Abstract
Several recent pharmacogenetic studies have investigated the variability in both outcome and toxicity in cisplatin-based therapies. These studies have focused on the genetic variability of therapeutic targets that could affect cisplatin response and toxicity in diverse type of cancer including lung, gastric, ovarian, testicular, and esophageal cancer. In this review, we seek to update the reader in this area of investigation, focusing primarily on DNA reparation enzymes and cisplatin metabolism through Glutathione S-Transferases (GSTs). Current evidence indicates a potential application of pharmacogenetics in therapeutic schemes in which cisplatin is the cornerstone of these treatments. Therefore, a collaborative effort is required to study these molecular characteristics in order to generate a genetic panel with clinical utility.
Keywords: pharmacogenetics, chemotherapy, cisplatin, polymorphisms, NER pathway, glutathione S-transferases
Introduction
Cisplatin is an alkylating agent used to treat several types of cancers that works by causing DNA lesions via the formation of intrastrand and interstrand crosslinks, resulting in the activation of various signal-transduction pathways that block cellular processes, such as replication and transcription. The action of cisplatin is cell cycle-independent, although in some cases, prolonged G2 phase cell-cycle arrest occurs (Siddik, 2003; Kelland, 2007). Cisplatin has a central role in cancer chemotherapy for testicular, ovarian/cervical, head and neck, and non-small-cell cancers. The side effects include nephrotoxicity (Wong and Giandomenico, 1999), hematogenesis and neurotoxicity (Decatris et al., 2004).
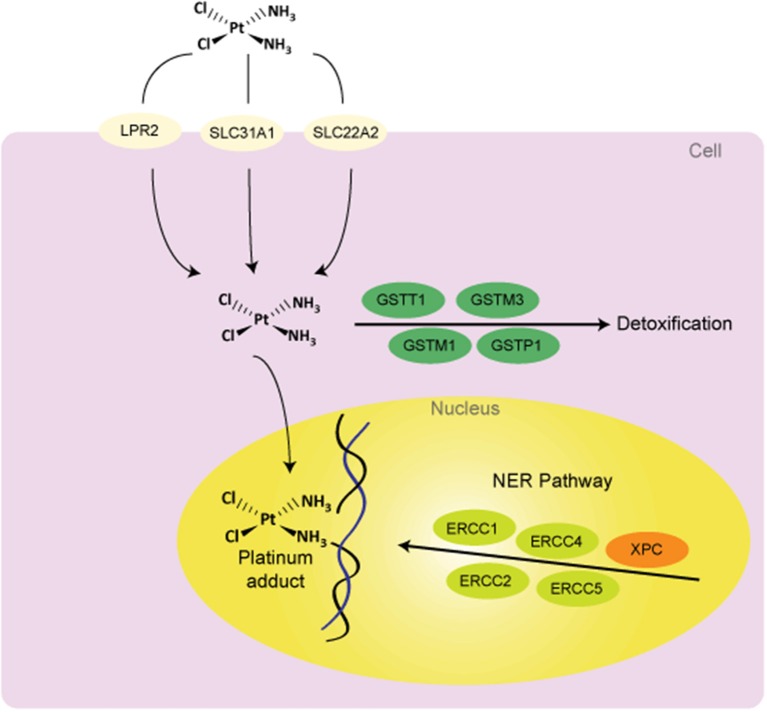
From the beginning, cisplatin has presented variations in therapeutic response. While some tumors are hypersensitive to anticancer therapy, other tumors have an intrinsic resistance. Investigations have sought an explanation of this variation and have suggested that the major resistance mechanisms include reduction in drug levels that reach the target DNA due to reduced uptake and/or increased efflux; increased cellular thiol levels; enhanced DNA repair and/or increased damage tolerance; and failure of cell-death pathways after the formation of platinum-DNA adducts (Fojo, 2001; Siddik, 2003; Wang and Lippard, 2005). In each of these processes there exist potential sites of pharmacogenetics variability (Figure 1). Changes at the genetic level causing modifications in cellular phenotype could explain some of the variability in response and toxicity to cisplatin-included chemotherapy. In this review, we discuss associations between genetic variants in the germ line and in outcomes following cisplatin-based chemotherapy. We mainly focus on DNA repair and cisplatin detoxification through Glutathione S-Transferases (GSTs).
Figure 1.
Potential sources of variability to clinical response to cisplatin treatment. Abbreviations: DNA, deoxyribonucleic acid; GSTs, glutathione S-Transferases; NER, nucleotide excision repair; LPR2, Low Phosphate Root2; SLC31A1 (CTR1), solute carrier family 31 (copper transporter), member 1; SLC22A2, solute carrier family 22 (organic cation transporter), member 2; ERCCs, Excision Repair Cross Complementing group of proteins; XPC, Xeroderma Pigmentosum Group C Protein.
Pharmacodynamic mechanisms
Cisplatin modulates several signal transduction pathways involving AKT (v-akt murine thymoma viral oncogene homolog), c-ABL (v-abl Abelson murine leukemia viral oncogene homolog 1), p53, and MAPK (mitogen-activated protein kinase)/JNK (c-Jun NH2-terminal kinase)/ERK (extracellular signal-regulated kinase). Cell death induced by cisplatin is concentration dependent and includes necrosis and apoptosis mechanisms (Gonzalez et al., 2001). Necrosis involves hyper-activation of Poly (ADP ribose) polymerase (PARP) (Nguewa et al., 2003) while apoptosis results from activation of CASP8, CASP9, CASP3, and CASP7 (Gonzalez et al., 2001).
Cisplatin distorts the structure of the DNA that generate intrastrand 1, 2—crosslinks binding proteins into shallow minor groove [high-mobility group (HMG) box proteins, repair proteins, transcription factors, histone H1] (Kartalou and Essigmann, 2001; Wozniak and Blasiak, 2002; Zdraveski et al., 2002). It covalently binds DNA and forms DNA adducts through intra- and interstrand crosslinks (ICLs). Intrastrand crosslinks are repaired by nucleotide excision repair (NER) using the other strand as a template. As both strands are compromised in ICLs, other enzymes are involved in their repair. Two major pathways of ICL repair exist; one is replication dependent and mainly involves homologous recombination, the second is replication independent and involves NER (Ho and Schärer, 2010). At the start of both of these pathways, translesion (TLS) polymerases are needed to bypass ICLs and restore one of the two DNA strands. Translesion synthesis is a mechanism used by cells to prevent common DNA damage from stalling replication forks and rising apoptosis levels. The most important TLS polymerases are Pol ζ (Polymerase zeta) and REV1 (Reversionless 1). Studies have shown that disruption or suppression of expression of both REV3L, the gene encoding the catalytic subunit of Pol ζ, or REV1 modifies sensitivity to cisplatin (Lin et al., 2006; Doles et al., 2010). Goricar et al. (2014) recently determined in patients with malignant mesothelioma that the mutant allele in REV1 rs3087403 and REV1 TGT haplotype associated with increased risk for leukopenia and neutropenia. REV3L rs465646, rs462779, and REV3L CCGG haplotype associated with longer overall survival (Goricar et al., 2014).
DNA repair enzymes
DNA damage repair mechanisms are as follows: direct repair of alkyl adducts; repair of base damage and single strand breaks by base excision repair; repair of double strand breaks by homologous recombination or by non-homologous end joining; repair of bulky DNA adducts by NER; and repair of mismatches and insertion/deletion loops by DNA mismatch repair (Camps et al., 2007). The NER pathway is one of the major DNA repair systems involved in the removal of platinum adducts. This pathway involves many proteins in lesion recognition, excision, DNA synthesis and ligation. Excision repair cross-complementary 1 (ERCC1) is a key protein involved in the process of NER and ERCC1-xeroderma pigmentosum (ERCC1-XPF) catalyzes incision on the incision 50 side to the site of DNA damage (Parker et al., 1991; Bessho, 1995). In addition to ERCC1, xeroderma pigmentosum complementary group D (XPD) encodes a helicase that participates in both NER and basal transcription as part of the transcription factor, IIH. Mutations destroying the enzymatic function of XPD protein are manifested clinically in combinations of three severe syndromes, including xeroderma pigmentosum, XP combined with Cockayne Syndrome and trichothiodystrophy (Lehmann, 2001; Clarkson and Wood, 2005). ERCC1 and ERCC2 (XPD) have pivotal roles in the NER pathway, this has been evidenced in studies where lower levels of intratumoral ERCC1 mRNA are significantly correlated with improved survival due to enhanced tumor cell sensitivity to cisplatin (Shirota et al., 2001). mRNA levels as well as the overexpression of ERCC1 and other enzymes have been implicated in the development of clinical resistance to platinum (Kirschner and Melton, 2010; Cheng et al., 2012).
Among these genes, the most studied is ERCC1 gene, mostly focused on the therapy of non-small cell lung cancer (NSCLC) and esophageal cancer. Polymorphisms in ERCC1 include mainly rs3212986 and rs11615. The polymorphism rs3212986 is located in the 3′ untranslated region and therefore may affect mRNA stability resulting in a decreased expression levels (Chen et al., 2000). In relation to rs3212986, the C allele leads to a change that results in an increase in overall survival (Zhou et al., 2004; Krivak et al., 2008; Takenaka et al., 2010), progression free survival (Krivak et al., 2008; Kim et al., 2009; Erčulj et al., 2012; Chen et al., 2013), treatment response (Li et al., 2010) and prognosis (Takenaka et al., 2010; Okuda et al., 2011). However, opposite associations have been reported in other studies related to reduced responses with the C allele (Bradbury et al., 2009; Kalikaki et al., 2009; Park et al., 2011; Wang et al., 2011), as well as increased toxicity (Khrunin et al., 2010; Tzvetkov et al., 2011; Erčulj et al., 2012). Wang et al. (2011) and Bradbury et al. (2009) showed that in esophageal cancer, patients with A/A or A/C genotype had improved outcomes compared with patients carrying wild-type genotypes. In addition, Park et al. (2011) have found similar results in metastatic cancer patients. On the contrary, opposite results have been found in NSCLC and ovarian cancer where the C allele relates to improved survival and treatment response. The variability in outcomes amongst these studies could be due to tumor characteristics (tissue-specific or organ-specific). The polymorphism C→T at codon 118 located on exon 4 of ERCC1 gene (rs11615) is expected to have the same effect. This polymorphism is associated with clinical response to platinum-based chemotherapy in NSCLC. The C allele is also related to an increase in overall survival (Isla et al., 2004; Ryu et al., 2004; Cheng et al., 2012; Joerger et al., 2012), progression free survival (Ryu et al., 2004; Cheng et al., 2012; Joerger et al., 2012), improved treatment response (Kalikaki et al., 2009) and prognosis (Okuda et al., 2011). Nevertheless, others authors detect opposite associations in larger-population studies, including amongst Chinese patients (Li et al., 2010; Ren et al., 2012): this should be considered in future research. Nephrotoxicity has been related to the C allele in rs3212986 ERCC1 (Tzvetkov et al., 2011), T allele in rs11615 ERCC1 (Tzvetkov et al., 2011) and C/T genotype in rs3212986 ERCC1 (Khrunin et al., 2010), independent of cancer type.
Another widely studied gene is ERCC2 (XPD). The presence of a variation in ERCC2 gene (rs13181 and rs1799793) reduces repair capacity, and results in greater efficacy of cisplatin treatment due to increased DNA damage and an enhanced cytotoxic effect. rs1799793 generates a positive effect in overall survival and progression free survival (Gurubhagavatula et al., 2004; Bradbury et al., 2009; Biason et al., 2012). Erčulj et al. (2012) found that G/G genotype is related to an increase in various types of toxicity (Erčulj et al., 2012) while nephrotoxicity has been shown by Joerger et al. (2012) (Joerger et al., 2012). The A allele in the mutation rs13181 increases overall survival (Park et al., 2001; Quintela-Fandino et al., 2006; Caronia et al., 2009; Chew et al., 2009). However, other authors have found the C allele related to increased overall survival (Bradbury et al., 2009) in esophageal cancer and progression free survival in pancreatic cancer (Avan et al., 2013). These discrepancies suggest that associations with C allele are not fully clear in these types of cancers, and that patients factors, treatment modalities and ethnic population could influence the outcome. Nonetheless, the majority of the results support an association between both rs1799793 and rs13181 and clinical outcomes in patients with NSCLC, osteosarcoma, breast cancer, ovarian cancer, and colorectal cancer. These significant associations in ERCC2 polymorphisms and clinical outcomes have included studies with a larger number of patients and differing patient populations.
Other studies found associations between ERCC5 mutations (rs1047768 and rs751402), PFS (progression free survival) (Sun et al., 2013) and OS (overall survival) (He et al., 2013). These studies have indicated that ERCC5 polymorphisms are involved in the efficacy of cisplatin neoadjuvant chemotherapy. Also, ototoxicity has related to rs2228001 mutation in the Xeroderma Pigmentosum Complementation group C (XPC) gene (Caronia et al., 2009). More information is needed about these associations to reach more powerful conclusions, including a greater number of patients and amongst different ethnic populations.
Additional DNA repair genes have also shown variability, including X-ray repair cross-complementing group 1 (XRCC1). This protein is involved in base excision repair. Among the mutations, we highlight rs25487 and rs1799782 mutations. In relation to rs25487, the mutant G variant has been associated with decreased treatment response (Gurubhagavatula et al., 2004; Giachino et al., 2007; Pacetti et al., 2009; Khrunin et al., 2010; Joerger et al., 2012; Ke et al., 2012; Miao et al., 2012), although opposite results exist (Quintela-Fandino et al., 2006; Sakano et al., 2006). Other evidence indicates associations between the G allele and neutropenia (Khrunin et al., 2010). T allele in rs1799782 mutation is related with an increase (Miao et al., 2012; Li and Li, 2013) and decrease in overall survival (Li et al., 2006; Shim et al., 2010). Li and Li (2013) and Miao et al. (2012) have performed studies in ovarian cancer with a large number of patients. Further data are required to confirm this association. Another finding is the relation between treatment response and the T allele (Wang et al., 2004; Yuan et al., 2006; Kim et al., 2009; Ke et al., 2012). This discrepancy may be due to cancer type or combined therapies. DNA repair enzymes might decrease the synergistic effects of combination of cisplatin and radiation and information from population should be added in future association specifics to subgroups (Li and Li, 2013). In addition, some studies have used cisplatin in combination with paclitaxel, gemcitabine, cyclophosphamide or 5-FU, depending on cancer type. Others factors that might affect variability in different populations are the stage of disease, patient status and period of follow-up in survival analysis.
With respect to X-ray repair cross complementing protein 3 (XRCC3), a protein involved in DNA double-strand breaks, the rs861539 mutation is the only one that relates to treatment outcome. Increased overall survival was associated with the T allele (De las Peñas et al., 2006; Chen et al., 2012) as was progression free survival (Font et al., 2008). However, Ren et al. (2012) have shown inverse results (Ren et al., 2012) including a large number of patient (n = 340) with NSCLC. More data are necessary to confirm these opposing results.
In summary, studies of association between genetic variants in the DNA repair system and clinical results show that these variants can be potential biomarkers for outcomes in the cisplatin-based therapies (Table 1). Despite race and treatment regimen, associations testing the polymorphism in ERCC1 appear to follow a consistent direction. rs3212986 and rs11615 polymorphisms should be considered in a future genetic panel because results were obtained in several researches with different treatment and demographic characteristics. Additional research should be performed in order to replicate results found with polymorphisms in ERCC2, XRCC1, and XRCC3. In additional studies, the later polymorphism should be used to evaluate clinical outcomes (overall survival and disease progression) considering different subgroups of patient. In relation to specific toxicities, associations with nephrotoxicity have been described and characterized, but likewise require confirmation.
Table 1.
Summary of association studies between genetic polymorphisms and outcomes in the cisplatin-based chemotherapy.
| Gene | Mutation | Cancer | References | Number of subjects | Results |
|---|---|---|---|---|---|
| ERCC1 | Gln504Lys | NSCL | Kalikaki et al., 2009 | 119 | C/C ↓ OS |
| rs3212986 | Nigro et al., 2010 | 7 | Related with survival | ||
| NC_000019.10:g.45409478C>A | Okuda et al., 2011 | 90 | C/C ↑ Prognosis | ||
| NG_015839.2:g.74351G>T | Takenaka et al., 2010 | 122 | C/C ↑ DFS and OS | ||
| Zhou et al., 2004 | 128 | C/C ↑ OS | |||
| Li et al., 2010 | 115 | C→ A ↓ Response | |||
| Advanced esophageal cancer | Wang et al., 2011 | 241 | C/C ↓ Remission rate and PFS | ||
| Bradbury et al., 2009 | 262 | Related with OS | |||
| Rumiato et al., 2013 | 143 | Related outcomes | |||
| Nasopharyngeal cancer | Chen et al., 2013 | 101 | C/C ↓ Risk of progression | ||
| Liu et al., 2013 | 104 | C/C ↓ PFS | |||
| Epithelial ovarian cancer | Kim et al., 2009 | 118 | C/A or A/A ↓ PFS and OS | ||
| Krivak et al., 2008 | 233 | C/C ↑ PFS and OS | |||
| Khrunin et al., 2010 | 104 | C/A ↑ Risk of nephrotoxicity | |||
| Malignant mesothelioma | Erčulj et al., 2012 | 133 | C/C ↑ PFS, Risk of toxicity | ||
| Cisplatin-treated cancer | Tzvetkov et al., 2011 | 79 | C allele ↓ eGFR (Nephrotoxicity) | ||
| Metastatic gastric cancer | Park et al., 2011 | 108 | C/C ↓ Response rate and Time to progression | ||
| Asn118Asn | NSCLC | Cheng et al., 2012 | 142 | C/C ↑ Response rate, PFS and OS | |
| rs11615 | Joerger et al., 2012 | 137 | C/C ↑ Response rate, PFS and OS | ||
| NC_000019.10:g.45420395A>G | Okuda et al., 2011 | 90 | C/C ↑ Prognosis | ||
| NG_015839.2:g.63434T>C | Ryu et al., 2004 | 109 | C/C ↑ Survival | ||
| Isla et al., 2004 | 62 | C allele ↑ Survival | |||
| Li et al., 2010 | 115 | C→ T ↑ Response | |||
| Su et al., 2007 | 230 | T allele ↑ Response | |||
| Ren et al., 2012 | 340 | C/C ↓ survival | |||
| Kalikaki et al., 2009 | 119 | C/C, C/T ↑ Response | |||
| Advanced esophageal cancer | Warnecke-eberz et al., 2009 | 52 | T/T ↑ Response | ||
| Epithelial ovarian cancer | Smith et al., 2007 | 103 | C/C ↑ Progression and death | ||
| Osteosarcoma | Hao et al., 2012 | 267 | T/T ↑ Event free survival | ||
| Esophageal adenocarcinoma | Metzger et al., 2012 | 217 | C/C ↓ Response | ||
| Melanoma | Liu et al., 2005 | 90 | C/C ↓ Response | ||
| Pancreatic cancer | Kamikozuru et al., 2008 | 67 | T allele ↑ PFS and OS | ||
| Cisplatin-treated cancer | Tzvetkov et al., 2011 | 79 | T allele ↓ eGFR (Nephrotoxicity) | ||
| Haplotype | Advanced gastric cancer | Goekkurt et al., 2009 | 156 | T allele/C allele ↑ grade 3-4 neutropenia | |
| rs3212986/rs11615 | |||||
| ERCC2 (XPD) | Asp312Asn | Esophageal cancer | Bradbury et al., 2009 | 262 | Related with OS |
| rs1799793 | |||||
| NC_000019.10:g.45364001C>T | |||||
| NG_007067.2:g.11587G>A | |||||
| Malignant mesothelioma | Erčulj et al., 2012 | 133 | G/G ↑ Risk of toxicity | ||
| Ovarian cancer | Khrunin et al., 2010 | 104 | G/G ↑ Severe neutropenia | ||
| NSCLC | Gurubhagavatula et al., 2004 | 103 | A allele ↓ OS | ||
| Joerger et al., 2012 | 137 | A allele related with OS | |||
| Squamous cell carcinoma of the head and neck | Quintela-Fandino et al., 2006 | 103 | A allele ↑ OS | ||
| Osteosarcoma | Biason et al., 2012 | 130 | G/A or A/A ↑ Response | ||
| Lys751Gln | Esophageal cancer | Bradbury et al., 2009 | 262 | Related with OS | |
| rs13181 | |||||
| NC_000019.10:g.45351661T>G | |||||
| NG_007067.2:g.23927A>C | |||||
| Pancreatic cancer | Avan et al., 2013 | 122 | Related with risk of death | ||
| Colorectal cancer | Park et al., 2001 | 73 | A/A ↑ response | ||
| NSCLC | Chen et al., 2012 | 355 | A/A ↑ OS | ||
| Ren et al., 2012 | 340 | A/A ↑ OS | |||
| Ludovini et al., 2011 | 192 | C/C ↑ PFS | |||
| Osteosarcoma | Caronia et al., 2009 | 91 | Allele G ↓ Response | ||
| Hao et al., 2012 | 267 | A/A ↑ Event free survival | |||
| Squamous cell carcinoma of the head and neck | Quintela-Fandino et al., 2006 | 103 | C allele ↑ OS | ||
| Breast cancer | Chew et al., 2009 | 55 | Related with clinical outcomes | ||
| Haplotype (rs1799793 /rs13181) | Advanced gastric cancer | Goekkurt et al., 2009 | 156 | Related with nephrotoxicity | |
| rs50872 | NSCLC | Kim et al., 2012 | 129 | A/A ↓ OS | |
| NC_000019.10:g.45359191A>G | |||||
| NG_007067.2:g.16397T>C | |||||
| Asp711Asp | NSCLC | Li et al., 2013 | 496 | C/T + T/T ↓ Response | |
| rs1052555 | |||||
| NC_000019.10:g.45352266G>A | |||||
| NG_007067.2:g.23322C>T | |||||
| XPC | Lys939Gln | Osteosarcoma | Caronia et al., 2009 | 91 | C/C association with ototoxicity |
| rs2228001 | |||||
| NC_000003.12:g.14145949G>T | |||||
| NG_011763.1:g.37724C>A | |||||
| ERCC5 | rs1047768 | Osteosarcoma | Sun et al., 2013 | 182 | T/T ↑ PFS and OS |
| NC_000013.11:g.102852167T>C | |||||
| NG_007146.1:g.11344T>C | |||||
| rs751402 | NSCLC | He et al., 2013 | 228 | A/A ↓ Response | |
| NC_000013.11:g.102845848A>G | |||||
| NG_007146.1:g.5025A>G | |||||
| XRCC1 | Gln399Arg | Ovarian cancer | Chung et al., 2006 | 36 | A allele ↓ Response |
| rs25487 | Khrunin et al., 2010 | 104 | G/G ↓ Severe neutropenia | ||
| NC_000019.10:g.43551574T>C | |||||
| NG_033799.1:g.29005A>G | NSCLC | Gurubhagavatula et al., 2004 | 103 | A allele ↓ OS | |
| Joerger et al., 2012 | 137 | G allele related with OS | |||
| Wang et al., 2004 | 105 | Gallele ↑ Response rate | |||
| Giachino et al., 2007 | 203 | A/A ↑ Median Survival Time | |||
| Ke et al., 2012 | 460 | A/A ↑ Survival | |||
| Lee et al., 2013 | 382 | A allele ↓ Response | |||
| Advanced gastric cancer | Goekkurt et al., 2009 | 156 | Related with OS | ||
| Ji et al., 2013 | 59 | A/A ↑ OS | |||
| Nasopharyngeal cancer | Zhai et al., 2013 | 60 | A/A related with remission | ||
| Ovarian cancer | Li and Li, 2013 | 335 | A/A ↑ Risk of death | ||
| Miao et al., 2012 | 195 | A/A ↑ Risk of death | |||
| Ovarian cancer | Khrunin et al., 2010 | 104 | A/A Severe neutropenia | ||
| Musculoskeletal cancer | Sakano et al., 2006 | 78 | G/A + A/A ↑ OS | ||
| Squamous cell carcinoma of the head and neck | Quintela-Fandino et al., 2006 | 103 | A allele ↑ OS | ||
| Biliary tract carcinoma | Pacetti et al., 2009 | 33 | G/G ↓ OS | ||
| Arg194Trp rs1799782 | Pancreatic cancer | Li et al., 2006 | 92 | T allele ↓ Survival | |
| NC_000019.10:g.43553422G>A | |||||
| NG_033799.1:g.27157C>T | |||||
| NSCLC | Sun et al., 2009 | 82 | C/T ↑ Response | ||
| Wang et al., 2004 | 105 | C/T or T/T ↑ Response | |||
| Hong et al., 2009 | 164 | C/T + T/T ↑ Response | |||
| Ke et al., 2012 | 460 | T/T ↓ Risk of death | |||
| Ovarian cancer | Li and Li, 2013 | 335 | T/T ↑ OS | ||
| Miao et al., 2012 | 195 | T/T ↑ Risk of death | |||
| Cervical cancer | Kim et al., 2008 | 66 | C/C ↓ Response | ||
| Gastric cancer | Shim et al., 2010 | 200 | C/T ↓ OS | ||
| Haplotype | NSCLC | Hong et al., 2009 | 164 | A - T haplotype ↑ Response | |
| (rs25487/rs1799782) | |||||
| XRCC3 | Thr241Met rs861539 | NSCLC | De las Peñas et al., 2006 | 135 | T/T ↑ OS |
| NC_000014.9:g.103699416G>A | Ren et al., 2012 | 340 | C/C ↑ OS | ||
| NG_011516.1:g.21071C>T NG_012307.1:g.75229G>A | Breast cancer | Chew et al., 2009 | 136 | C/C ↑ Response rate and PFS | |
| Advanced esophageal cancer | Font et al., 2008 | 28 | T/T ↑ TTP | ||
| Adenocarcinoma of esophageal and stomach | Ott et al., 2011 | 258 | T allele ↑ OS |
Polymorphisms on DNA repair.
OS, Overall survival; PFS, Progression free survival; TTP, Time to progression; EFS, Event free survival; DFS, Disease free survival; G-CSF, Granulocyte-Colony Stimulating Factor; eGFR, Glomerular filtration rate.
Pharmacokinetic mechanisms
Evidence indicates that reduced drug accumulation is a significant mechanism of cisplatin resistance (Kelland, 1993). The cause may be an inhibition in drug uptake, an increase in drug efflux, or both. Studies concerning the mechanisms of cisplatin uptake into the cell have focused on both passive diffusion (Hromas et al., 1987; Binks and Dobrota, 1990; Mann et al., 1991) and copper transporters (Katano et al., 2002; Ohashi et al., 2003; Safaei et al., 2004).
Recent studies have demonstrated that mutation or deletion of the CTR1 gene results in increased cisplatin resistance and reduction of platinum levels (Ishida et al., 2002). Copper-transporting P-type adenosine triphosphate (ATP7B) is associated with cisplatin resistance in vitro (Komatsu et al., 2000), and in various cancers (Nakayama et al., 2002, 2004; Ohbu et al., 2003). ATP-binding cassette sub-family C2 (ABCC2), another transporter protein, also has a role in cisplatin resistance, probably promoting drug efflux (Koike et al., 1997; Kool et al., 1997; Cui et al., 1999). ABCC3 is a member of the multidrug resistance protein (MRP) family. Caronia et al. (2011) found that rs4148416 was associated with low survival. In addition, the ABCB1 gene that is well-known and encodes P-glycoprotein, contains three polymorphisms (rs2032582, rs1045642, and rs1128503) that have been studied individually and as a haplotype, however, the results have been inconsistent (Caronia et al., 2011).
Detoxification
Cisplatin is inactivated by conjugation with glutathione through the GSTs. This phase II enzyme catalyzes the conjugation of reactive metabolites with negatively charged hydrophilic molecules for disposal in excretion processes. Genetic variations in GSTs have been implicated in cellular resistance to cancer chemotherapy and in outcomes of cisplatin-based treatments. When GSTs enzymes with reduced activity are present, the available concentration in the drug in tumor tissue increases. In these patients therapy might be more effective, but might also be severely toxic (Strange et al., 2000; Siddik, 2003; Quiñones et al., 2006). Several studies have shown significant association between polymorphic GSTs genes and cisplatin treatment response suggesting these polymorphisms as potential biomarkers (Table 2).
Table 2.
Summary of association studies between genetic polymorphisms on Glutathione-S-Transferases and outcomes in the cisplatin-based chemotherapy.
| Gene | Mutation | Cancer | References | Number of subjects | Results |
|---|---|---|---|---|---|
| GSTP1 | Ile105Val | Testicular cancer | Oldenburg et al., 2007b | 173 | G/G ↓ Ototoxicity |
| rs1695 | Testicular cancer | Oldenburg et al., 2007a | 238 | G/G ↓ neurotoxicity | |
| NC_000011.10:g.67585218A>G | Ovarian cancer | Khrunin et al., 2010 | 104 | A/A ↑ OS and PFS | |
| NG_012075.1:g.6624A>G | Urothelial cancer | Yokomizo et al., 2007 | 179 | G allele ↑ myelosuppression | |
| Epithelial ovarian cancer | Kim et al., 2009 | 118 | A/A ↑ Risk for grade 3 or 4 Hematological Toxicity | ||
| Advanced gastric cancer | Ji et al., 2013 | 59 | G/G ↑ Survival | ||
| Goekkurt et al., 2009 | 156 | A/A ↑ Grade 3-4 neutropenia and neurotoxicity | |||
| Ruzzo et al., 2006 | 175 | A/A ↓ Survival | |||
| Osteosarcoma | Windsor et al., 2012 | 60 | G Allele ↑ Myelosuppression | ||
| Yang et al., 2012 | 187 | G Allele ↑ Rates of response | |||
| NSCLC | Joerger et al., 2012 | 137 | G/G ↑ Risk of polyneuropathy | ||
| Sun et al., 2010 | 113 | G Allele ↑ Response | |||
| Medulloblastoma | Rednam et al., 2013 | 106 | G Allele ↑ ototoxicity | ||
| Gastric cancer | Goekkurt et al., 2006 | 52 | G/G ↑ survival | ||
| GSTA1 | rs3957357 | Ovarian cancer | Khrunin et al., 2010 | 104 | T/T ↑ Survival vs. C/C |
| NC_000006.12:g.52803889A>G | |||||
| NM_145740.3:c.-135T>C | |||||
| GSTT1 | Null | Epithelial ovarian cancer | Kim et al., 2009 | 118 | Non-null ↓ OS, PFS |
| Advanced gastric cancer | Goekkurt et al., 2009 | 156 | Non-null ↑ OS and PFS | ||
| Platinum chemotherapy | Jurajda et al., 2012 | 55 | Null allele ↑ onset of ototoxicity | ||
| Pediatric solid tumor | Choeyprasert et al., 2013 | 68 | Non-null related with ototoxicity | ||
| GSTM1 | Null | Ovarian cancer | Beeghly et al., 2006 | 215 | Null allele ↑ OS |
| Khrunin et al., 2010 | 104 | Null allele ↓ Thrombocytopenia, anemia and neuropathy | |||
| Neck and head cancer | Dhawan et al., 2013 | 23 | Null allele ↑ Toxicity | ||
| Breast cancer | Petros et al., 2005 | 85 | Null allele ↑ OS | ||
| Testicular cancer | Oldenburg et al., 2007b | 173 | Non-null ↑ ototoxicity | ||
| Oldenburg et al., 2007a | 238 | Non-null ↑ ototoxicity | |||
| Null allele ↓ ototoxicity | |||||
| Advanced ovarian cancer | Medeiros et al., 2003 | 24 | Null allele ↑ PFS and OS | ||
| Ott et al., 2008 | 139 | Null allele ↑ OS | |||
| GSTM3 | rs1799735 | Cisplatin-based chemotherapy | Peters et al., 2000 | 19 | Deletion in intron 6 ↓ ototoxicity |
| NC_000001.10:g.110280254delC, NC_000001.10:g.110280254delCinsCCT | |||||
| Cisplatin-based chemotherapy | Khrunin et al., 2010 | 104 | AGG/AGG ↓ Thrombocytopenia, anemia and neuropathy |
OS, Overall survival; PFS, Progression free survival; TTP, Time to progression; EFS, Event free survival; G-CSF, Granulocyte-Colony Stimulating Factor.
In the GSTs superfamily there are eight cytosolic classes (Alpha, kappa, mu, omega, pi, sigma, theta, and zeta) (Katoh et al., 2008; Luo et al., 2011). GSTP1, GSTM1, and GSTT1 genes, have been the most widely studied in relation to the functional polymorphisms. GSTP1 is widely expressed in normal human epithelial tissues. A single nucleotide substitution (A→G) at position 313 (rs1695) of the GSTP1 gene, results in replacement of isoleucine with valine at codon 105 of the enzyme, substantially diminishes GSTP1 enzyme activity. On the contrary, GSTM1 and GSTP1 genetically delected (homozygous null allele) will lead to an absence of enzymatic activity (Stoehlmacher et al., 2002).
The GSTP1 gene has been the most studied in a wide number of cancers with controversial results related to cisplatin-based therapy. Some investigations have shown that patients with G/G genotype present less toxicity (Oldenburg et al., 2007a,b; Goekkurt et al., 2009; Kim et al., 2009) with more survival (Goekkurt et al., 2006; Ruzzo et al., 2006; Ji et al., 2013) and better therapy response (Sun et al., 2010; Yang et al., 2012). On the other hand, the G allele has been associated with a risk of myelosuppression, polyneuropathy, and toxicity (Yokomizo et al., 2007; Joerger et al., 2012; Windsor et al., 2012; Rednam et al., 2013). In ovarian cancer, the A allele is related to better PFS and OS (Khrunin et al., 2010). GSTP1 A/A genotype has been found to predict suboptimal response to flurouracil/cisplatin chemotherapy and poor survival in patients with advanced gastric cancer (Ruzzo et al., 2006). The influence of rs1695 GSTP1 on toxicity to taxane-and platinum-based chemotherapy is in debate (Kim et al., 2009).
Polymorphism of GSTM1 and GSTT1 genes is associated with cisplatin-based treatments. GSTM1 null has been specifically related to an increase of OS and PFS (Medeiros et al., 2003; Petros et al., 2005; Beeghly et al., 2006; Ott et al., 2008). Concerning toxicity, it has been associated with a decrease in toxicity (Oldenburg et al., 2007a,b; Khrunin et al., 2010), although Dhawan et al. (2013) showed the opposite but with a small sample (n = 23) (Dhawan et al., 2013). On the GSTT1 gene, the non-null allele relates to an increase in overall survival and progression free survival (Goekkurt et al., 2009), however, Kim et al. (2009) showed the opposite but this contradiction apparently is caused by different definitions of patient response. Moreover, the null allele has also associated with an increase in ototoxicity (Jurajda et al., 2012; Choeyprasert et al., 2013). Finally, additional studies examining the GSTA1 gene showed the T/T genotype (rs3957357) associates with an increase of overall survival (Khrunin et al., 2010). Regarding to GSTM3 gene, the AGG/AGG haplotype (rs1799735) is related to less thrombocytopenia, anemia and neuropathy (Khrunin et al., 2010). Nevertheless, more evidence is needed in order to determine a clear role of GSTA1 and GSTM3 genes on cisplatin-based therapy.
Polymorphisms in the GSTP1 gene have shown controversial results among different types of cancer. Some studies found the polymorphic allele related to less toxicity, better therapy response and more survival but others found the opposite regarding to toxicity (Rednam et al., 2013). The results obtained by several authors demonstrate that the GSTM1 null allele is consistently related to overall survival in different types of cancer. Concerning toxicity, few investigation have found associations, therefore the role of this polymorphism on toxicity is not clear. On the other hand, the GSTT1 null allele associates with toxicity in patients carrying this polymorphism. Regarding OS and PFS it appears that null allele is related to decreased OS and PFS, although one author showed the opposite (Ruzzo et al., 2006; Goekkurt et al., 2009). This contradiction apparently is caused by different definitions of patient response.
Together, the evidence appears to indicate a strong association between GSTs polymorphisms and clinical response (overall survival and disease progression). However, the effects on toxicity do not appear to have a clear and dominant trend, and may be related to differing treatment modalities in each of the studies. Despite this, with the data presented we can conclude that the GSTP1 polymorphic allele and the GSTM1 and GSTT1 null alleles appear to result in enhanced overall survival and progression free survival, particularly in gastric cancer where the data have been more consistent. Lack of activity in GSTs enzymes appear to lead to a better treatment response.
Conclusion
Personalized therapy promises improved outcomes to treatment with respect to efficacy and toxicity of treatment. Ideally, sub-groups of patients that would require adjustment to therapy based on genetic information could be detected prior to commencing treatment, and therapy accordingly optimized. Pharmacogenetics, the study of the role of inheritance in individual variation in drug response, can address cisplatin cellular resistance, providing tools to achieve the modification of current treatments in different types of cancer, including lung, gastric, ovarian, testicular and, esophageal cancers (Weinshilboum, 2003).
Variable responses to different treatments, including cisplatin, have been seen from different points of view. When looking into the genetic variability in processes where cisplatin is involved, including pharmacokinetics and pharmacodynamics, efforts have delivered evidence regarding DNA repair systems and metabolization systems. Within the variability in DNA repair processes, key genes involved include ERCC1, ERCC2 (XPD), ERCC5, XRCC1, XRCC3, and XPC genes. Studies examining the genetic variability of cisplatin metabolism have shown that the main genes involved are GSTP1, GSTM3, GSTM1, and GSTT1. Currently there appears to be a group of genes that would influence variability in response and toxicity in cisplatin-based therapies which we present here in this up-dated review.
Diverse results have been found among the polymorphisms analyzed in both DNA repair enzymes and detoxification enzymes. These contradictions and variations are primarily due to the heterogeneity amongst studies (patient population, treatment and number of subjects). Another possibility is with the inclusion of a large number of candidate genes, there is always a risk of false positive associations. For example, recent studies showed a relationship between rs12201199 in thiopurine S-methyltransferase gene (TPMT) and rs9332377 in the catechol-O-methyltransferase gene (COMPT) with cisplatin-induced hearing loss in children (Ross et al., 2009). Our opinion is that future studies in this line should include the genes we have highlighted, and that a collaborative effort is required to improve the quality and strength of evidence in order to achieve a validated panel of polymorphisms that guides therapeutic decisions.
Finally, prospective clinical studies employing polymorphism panels in these treatment procedures are required to determine whether adjustment of therapy based on genetic information can influence outcomes in these scenarios.
Author contributions
Ángela Roco: Review of intellectual content and Final approval, Juan Cayún: Substantial contributions, Stephania Contreras: Substantial contributions, Jana Stojanova: Substantial contributions, Luis Quiñones: Review of intellectual content and Final approval.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work in the author's laboratory has been financed by Grants FONDECYT 1140434, Chile.
References
- Avan A., Pacetti P., Reni M., Milella M., Vasile E., Mambrini A., et al. (2013). Prognostic factors in gemcitabine-cisplatin polychemotherapy regimens in pancreatic cancer: XPD-Lys751Gln polymorphism strikes back. Int. J. Cancer. 133, 1016–1022. 10.1002/ijc.28078 [DOI] [PubMed] [Google Scholar]
- Beeghly A., Katsaros D., Chen H., Fracchioli S., Zhang Y., Massobrio M., et al. (2006). Glutathione S-transferase polymorphisms and ovarian cancer treatment and survival. Gynecol. Oncol. 100, 330–337. 10.1016/j.ygyno.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Bessho T. (1995). Purification and characterization of the XPF-ERCC1 complex of human DNA repair excision nuclease. J. Biol. Chem. 270, 22657–22660. 10.1074/jbc.270.39.22657 [DOI] [PubMed] [Google Scholar]
- Biason P., Hattinger C. M., Innocenti F., Talamini R., Alberghini M., Scotlandi K., et al. (2012). Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 12, 476–483. 10.1038/tpj.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binks S. P., Dobrota M. (1990). Kinetics and mechanism of uptake of platinum-based pharmaceuticals by the rat small intestine. Biochem. Pharmacol. 40, 1329–1336. 10.1016/0006-2952(90)90400-F [DOI] [PubMed] [Google Scholar]
- Bradbury P. A., Kulke M. H., Heist R. S., Zhou W., Ma C., Xu W., et al. (2009). Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet. Genomics 19, 613–625. 10.1097/FPC.0b013e32832f3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C., Sirera R., Iranzo V., Tarón M., Rosell R. (2007). Gene expression and polymorphisms of DNA repair enzymes: cancer susceptibility and response to chemotherapy. Clin. Lung Cancer 8, 369–375. 10.3816/CLC.2007.n.017 [DOI] [PubMed] [Google Scholar]
- Caronia D., Patiño-García A., Milne R. L., Zalacain-Díez M., Pita G., Alonso M. R., et al. (2009). Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 9, 347–353. 10.1038/tpj.2009 [DOI] [PubMed] [Google Scholar]
- Caronia D., Patiño-Garcia A., Peréz-Martínez A., Pita G., Moreno L. T., Zalacain-Díez M., et al. (2011). Effect of ABCB1 and ABCC3 polymorphisms on osteosarcoma survival after chemotherapy: a pharmacogenetic study. PLoS ONE 6:e26091. 10.1371/journal.pone.0026091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wang F., Wang Z., Li C., Luo H., Liang Y., et al. (2013). Polymorphisms in ERCC1 C8092A predict progression-free survival in metastatic/recurrent nasopharyngeal carcinoma treated with cisplatin-based chemotherapy. Cancer Chemother. Pharmacol. 72, 315–322. 10.1007/s00280-013-2196-8 [DOI] [PubMed] [Google Scholar]
- Chen P., Wiencke J., Aldape K., Kesler-Diaz A., Miike R., Kelsey K., et al. (2000). Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol. Biomarkers Prev. 9, 843–847. [PubMed] [Google Scholar]
- Chen X., Sun H., Ren S., Kim Curran V., Zhang L., Zhou S., et al. (2012). Association of XRCC3 and XPD751 SNP with efficacy of platinum-based chemotherapy in advanced NSCLC patients. Clin. Transl. Oncol. 14, 207–213. 10.1007/s12094-012-0785-3 [DOI] [PubMed] [Google Scholar]
- Cheng J., Ha M., Wang Y., Sun J., Chen J., Wang Y., et al. (2012). A C118T polymorphism of ERCC1 and response to cisplatin chemotherapy in patients with late-stage non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 138, 231–238. 10.1007/s00432-011-1090-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew H. K., Doroshow J. H., Frankel P., Margolin K. A., Somlo G., Lenz H., et al. (2009). Phase II studies of gemcitabine and cisplatin in heavily and minimally pretreated metastatic breast cancer. J. Clin. Oncol. 27, 2163–2169. 10.1200/JCO.2008.17.4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choeyprasert W., Sawangpanich R., Lertsukprasert K., Udomsubpayakul U., Songdej D., Unurathapan U., et al. (2013). Cisplatin-induced ototoxicity in pediatric solid tumors: the role of glutathione S-transferases and megalin genetic polymorphisms. J. Pediatr. Hematol. Oncol. 35, e138–e143. 10.1097/MPH.0b013e3182707fc5 [DOI] [PubMed] [Google Scholar]
- Chung H. H., Kim M.-K., Kim J. W., Park N.-H., Song Y.-S., Kang S.-B., et al. (2006). XRCC1 R399Q polymorphism is associated with response to platinum-based neoadjuvant chemotherapy in bulky cervical cancer. Gynecol. Oncol. 103, 1031–1037. 10.1016/j.ygyno.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Wood R. D. (2005). Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair 4, 1068–1074. 10.1016/j.dnarep.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Cui Y., König J., Buchholz J. K., Spring H., Leier I., Keppler D. (1999). Drug resistance and ATP-dependent conjugate transport mediated by the apical multidrug resistance protein, MRP2, permanently expressed in human and canine cells. Mol. Pharmacol. 55, 929–937. [PubMed] [Google Scholar]
- Decatris M. P., Sundar S., O'Byrne K. J. (2004). Platinum-based chemotherapy in metastatic breast cancer: current status. Cancer Treat. Rev. 30, 53–81. 10.1016/S0305-7372(03)00139-7 [DOI] [PubMed] [Google Scholar]
- De las Peñas R., Sanchez-Ronco M., Alberola V., Taron M., Camps C., Garcia-Carbonero R., et al. (2006). Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann. Oncol.? 17, 668–675. 10.1093/annonc/mdj135 [DOI] [PubMed] [Google Scholar]
- Dhawan D., Panchal H., Shukla S., Padh H. (2013). Genetic variability & chemotoxicity of 5-fluorouracil & cisplatin in head & neck cancer patients: a preliminary study. Indian J. Med. Res. 137, 125–129. [PMC free article] [PubMed] [Google Scholar]
- Doles J., Oliver T. G., Cameron E. R., Hsu G., Jacks T., Walker G. C., et al. (2010). Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 107, 20786–20791. 10.1073/pnas.1011409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erčulj N., Kovač V., Hmeljak J., Dolžan V. (2012). The influence of platinum pathway polymorphisms on the outcome in patients with malignant mesothelioma. Ann. Oncol. 23, 961–967. 10.1093/annonc/mdr324 [DOI] [PubMed] [Google Scholar]
- Fojo T. (2001). Cancer, DNA repair mechanisms, and resistance to chemotherapy. J. Natl. Cancer Inst. 93, 1434–1436. 10.1093/jnci/93.19.1434 [DOI] [PubMed] [Google Scholar]
- Font A., Salazar R., Maurel J., Taron M., Ramirez J. L., Tabernero J., et al. (2008). Cisplatin plus weekly CPT-11/docetaxel in advanced esophagogastric cancer: a phase I study with pharmacogenetic assessment of XPD, XRCC3 and UGT1A1 polymorphisms. Cancer Chemother. Pharmacol. 62, 1075–1083. 10.1007/s00280-008-0700-3 [DOI] [PubMed] [Google Scholar]
- Giachino D. F., Ghio P., Regazzoni S., Mandrile G., Novello S., Selvaggi G., et al. (2007). Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin. Cancer Res. 13, 2876–2881. 10.1158/1078-0432.CCR-06-2543 [DOI] [PubMed] [Google Scholar]
- Goekkurt E., Al-Batran S.-E., Hartmann J. T., Mogck U., Schuch G., Kramer M., et al. (2009). Pharmacogenetic analyses of a phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil and leucovorin plus either oxaliplatin or cisplatin: a study of the arbeitsgemeinschaft internistische onkologie. J. Clin. Oncol. 27, 2863–2873 10.1200/JCO.2008.19.1718 [DOI] [PubMed] [Google Scholar]
- Goekkurt E., Hoehn S., Wolschke C., Wittmer C., Stueber C., Hossfeld D. K., et al. (2006). Polymorphisms of glutathione S-transferases (GST) and thymidylate synthase (TS)–novel predictors for response and survival in gastric cancer patients. Br. J. Cancer 94, 281–286. 10.1038/sj.bjc.6602891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V. M., Fuertes M. A., Alonso C., Perez J. M. (2001). Is cisplatin-induced cell death always produced by apoptosis? Mol. Pharmacol. 59, 657–663. 10.1124/mol.59.4.657 [DOI] [PubMed] [Google Scholar]
- Goricar K., Kovac V., Dolzan V. (2014). Polymorphisms in translesion polymerase genes influence treatment outcome in malignant mesothelioma. Pharmacogenomics 15, 941–950. 10.2217/pgs.14.14 [DOI] [PubMed] [Google Scholar]
- Gurubhagavatula S., Liu G., Park S., Zhou W., Su L., Wain J. C., et al. (2004). XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J. Clin. Oncol. 22, 2594–2601. 10.1200/JCO.2004.08.067 [DOI] [PubMed] [Google Scholar]
- Hao T., Feng W., Zhang J., Sun Y. J., Wang G. (2012). Association of four ERCC1 and ERCC2 SNPs with survival of bone tumour patients. Asian Pac. J. Cancer Prev. 13, 3821–3824. 10.7314/APJCP.2012.13.8.3821 [DOI] [PubMed] [Google Scholar]
- He C., Duan Z., Li P., Xu Q., Yuan Y. (2013). Role of ERCC5 promoter polymorphisms in response to platinum-based chemotherapy in patients with advanced non-small-cell lung cancer. Anticancer Drugs 24, 300–305. 10.1097/CAD.0b013e32835bd6ce [DOI] [PubMed] [Google Scholar]
- Ho T. V., Schärer O. D. (2010). Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 51, 552–566. 10.1002/em.20573 [DOI] [PubMed] [Google Scholar]
- Hong C. Y., Xu Q., Yue Z., Zhang Y., Yuan Y. (2009). Correlation of the sensitivity of NP chemotherapy in non-small lung cancer with DNA repair gene XRCC1 polymorphism. Chin. J. Cancer 28, 1291–1297. 10.5732/cjc.009.10139 [DOI] [PubMed] [Google Scholar]
- Hromas R. A., North J. A., Burns C. P. (1987). Decreased cisplatin uptake by resistant L1210 leukemia cells. Cancer Lett. 36, 197–201. 10.1016/0304-3835(87)90091-7 [DOI] [PubMed] [Google Scholar]
- Ishida S., Lee J., Thiele D. J., Herskowitz I. (2002). Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc. Natl Acad. Sci. U.S.A. 99, 14298–14302. 10.1073/pnas.162491399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isla D., Sarries C., Rosell R., Alonso G., Domine M., Taron M., et al. (2004). Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann. Oncol. 15, 1194–1203. 10.1093/annonc/mdh319 [DOI] [PubMed] [Google Scholar]
- Ji M., Xu B., Jiang J. T., Wu J., Li X.-D., Zhao W. Q., et al. (2013). Relationship between glutathione S-transferase P1 (GSTP1), X-ray repair cross complementing group 1 (XRCC1) and 5,10-methylenetetrahydrofolate reductase (5,10-MTHFR) gene polymorphisms and response to chemotherapy in advanced gastric cancer. Onkologie 36, 335–340. 10.1159/000351260 [DOI] [PubMed] [Google Scholar]
- Joerger M., Burgers S. A., Baas P., Smit E. F., Haitjema T. J., Bard M. P. L., et al. (2012). Germline polymorphisms in patients with advanced nonsmall cell lung cancer receiving first-line platinum-gemcitabine chemotherapy: a prospective clinical study. Cancer 118, 2466–2475. 10.1002/cncr.26562 [DOI] [PubMed] [Google Scholar]
- Jurajda M., Talach T., Kostřica R., Lakomý R., Kocák I., Cvanová M. (2012). Genetic background of cisplatin induced ototoxicity. Klin. Onkol. 25, 184–187. [PubMed] [Google Scholar]
- Kalikaki A., Kanaki M., Vassalou H., Souglakos J., Voutsina A., Georgoulias V., et al. (2009). DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin. Lung Cancer 10, 118–123. 10.3816/CLC.2009.n.015 [DOI] [PubMed] [Google Scholar]
- Kamikozuru H., Kuramochi H., Hayashi K., Nakajima G., Yamamoto M. (2008). ERCC1 codon 118 polymorphism is a useful prognostic marker in patients with pancreatic cancer treated with platinum-based chemotherapy. Int. J. Oncol. 32, 1091–1096. 10.3892/ijo.32.5.1091 [DOI] [PubMed] [Google Scholar]
- Kartalou M., Essigmann J. M. (2001). Recognition of cisplatin adducts by cellular proteins. Mutat. Res. 478, 1–21. 10.1016/S0027-5107(01)00142-7 [DOI] [PubMed] [Google Scholar]
- Katano K., Kondo A., Safaei R., Holzer A., Samimi G., Mishima M., et al. (2002). Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 62, 6559–6565. [PubMed] [Google Scholar]
- Katoh T., Yamano Y., Tsuji M., Watanabe M. (2008). Genetic polymorphisms of human cytosol glutathione S-transferases and prostate cancer. Pharmacogenomics 9, 93–104. 10.2217/14622416.9.1.93 [DOI] [PubMed] [Google Scholar]
- Ke H. G., Li J., Shen Y., You Q.-S., Yan Y., Dong H.-X., et al. (2012). Prognostic significance of GSTP1, XRCC1 and XRCC3 polymorphisms in non-small cell lung cancer patients. Asian Pac. J. Cancer Prev. 13, 4413–4416. 10.7314/APJCP.2012.13.9.4413 [DOI] [PubMed] [Google Scholar]
- Kelland L. (2007). The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7, 573–584. 10.1038/nrc2167 [DOI] [PubMed] [Google Scholar]
- Kelland L. R. (1993). New platinum antitumor complexes. Crit. Rev. Oncol. Hematol. 15, 191–219. 10.1016/1040-8428(93)90042-3 [DOI] [PubMed] [Google Scholar]
- Khrunin A. V., Moisseev A., Gorbunova V., Limborska S. (2010). Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharmacogenomics J. 10, 54–61. 10.1038/tpj.2009.45 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Kim M.-K., Chung H. H., Kim J. W., Park N. H., Song Y. S., et al. (2009). Genetic polymorphisms affecting clinical outcomes in epithelial ovarian cancer patients treated with taxanes and platinum compounds: a Korean population-based study. Gynecol. Oncol. 113, 264–269. 10.1016/j.ygyno.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Kim K., Kang S. B., Chung H. H., Kim J. W., Park N. H., Song Y. S. (2008). XRCC1 Arginine194Tryptophan and GGH-401Cytosine/Thymine polymorphisms are associated with response to platinum-based neoadjuvant chemotherapy in cervical cancer. Gynecol. Oncol. 111, 509–515. 10.1016/j.ygyno.2008.08.034 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee G.-W., Lee M. J., Cho Y. J., Jeong Y. Y., Kim H. C., et al. (2012). Clinical significance of ERCC2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy. Lung Cancer 77, 578–584. 10.1016/j.lungcan.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Kirschner K., Melton D. W. (2010). Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 30, 3223–3232. [PubMed] [Google Scholar]
- Koike K., Kawabe T., Tanaka T., Toh S., Uchiumi T., Wada M., et al. (1997). A canalicular multispecific organic anion transporter (cMOAT) antisense cDNA enhances drug sensitivity in human hepatic cancer cells. Cancer Res. 57, 5475–5479. [PubMed] [Google Scholar]
- Komatsu M., Sumizawa T., Mutoh M., Chen Z. S., Terada K., Furukawa T., et al. (2000). Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 60, 1312–1316. [PubMed] [Google Scholar]
- Kool M., de Haas M., Scheffer G. L., Scheper R. J., van Eijk M. J., Juijn J. A., et al. (1997). Analysis of expression of cMOAT (MRP2), MRP3, MRP4, and MRP5, homologues of the multidrug resistance-associated protein gene (MRP1), in human cancer cell lines. Cancer Res. 57, 3537–3547. [PubMed] [Google Scholar]
- Krivak T. C., Darcy K. M., Tian C., Armstrong D., Baysal B. E., Gallion H., et al. (2008). Relationship between ERCC1 polymorphisms, disease progression, and survival in the Gynecologic Oncology Group Phase III Trial of intraperitoneal versus intravenous cisplatin and paclitaxel for stage III epithelial ovarian cancer. J. Clin. Oncol. 26, 3598–3606. 10.1200/JCO.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Kang H. G., Yoo S. S., Kang Y. R., Choi Y. Y., Lee W. K., et al. (2013). Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer 82, 330–339. 10.1016/j.lungcan.2013.07.024 [DOI] [PubMed] [Google Scholar]
- Lehmann A. R. (2001). The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15, 15–23. 10.1101/gad.859501 [DOI] [PubMed] [Google Scholar]
- Li D., Frazier M., Evans D. B., Hess K. R., Crane C. H., Abbruzzese J. L. (2006). Genes are associated with reduced survival of pancreatic. J. Clin. Oncol. 24, 1720–1728. 10.1200/JCO.2005.04.4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Sun X., Sun N., Qin S., Cheng H., Feng J., et al. (2010). Association between polymorphisms of ERCC1 and XPD and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer. Am. J. Clin. Oncol. 33, 489–494. 10.1097/COC.0b013e3181b9cedc [DOI] [PubMed] [Google Scholar]
- Li K., Li W. (2013). Association between polymorphisms of XRCC1 and ADPRT genes and ovarian cancer survival with platinum-based chemotherapy in Chinese population. Mol. Cell. Biochem. 372, 27–33. 10.1007/s11010-012-1442-4 [DOI] [PubMed] [Google Scholar]
- Li X. D., Han J. C., Zhang Y. J., Li H. B., Wu X. Y. (2013). Common variations of DNA repair genes are associated with response to platinum-based chemotherapy in NSCLCs. Asian Pac. J. Cancer Prev. 14, 145–148. 10.7314/APJCP.2013.14.1.145 [DOI] [PubMed] [Google Scholar]
- Lin X., Okuda T., Trang J., Howell S. B. (2006). Human REV1 modulates the cytotoxicity and mutagenicity of cisplatin in human ovarian carcinoma cells. Mol. Pharmacol. 69, 1748–1754. 10.1124/mol.105.020446 [DOI] [PubMed] [Google Scholar]
- Liu D., Day S. J. O., Yang D., Boasberg P., Milford R., Kristedja T., et al. (2005). Impact of gene polymorphisms on clinical outcome for stage IV melanoma patients treated with biochemotherapy: an exploratory study. Clin. Cancer Res. 11, 1237–1246. [PubMed] [Google Scholar]
- Liu H., Qi B., Guo X., Tang L. Q., Chen Q. Y., Zhang L., et al. (2013). Genetic variations in radiation and chemotherapy drug action pathways and survival in locoregionally advanced nasopharyngeal carcinoma treated with chemoradiotherapy. PLoS ONE 8:e82750. 10.1371/journal.pone.0082750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludovini V., Floriani I., Pistola L., Minotti V., Meacci M., Chiari R., et al. (2011). Association of cytidine deaminase and xeroderma pigmentosum group D polymorphisms with response, toxicity, and survival in cisplatin/gemcitabine-treated advanced non-small cell lung cancer patients. J. Thorac. Oncol. 6, 2018–2026. 10.1097/JTO.0b013e3182307e1f [DOI] [PubMed] [Google Scholar]
- Luo W., Kinsey M., Schiffman J. D., Lessnick S. L. (2011). Glutathione S-transferases in pediatric cancer. Front Oncol. 1:39. 10.3389/fonc.2011.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S. C., Andrews P. A., Howell S. B. (1991). Modulation of cis-diamminedichloroplatinum(II) accumulation and sensitivity by forskolin and 3-isobutyl-1-methylxanthine in sensitive and resistant human ovarian carcinoma cells. Int. J. Cancer 48, 866–872. 10.1002/ijc.2910480613 [DOI] [PubMed] [Google Scholar]
- Medeiros R., Pereira D., Afonso N., Palmeira C., Faleiro C., Afonso-Lopes C., et al. (2003). Platinum/paclitaxel-based chemotherapy in advanced ovarian carcinoma: glutathione S-transferase genetic polymorphisms as predictive biomarkers of disease outcome. Int. J. Clin. Oncol. 8, 156–161. 10.1007/s10147-003-0318-8 [DOI] [PubMed] [Google Scholar]
- Metzger R., Warnecke-Eberz U., Alakus H., Kütting F., Brabender J., Vallböhmer D., et al. (2012). Neoadjuvant radiochemotherapy in adenocarcinoma of the esophagus: ERCC1 gene polymorphisms for prediction of response and prognosis. J. Gastrointest. Surg.? 16, 26–34. discussion 34. 10.1007/s11605-011-1700-x [DOI] [PubMed] [Google Scholar]
- Miao J., Zhang X., Tang Q.-L., Wang X.-Y., Kai L. (2012). Prediction value of XRCC 1 gene polymorphism on the survival of ovarian cancer treated by adjuvant chemotherapy. Asian Pac. J. Cancer Prev. 13, 5007–5010. 10.7314/APJCP.2012.13.10.5007 [DOI] [PubMed] [Google Scholar]
- Nakayama K., Kanzaki A., Ogawa K., Miyazaki K., Neamati N., Takebayashi Y. (2002). Copper-transporting P-type adenosine triphosphatase (ATP7B) as a cisplatin based chemoresistance marker in ovarian carcinoma: comparative analysis with expression of MDR1, MRP1, MRP2, LRP and BCRP. Int. J. Cancer 101, 488–495. 10.1002/ijc.10608 [DOI] [PubMed] [Google Scholar]
- Nakayama K., Kanzaki A., Terada K., Mutoh M., Ogawa K., Sugiyama T., et al. (2004). Prognostic value of the Cu-transporting ATPase in ovarian carcinoma patients receiving cisplatinbased chemotherapy. Clin. Cancer Res. 10, 2804–2811. 10.1158/1078-0432.CCR-03-0454 [DOI] [PubMed] [Google Scholar]
- Nguewa P. A., Fuertes M. A., Alonso C., Peréz J. M. (2003). Pharmacological modulation of Poly (ADP-ribose) polymerase-mediated cell death: exploitation in cancer chemotherapy. Mol. Pharmacol. 64, 1007–1014. 10.1124/mol.64.5.1007 [DOI] [PubMed] [Google Scholar]
- Nigro C. L. O., Monteverde M., Riba M., Lattanzio L., Tonissi F., Garrone O., et al. (2010). Expression profiling and long lasting responses to chemotherapy in metastatic gastric cancer. Int. J. Oncol. 37, 1219–1228. 10.3892/ijo_00000773 [DOI] [PubMed] [Google Scholar]
- Ohashi K., Kajiya K., Inaba S., Hasegawa T., Seko Y., Furuchi T., et al. (2003). Copper(II) protects yeast against the toxicity of cisplatin independently of the induction of metallothionein and the inhibition of platinum uptake. Biochem. Biophys. Res. Comm. 310, 148–152. 10.1016/j.bbrc.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Ohbu M., Ogawa K., Konno S., Kanzaki A., Terada K., Sugiyama T., et al. (2003). Copper-transporting P-type adenosine triphosphatase (ATP7B) is expressed in human gastric carcinoma. Cancer Lett. 189, 33–38. 10.1016/S0304-3835(02)00462-7 [DOI] [PubMed] [Google Scholar]
- Okuda K., Sasaki H., Hikosaka Y., Kawano O., Yukiue H., Yano M., et al. (2011). Excision repair cross complementation group 1 polymorphisms predict overall survival after platinum-based chemotherapy for completely resected non-small-cell lung cancer. J. Surg. Res. 168, 206–212. 10.1016/j.jss.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Oldenburg J., Kraggerud S. M., Brydøy M., Cvancarova M., Lothe R. A., Fossa S. D. (2007a). Association between long-term neuro-toxicities in testicular cancer survivors and polymorphisms in glutathione-s-transferase-P1 and -M1, a retrospective cross sectional study. J. Transl. Med. 5:70. 10.1186/1479-5876-5-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg J., Kraggerud S. M., Cvancarova M., Lothe R. A., Fossa S. D. (2007b). Cisplatin-induced long-term hearing impairment is associated with specific glutathione s-transferase genotypes in testicular cancer survivors. J. Clin. Oncol. 25, 708–714. 10.1200/JCO.2006.08.9599 [DOI] [PubMed] [Google Scholar]
- Ott K., Lordick F., Becker K., Ulm K., Siewert J., Höfler H., et al. (2008). Glutathione-S-transferase P1, T1 and M1 genetic polymorphisms in neoadjuvant-treated locally advanced gastric cancer: GSTM1-present genotype is associated with better prognosis in completely resected patients. Int. J. Colorectal Dis. 23, 773–782. 10.1007/s00384-008-0490-4 [DOI] [PubMed] [Google Scholar]
- Ott K., Rachakonda P. S., Panzram B., Keller G., Lordick F., Becker K., et al. (2011). DNA repair gene and MTHFR gene polymorphisms as prognostic markers in locally advanced adenocarcinoma of the esophagus or stomach treated with cisplatin and 5-fluorouracil-based neoadjuvant chemotherapy. Ann. Surg. Oncol. 18, 2688–2698. 10.1245/s10434-011-1601-y [DOI] [PubMed] [Google Scholar]
- Pacetti P., Giovannetti E., Mambrini A., Nannizzi S., Orlandi M., Tartarini R., et al. (2009). Single nucleotide polymorphisms and clinical outcome in patients with biliary tract carcinoma treated with epirubicin, cisplatin and capecitabine. Anticancer Res. 29, 1835–1840. [PubMed] [Google Scholar]
- Park D. J., Stoehlmacher J., Zhang W., Tsao-wei D. D., Groshen S., Lenz H. (2001). A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 61, 8654–8658. [PubMed] [Google Scholar]
- Park S. R., Kong S.-Y., Nam B.-H., Choi I. J., Kim C. G., Lee J. Y., et al. (2011). CYP2A6 and ERCC1 polymorphisms correlate with efficacy of S-1 plus cisplatin in metastatic gastric cancer patients. Br. J. Cancer 104, 1126–1134. 10.1038/bjc.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. J., Gill I., Tarone R., Vionnet J. A., Grunberg S., Muggia F. M., et al. (1991). Platinum-DNA damage in leukocyte DNA of patients receiving carboplatin and cisplatin chemotherapy, measured by atomic absorption spectrometry. Carcinogenesis 12, 1253–1258. 10.1093/carcin/12.7.1253 [DOI] [PubMed] [Google Scholar]
- Peters U., Preisler-Adams S., Hebeisen A., Hahn M., Seifert E., Lanvers C., et al. (2000). Glutathione S-transferase genetic polymorphisms and individual sensitivity to the ototoxic effect of cisplatin. Anticancer Drugs 11, 639–643. [DOI] [PubMed] [Google Scholar]
- Petros W. P., Hopkins P. J., Spruill S., Broadwater G., Vredenburgh J. J., Colvin O. M., et al. (2005). Associations between drug metabolism genotype, chemotherapy pharmacokinetics, and overall survival in patients with breast cancer. J. Clin. Oncol. 23, 6117–6125. 10.1200/JCO.2005.06.075 [DOI] [PubMed] [Google Scholar]
- Quiñones L., Lee K., Varela F. N., Escala M., García K., Godoy L., et al. (2006). Cancer pharmacogenetics: Study of genetically determined variations on cancer susceptibility due to xenobiotic exposure. Rev. Méd. Chil. 134, 499–515. 10.4067/S0034-98872006000400015 [DOI] [PubMed] [Google Scholar]
- Quintela-Fandino M., Hitt R., Medina P. P., Gamarra S., Manso L., Cortes-Funes H., et al. (2006). DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J. Clin. Oncol. 24, 4333–4339. 10.1200/JCO.2006.05.8768 [DOI] [PubMed] [Google Scholar]
- Rednam S., Scheurer M. E., Adesina A. (2013). Glutathione S-transferase P1 single nucleotide polymorphism predicts permanent ototoxicity in children with medulloblastoma. Pediatr Blood Cancer 60, 593–598. 10.1002/pbc.24366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Zhou S., Wu F., Zhang L., Li X., Zhang J., et al. (2012). Association between polymorphisms of DNA repair genes and survival of advanced NSCLC patients treated with platinum-based chemotherapy. Lung Cancer 75, 102–109. 10.1016/j.lungcan.2011.05.023 [DOI] [PubMed] [Google Scholar]
- Ross C. J. D., Katzov-Eckert H., Dubé M.-P., Brooks B., Rassekh S. R., Barhdadi A., et al. (2009). Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat. Genet. 41, 1345–1349. 10.1038/ng.478 [DOI] [PubMed] [Google Scholar]
- Rumiato E., Cavallin F., Boldrin E., Cagol M., Alfieri R., Basso D., et al. (2013). ERCC1 C8092A (rs3212986) polymorphism as a predictive marker in esophageal cancer patients treated with cisplatin/5-FU-based neoadjuvant therapy. Pharmacogenet. Genomics 23, 597–604. 10.1097/FPC.0b013e3283653afc [DOI] [PubMed] [Google Scholar]
- Ruzzo A., Graziano F., Kawakami K., Watanabe G., Santini D., Catalano V., et al. (2006). Pharmacogenetic profiling and clinical outcome of patients with advanced gastric cancer treated with palliative chemotherapy. J. Clin. Oncol. 24, 1883–1891. 10.1200/JCO.2005.04.8322 [DOI] [PubMed] [Google Scholar]
- Ryu J. S., Hong Y.-C., Han H.-S., Lee J.-E., Kim S., Park Y.-M., et al. (2004). Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer 44, 311–316. 10.1016/j.lungcan.2003.11.019 [DOI] [PubMed] [Google Scholar]
- Safaei R., et al. (2004). Cross-resistance to cisplatin in cells with acquired resistance to copper. Cancer Chemother. Pharmacol. 53, 239–246. 10.1007/s00280-003-0736-3 [DOI] [PubMed] [Google Scholar]
- Sakano S., Wada T., Matsumoto H., Sugiyama S., Inoue R., Eguchi S., et al. (2006). Single nucleotide polymorphisms in DNA repair genes might be prognostic factors in muscle-invasive bladder cancer patients treated with chemoradiotherapy. Br. J. Cancer 95, 561–570. 10.1038/sj.bjc.6603290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H. J., Yun J. Y., Hwang J. E., Bae W. K., Cho S. H., Lee J. H., et al. (2010). BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci. 101, 1247–1254. 10.1111/j.1349-7006.2010.01514.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirota Y., Stoehlmacher J., Brabender J., Xiong Y. P., Uetake H., Danenberg K. D., et al. (2001). ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J. Clin. Oncol. 19, 4298–4304. [DOI] [PubMed] [Google Scholar]
- Siddik Z. H. (2003). Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279. 10.1038/sj.onc.1206933 [DOI] [PubMed] [Google Scholar]
- Smith S., Su D., Rigault de la Longrais I. A., Schwartz P., Puopolo M., Rutherford T. J., et al. (2007). ERCC1 genotype and phenotype in epithelial ovarian cancer identify patients likely to benefit from paclitaxel treatment in addition to platinum-based therapy. J. Clin. Oncol. 25, 5172–5179. 10.1200/JCO.2007.11.8547 [DOI] [PubMed] [Google Scholar]
- Stoehlmacher J., Park D. J., Zhang W., Groshen S., Tsao-wei D. D., Yu M. C., et al. (2002). Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J. Natl. Cancer Inst. 94, 936–942. 10.1093/jnci/94.12.936 [DOI] [PubMed] [Google Scholar]
- Strange R. C., Jones P. W., Fryer A. (2000). Glutathione S-transferase: genetics and role in toxicology. Toxicology Lett. 112–113, 357–363. 10.1016/S0378-4274(99)00230-1 [DOI] [PubMed] [Google Scholar]
- Su D., Ma S., Liu P., Jiang Z., Lv W., Zhang Y., et al. (2007). Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer 56, 281–288. 10.1016/j.lungcan.2006.12.002 [DOI] [PubMed] [Google Scholar]
- Sun N., Sun X., Chen B., Cheng H., Feng J., Cheng L., et al. (2010). MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non-small cell lung cancer. Cancer Chemother. Pharmacol. 65, 437–446. 10.1007/s00280-009-1046-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.-H., Hou W.-G., Zhao H.-X., Zhao Y.-L., Ma C., Liu Y. (2013). Single nucleotide polymorphisms in the NER pathway and clinical outcome of patients with bone malignant tumors. Asian Pac. J. Cancer Prev. 14, 2049–2052. 10.7314/APJCP.2013.14.3.2049 [DOI] [PubMed] [Google Scholar]
- Sun X., Li F., Sun N., Shukui Q., Baoan C., Jifeng F., et al. (2009). Lung cancer polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer 65, 230–236. 10.1016/j.lungcan.2008 [DOI] [PubMed] [Google Scholar]
- Takenaka T., Yano T., Kiyohara C., Miura N., Kouso H., Ohba T., et al. (2010). Effects of excision repair cross-complementation group 1 (ERCC1) single nucleotide polymorphisms on the prognosis of non-small cell lung cancer patients. Lung Cancer 67, 101–107. 10.1016/j.lungcan.2009.03.007 [DOI] [PubMed] [Google Scholar]
- Tzvetkov M. V., Behrens G., O'Brien V. P., Hohloch K., Brockmöller J., Benöhr P. (2011). Pharmacogenetic analyses of cisplatin-induced nephrotoxicity indicate a renoprotective effect of ERCC1 polymorphisms. Pharmacogenomics 12, 1417–1427. 10.2217/pgs.11.93 [DOI] [PubMed] [Google Scholar]
- Wang D., Lippard S. J. (2005). Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4, 307–320. 10.1038/nrd1691 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen J., Li X., He Y., Hu B., Ji C., et al. (2011). Genetic polymorphisms of ERCC1 and their effects on the efficacy of cisplatin-based chemotherapy in advanced esophageal carcinoma. Oncol. Rep. 25, 1047–1052. 10.3892/or.2011.1170 [DOI] [PubMed] [Google Scholar]
- Wang Z. H., Miao X. P., Tan W., Zhang X. R., Xu B. H., Lin D. X. (2004). Single nucleotide polymorphisms in XRCC1 and clinical response to platin-based chemotherapy in advanced non-small cell lung cancer. Chin. J. Cancer 23, 865–868. [PubMed] [Google Scholar]
- Warnecke-eberz U., Vallböhmer D., Alakus H., Kütting F., Lurje G., Bollschweiler E., et al. (2009). ERCC1 and XRCC1 gene polymorphisms predict response to neoadjuvant radiochemotherapy in esophageal cancer. J. Gastrointest. Surg. 13, 1411–1421. 10.1007/s11605-009-0881-z [DOI] [PubMed] [Google Scholar]
- Weinshilboum R. (2003). Inheritance and drug response. N Engl. J. Med. 348, 529–537. 10.1056/NEJMra020021 [DOI] [PubMed] [Google Scholar]
- Windsor R. E., Strauss S. J., Kallis C., Wood N. E., Whelan J. S. (2012). Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: a pilot study. Cancer 118, 1856–1867. 10.1002/cncr.26472 [DOI] [PubMed] [Google Scholar]
- Wong E., Giandomenico C. M. (1999). Current status of platinum-based antitumor drugs. Chem. Rev. 99, 2451–2466 10.1021/cr980420v [DOI] [PubMed] [Google Scholar]
- Wozniak K., Blasiak J. (2002). Recognition and repair of DNA–cisplatin adducts. Acta Biochim. Pol. 49, 583–596. [PubMed] [Google Scholar]
- Yang L.-M., Li X.-H., Bao C.-F. (2012). Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac. J. Cancer Prev. 13, 5883–5886. 10.7314/APJCP.2012.13.11.5883 [DOI] [PubMed] [Google Scholar]
- Yokomizo A., Yamamoto K., Kinukawa N., Tsunoda T., Koga H., Naito S. (2007). Association analysis of glutathione-S-transferase P1 (GSTP1) polymorphism with urothelial cancer susceptibility and myelosuppression after M-VAC chemotherapy. Int. J. Urol. 14, 500–504. 10.1111/j.1442-2042.2007.01769.x [DOI] [PubMed] [Google Scholar]
- Yuan P., Miao X., Zhang X., Wang Z., Tan W., Sun Y., et al. (2006). [XRCC1 and XPD genetic polymorphisms predict clinical responses to platinum-based chemotherapy in advanced non-small cell lung cancer]. Zhonghua Zhong Liu Za Zhi. 28, 196–199. [PubMed] [Google Scholar]
- Zdraveski Z. Z., Mello J. A., Farinelli C. K., Essigmann J. M., Marinus M. G. (2002). MutS preferentially recognizes cisplatin-over oxaliplatin-modified DNA. J. Biol. Chem. 277, 1255–1260 10.1074/jbc.M105382200 [DOI] [PubMed] [Google Scholar]
- Zhai X., Hu Q., Gu K., Wang J., Zhang J., Wu Y. (2013). Significance of XRCC1 Codon399 polymorphisms in Chinese patients with locally advanced nasopharyngeal carcinoma treated with radiation therapy. Asia Pac. J. Clin. Oncol. [Epub ahead of print]. 10.1111/ajco.12117 [DOI] [PubMed] [Google Scholar]
- Zhou W., Gurubhagavatula S., Liu G., Park S., Neuberg D. S., Wain J. C., et al. (2004). Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin. Cancer Res. 10, 4939–4943. 10.1158/1078-0432.CCR-04-0247 [DOI] [PubMed] [Google Scholar]