antibody (9E10, Covance) at 1:1000, mouse anti-HA antibody (16B12, Covance) at 1:1000, rabbit anti-Clb2p antibody (Santa Cruz Biotechnology) at 1:1000, and goat anti-Cdc20p antibody yC-20 (Santa Cruz Biotechnology) at 1:200, all in PBS containing 2% BSA and 0.2% Tween 20.
Northern blotting
Total RNA was prepared with the RiboPure-Yeast kit (Ambion) and separated on a glyoxal gel with reagents from the NorthernMax-Gly kit (Ambion). Probes for CDC20 and ACT1 mRNA were prepared with the DECAprime II kit (Ambion). Hybridization was performed as previously described (Goh et al. 2000). Briefly, hybridization was carried out in pH 6.2 buffer (0.6 M NaCl, 0.18 M Na2HPO4, 0.006 M EDTA, 10% Dextran sulfate [500,000], 1% lauroyl sarcosinate) and 46 μg/mL salmon sperm DNA overnight at 65°C. The membrane was first washed at room temperature in 2× SSC plus 0.1% SDS, followed by extensive washes at 55°C in 0.1× SSC plus 0.1% SDS. The blot was then exposed to a storage phosphor screen (Amersham), and signals were quantified with NIH image software.
Acknowledgments
We thank Marion Shonn for providing the plasmid for PGAL-pds1Δdb, Andrew Murray for cdc20 mutant plasmids and yeast strain cdc16-1, Wolfgang Seufert for the CDH1 disruption plasmid, and Kevin Hardwick for MAD1 and MAD3 disruption plas-mids, as well as anti-Mad1p and anti-Mad3p antibodies. We thank Hong Hwa Lim for advice on CDC20 Northern blotting. We also thank Tim Huffaker for critically reading the manuscript. This work was supported by grants from the National Institutes of Health (NIH) and the David and Lucile Packard Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1184204.
References
- Ausubel F.M., Brent, R., Kingston, B.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 2000. Current protocols in molecular biology, John Wiley & Sons, Inc., New York.
- Chen R.H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158: 487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady, D.M., Smith, D., Murray, A.W., and Hardwick, K.G. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell 10: 2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E. and Chen, R.H. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell 13: 1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.J., Segal, M., Andrews, C.A., Rudyak, S.G., Jensen, S., Smith, K., and Reed, S.I. 2003. S-Phase checkpoint controls mitosis via an APC-independent Cdc20p function. Nat. Cell Biol. 5: 928-935. [DOI] [PubMed] [Google Scholar]
- Cleveland D.W., Mao, Y., and Sullivan, K.F. 2003. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112: 407-421. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O. and Koshland, D. 1999. Pds1p of budding yeast has dual roles: Inhibition of anaphase initiation and regulation of mitotic exit. Genes & Dev. 13: 1950-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson, V.L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N., and Taylor, S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161: 267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal, V., Scott, M.L., Benezra, R., and Sorger, P.K. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635-645. [DOI] [PubMed] [Google Scholar]
- Fang G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 13: 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu, H., and Kirschner, M.W. 1998a. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 12: 1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998b. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2: 163-171. [DOI] [PubMed] [Google Scholar]
- Fraschini R., Beretta, A., Sironi, L., Musacchio, A., Lucchini, G., and Piatti, S. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20: 6648-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D., Colaluca, I.N., Vodermaier, H.C., Biamonti, G., Giacca, M., Falaschi, A., Riva, S., and Peverali, F.A. 2003. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 22: 3715-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber P.M. and Rine, J. 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161: 521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S., Kramer, E., Gieffers, C., Gannon, J., Peters, J.M., and Hunt, T. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153: 137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.Y., Lim, H.H., and Surana, U. 2000. Cdc20 protein contains a destruction-box but, unlike Clb2, its proteolysisis not acutely dependent on the activity of anaphase-promoting complex. Eur. J. Biochem. 267: 434-449. [DOI] [PubMed] [Google Scholar]
- Hames R.S., Wattam, S.L., Yamano, H., Bacchieri, R., and Fry, A.M. 2001. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 20: 7117-7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G. and Murray, A.W. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131: 709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Johnston, R.C., Smith, D.L., and Murray, A.W. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148: 871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Burton, J.L., and Solomon, M.J. 2002. The anaphase-promoting complex: It's not just for mitosis any more. Genes & Dev. 16: 2179-2206. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z., Chung, Y.S., Mochizuki, Y., Hardy, C.F., and Cohen-Fix, O. 2001. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 11: 1347-1352. [DOI] [PubMed] [Google Scholar]
- Howell B.J., Hoffman, D.B., Fang, G., Murray, A.W., and Salmon, E.D. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150: 1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis, L., and Roberts, B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtu-bule function. Cell 66: 507-517. [DOI] [PubMed] [Google Scholar]
- Huang J.N., Park, I., Ellingson, E., Littlepage, L.E., and Pellman, D. 2001. Activity of the APCCdh1 form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154: 85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L.H., Lau, L.F., Smith, D.L., Mistrot, C.A., Hardwick, K.G., Hwang, E.S., Amon, A., and Murray, A.W. 1998. Budding yeast Cdc20: A target of the spindle checkpoint. Science 279: 1041-1044. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Earle, E., Fowler, K.J., and Choo, K.H. 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes & Dev. 14: 2277-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M.J., Beardmore, V.A., Weinstein, J., and Gorbsky, G.J. 2002. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158: 841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin, D.P., Matsumoto, S., Kitazono, A., and Matsumoto, T. 1998. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science 279: 1045-1047. [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer, N., Podtelejnikov, A.V., Mann, M., and Peters, J.M. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell 11: 1555-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. and Murray, A.W. 1991. Feedback control of mitosis in budding yeast. Cell 66: 519-531. [DOI] [PubMed] [Google Scholar]
- Li X. and Nicklas, R.B. 1995. Mitotic forces control a cell-cycle checkpoint. Nature 373: 630-632. [DOI] [PubMed] [Google Scholar]
- Michel L.S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P.K., Murty, V.V., and Benezra, R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409: 355-359. [DOI] [PubMed] [Google Scholar]
- Millband D.N. and Hardwick, K.G. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22: 2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M.C., Kaiser, P., Rudyak, S., Baskerville, C., Watson, M.H., and Reed, S.I. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 424: 1009-1013. [DOI] [PubMed] [Google Scholar]
- Musacchio A. and Hardwick, K.G. 2002. The spindle checkpoint: Structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 3: 731-741. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: The molecular biology of chromosome separation. Science 297: 559-565. [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Waters, J.C., Salmon, E.D., and Ward, S.C. 2001. Checkpoint signals in grasshopper meiosis are sensitive to microtubule attachment, but tension is still essential. J. Cell Sci. 114: 4173-4183. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M. and Kirschner, M.W. 2000. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 14: 655-665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Lee, E., and Kirschner, M.W. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes & Dev. 15: 2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Hwang, E.S., Visintin, R., and Amon, A. 1998. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 8: 750-760. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Cole, R.W., Khodjakov, A., and Sluder, G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130: 941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott E.J. and Hoyt, M.A. 1998. Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics 148: 599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Lutum, A.S., and Seufert, W. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683-693. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner, M., Mocker, D., and Seufert, W. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20: 5165-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Zachariae, W., Ciosk, R., and Nasmyth, K. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17: 1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., Andreassen, P.R., Lacroix, F.B., Wilson, L., and Margolis, R.L. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. 98: 4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen C.S., Lukas, C., Kramer, E.R., Peters, J.M., Bartek, J., and Lukas, J. 2000. Nonperiodic activity of the human anaphase-promoting complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Mol. Cell. Biol. 20: 7613-7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B.M. and Murray, A.W. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11: 1462-1467. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan, G.K., and Yen, T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154: 925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj, R., Li, B., and Yu, H. 2001. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1: 227-237. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg R.L. and Morgan, D.O. 1999. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes & Dev. 13: 1936-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C.D., Brady, D.M., Johnston, R.C., Hanna, J.S., Hardwick, K.G., and Spencer, F.A. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13: 3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. 1997. Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J. Biol. Chem. 272: 28501-28511. [DOI] [PubMed] [Google Scholar]
- Weiss E. and Winey, W. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132: 111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14: 706-714. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Lees, E. 2001. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: Model for spindle checkpoint regulation. Mol. Cell. Biol. 21: 5190-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]