Abstract
The hypothesis that interspecific hybridisation promotes invasiveness has received much recent attention, but tests of the hypothesis can suffer from important limitations. Here, we provide the first systematic review of studies experimentally testing the hybridisation-invasion (H-I) hypothesis in plants, animals and fungi. We identified 72 hybrid systems for which hybridisation has been putatively associated with invasiveness, weediness or range expansion. Within this group, 15 systems (comprising 34 studies) experimentally tested performance of hybrids vs. their parental species and met our other criteria. Both phylogenetic and non-phylogenetic meta-analyses demonstrated that wild hybrids were significantly more fecund and larger than their parental taxa, but did not differ in survival. Resynthesised hybrids (which typically represent earlier generations than do wild hybrids) did not consistently differ from parental species in fecundity, survival or size. Using meta-regression, we found that fecundity increased (but survival decreased) with generation in resynthesised hybrids, suggesting that natural selection can play an important role in shaping hybrid performance – and thus invasiveness – over time. We conclude that the available evidence supports the H-I hypothesis, with the caveat that our results are clearly driven by tests in plants, which are more numerous than tests in animals and fungi.
Keywords: Adaptive evolution, colonisation, hybridisation, introgression, invasion genetics, phylogenetic meta-analysis, polyploidy, range expansion, weeds
INTRODUCTION
The study of what determines invasion success in non-native organisms has a long history (Elton 1958; Baker 1974), although not always a successful one (Perrins et al. 1992; Mack 1996). Genetic and evolutionary factors have been considered important (Baker & Stebbins 1965; Crawford & Whitney 2010), including the hypothesis that interspecific hybridisation may promote invasiveness (Stebbins 1985; Abbott 1992). This Hybridisation-Invasion (H-I) hypothesis has received particular attention in the past decade-and-a-half following the publication of Ellstrand & Schierenbeck (2000). In addition to summarising the mechanisms by which hybridisation could enhance invasiveness, Ellstrand and Schierenbeck compiled preliminary support for the hypothesis via a list of 28 taxa in which hybridisation has preceded invasiveness. This original list has since been expanded to 35 hybrid taxa (Schierenbeck & Ellstrand 2009), contributing to the increasing acceptance of the idea that hybridisation can be a driver of biological invasions (e.g. Darling et al. 2008; Le Roux & Wieczorek 2009; but see Whitney et al. 2009). As species are transported globally with increasing frequency, invasive hybrids will continue being introduced to new regions, and new opportunities for interspecific mating will undoubtedly result in the formation of novel hybrid taxa. The hypothesised connection between hybridisation and invasiveness is, therefore, likely to remain a critical issue, both for conservation and for better understanding the evolutionary ecology of colonising species. However, despite rapidly growing interest in this topic, we currently lack a comprehensive assessment of the evidence either supporting or refuting a causal link between hybridisation and invasiveness.
Ellstrand & Schierenbeck (2000) outlined several, non-mutually exclusive, mechanisms by which hybridisation could enhance invasiveness; we highlight them briefly here (see also Rieseberg et al. 2007). First, hybridisation can create novel phenotypes relative to the parental taxa, increasing the likelihood of survival and establishment success in novel habitats. Such novelty includes transgressive phenotypes, where hybrids exhibit trait values that fall outside the range of their parents (Rieseberg et al. 1999), as well as novel combinations of parental phenotypic traits (Hovick et al. 2012). Second, hybridisation can lead to increased phenotypic and genetic variation relative to the parental taxa, which may help hybrids better cope with environmental stochasticity and increase their evolutionary potential (Anderson & Stebbins 1954; Stebbins 1959). Heterosis is a special case of increased genetic variation, where hybrids (particularly F1s) experience performance gains due to increased heterozygosity. When hybridisation is accompanied by mechanisms that stabilise heterotic lineages (i.e. polyploidy, clonal growth or agamospermy), the resulting hybrids may experience increased invasiveness (e.g. Parepa et al. 2014). Finally, if the parental taxa are relatively isolated and occur in small populations, hybridisation could lead to the purging of genetic load, and the resulting fitness boost could increase invasiveness (Ellstrand & Schierenbeck 2000). To our knowledge, this final mechanism has not yet been demonstrated empirically.
Establishing a causal relationship between hybridisation and invasiveness can be challenging. A first challenge is that, because many hybrid taxa are sterile, performance assessments cannot always rely on fecundity as an indicator of potential population growth rates. Second, the outcomes of performance comparisons between hybrids and parents frequently depend on which hybrid class (F1, BC1, etc.) is examined (Arnold & Hodges 1995; Arnold & Martin 2010). For example, hybrids experiencing heterosis in the F1 generation may experience hybrid breakdown as segregating genes in subsequent generations lead to decreasing fitness (Hooftman et al. 2007). Even within the same hybrid class, hybrid individuals can be highly variable, resulting in biologically meaningful differences among lineages with differing parental backgrounds (Pyšek et al. 2003; Hartman et al. 2013). Third, the environmental context can determine relative performance of hybrids vs. parents (Arnold & Martin 2010), and in at least one case, hybrids outperform their parental taxa by way of different traits in different environments (Hovick et al. 2012). These considerations suggest that the performance metric, the choice of hybrid material and the environmental setting all need to be carefully considered to provide meaningful tests of the H-I hypothesis.
A further challenge in evaluating the H-I hypothesis is that observational studies documenting increasing frequencies of hybrids relative to their parental taxa tell us nothing definitive about relative hybrid performance and therefore invasiveness. Even in the absence of a hybrid advantage, interspecific mating will often lead to at least low levels of introgression of selectively neutral alleles, creating hybrid swarms (e.g. Hasselman et al. 2014). Yet, some studies of hybrid invaders interpret high abundances of hybrids relative to their parental taxa as evidence that hybridisation has promoted invasiveness in that taxon (e.g. Urbanska et al. 1997; Moody & Les 2002; Tavalire et al. 2012; Wu et al. 2013). This practice can result in unsubstantiated claims regarding the role of hybridisation being propagated throughout the literature.
Because of these challenges, testing the H-I hypothesis clearly requires an experimental approach. At the most basic level, these experiments must compare hybrid performance with that of the parental taxa in a common environment in order to remove the effects of environmental variation and historical chance while focusing on genetically based performance differences among taxa. In any such experiment, the proper parental taxa must be used as benchmarks for comparison with hybrid performance. Ideally, both parents would be included in any such tests, but at a minimum the more invasive parental taxon must be. For example, if an invasive hybrid results from hybridisation between a relatively benign native species and an introduced invasive species, then performance assessments that exclude the invasive parent are unlikely to be informative: the invasive parent may have been capable of spreading just as rapidly and achieving the same densities even if hybridisation had not occurred. Unfortunately, a surprising number of studies fail to include comparisons to the more invasive parent (see Discussion, below).
Previous compilations of taxa that are both invasive and hybrid-derived (Ellstrand & Schierenbeck 2000; Schierenbeck & Ellstrand 2009) represent a vital first step in determining whether hybridisation and invasiveness are causally related. However, these compilations were not intended to be exhaustive. More importantly, given that hybridisation is not rare (animals: Schwenk et al. 2008; plants: Whitney et al. 2010a; fungi: Giraud et al. 2008), we should expect some fraction of invasive taxa to be hybrids, even in the absence of an underlying causal relationship. Our aims are to clarify the criteria needed for evaluating the H-I hypothesis and to evaluate tests meeting these criteria that have been conducted to date. We perform a systematic review to address three questions: (1) based on a comprehensive evaluation of the literature, how frequently has hybridisation been associated with invasiveness in plants, animals, and fungi? (2) to what extent has the H-I hypothesis been evaluated experimentally? and (3) do those evaluations support or refute the H-I hypothesis? We expand earlier compilations of invasive hybrids (Schierenbeck & Ellstrand 2009) by including a number of new hybrid taxa. In addition, we present recommendations for designing future tests of whether hybridisation promotes invasiveness.
METHODS
To investigate the H-I hypothesis, we carried out a systematic review (sensu Liberati et al. 2009) that included meta-analyses. In doing so, we followed the PRISMA statement (Liberati et al. 2009) to the extent possible; note that PRISMA was developed for systematic reviews in medicine and thus not all provisions are relevant here. Our first step was to conduct an exhaustive literature review in an attempt to identify all plant, animal, and fungal taxa for which interspecific hybridisation is postulated to have enhanced invasiveness or contributed to geographic range expansions. We then assessed the extent to which this hypothesis has been tested experimentally, and then quantified the effects of hybridisation on invasiveness using meta-analysis. Following Richardson et al. (2000), we define invasiveness as a measure of the ability of a species, without human assistance, to increase in population size and spatial distribution following introduction. However, here we have relaxed the ‘introduction’ criterion to also include a handful of systems where both parents are native to the region from which a hybrid has subsequently spread. Thus, our dataset includes putatively invasive hybrids originating via a number of possible scenarios: (1) hybridisation in the native range, followed by local or regional range expansion, (2) hybridisation in the native range, followed by introduction to a novel region, (3) hybridisation in the novel range between two (or more) introduced parents, and (4) hybridisation in the novel range between one introduced and one native parent. These scenarios are not mutually exclusive because in some systems hybrids have been introduced repeatedly to novel regions and/or the parental taxa recurrently hybridise.
Literature search criteria
On May 10, 2012 (and again on August 5, 2013 to update our dataset), we searched Thomson Reuters Web of Science (http://apps.webofknowledge.com) for the keywords ‘hybrid*’ and ‘inva*’, subsequently limiting our records to those in the following categories (to minimise references from the medical literature): Ecology, Evolutionary Biology, Plant Sciences, Marine and Freshwater Biology, Entomology, Zoology, Fisheries and Agronomy. This search yielded 945 references. On the same days, we also conducted a forward citation search on three seminal papers that address the implications of hybridisation for biological invasions (Stebbins 1985; Abbott 1992; Ellstrand & Schierenbeck 2000); this added 593 references to our list. We read all 1538 abstracts and every study in which an invasive or weedy species had been determined to be a hybrid, retaining studies in which hybridisation was linked to invasiveness. Additional relevant references cited by these studies were added to our database as we encountered them. We kept a running list of hybrid systems that fit our criteria (see following paragraph), and once we had finished reviewing the studies in our database we conducted one additional taxon-specific search for each of the 72 hybrid systems identified (last updated August 5, 2013) to ensure we had not overlooked any experimental parent-hybrid comparisons in those systems. These study selection steps are summarised in Appendix 1, and Appendix 2 presents the resulting list of 72 systems in which hybridisation has a presumed link to invasiveness. We then confirmed that the use of a single search engine did not result in appreciable numbers of relevant studies being missed (Appendix 3).
Our search included hybrid plant, animal and fungal taxa (including the ‘fungoid’ stramenopiles; bacteria were omitted). We excluded hybrids that have only been synthesised experimentally (i.e. have no analogues in nature), and we excluded transgenic hybrids where hybrid advantage is linked to transformed genes such as those providing herbicide resistance. While there are many fascinating systems where intraspecific hybridisation (i.e. ‘wide crossing’) may have enhanced invasiveness (Lavergne & Molofsky 2007; Culley & Hardiman 2009; Geiger et al. 2011), we limited our review to interspecific hybridisation. Finally, one interspecific hybrid system identified previously (Schierenbeck & Ellstrand 2009) as a potential example of the H-I process was excluded because recent evidence argues against its hybrid origin (‘strawhull’ rice: Reagon et al. 2011; Sun et al. 2013).
Assessing the literature: what do we know about hybrid invaders?
For each hybrid system we recorded basic biological information (see Appendix 2) including growth form (for plants: tree, herb, grass), life history (e.g. annual, perennial) and ploidy of the parental and hybrid taxa. We recorded whether the parental taxa had commercial origins (i.e. horticulture, agriculture or the pet trade), the provenance of parental taxa (native or introduced) and whether hybridisation occurred before or after introduction to a novel region.
Lastly, for each system we collected information about the strength of evidence supporting the H-I hypothesis. We noted whether such inferences were based on ad hoc observations, observational studies (measurements taken in naturally occurring populations), lab- or greenhouse-based experimental studies, or field-based experimental studies conducted in a common environment (including common garden and mesocosm experiments). We recorded the metrics by which hybrid performance was assessed and the parental taxa to which hybrid performance was compared. Finally, we recorded which hybrid class(es) were investigated.
Meta-analysis: do hybrids outperform parents?
For the meta-analysis, we identified every study in our dataset conducting an experimental hybrid-parent performance comparison, including field-, greenhouse-, and lab-based experiments. We excluded non-experimental observations from natural populations, as this approach does not permit taxon-specific (i.e. genetic) effects to be separated from environmental effects. We also excluded performance comparisons that were unable to test the H-I hypothesis because of uninformative benchmarks (e.g. studies failing to compare a hybrid with its most invasive parental taxon). We extracted performance data for hybrids and their parental taxa, grouping performance metrics into three categories: fecundity (including for Lactuca, the product of germination rate, survival rate and seed output plant−1; see Hooftman et al. 2005), survival and organism size. We consider these individual performance metrics to be potential components of population growth rates and thus proxies for invasiveness, while acknowledging that they can be imperfect indicators (see Discussion). For plants, roughly half of the reported fecundity estimates included values of zero for individuals that died before setting seed (Helianthus, Lactuca and Sorghum), while other estimates excluded pre-reproduction mortality (Reynoutria, Senecio vulgaris and Senecio squalidus); Raphanus fecundity estimates in our database represent a combination of both approaches. Estimates including pre-reproduction mortality are likely to be more robust indicators of population growth rates (see Discussion), but we included both types of estimates for meta-analyses. For the two hybrid fungal pathogens in our dataset only pathogenicity was reported, and we considered this a fecundity proxy for our meta-analysis. We extracted means and standard deviations (or the data needed to calculate them) from tables and figures, using in the latter case the object measurement tool in Adobe Acrobat Professional version 7 (Adobe Systems Inc., San Jose, CA, USA).
We used Hedges’d as our effect size metric:
where
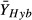 and
and 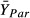 are
mean performance of the hybrid and parental taxa, s is the pooled standard
deviation and J is a small-sample correction factor (Rosenberg
et al. 2013). Positive values for
d thus indicate hybrids that outperform their parents, as predicted by the H-I
hypothesis. If multiple hybrid classes were considered in a single experiment, we extracted data
separately by hybrid class (e.g. resynthesised F1 vs. wild hybrids). Some of the studies
in our meta-analysis reported data from both parental taxa, in which case we calculated
Hedges’ d twice, once for the hybrid relative to the more ‘invasive’ parent
(i.e. that with higher fecundity, larger size or higher survival), and a second time for the hybrid
relative to mean performance of both parents.
are
mean performance of the hybrid and parental taxa, s is the pooled standard
deviation and J is a small-sample correction factor (Rosenberg
et al. 2013). Positive values for
d thus indicate hybrids that outperform their parents, as predicted by the H-I
hypothesis. If multiple hybrid classes were considered in a single experiment, we extracted data
separately by hybrid class (e.g. resynthesised F1 vs. wild hybrids). Some of the studies
in our meta-analysis reported data from both parental taxa, in which case we calculated
Hedges’ d twice, once for the hybrid relative to the more ‘invasive’ parent
(i.e. that with higher fecundity, larger size or higher survival), and a second time for the hybrid
relative to mean performance of both parents.
We performed a series of data-aggregation steps to minimise non-independence in our dataset. When multiple performance metrics fitting one of our categories were reported in a single study, we selected the most comprehensive (e.g. total biomass instead of aboveground biomass). For studies conducted using multiple experimental treatments, we selected the treatment most closely approximating natural conditions in the field (i.e. herbivores present); lacking an obvious selection on that basis, we chose one at random. We used fixed-effect meta-analysis with inverse variance weighting to aggregate effect sizes from multiple sites or habitats within a single study (Mengersen et al. 2013), doing so separately for each parental taxon and hybrid class. We used weighted means to aggregate data from multiple hybrid classes within an experiment, using two alternative aggregation approaches. In the first, we simply combined data from all hybrid classes; this can be used to calculate an overall hybrid effect, but it ignores meaningful distinctions between the types of hybrids used across studies. Thus, in the second approach we distinguished three hybrid categories: (1) naturally occurring, or ‘wild’ hybrids usually assumed to be advanced-generation hybrids, (2) resynthesised F1 hybrids from experimental crosses, and (3) resynthesised post-F1 hybrids from experimental crosses (a broad category, ranging from F2 and BC1 to F10 hybrids). Finally, for hybrid taxa that had been studied in multiple studies, we aggregated effect sizes using one final fixed-effects meta-analysis, yielding no more than one effect size per species in every analysis. We conducted random-effect meta-analyses with inverse variance weighting on these data in R (version 3.0.2; R Development Core Team 2013) using the metafor package (version 1.9-3; Viechtbauer 2010). As a measure of heterogeneity in our analyses we present I2 values for each model (Higgins & Thompson 2002). I2 ranges from 0 to 100% and can be interpreted as the proportion of variability in a given effect size estimate due to between-study heterogeneity.
Because we expected the group of resynthesised post-F1 hybrids to be a heterogeneous sample, we also created a subset of our full dataset for more detailed analyses using meta-regression (Mengersen et al. 2013). For the few hybrid taxa in which performance data from multiple hybrid classes were reported, we re-aggregated effect sizes within and among studies (as above) to yield a single effect size per taxon and per hybrid class. This gave distinct effect sizes for wild hybrids, F1s, generation 2 hybrids (including F2, BC1 and progeny of selfed F1s), generation 3 hybrids (including F3, BC2 and progeny of selfed F2s) and so on. We included hybrid class as a continuous, fixed effect in meta-regressions, which tested for significant increases or decreases in effect size across generations. Because hybrid class is unknown for wild hybrids in the dataset, we omitted them from these analyses. To account for multiple effect sizes per species, we used a multivariate approach in metafor (the rma.mv command), specifying hybrid taxon as a random factor and adding a study-level random variable to achieve a random-effects meta-regression. Where the hybrid class effect was significant it was sometimes inconsistent across taxa, so in these cases we re-ran random-effects meta-regressions separately for each hybrid taxon in the dataset (note that data are lacking to assess taxon × class interactions statistically). All meta-regressions included an intercept, although we report only on inferences regarding significance of the slopes.
Because all species have a shared evolutionary history, non-independence among species is an important consideration for meta-analyses where effect sizes can be mapped onto phylogenies (Lajeunesse 2009; Chamberlain et al. 2012). We conducted phylogenetic meta-analyses using PhyloMeta version 1.3 (Lajeunesse 2011) to account for this non-independence. A phylogeny was constructed as follows: a base topology of plant taxa was derived from Phylomatic v.3 (Webb & Donoghue 2005) using tree R20120829; resolution within Asteraceae was added using topologies in Funk et al. (2009); animal and fungal taxa were added based on Maddison & Schulz (2007). We aged internal nodes for the phylogeny of plant taxa based on Wikström et al. (2001), using the bladj algorithm in Phylocom to interpolate ages of undated nodes (Webb et al. 2008). We added ages to all remaining nodes using the Expert Result dates from Hedges et al. (2006). This base tree with all taxa and node ages is presented in Appendix 4. Phylogenetic meta-analysis results did not differ qualitatively from non-phylogenetic results. Because of this concordance and because PhyloMeta does not support meta-regression, for the sake of consistency and ease of interpretation we present results only from non-phylogenetic meta-analyses and meta-regressions in the main text; see Appendix 4 for phylogenetic meta-analysis results.
The results we present make no distinction between experiments conducted in the field vs. lab or greenhouse experiments. Where sample sizes were large enough to run analyses using only field-collected data, outcomes were similar to those using the more complete dataset (data not shown); heterogeneity tests also indicated similar effect sizes in field- vs. lab-based investigations (Appendix 5; see Table1 for Field/Lab designations). Because of limited sample sizes, we present comparisons between hybrids and their ‘more invasive’ parent only. Comparisons of hybrid performance to the average performance of both parents yielded similar results, although with slightly larger effect sizes (data not shown); the results presented below can therefore be considered conservative.
Table 1.
Experimental studies included in meta-analyses
| Hybrid Taxon | Parental Taxa | Field/Lab | Hybrid Classes§ | Fecundity | Survival | Size | Reference |
|---|---|---|---|---|---|---|---|
| Plant taxa | |||||||
| Carpobrotus hybrids¶ | C. edulisE × C. chilensisE | Field | Wild | W>P1>P2† | P2=P1=W | Vila & D'Antonio 1998b | |
| Field | Wild | W=P2=P1 | W=P1=P2 | Vila & D'Antonio 1998a | |||
| Field | Wild | P2a=P1ab=Wb | Weber & D'Antonio 1999 | ||||
| Helianthus annuus texanus | H. a. annuusN × H. debilisN | Field | Wild, BC1 | W>P1a=P2ab=G2b | W=G2=P2>P1 | P1=W=G2>P2† | Whitney et al. 2006, 2010bb |
| Field | Wild, BC1, BC7 | G7=W=P1>P2>G2 | Wa=P2ab=G2ab=P1b>G7 | W=P1>G7>G2>P2 | Hovick et al. unpub. (see Appendix 6) | ||
| Field | Wild, BC1, BC4 | W=P1=G4>G2 | P1a=G2a=G4ab=Wb | P1a=G4ab= G2b=Wb | Whitney et al. unpub.(see Appendix 6) | ||
| Lactuca hybrids | L. serriolaN × L. sativaE | Field | BC1, S1 | G2>P2=P1 | G2>P2>P1 | Hooftman et al. 2005 | |
| Field | F1, S1-3, BC1S1, BC1-3 | G1a=G2ab=G3bc=P1bc=G4c>P2 | G1a=G2ab=G3b=G4b=P1b>P2 | Hooftman et al. 2007; | |||
| Field | BC1S1 | P1=G3>P2 | G3=P1>P2 | P2=G3=P1 | Hartman et al. 2013 | ||
| Myriophyllum hybrids¶ | M. spicatumE × M. sibiricumE | Lab | Wild | W>P1 | LaRue et al. 2013 | ||
| Raphanus hybrids | R. raphanistrumE × R. sativusE | Field | G3 | P1>G3* | G3>P1* | Campbell & Snow 2007 | |
| Field | G4 | G4>P1 | G4>P1 | G4>P1 | Campbell et al. 2006; | ||
| Field | G4 | G4>P1* | P1=G4* | Hovick et al. 2012 | |||
| Field | Wild | W>P1>P2 | W>P1>P2† | W=P2>P1 | Ridley & Ellstrand 2009 | ||
| Field | F1 | P1>G1* | P1>G1*,† | Snow et al. 2001 | |||
| Field | F2, F10 | P1=G2; G10=P1 | Snow et al. 2010‡ | ||||
| Reynoutria × bohemica¶ | R. japonicaE × R. sachalinensisE | Field | Wild | P1=W=P2*,† | P2=W=P1 | Pyšek et al. 2003 | |
| Field | Wild | P1=W>P2* | Brabec & Pyšek 2000 | ||||
| Field | F1, BC1, F2 | G2=G1=P1 | Gammon et al. 2010 | ||||
| Field | Wild | W=P1=P2*,† | W>P2=P1* | Parepa et al. 2014 | |||
| Lab | Wild | W=P1* | Richards et al. 2008 | ||||
| Lab | Wild | P1=W* | Rouifed et al. 2011 | ||||
| Senecio vulgaris var. hibernicus | S. squalidusE × S. vulgaris var. vulgarisN | Field | Wild | P1=W=P2 | W=P1=P2* | P2=W=P1 | Hawkes et al. 2010 |
| Senecio squalidus | S. aethnensisE × S. chrysanthemumifoliusE | Lab | Wild | P2=W>P1 | P2a=Wab=P1b | Brennan et al. 2012 | |
| Sorghum almum | S. halepenseE × S. bicolorE | Field | F1 | G1=P1 | G1=P1* | Arriola & Ellstrand 1997 | |
| Spartina hybrids¶ | S. alternifloraE × S. foliosaN | Lab | Wild | W=P1>P2 | Ayres et al. 2004 | ||
| Typha × glauca¶ | T. angustifoliaE × T. latifoliaN | Field | Wild | W>P2=P1 | Sullivan et al. 2010 | ||
| Fungal taxa | |||||||
| Melampsora × columbiana | M. medusaeN × M. occidentalisN | Lab | Wild | P2>W>P1 | Newcombe et al. 2000 | ||
| Lab | Wild | W=P1>P2 | Newcombe et al. 2001 | ||||
| Pythium hybrids | P. arrhenomanesE (or similar) × P. phragmitisN | Lab | Wild | W=P1>P2* | Nechwatal & Mendgen 2009 | ||
| Animal taxa | |||||||
| Ambystoma hybrids | A. tigrinum mavortiumE × A. californienseN | Field | F1, F2, BC1 | P2=P1=G1=G2 | P1=G1=G2>P2 | Ryan et al. 2009 | |
| Lab | Wild, F1, F2, BC1 | P1>G1=G2>P2>W | P1>G2a=Wab=G1b>P2 | Johnson et al. 2010 | |||
| Cyprinella hybrids | C. lutrensisE × C. venustaN | Lab | F1 | G1>P1=P2* | Blum et al. 2010 | ||
Significant differences as reported in references; otherwise, significance reflects means separated by >2 SEM, based on our aggregated data.
Excluded from meta-analysis because variability not reported.
G2 and G10 hybrids assessed in separate experiments.
All hybrid classes used (Sx = selfed Fxs; BCx = backcrossed Fxs; BC1S1 = selfed BC1s).
Capable of clonal growth (Flora of North America Editorial Committee 1993+).
For each entry, significant differences are indicated by inequality signs and superscript letters, where necessary (taxa sharing the same letter do not differ). Differences are from referenced studies or our data aggregations (see footnote) and were not used in meta-analysis. Parental taxa are denoted P1 and P2, reflecting their order in the ‘Parental Taxa’ column; P1 is the more invasive/higher performing parent used for meta-analyses. Parents are denoted as native (N) or exotic (E), relative to the region where hybrids are invasive.
Results
Including those that have been identified in previous reviews (Ellstrand & Schierenbeck 2000; Schierenbeck & Ellstrand 2009), we found 72 hybrid systems (i.e. a hybrid taxon and its two or more parental species) for which hybridisation has been putatively associated with invasiveness, weediness or range expansion (Appendix 2). The vast majority are plants (n = 59 systems), followed by fungal pathogens (including fungi and stramenopiles; n = 8) and animals (n = 5). Plant hybrids in our database come from 20 families, primarily Asteraceae (n = 16), Poaceae (n = 8) and Brassicaceae (n = 6).
Experimental tests of the H-I hypothesis
From our full list of 72 hybrids, we found 18 hybrid taxa for which experimental hybrid-parent performance comparisons utilizing the relevant parent(s) have been conducted (reported in 40 studies and two unpublished datasets), either in the field (24 studies plus our unpublished datasets) or in the lab or greenhouse (16 studies). Only 14 of the 59 plant hybrids in our dataset (23.7%) have been assessed experimentally in such a way as to test the H-I hypothesis (Appendix 2). Nine of the 14 hybrid systems have been assessed in the field, with the remainder tested only in the lab or greenhouse. These experiments compare hybrid performance to that of the relevant parental taxa, either both parents (e.g. Vila & D'Antonio 1998b; Hooftman et al. 2005; Sullivan et al. 2010) or, if one parent is clearly more invasive or a superior competitor (i.e. if the inferior parent has not become naturalised despite opportunity), the superior parent (e.g. Hovick et al. 2012). For 11 of the remaining 45 taxa that have not been adequately tested, observational or incomplete experimental data have been reported that are suggestive of hybrid performance gains; these include some studies comparing hybrids with their inferior competitor parent only (e.g. Grosholz 2010) and some studies comparing hybrids with only one parent, either because the second is not clearly inferior (Van Grunsven et al. 2009), or because the second parent is not yet known (Henery et al. 2010; Hahn et al. 2012). For the remaining 34 of these 45 plant hybrids, hybridisation has been confirmed and also suggested as the underlying driver of weediness or invasiveness, but we have found no data on which to judge the latter claim.
For fungal pathogens, two hybrids have been assessed sufficiently to yield experimental tests of the H-I hypothesis: the rust Melampsora x columbiana (Newcombe et al. 2000, 2001) and a Pythium hybrid found on European Phragmites australis (Nechwatal & Mendgen 2009). All such investigations have been conducted in the lab, thus the relative performance of hybrid pathogens in the field is not known.
Of the five invasive animal hybrids in our dataset, two systems have been investigated experimentally. Ambystoma hybrids have been assessed both in field (Ryan et al. 2009) and lab tests (Johnson et al. 2010) and Cyprinella hybrids have been assessed in the lab only (Blum et al. 2010).
For our meta-analyses, we excluded eight of the 42 studies reporting experimental data, either because they failed to report some measure of variability or because they only reported data (percent germination, photosynthesis-related traits and walking speed) not fitting with our three performance categories; this exclusion eliminated three hybrid plant taxa from subsequent analyses (Sarcocornia hybrids, Sphagneticola hybrids and Viola x tatrae), cutting our final list to 15 hybrid taxa (11 plant, two animal and two fungal hybrids; see Table1 and Appendix 4).
Meta-analysis: does hybridisation lead to hybrid performance gains?
Many of the 34 studies included in our meta-analyses used multiple hybrid classes, reported data from multiple unique experiments or established experiments across multiple locations. Thus, we calculated a total of 47 fecundity-based, 52 size-based and 42 survival-based effect sizes from experimental hybrid-parent performance comparisons. In all analyses, effect-size heterogeneity (I2) among taxa was substantial, which likely contributed to non-significant differences in heterogeneity tests comparing F1, post-F1 and wild hybrids (fecundity: Q = 3.32, P = 0.190, I2 = 86.1%; survival: Q = 1.18, P = 0.553, I2 = 93.3%; size: Q = 2.54, P = 0.280, I2 = 96.2%; all d.f.=2). We thus conducted analyses on all hybrid classes combined; however, because of an a priori interest in relative performance by each group and because these categories represent hybrid populations with distinct histories of genotypic filtering and selective pressures, we also conducted analyses separately for each hybrid class. This approach is also supported by (less conservative) fixed-effects analyses, which found significant differences between hybrid classes (results not shown). We found no indication of publication bias based on funnel plots and rank correlation tests for funnel plot asymmetry (see Appendix 5).
Do hybrids out-reproduce their parental taxa?
Meta-analyses of fecundity indicated that naturally occurring wild hybrids significantly outperformed their ‘more invasive’ parental taxon (Z = 2.79, P = 0.005, n = 6, I2 = 72.1%; see ‘Overall M-A results’ in Fig.1a). Within individual taxa, wild hybrids were uniformly more fecund than their more invasive parents, with the effect reaching statistical significance in Raphanus and Senecio squalidus (and nearly reaching statistical significance in Helianthus, with a 95% confidence interval of [−0.034, 0.281], Fig.1a). For all hybrid classes considered together, hybrids were also significantly more fecund than their more invasive parent (Z = 2.04, P = 0.042, n = 9, I2 = 64.1%; Fig.1a). However, resynthesised F1 and later-generation (post-F1) hybrids did not have significantly enhanced fecundity (F1: Z = −0.73, P = 0.468, n = 4, I2 = 92.7%; post-F1: Z = 0.72, P = 0.470, n = 4, I2 = 85.2%; Fig.1a).
Figure 1.
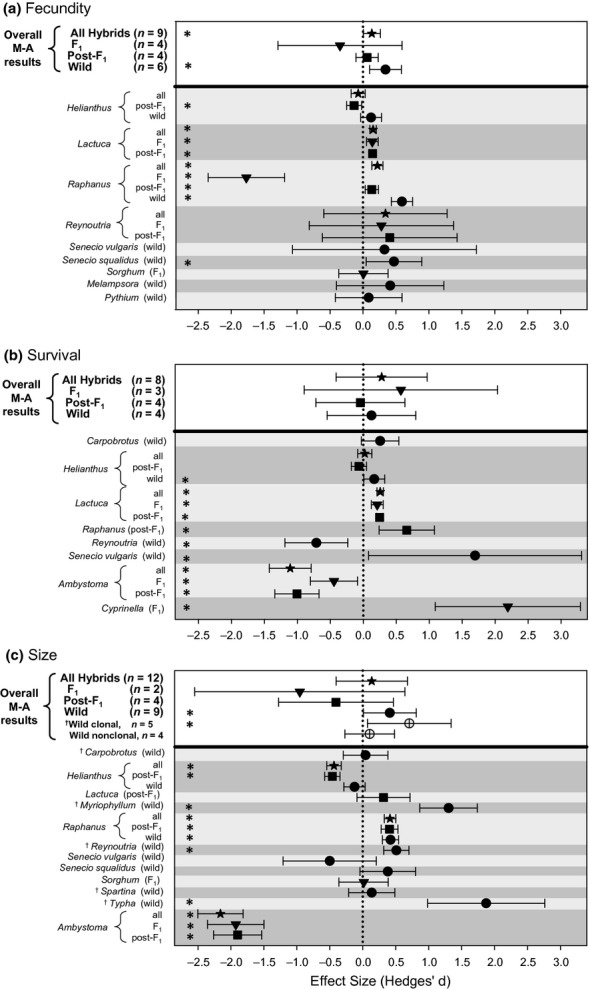
Meta-analysis results, showing effect sizes and 95% confidence intervals for hybrid vs. parental performance for (a) fecundity, (b) survival and (c) size. Effects greater than zero (dotted vertical line) indicate greater hybrid performance relative to their ‘more invasive’ parental taxon. Confidence intervals excluding zero are noted with an asterisk along the left side of the panel. Overall effect sizes shown in the non-shaded region at the top of each panel are from random effect meta-analyses conducted separately for (1) all hybrid classes combined, (2) resynthesised F1 hybrids, (3) resynthesised post-F1 hybrids and (4) wild hybrids. In panel C, separate overall effect sizes are also shown for wild hybrids that are capable vs. incapable of clonal growth. Individual effect sizes are shown beneath the solid horizontal line, with taxa separated by alternate shading and symbols indicating different hybrid classes.
Our meta-regression of fecundity responses indicated a marginally significant generation effect across all three taxa, with relative hybrid fecundity increasing over subsequent generations (βgen ± SE = 0.106 ± 0.058, Z = 1.83, P = 0.067, I2 = 95.6%), although this relationship varied across taxa (Fig.2a). When taxa were assessed separately with linear models, the generation effect was marginally significant and positive for Helianthus, not significant for Raphanus, and significantly negative for Lactuca (Helianthus: βgen = 0.218 ± 0.117, Z = 1.87, P = 0.062, I2 = 93.2%; Raphanus: βgen = 0.132 ± 0.108, Z = 1.223, P = 0.221, I2 = 93.7%; Lactuca: βgen = −0.067 ± 0.032, Z = −2.13, P = 0.033, I2 = 74.6%). Despite apparent nonlinearity in the scatterplot (Fig.2a), a polynomial relationship was not supported for all three taxa combined (polynomial model P = 0.101, βgen P = 0.133 and βgen2 P = 0.286). In contrast, a polynomial model did fit the combined Helianthus and Raphanus data well (polynomial model P = 0.004; βgen=0.688 ± 0.259, Z = 2.66, P = 0.008; βgen2=−0.049 ± 0.023, Z = −2.162, P = 0.031, I2 = 93.9%), indicating that hybrid relative fecundity for these two taxa increased rapidly over the first few generations before reaching a point where hybrids outperformed their parents (Fig.2a).
Figure 2.
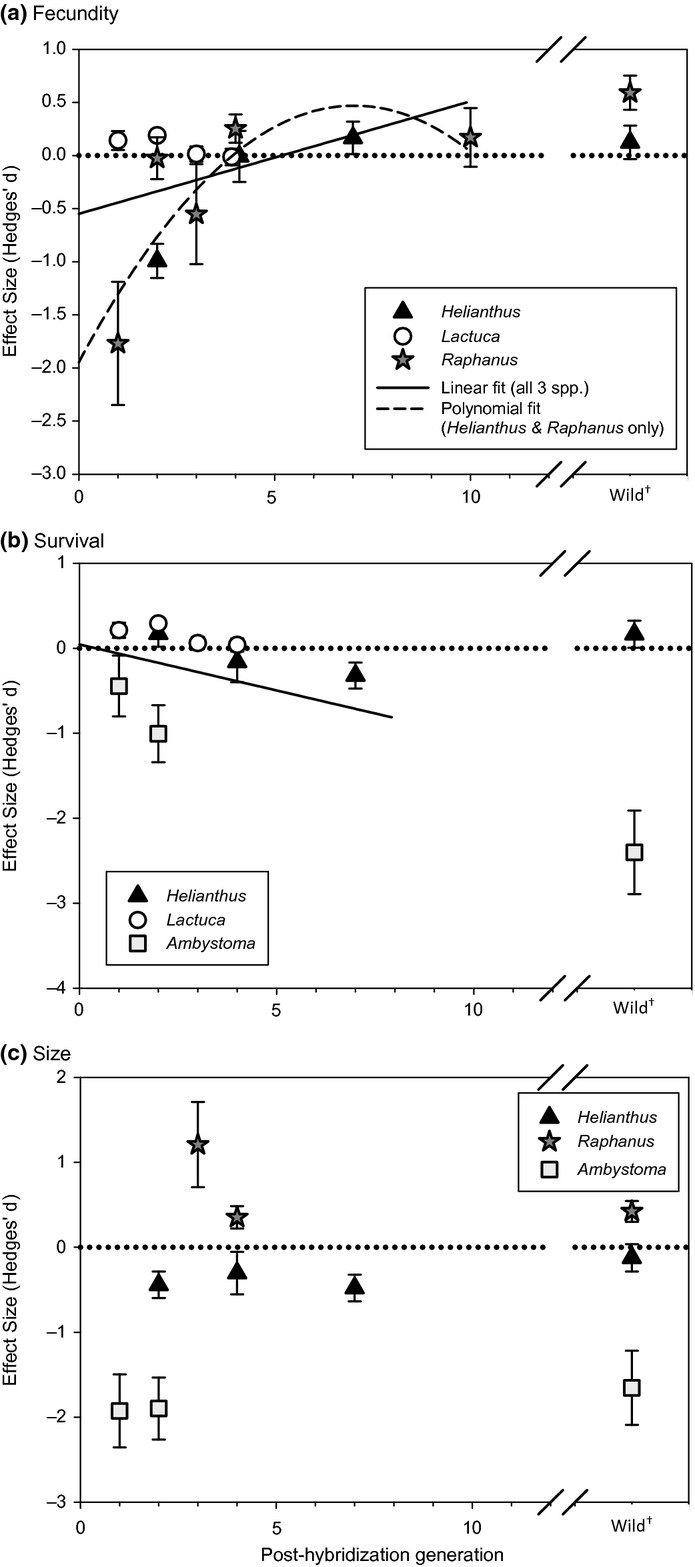
Meta-regression results, showing effect sizes (hybrid vs. parental performance) by hybrid generation for (a) fecundity, (b) survival and (c) size. Effects greater than zero (above the dotted horizontal line) indicate greater hybrid performance relative to their ‘more invasive’ parental taxa. Error bars indicate 95% confidence intervals. The best-fit lines in panels A and B show significant relationships between hybrid generation and Hedges’ d for fecundity and survival based on meta-regression parameter estimates; the meta-regression for size in panel C was not statistically significant. In addition to the linear best-fit line, panel A also depicts the nonlinear relationship between hybrid generation and relative fecundity supported for Helianthus and Raphanus (considered apart from Lactuca; see Results). Note that effect sizes for naturally occurring wild hybrids were not included in the analysis, but are shown in the figure for comparison.
Do hybrids outlive their parental taxa?
Meta-analyses of survival indicated that none of our three hybrid classes differed from their parental taxa (wild: Z = 0.36, P = 0.718, n = 4, I2 = 81.9%; F1: Z = 0.76, P = 0.446, n = 3, I2 = 92.0%; post-F1: Z = −0.12, P = 0.901, n = 4, I2 = 96.2%; Fig.1b), but we note that small sample sizes for these analyses limit our power to detect any true effects. The combination of all hybrid classes gave the same non-significant result (Z = 0.79, P = 0.429, n = 8, I2 = 93.9%). Unlike our other performance metrics, relative survival of hybrids tended to vary more among taxa than among hybrid classes within taxa. Regardless of hybrid class, all Lactuca, Raphanus, Senecio vulgaris and Cyprinella hybrids had significantly higher survival than their parents, whereas all Reynoutria and Ambystoma hybrids had significantly lower survival (Fig.1b).
Across the three systems for which data are available, relative hybrid survival significantly decreased across generations (βgen = −0.122 ± 0.048, Z = −2.55, P = 0.011; I2 = 93.3%; Fig.2b). This pattern was similar for Helianthus and Lactuca when analysed separately (Helianthus: βgen = −0.097 ± 0.022, Z = −4.41, P < 0.001, I2 = 0.0%; Lactuca: βgen = −0.077 ± 0.042, Z = −1.82, P = 0.068, I2 = 86.8%; note that Ambystoma had too few data points to be assessed separately).
Are hybrids larger than their parental taxa?
In addition to being more fecund, wild hybrids were also significantly larger than their more invasive parent (Z = 2.00, P = 0.046, n = 9, I2 = 89.4%; Fig.1c). In contrast to the fecundity data, effect size variation among wild hybrid taxa for organism size was much greater (89.4 vs. 72.1%), with a broader range of effect size estimates (Fig.1c) and two influential outlier genera with large wild hybrids relative to their parents (the plants Typha and Myriophyllum; Cook's D > 0.34 and Dffits > 0.65 for both). Because relative size advantages may be particularly relevant for species that persist and spread by clonal growth, we re-analysed the wild hybrid size data by clonal growth category (present vs. absent; see Table1). Although effect sizes for these two groups were not significantly different from each other (Q = 2.36, d.f.=1, P = 0.124), only wild hybrids capable of clonal growth were larger than their parental taxa (clonal: Z = 2.18, P = 0.029, n = 5, I2 = 88.0%; non-clonal: Z = 0.55, P = 0.581, n = 4, I2 = 90.8%; Fig.1c). At the level of individual clonal taxa, hybrids were significantly larger than parents in Myriophyllum, Reynoutria and Typha but not in Carpobrotus or Spartina. For resynthesised F1s and post-F1s, hybrids were smaller than their parental taxa on average, but with large confidence intervals that overlapped zero (F1: Z = −0.99, P = 0.325, n = 2, I2 = 97.8%; post-F1: Z = −0.77, P = 0.444, n = 4, I2 = 98.5%). When all hybrid classes were considered together, hybrid and parental size did not differ (Z = 0.50, P = 0.620, n = 12, I2 = 97.1%).
Among the three resynthesised hybrid taxa that have been assessed, relative hybrid size did not vary across generations (βgen = −0.023 ± 0.078, Z = −0.30, P = 0.768, I2 = 98.1%; Fig.2c).
Discussion
We found moderate support for the hypothesised connection between interspecific hybridisation and increased invasiveness. Quantitative data from experimental performance comparisons that could be used to test the H-I hypothesis were available for roughly 21% of all invasive or weedy hybrid taxa in our dataset. Not surprisingly, among-study heterogeneity was high in all cases, reflecting species-specific responses to hybridisation in addition to likely variation in the design of experiments and performance assessments. The strongest evidence supporting the H-I hypothesis was found in fecundity-based performance assessments involving wild (as opposed to resynthesised) hybrids; evidence was strongest in plants but weaker in the few animal and fungal taxa for which data were available. Although fecundity relative to parents was on average low in resynthesised hybrids, it increased significantly over time in two of the three systems for which we could assess the pattern; in these systems, resynthesised hybrids outperformed their parents by approximately the fifth generation post-hybridisation. Hybrid survival was more variable among taxa, and although no overall effect was seen in the meta-analysis, hybrid survival relative to parents tended to be high for plants and inconsistent between the two animal systems evaluated (note that tests in animal systems have only focused on resynthesised, instead of wild, hybrids). For resynthesised hybrids, there was an overall pattern of decreasing relative survival over time post-hybridisation. Finally, with respect to size, the meta-analysis revealed that wild hybrids were larger than their parental taxa, a pattern that was particularly strong for taxa in which hybrids are capable of clonal growth.
Designing experiments to test the H-I hypothesis: two basic criteria
One of the clearest patterns from our review is that many studies appearing to test the H-I hypothesis have not adequately done so. At a minimum, we argue that an informative test must meet two basic criteria. First, it must compare the performance of hybrids to that of the most invasive parent to ensure that hybrid performance is assessed relative to the correct benchmark. Where two introduced or two native species have hybridised, typically both parents must be included in the assessment to determine whether hybridisation has enhanced invasiveness. Where hybrids result from crossing one native taxon with one introduced and invasive taxon, the introduced parent must be included in any assessment for meaningful comparisons. If the most relevant parent is omitted, then inferences regarding the H-I hypothesis are impossible. For example, California Spartina is often cited as a case where hybridisation is associated with invasiveness (e.g. Ayres et al. 2008; Grosholz 2010). However, we found no experimental tests comparing fecundity or survival of the hybrid to the invasive parent, and a test of size found no difference between the taxa (Table1). A test of resistance to goose herbivory found decreased palatability in the hybrid relative to the native parent S. foliosa (Grosholz 2010). This pattern was interpreted as support for the H-I hypothesis, via the suggestion that hybrids ‘may pose a greater risk to natural systems than the parent species.’ Again, however, no comparison was made to the invasive parent S. alterniflora. It could be that the invasive parent has equal (or even lower) palatability relative to the hybrid, in which case hybridisation could have nothing to do with performance or invasiveness in this system.
In some cases, accurate identification of both parental taxa is a barrier to our first criterion for testing the H-I hypothesis. For example, a tetraploid cytotype of spotted knapweed (Centaurea stoebe s.l.) has been recorded as invasive in North America, with molecular analyses pointing to an allopolyploid origin between the diploid Centaurea stoebe s. str. and an unknown parent (Mraz et al. 2012). Robust common garden experiments have been conducted in this system (Henery et al. 2010; Hahn et al. 2012), but without knowing the second parent's identity inferences regarding the H-I hypothesis are moot. Similar challenges exist in other systems (e.g. Kim et al. 2008; Inderbitzin et al. 2011), emphasising the continued importance of phylogenetic and taxonomic research in putative cases of hybridisation-induced invasiveness.
We suggest that a second criterion for testing the H-I hypothesis is that performance assessments be conducted experimentally in a common environment, as observations of hybrid performance or abundance in naturally occurring populations are not unambiguously interpretable. This criterion is not satisfied by a handful of key studies often cited in support of the H-I hypothesis that document increased abundances of hybrids relative to their parental taxa (Urbanska et al. 1997; Moody & Les 2002). Abundant hybrids could indeed result from enhanced invasiveness, but the same patterns could also result from chance dispersal events or simply from the introgression of neutral alleles from one parental genome into the other.
Fitness components vs. population growth rates as measures of invasiveness
In our meta-analyses, the degree to which the H-I hypothesis was supported varied with the fitness component (fecundity, survival or size) examined. In some cases, this pattern may reflect expected life-history tradeoffs: for example, resynthesised Helianthus hybrids displayed an increase in relative fecundity but a decrease in relative survival over time post-hybridisation (Fig.2a,b). This issue highlights the importance of knowing how individual fitness components are related to the population growth rate (λ), which determines invasiveness. While fitness components are expected to generally be positively correlated with λ, the details matter. For instance, if populations are establishment (microsite) limited rather than propagule limited (Poulsen et al. 2007), then beyond a certain point, increased fecundity will not necessarily result in increased λ.
Female fecundity alone could be misleading in tests of the H-I hypothesis if male fitness (i.e. pollen or sperm viability) is severely depressed in hybrids relative to their parents. Because we only found pollen viability data for four taxa representing a variety of hybrid classes (Sorghum, Reynoutria, Raphanus and Senecio squalidus), a meaningful meta-analysis was not possible. However, for resynthesised Raphanus hybrids the change in pollen viability over time is similar to the changes in Raphanus fecundity we identified using meta-regression; that is, pollen viability is depressed in the F1 generation but then recovers by approximately generation 3–6, eventually reaching values equal to or greater than that of its parental taxon (Snow et al. 2010). It remains unknown whether male fitness responds similarly in other invasive hybrids and to what extent such patterns might influence population growth rates.
Four studies (of two hybrid systems) included in our meta-analyses do report λ for hybrids vs. parent populations (Hooftman et al. 2005, 2007; Campbell et al. 2006; Hartman et al. 2013). In Raphanus, λ did not differ between paired field populations of resynthesised hybrids and their better performing parental taxon over the second through fourth years post-hybridisation in Michigan (Campbell et al. 2006). From the perspective of testing the H-I hypothesis, this finding concurs with a common garden experiment in the same region where fourth-generation hybrids did not outperform their parental taxon (hybrids were significantly less fecund and had similar survival to the parent; Campbell et al. 2006). In Lactuca, λ was estimated from experimental populations of hybrids and both parental taxa by multiplying germination and survival rates by seed output plant−1 (Hooftman et al. 2005, 2007; Hartman et al. 2013); in all cases, differences in λ between taxa reflected differences in survival but not in seed output, suggesting that for Lactuca, survival is a better indicator of λ than is fecundity. Unfortunately, without estimates of λ and individual fitness components from other systems, questions remain about the degree to which these alternative performance metrics may indicate differences in population growth rates. Overall, future tests of the H-I hypothesis for sexually-reproducing species would be better served if λ (rather than fitness components alone) were estimated as the measure of invasiveness.
For some taxa, size (or vegetative growth rate) may be the most relevant measure of invasiveness. Many invasive plants reproduce primarily via clonal spread (Pyšek & Richardson 2007); and indeed clonality has been identified as an important mechanism by which heterosis achieved in the F1 (which is normally broken down by recombination in sexual species) can be stabilised in hybrid lineages, potentially increasing invasiveness (Ellstrand & Schierenbeck 2000). Invasive populations of hybrid Myriophyllum, Reynoutria and Typha can reproduce clonally, have been shown to be largely composed of F1 individuals (Kirk et al. 2011; Bailey 2013; LaRue et al. 2013), and in our analyses were all significantly larger than their parents (Fig.1c). For taxa such as these, size may be a relevant measure of invasive potential; thus a pattern in which hybrids are larger than parents can be interpreted as support for the H-I hypothesis. Conversely, although Carpobrotus and Spartina hybrids also exhibit clonal growth, sexual reproduction (and thus recombination) appears to be common in these systems (Gallagher et al. 1997; Ayres et al. 2008), and hybrids were not larger than their parental taxa (Fig.1c). The contrasting patterns expressed by these two groups of hybrid taxa illustrate the critical importance of measuring performance in a way that captures invasive potential appropriately for each particular system.
How does within-taxon variation in relative hybrid performance affect our understanding of the H-I hypothesis?
Hybrid taxa are often highly variable, and our review highlights three key aspects of this variability that have direct bearing on how the H-I hypothesis is conceived, interpreted and tested. These non-mutually exclusive sources of variation can be distinguished as habitat effects, generation effects and lineage effects.
Habitat effects: genotype by environment (G × E) interactions influence hybrid invasiveness
Habitat-specific performance advantage can lead to a complicated mosaic of peaks in hybrid relative performance. For example, Carpobrotus hybrids had higher relative growth rates than both of their parental taxa in backdune (but not bluff scrub) habitats, yet hybrid survival in response to herbivory was greatest in the bluff scrub (Vila & D'Antonio 1998b). At a larger geographic scale (North America), Raphanus hybrids have been shown to outperform their weedy parental taxa in California, where they are currently invasive (Campbell et al. 2006; Ridley & Ellstrand 2009), and in Texas, where they do not yet occur naturally (Hovick et al. 2012), but not in Michigan (Campbell et al. 2006). Although hybrids were traditionally thought to occur only in intermediate hybrid zones (Anderson 1948; Barton & Hewitt 1985), mounting evidence suggests that genome shuffling following hybridisation can result in phenotypes that are able to thrive in non-intermediate and even non-parental habitats (Rieseberg et al. 1999, 2007). Site-specific variation in hybrid relative performance will make it more difficult to reject the null hypothesis of no difference between hybrids and their parents if some common (but not ubiquitous) combination of biotic and abiotic factors exists that favours hybrid invaders. These considerations suggest there may be great value in assessing performance across a range of conditions.
Generation effects: not all hybrid generations are created equal
In most cases, assessments of hybrid relative performance depend greatly on which hybrid class(es) are investigated. Our meta-analyses indicate that wild (presumably later-generation) hybrids are more likely to outperform their parental taxa than are resynthesised (mostly early-generation) hybrids, on average. This pattern conforms to long-held expectations that hybrid performance should increase in post-F1 hybrid classes as poorly adapted genotypes are filtered from the population by natural selection (reviewed by Arnold & Hodges 1995). Similarly, based on our meta-regressions, relative fecundity increased over subsequent generations post-hybridisation (Fig.2a), and in many cases, the earliest-generation (F1) hybrids performed worse than their parents and all other hybrid classes (Fig.1a–c). Hybrid survival also varied by generation but in the opposite direction, decreasing over time post-hybridisation (Fig.2b). This pattern may reflect a survival-fecundity life history tradeoff (cf. Helianthus from Fig.2a and b).
These considerations mean that the choice of hybrid class used in testing the H-I hypothesis is a crucial one. On one hand, resynthesising hybrid lineages for testing the H-I hypothesis is the cleanest way to isolate the effects of hybridisation from other evolutionary factors such as population admixture and bottlenecks. On the other, tests using early-generation hybrid populations will often include poorly adapted genotypes (e.g. Snow et al. 2010) and thus may give misleading answers about whether hybrids are capable of outperforming the parental species. Early-generation hybrid populations often have low average fitness but with high variance and a small number of well-adapted individuals (e.g. Whitney et al. 2006). The composition of these populations can therefore change substantially in only a few generations, making tests with only early-generation hybrids suspect. These problems are avoided if resynthesised lineages are first allowed to evolve in natural conditions post-hybridisation (Campbell et al. 2006; Whitney et al. 2006), thus letting natural selection filter out poorly adapted genotypes and mimicking naturally occurring hybridisation/invasion events.
Lineage effects: variable outcomes among hybridisation events
Hybrid lineages derived from the same pair of parental species, but formed with input from different parental individuals, can differ substantially in traits and performance (Pyšek et al. 2003; Hartman et al. 2013). Such variability can also play out at the population level, which means that source populations must be chosen carefully for experimental comparisons (or for resynthesising hybrid lineages). For instance, a hybrid-parent comparison that draws parental individuals from far beyond the supposed range of hybrid formation may be uninformative, particularly where parental taxa are themselves highly variable.
Strong associations between hybridisation and polyploidy for weedy and invasive hybrids
In many systems where the H-I hypothesis has been invoked, hybridisation is also closely associated with polyploidy (Stebbins 1985). In our dataset, 37 of 59 plant hybrids (63%) are reported to be polyploid, and of these nearly half have increased ploidy relative to their parental taxa (16 of 37; see Appendix 2). Polyploidy is not as well studied in fungal pathogens (Albertin & Marullo 2012), but two fungal hybrids from our database are also reported to be polyploid (Verticillium longisporum and Phytophthora alni alni). Because polyploidy alone may contribute to invasiveness (Pandit et al. 2011; te Beest et al. 2012), an important future research direction is disentangling the effects of hybridisation from those of polyploidy.
A robust ‘gold standard’ design for experimentally separating the effects of hybridisation from those of polyploidy in systems experiencing both phenomena would be to create resynthesised hybrid lineages in which both effects are manipulated independently. That is, experimental crosses would be exposed (or not) to chromosome doubling agents such as colchicine to create homoploid hybrid descendants, allopolyploid (hybrid) descendants, and autopolyploid (non-hybrid) descendants from the same set of parents. Performance could then be compared among parental taxa and their descendant lineages to disentangle the phenotypic effects of hybridisation and polyploidy. Such experiments with invasive polyploid taxa would benefit by considering the same sources of variation discussed above (habitat, generation and lineage effects), making strong inferences on the roles of hybridisation and polyploidy for polyploid invaders possible.
The H-I hypothesis: conclusions and future directions
Based on experiments that have been reported to date, the H-I hypothesis is moderately supported across taxa, with strong support in some systems (particularly plants) and little support in others for which it has been postulated (e.g. Senecio vulgaris var. hibernicus). Overall, our results suggest an important but variable effect of hybridisation in triggering or allowing invasions, although we note that these inferences rely on estimates of fecundity, survival and size as proxies of population growth rates and thus of invasiveness. Variability in outcomes (i.e. whether hybridisation triggers invasiveness or not) is consistent with a main theme of the current invasion literature emphasising that invasion is multicausal (Rejmánek et al. 2005); as historic, species-specific and environment-specific factors can predominate, different invasions across different taxa and locations are unlikely to be linked to a single ‘smoking gun.’
The list of invasive hybrids initially compiled by Ellstrand & Schierenbeck (2000) continues to grow (see Appendix 2); however, in most systems we still lack well-designed experiments to test the H-I hypothesis. Critical components of future tests include ensuring that: (1) hybrid performance is compared to the correct benchmark, i.e. the more invasive parent, (2) tests are experimental and carried out in a common environment, (3) species-specific performance metrics are used that accurately reflect the invasiveness of species with different life histories, and (4) more tests in animal and fungal taxa are attempted.
Acknowledgments
Thanks to Allison Snow, Loren Rieseberg, Jim Grover, J. Chris Pires, the Rudgers-Whitney lab group and anonymous reviewers for comments on the manuscript, advice and discussion. Sincere thanks to the authors of the original studies included in the meta-analyses. This work was supported by NSF grants DEB 0716868 and DEB 1257965 (to KDW) and DEB 1146203 (to SMH and KDW).
Authorship
Designed the study, reviewed the literature, reconstructed phylogenies, and wrote the manuscript: SMH and KDW. Compiled data for the meta-analyses and performed all statistical analyses: SMH.
Supporting Information
Additional Supporting Information may be downloaded via the online version of this article at Wiley Online Library (www.ecologyletters.com).
Supplementary Information
References
- 1.Abbott RJ. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- 2.Albertin W. Marullo P. Polyploidy in fungi: evolution after whole-genome duplication. Proc. Biol. Sci. 2012;279:2497–2509. doi: 10.1098/rspb.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson E. Hybridization of the habitat. Evolution. 1948;2:1–9. [Google Scholar]
- 4.Anderson E. Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- 5.Arnold ML. Hodges SA. Are natural hybrids fit or unfit relative to their parents? Trends Ecol. Evol. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- 6.Arnold ML. Martin NH. Hybrid fitness across time and habitats. Trends Ecol. Evol. 2010;25:530–536. doi: 10.1016/j.tree.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Arriola PE. Ellstrand NC. Fitness of interspecific hybrids in the genus Sorghum: Persistence of crop genes in wild populations. Ecol. Appl. 1997;7:512–518. [Google Scholar]
- 8.Ayres DR, Smith DL, Zaremba K, Klohr S. Strong DR. Spread of exotic cordgrasses and hybrids (Spartina sp.) in the tidal marshes of San Francisco Bay, California, USA. Biol. Invasions. 2004;6:221–231. [Google Scholar]
- 9.Ayres DR, Zaremba K, Sloop CM. Strong DR. Sexual reproduction of cordgrass hybrids (Spartina foliosa x alterniflora) invading tidal marshes in San Francisco Bay. Divers. Distrib. 2008;14:187–195. [Google Scholar]
- 10.Bailey J. The Japanese knotweed invasion viewed as a vast unintentional hybridisation experiment. Heredity. 2013;110:105–110. doi: 10.1038/hdy.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker HG. The evolution of weeds. Annu. Rev. Ecol. Syst. 1974;5:1–24. [Google Scholar]
- 12.Baker HG. Stebbins GL. The Genetics of Colonizing Species. NY: Academic Press New York; 1965. [Google Scholar]
- 13.Barton NH. Hewitt GM. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- 14.te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum MJ, Walters DM, Burkhead NM, Freeman BJ. Porter BA. Reproductive isolation and the expansion of an invasive hybrid swarm. Biol. Invasions. 2010;12:2825–2836. [Google Scholar]
- 16.Brabec J. Pyšek P. Establishment and survival of three invasive taxa of the genus Reynoutria (Polygonaceae) in mesic mown meadows: a field experimental study. Folia Geobotanica. 2000;35:27–42. [Google Scholar]
- 17.Brennan AC, Barker D, Hiscock SJ. Abbott RJ. Molecular genetic and quantitative trait divergence associated with recent homoploid hybrid speciation: a study of Senecio squalidus (Asteraceae) Heredity. 2012;108:87–95. doi: 10.1038/hdy.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell LG. Snow AA. Competition alters life history and increases the relative fecundity of crop-wild radish hybrids (Raphanus spp.) New Phytol. 2007;173:648–660. doi: 10.1111/j.1469-8137.2006.01941.x. [DOI] [PubMed] [Google Scholar]
- 19.Campbell LG, Snow AA. Ridley CE. Weed evolution after crop gene introgression: greater survival and fecundity of hybrids in a new environment. Ecol. Lett. 2006;9:1198–1209. doi: 10.1111/j.1461-0248.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 20.Chamberlain SA, Hovick SM, Dibble CJ, Rasmussen NL, Van Allen BG, Maitner BS, et al. Does phylogeny matter? Assessing the impact of phylogenetic information in ecological meta-analysis. Ecol. Lett. 2012;15:627–636. doi: 10.1111/j.1461-0248.2012.01776.x. [DOI] [PubMed] [Google Scholar]
- 21.Crawford KM. Whitney KD. Population genetic diversity influences colonization success. Mol. Ecol. 2010;19:1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x. [DOI] [PubMed] [Google Scholar]
- 22.Culley TM. Hardiman NA. The role of intraspecific hybridization in the evolution of invasiveness: a case study of the ornamental pear tree Pyrus calleryana. Biol. Invasions. 2009;11:1107–1119. [Google Scholar]
- 23.Darling JA, Bagley MJ, Roman J, Tepolt CK. Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus. Mol. Ecol. 2008;17:4992–5007. doi: 10.1111/j.1365-294X.2008.03978.x. [DOI] [PubMed] [Google Scholar]
- 24.Ellstrand NC. Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl Acad. Sci. USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elton CE. The Ecology of Invasions by Animals and Plants. London, UK: Methuen & Company Ltd; 1958. [Google Scholar]
- 26.Flora of North America Editorial Committee. 1993. +). Flora of North America North of Mexico. New York and Oxford.
- 27.Funk VA, Susanna A, Stuessey FT. Bayer RJ. Systematics, Evolution, and Biogeography of Compositae. Austria: International Association for Plant Taxonomy Vienna; 2009. [Google Scholar]
- 28.Gallagher KG, Schierenbeck KA. Dantonio CM. Hybridization and introgression in Carpobrotus spp. (Aizoaceae) in California. 2. Allozyme evidence. Am. J. Bot. 1997;84:905–911. [PubMed] [Google Scholar]
- 29.Gammon MA, Baack E, Orth JF. Kesseli R. Viability, Growth, and Fertility of Knotweed Cytotypes in North America. Invasive Plant Sci. Manage. 2010;3:208–218. [Google Scholar]
- 30.Geiger JH, Pratt PD, Wheeler GS. Williams DA. Hybrid vigor for the invasive exotic Brazilian peppertree (Schinus terebinthifolius Raddi., Anacardiaceae) in Florida. Int. J. Plant Sci. 2011;172:655–663. [Google Scholar]
- 31.Giraud T, Refregier G, Le Gac M, de Vienne DM. Hood ME. Speciation in fungi. Fungal Genet. Biol. 2008;45:791–802. doi: 10.1016/j.fgb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Grosholz E. Avoidance by grazers facilitates spread of an invasive hybrid plant. Ecol. Lett. 2010;13:145–153. doi: 10.1111/j.1461-0248.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- 33.Hahn MA, Buckley YM. Muller-Scharer H. Increased population growth rate in invasive polyploid Centaurea stoebe in a common garden. Ecol. Lett. 2012;15:947–954. doi: 10.1111/j.1461-0248.2012.01813.x. [DOI] [PubMed] [Google Scholar]
- 34.Hartman Y, Uwimana B, Hooftman DAP, Schranz ME, van de Wiel CCM, Smulders MJM, et al. Genomic and environmental selection patterns in two distinct lettuce crop-wild hybrid crosses. Evol. Appl. 2013;6:569–584. doi: 10.1111/eva.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasselman DJ, Argo EE, McBride MC, Bentzen P, Schultz TF, Perez-Umphrey AA, et al. Human disturbance causes the formation of a hybrid swarm between two naturally sympatric fish species. Mol. Ecol. 2014;23:1137–1152. doi: 10.1111/mec.12674. [DOI] [PubMed] [Google Scholar]
- 36.Hawkes CV, Douglas AE. Fitter AH. Origin, local experience, and the impact of biotic interactions on native and introduced Senecio species. Biol. Invasions. 2010;12:113–124. [Google Scholar]
- 37.Hedges SB, Dudley J. Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 38.Henery ML, Bowman G, Mraz P, Treier UA, Gex-Fabry E, Schaffner U, et al. Evidence for a combination of pre-adapted traits and rapid adaptive change in the invasive plant Centaurea stoebe. J. Ecol. 2010;98:800–813. [Google Scholar]
- 39.Higgins JPT. Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 40.Hooftman DAP, Oostermeijer JGB, Jacobs MMJ. Den Nijs HCM. Demographic vital rates determine the performance advantage of crop-wild hybrids in lettuce. J. Appl. Ecol. 2005;42:1086–1095. [Google Scholar]
- 41.Hooftman DAP, Jong MJD, Oostermeijer JGB. Den Nijs H. Modelling the long-term consequences of crop-wild relative hybridization: a case study using four generations of hybrids. J. Appl. Ecol. 2007;44:1035–1045. [Google Scholar]
- 42.Hovick SM, Campbell LG, Snow AA. Whitney KD. Hybridization Alters Early Life-History Traits and Increases Plant Colonization Success in a Novel Region. Am. Naturalist. 2012;179:192–203. doi: 10.1086/663684. [DOI] [PubMed] [Google Scholar]
- 43.Inderbitzin P, Davis RM, Bostock RM. Subbarao KV. The Ascomycete Verticillium longisporum Is a Hybrid and a Plant Pathogen with an Expanded Host Range. PLoS ONE. 2011;6:e18260. doi: 10.1371/journal.pone.0018260. & (3), [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson JR, Fitzpatrick BM. Shaffer HB. Retention of low-fitness genotypes over six decades of admixture between native and introduced tiger salamanders. BMC Evol. Biol. 2010;10:147. doi: 10.1186/1471-2148-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim ST, Sultan SE. Donoghue MJ. Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proc. Natl Acad. Sci. USA. 2008;105:12370–12375. doi: 10.1073/pnas.0805141105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirk H, Connolly C. Freeland JR. Molecular genetic data reveal hybridization between Typha angustifolia and Typha latifolia across a broad spatial scale in eastern North America. Aquat. Bot. 2011;95:189–193. [Google Scholar]
- 47.Lajeunesse MJ. Meta-Analysis and the Comparative Phylogenetic Method. Am. Naturalist. 2009;174:369–381. doi: 10.1086/603628. [DOI] [PubMed] [Google Scholar]
- 48.Lajeunesse MJ. phyloMeta: a program for phylogenetic comparative analyses with meta-analysis. Bioinformatics. 2011;27:2603–2604. doi: 10.1093/bioinformatics/btr438. [DOI] [PubMed] [Google Scholar]
- 49.LaRue EA, Grimm D. Thum RA. Laboratory crosses and genetic analysis of natural populations demonstrate sexual viability of invasive hybrid watermilfoils (Myriophyllum spicatum x M. sibiricum) Aquat. Bot. 2013;109:49–53. [Google Scholar]
- 50.Lavergne S. Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Roux J. Wieczorek AM. Molecular systematics and population genetics of biological invasions: towards a better understanding of invasive species management. Ann. Appl. Biol. 2009;154:1–17. [Google Scholar]
- 52.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JPA, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. Bmj-British Medical Journal. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mack RN. Predicting the identity and fate of plant invaders: emergent and emerging approaches. Biol. Conserv. 1996;78:107–121. [Google Scholar]
- 54.Maddison DR. Schulz K-S. 2007. & The Tree of Life Web Project http://tolweb.org.
- 55.Mengersen K, Jennions MD. Schmid CH. Statistical models for the meta-analysis of nonindependent data. In: Koricheva J, Gurevitch J, Mengersen K, editors; Handbook of Meta-Analysis in Ecology and Evolution. Princeton, NJ: Princeton University Press; 2013. pp. 255–283. [Google Scholar]
- 56.Moody ML. Les DH. Evidence of hybridity in invasive watermilfoil (Myriophyllum) populations. Proc. Natl Acad. Sci. USA. 2002;99:14867–14871. doi: 10.1073/pnas.172391499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mraz P, Garcia-Jacas N, Gex-Fabry E, Susanna A, Barres L. Muller-Scharer H. Allopolyploid origin of highly invasive Centaurea stoebe s.l. (Asteraceae) Mol. Phylogenet. Evol. 2012;62:612–623. doi: 10.1016/j.ympev.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Nechwatal J. Mendgen K. Evidence for the occurrence of natural hybridization in reed-associated Pythium species. Plant. Pathol. 2009;58:261–270. [Google Scholar]
- 59.Newcombe G, Stirling B, McDonald S. Bradshaw HD. Melampsora xcolumbiana, a natural hybrid of M-medusae and M-occidentalis. Mycol. Res. 2000;104:261–274. [Google Scholar]
- 60.Newcombe G, Stirling B. Bradshaw HD. Abundant pathogenic variation in the new hybrid rust Melampsora xcolumbiana on hybrid poplar. Phytopathology. 2001;91:981–985. doi: 10.1094/PHYTO.2001.91.10.981. [DOI] [PubMed] [Google Scholar]
- 61.Pandit MK, Pocock MJO. Kunin WE. Ploidy influences rarity and invasiveness in plants. J. Ecol. 2011;99:1108–1115. [Google Scholar]
- 62.Parepa M, Fischer M, Krebs C. Bossdorf O. Hybridization increases invasive knotweed success. Evol. Appl. 2014;7:413–420. doi: 10.1111/eva.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perrins J, Williamson M. Fitter A. Do annual weeds have predictable characters? Acta Oecol.-Int. J. Ecol. 1992;13:517–533. [Google Scholar]
- 64.Poulsen JR, Osenberg CW, Clark CJ, Levey DJ. Bolker BM. Plants as reef fish: fitting the functional form of seedling recruitment. Am. Naturalist. 2007;170:167–183. doi: 10.1086/518945. [DOI] [PubMed] [Google Scholar]
- 65.Pyšek P. Richardson DM. Traits associated with invasiveness in alien plants: where do we stand? In: Nentwig W, editor; Biological Invasions. Germany: Springer-Verlag Berlin; 2007. pp. 97–125. [Google Scholar]
- 66.Pyšek P, Brock JH, Bimova K, Mandak B, Jarosik V, Koukolikova I, et al. Vegetative regeneration in invasive Reynoutria (Polygonaceae) taxa: the determinant of invasibility at the genotype level. Am. J. Bot. 2003;90:1487–1495. doi: 10.3732/ajb.90.10.1487. [DOI] [PubMed] [Google Scholar]
- 67.R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 68.Reagon M, Thurber CS, Olsen KM, Jia YL. Caicedo AL. The long and the short of it: SD1 polymorphism and the evolution of growth trait divergence in U.S. weedy rice. Mol. Ecol. 2011;20:3743–3756. doi: 10.1111/j.1365-294X.2011.05216.x. [DOI] [PubMed] [Google Scholar]
- 69.Rejmánek M, Richardson DM. Pyšek P. Plant invasions and invasibility of plant communities. In: van der Maarel E, editor; Vegetation Ecology. Oxford: Blackwell; 2005. pp. 332–355. [Google Scholar]
- 70.Richards CL, Walls RL, Bailey JP, Parameswaran R, George T. Pigliucci M. Plasticity in salt tolerance traits allows for invasion of novel habitat by Japanese knotweed s. l. (Fallopia japonica and F-bohemica, Polygonaceae) Am. J. Bot. 2008;95:931–942. doi: 10.3732/ajb.2007364. [DOI] [PubMed] [Google Scholar]
- 71.Richardson DM, Pyšek P, Rejmanek M, Barbour MG, Panetta FD. West CJ. Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib. 2000;6:93–107. [Google Scholar]
- 72.Ridley CE. Ellstrand NC. Evolution of enhanced reproduction in the hybrid-derived invasive, California wild radish (Raphanus sativus) Biol. Invasions. 2009;11:2251–2264. [Google Scholar]
- 73.Rieseberg LH, Archer MA. Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- 74.Rieseberg LH, Kim SC, Randell RA, Whitney KD, Gross BL, Lexer C, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenberg MS, Rothstein HR. Gurevitch J. Effect sizes: conventional choices and calculations. In: Koricheva J, Gurevitch J, Mengersen K, editors; Handbook of Meta-Analysis in Ecology and Evolution. NJ: Princeton University Press Princeton; 2013. pp. 61–71. [Google Scholar]
- 76.Rouifed S, Puijalon S, Viricel MR. Piola F. Achene buoyancy and germinability of the terrestrial invasive Fallopia x bohemica in aquatic environment: a new vector of dispersion? Ecoscience. 2011;18:79–84. [Google Scholar]
- 77.Ryan ME, Johnson JR. Fitzpatrick BM. Invasive hybrid tiger salamander genotypes impact native amphibians. Proc. Natl Acad. Sci. USA. 2009;106:11166–11171. doi: 10.1073/pnas.0902252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schierenbeck KA. Ellstrand NC. Hybridization and the evolution of invasiveness in plants and other organisms. Biol. Invasions. 2009;11:1093–1105. [Google Scholar]
- 79.Schwenk K, Brede N. Streit B. Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philos Trans R Soc B: Biol Sci. 2008;363:2805–2811. doi: 10.1098/rstb.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snow AA, Uthus KL. Culley TM. Fitness of hybrids between weedy and cultivated radish: implications for weed evolution. Ecol. Appl. 2001;11:934–943. [Google Scholar]
- 81.Snow AA, Culley TM, Campbell LG, Sweeney PM, Hegde SG. Ellstrand NC. Long-term persistence of crop alleles in weedy populations of wild radish (Raphanus raphanistrum) New Phytol. 2010;186:537–548. doi: 10.1111/j.1469-8137.2009.03172.x. [DOI] [PubMed] [Google Scholar]
- 82.Stebbins GL. The role of hybridization in evolution. Proc. Am. Philos. Soc. 1959;103:231–251. [Google Scholar]
- 83.Stebbins GL. Polyploidy, hybridization, and the invasion of new habitats. Ann. Mo. Bot. Gard. 1985;72:824–832. [Google Scholar]
- 84.Sullivan L, Wildova R, Goldberg D. Vogel C. Growth of three cattail (Typha) taxa in response to elevated CO(2) Plant Ecol. 2010;207:121–129. [Google Scholar]
- 85.Sun J, Qian Q, Ma DR, Xu ZJ, Liu D, Du HB, et al. Introgression and selection shaping the genome and adaptive loci of weedy rice in northern China. New Phytol. 2013;197:290–299. doi: 10.1111/nph.12012. [DOI] [PubMed] [Google Scholar]
- 86.Tavalire HF, Bugbee GE, LaRue EA. Thum RA. Hybridization, cryptic diversity, and invasiveness in introduced variable-leaf watermilfoil. Evol. Appl. 2012;5:892–900. doi: 10.1111/j.1752-4571.2012.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Urbanska KM, Hurka H, Landolt E, Neuffer B. Mummenhoff K. Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, central Switzerland: biosystematic and molecular evidence. Plant Syst. Evol. 1997;204:233–256. [Google Scholar]
- 88.Van Grunsven RHA, Bos F, Ripley BS, Suehs CM. Veenendaal EM. Release from soil pathogens plays an important role in the success of invasive Carpobrotus in the Mediterranean. South African J. Botany. 2009;75:172–175. [Google Scholar]
- 89.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–49. [Google Scholar]
- 90.Vila M. D'Antonio CM. Fitness of invasive Carpobrotus (Aizoaceae) hybrids in coastal California. Ecoscience. 1998a;5:191–199. [Google Scholar]
- 91.Vila M. D'Antonio CM. Hybrid vigor for clonal growth in Carpobrotus (Aizoaceae) in coastal California. Ecol. Appl. 1998b;8:1196–1205. [Google Scholar]
- 92.Webb CO. Donoghue MJ. Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes. 2005;5:181–183. & http://phylodiversity.net/phylomatic/ [Google Scholar]
- 93.Webb CO, Ackerly DD. Kembel SW. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 94.Weber E. D'Antonio CM. Phenotypic plasticity in hybridizing Carpobrotus spp. (Aizoaceae) from coastal California and its role in plant invasion. Can. J. Bot.-Revue Canadienne De Botanique. 1999;77:1411–1418. [Google Scholar]
- 95.Whitney KD, Randell RA. Rieseberg LH. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am. Naturalist. 2006;167:794–807. doi: 10.1086/504606. [DOI] [PubMed] [Google Scholar]
- 96.Whitney KD, Ahern JR. Campbell LG. Hybridization-prone plant families do not generate more invasive species. Biol. Invasions. 2009;11:1205–1215. [Google Scholar]
- 97.Whitney KD, Ahern JR, Campbell LG, Albert LP. King MS. Patterns of hybridization in plants. Perspectives in Plant Ecol. Evol. Systematics. 2010a;12:175–182. [Google Scholar]
- 98.Whitney KD, Randell RA. Rieseberg LH. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytol. 2010b;187:230–239. doi: 10.1111/j.1469-8137.2010.03234.x. [DOI] [PubMed] [Google Scholar]
- 99.Wikström N, Savolainen V. Chame MW. Evolution of angiosperms: calibrating the family tree. Proc. Biol. Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu W, Zhou RC, Ni GY, Shen H. Ge XJ. Is a new invasive herb emerging? Molecular confirmation and preliminary evaluation of natural hybridization between the invasive Sphagneticola trilobata (Asteraceae) and its native congener S-calendulacea in South China. Biol. Invasions. 2013;15:75–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


