Abstract
C4 photosynthesis depends on the strict compartmentalization of CO2 assimilatory enzymes. cis-regulatory mechanisms are described that ensure mesophyll-specific expression of the gene encoding the C4 isoform of phosphoenolpyruvate carboxylase (ppcA1) of the C4 dicot Flaveria trinervia. To elucidate and understand the anatomy of the C4 ppcA1 promoter, detailed promoter/reporter gene studies were performed in the closely related C4 species F. bidentis, revealing that the C4 promoter contains two regions, a proximal segment up to −570 and a distal part from −1566 to −2141, which are necessary but also sufficient for high mesophyll-specific expression of the β-glucuronidase reporter gene. The distal region behaves as an enhancer-like expression module that can direct mesophyll-specific expression when inserted into the ppcA1 promoter of the C3 plant F. pringlei. Mesophyll expression determinants were restricted to a 41-bp segment, referred to as mesophyll expression module 1 (Mem1). Evolutionary and functional studies identified the tetranucleotide sequence CACT as a key component of Mem1.
INTRODUCTION
C4 plants are characterized by high rates of photosynthesis as well as an efficient use of water and nitrogen resources. This is because of their unique mode of carbon assimilation that concentrates CO2 at the site of ribulose bisphosphate carboxylase/oxygenase. The functioning of C4 photosynthesis is dependent upon the strict compartmentation of the CO2 assimilatory enzymes into two distinct cell types, mesophyll and bundle-sheath cells. The primary carboxylating enzyme, phosphoenolpyruvate carboxylase, accumulates exclusively in the mesophyll cells, and the secondary carboxylase, ribulose bisphosphate carboxylase/oxygenase, and the decarboxylating enzymes, such as NADP-dependent malic enzyme, are restricted to the bundle-sheath cells (Hatch, 1987).
This division of labor between mesophyll and bundle-sheath cells is the result of differential gene expression. In NADP-malic enzyme-type C4 species, for instance, transcripts for phosphoenolpyruvate carboxylase, pyruvate phosphate dikinase, NADP-malic enzyme, and the small subunit of ribulose bisphosphate carboxylase/oxygenase, accumulate differentially in the two cell types. This differential accumulation is largely because of transcriptional control (Sheen, 1999).
C4 plants occur in at least 18 families of monocotyledonous and dicotyledonous plants. This indicates that C4 plants must have evolved several times independently from C3 ancestors during the evolution of angiosperms (Kellogg, 1999; Sage et al., 1999). The multiple independent origin of C4 photosynthesis suggests that the evolution of a C3 into a C4 species must have been relatively easy in genetic terms. The available molecular data on the C4 cycle enzymes support this point of view. None of the C4 enzymes, phosphoenolpyruvate carboxylase (PEPC), pyruvate orthophosphate dikinase, or NADP-dependent malic enzyme, are unique to C4 plants. Nonphotosynthetic isoforms of these enzymes are also present in C3 species and in the nonphotosynthetic tissues of C4 species. The ubiquitous presence of these nonphotosynthetic isoforms of the C4 cycle enzymes in C3 plants indicates that these C3 isoforms served as the starting point for the evolution of the C4 genes (reviewed in Monson, 1999).
As a starting point to understanding the molecular basis of the evolution of C4 genes, we are focusing on the C4 gene for PEPC and are using the genus Flaveria (Asteraceae) (Powell, 1978) as an experimental system. Flaveria has C3 and C4 species and a large number of C3-C4 photosynthetic intermediates (reviewed in Edwards and Ku, 1987). These intermediates differ in the expression of the C4 photosynthetic traits, and there is convincing evidence that at least some of these species are true evolutionary intermediates (Monson and Moore, 1989).
Three major changes must have occurred during C3-to-C4 evolution to transform a C3 PEPC gene into a C4 gene (reviewed in Westhoff and Gowik, 2004). C4 PEPC genes are highly expressed (Hermans and Westhoff, 1990; Crétin et al., 1991), whereas C3 PEPC transcripts generally occur only in moderate amounts in plant tissues (Crétin et al., 1991; Ernst and Westhoff, 1996). First, therefore, promoter strength had to increase. Second, a mesophyll-specific expression pattern had to evolve because the strict compartmentation of PEPC is imperative for an effectively functioning C4 cycle (Hatch, 1987). Finally, the metabolic context of C3 and C4 PEPCs differs; therefore, the PEPC protein had to change its kinetic and regulatory enzyme properties to meet the metabolic requirements of C4 photosynthesis (Svensson et al., 2003).
The C4 PEPCs of C4 Flaveria species are encoded by the phosphoenolpyruvate carboxylase A (ppcA) gene class (Hermans and Westhoff, 1992). ppcA orthologous PEPC genes are found in all C3 and C3-C4 intermediate Flaveria species, indicating that this PEPC gene class was already present in the last common ancestor of present C3 and C4 Flaveria species (Westhoff and Gowik, 2004). The comparative enzymatic analysis of ppcA PEPC proteins from C3, C3-C4 intermediate, and C4 Flaveria species revealed that the ppcA PEPCs of F. pringlei (C3) and F. trinervia (C4) are typical C3 and C4 PEPCs, respectively, and that only a few amino acid changes, most notably a C4 invariant Ser residue in the vicinity of the catalytic site, were responsible for the observed differences in kinetic and regulatory behavior (Svensson et al., 1997; Bläsing et al., 2000). The ppcA PEPCs from the C3-C4 species F. pubescens and F. brownii were found to be intermediate, indicating that the ppcA PEPCs changed gradually from C3 to C4 (Engelmann et al., 2003), and this PEPC gene class could serve as an evolutionary model to unravel the C4-associated changes in enzyme and gene expression characteristics (Svensson et al., 2003; Westhoff and Gowik, 2004).
Analysis of ppcA1 promoter/β-glucuronidase (GUS) reporter gene fusions in the C4 plant F. bidentis revealed that the ppcA1 promoter of F. trinervia directs high levels of reporter gene expression in the mesophyll cells (Stockhaus et al., 1997). This demonstrated that the expression of the corresponding gene is largely determined by transcription and that the 2188 bp (with reference to the AUG start codon of the ppcA1 reading frame) of the 5′ flanking sequences contain all the essential cis-regulatory elements for a high and mesophyll-specific expression. By contrast, the 2538 bp (with reference to the AUG start codon) of the 5′ flanking sequences of the ppcA1 gene of F. pringlei were found to be a weak promoter and did not direct any organ- or cell-specific expression (Stockhaus et al., 1997). Both promoters thus exhibited all the attributes that were expected from the accumulation patterns of their corresponding RNAs and proteins (Höfer et al., 1992; Ernst and Westhoff, 1996).
To fully understand the anatomy of the C4 ppcA1 promoter and to identify the cis-regulatory elements that are essential for its mesophyll specificity, detailed promoter reporter gene analyses with transgenic F. bidentis were performed. These experiments revealed that the C4 promoter contains two regions, a proximal region up to −570 (PR) and a distal region from −1566 to −2141 (DR), which are necessary and sufficient for a high mesophyll-specific expression. The DR behaves as an enhancer-like expression module and is able to confer a mesophyll expression component to the ppcA1 promoter of F. pringlei. Further dissection of the DR identified a 41-bp module (mesophyll expression module 1 [Mem1]) that in conjunction with the PR, is sufficient for mesophyll-specific expression. Evolutionary and functional analyses identified the tetranucleotide CACT as a key element of Mem1.
RESULTS
The Distal Segment of the C4 ppcA1 Promoter Is Required for both Expression Specificity and Quantity and Behaves as an Enhancer-Like Expression Module
In the C3 plant tobacco (Nicotiana tabacum), the C4 ppcA1 promoter behaved essentially as a palisade parenchyma–specific promoter. The expression in the palisade parenchyma was lost when the 5′ distal 1618 bp of 5′ distal sequences were removed and the remaining 570 bp of proximal sequences were analyzed for promoter activity (Stockhaus et al., 1994). This finding suggested that the 5′ DR of the promoter contains cis-regulatory elements that are absolutely essential for a high level of expression and for mesophyll specificity. To define this distal promoter region precisely, a systematic deletion analysis was performed using the high level of expression in the palisade parenchyma cells of the C3 plant tobacco as a test system.
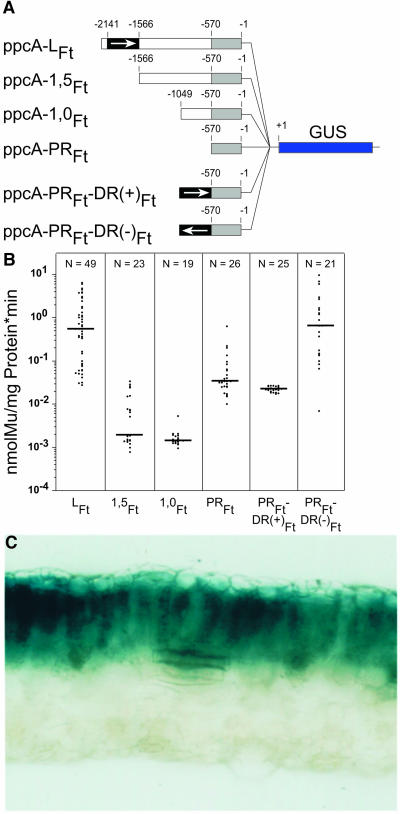
When the ppcA1 starting promoter of 2188 bp (named ppcA-LFt; Figure 1) was shortened by 623 bp (construct ppcA-1,5Ft; Figure 2A), the expression activity was almost entirely lost. It is highly significant that no palisade parenchyma expression was detected by histochemical staining (data not shown). Further deletion of 5′ promoter sequences (construct ppcA-1,0Ft; Figure 2A) influenced the resulting promoter activity as compared with the ppcA-1,5Ft promoter construct only marginally (Figure 2B). Both the ppcA-1,5Ft and ppcA-1,0Ft constructs showed a lower promoter activity than the ppcA-PRFt promoter fragment (Figures 2A and 2B). These observations suggested that the region between base pairs −570 and −1565 appears to contain sequences that reduce promoter activity but that the DR between base pairs −1565 and −2188 is absolutely essential for the C4 ppcA1 promoter activity in tobacco.
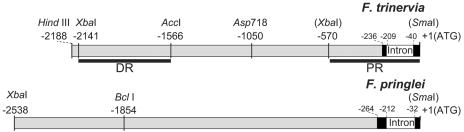
Figure 1.
Restriction Maps of the 5′ Flanking Regions of the ppcA1 Genes of F. trinervia and F. pringlei.
Nucleotide positions are numbered with respect to the AUG translational initiation codon. The DR and PR of the ppcA1 promoter of F. trinervia are labeled by black bars.
Figure 2.
Deletion Analysis of the ppcA1 Promoter of F. trinervia in the C3 Plant Tobacco.
(A) Structures of the ppcA1/GUS chimerical genes used for tobacco transformation. The nucleotide numbers refer to the translation initiation codon. The DR of the ppcA1-promoter from the C4 plant F. trinervia is indicated by a black box, and the PR is indicated by a gray box.
(B) GUS activities in leaves of transgenic tobacco plants. The median value of the GUS activities is expressed in nanomoles of the reaction product 4-methylumbelliferone (Mu) generated per milligram of protein per minute. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column.
(C) Histochemical localization of GUS activity in a leaf section of a transgenic tobacco plant transformed with the ppcA-PRFt-DR(−)Ft construct. Incubation for 6 h.
This fact was tested directly by fusing the distal part between base pairs −1566 and −2141 in direct and inverse orientation with the proximal −570 bp of promoter sequences (constructs ppcA-PRFt-DR(+)Ft and ppcA-PRFt-DR(−)Ft; Figure 2A) and by analyzing the promoter activities in transgenic tobacco. The GUS activity of the ppcA-PRFt-DR(−)Ft construct in the leaf was comparable to that of the LFt chimerical gene, but the activity of the ppcA-PRFt-DR(+)Ft construct was drastically reduced (Figure 2B). For the ppcA-PRFt-DR(−)Ft construct, histochemical analyses showed that this promoter directed a palisade parenchyma–specific expression of the GUS reporter gene (Figure 2C). It follows from these expression analyses with the C3 plant tobacco that the distal and proximal parts of the promoter will be sufficient for the ppcA1 promoter activity and that the nucleotide sequences between −570 and −1566 are probably not necessary for its expression specificity.
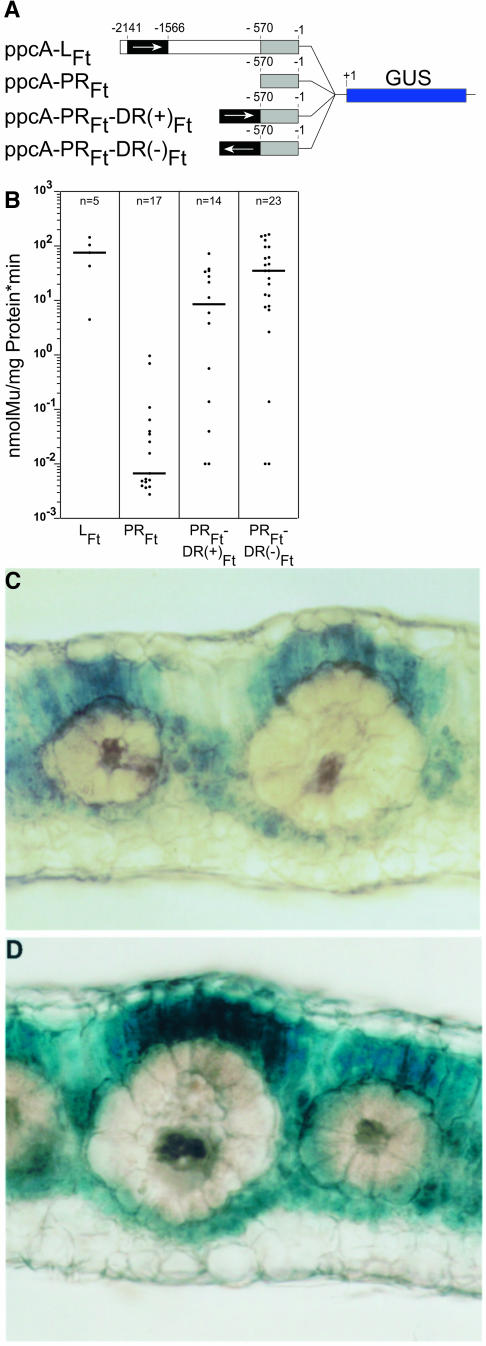
To verify these conclusions, transformation experiments with the C4 plant F. bidentis were performed. F. bidentis is very closely related to F. trinervia and is used for these experiments (Stockhaus et al., 1997) because this species, in contrast with F. trinervia, may be transformed by Agrobacterium tumefaciens (Chitty et al., 1994). Quantitative measurements of GUS activity showed that ppcA-PRFt-DR(−)Ft promoter was approximately half as active as the full C4 ppcA1 promoter, whereas the ppcA-PRFt-DR(+)Ft promoter activity was reduced to approximately one-quarter (Figure 3B). Both promoters directed a mesophyll-specific expression of the GUS reporter gene and showed the same expression pattern as the full C4 ppcA1 promoter (Stockhaus et al., 1997). Two conclusions were drawn from these experiments. First, the DR and PR of the promoter (Figure 1) are sufficient for an elevated and mesophyll-specific promoter activity (i.e., the nucleotide sequences between −570 and −1566 are essentially dispensable). Second, because the C4-DR functions both in the correct and the inverse orientation, this cis-regulatory region shows the typical features of a transcriptional enhancer (Blackwood and Kadonaga, 1998).
Figure 3.
Analysis of the ppcA1 GUS Reporter Gene Constructs ppcA-PRFt-DR(+)Ft and ppcA-PRFt-DR(−)Ft in Transgenic F. bidentis.
(A) Structures of the ppcA1/GUS chimerical genes.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (n) are indicated at the top of each column. Median values are shown. Mu, 4-methylumbelliferone.
(C) and (D) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with the ppcA-PRFt-DR(+)Ft (C) or the ppcA-PRFt-DR(−)Ft construct (D). Incubation time was 20 min in case of the ppcA-PRFt-DR(+)Ft plant and 10 min in case of the ppcA-PRFt-DR(−)Ft plant.
The DR of the C4 ppcA1 Promoter Provides Mesophyll Specificity but No Raised Expression Quantity in the Context of the C3 ppcA1 Promoter
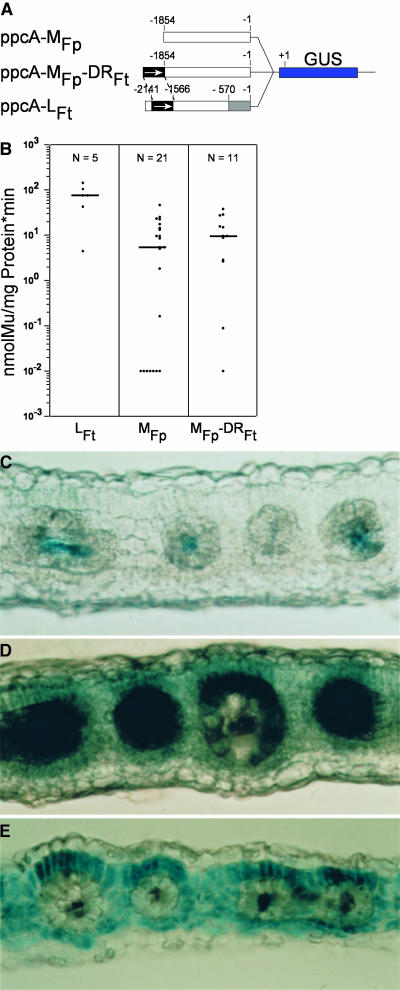
The C4-DR could function as a C4 expression module that confers both height and specificity of expression. If this were true, one should expect that upon transfer of the C4-DR into the ppcA1 promoter of the C3 plant F. pringlei, the C4-C3 hybrid promoter would behave like a C4 ppcA1 promoter and show a high level of expression in the mesophyll cells. To test this, the DR of the C3 ppcA1 promoter from nucleotides −2538 to −1854 (Figure 1) was removed, giving rise to ppcA-MFp, and replaced by the C4-DR in correct orientation (Figure 4A).
Figure 4.
Analysis of the ppcA1 GUS Reporter Gene Constructs ppcA-MFp and ppcA-MFp-DR(+)Ft in Transgenic F. bidentis.
(A) Structures of the ppcA1/GUS chimerical genes.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values are shown. Mu, 4-methylumbelliferone.
(C) to (E) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with ppcA-MFp ([C] and [D]) or ppcA-MFp-DR(+)Ft (E). Incubation times were 22 min (C), 48 min (D), and 43 min (E).
It is known from previous work (Stockhaus et al., 1997) that the ppcA1 promoter of F. pringlei (ppcA-LFp) is relatively weak when compared with the C4 ppcA1 promoter and directs expression in all cells of the leaf, including the vascular bundle. The shortened promoter version ppcA-MFp behaves similarly (Figures 4B and 4C). The addition of the C4-DR to the ppcA-MFp (resulting in ppcA-MFp-DRFt) causes just a small increase in the expression strength (Figure 4B). However, the in situ analysis of the transgenic plants revealed that the C4-C3 chimerical promoter had acquired a mesophyll expression component that was not detectable with the ppcA-MFp construct (Figure 4E). A visual comparison of the in situ promoter activities of ppcA-MFp and ppcA-MFp-DRFt also suggested that the C4-DR did not only add a mesophyll expression component to the ppcA1 promoter part of F. pringle but reduced its original activity in bundle-sheath cells and vascular tissue. We concluded from these experiments that the C4-DR contains mesophyll expression components that are not able to increase the strength of the C3 ppcA1 promoter substantially.
Mapping of cis-Regulatory Elements in the C4 DR of the C4 ppcA1 Promoter
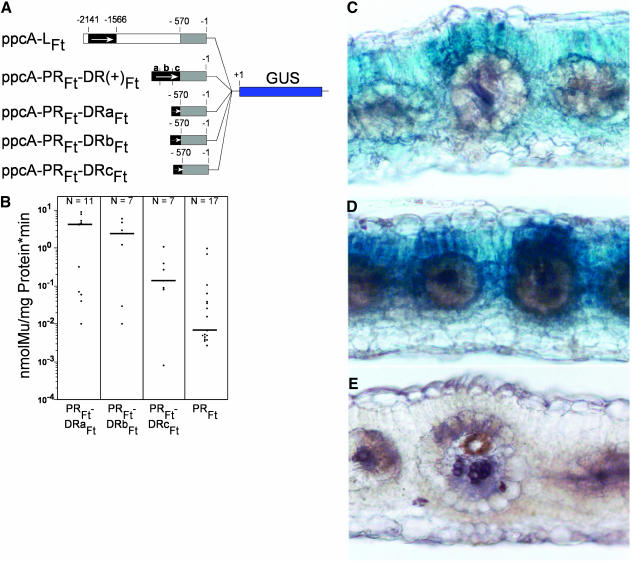
The C4-DR consists of 575 bp. To identify the cis-regulatory element(s) within the C4-DR more precisely, this region was dissected into three pieces of approximately equal size that overlap by 50 bp (Figure 5). Each fragment of the C4-DR, named a to c, was fused with the C4-PR of the ppcA1 promoter of F. trinervia in the correct orientation, and the resulting constructs, ppcA-PRFt-DRaFt, ppcA-PRFt-DRbFt, and ppcA-PRFt-DRcFt (Figure 6A), were transformed into F. bidentis.
Figure 5.
Nucleotide Sequence of the DR of the ppcA1 Promoter of F. trinervia Showing the Location of the Three Subfragments a, b, and c.
Figure 6.
Analysis of the ppcA1 GUS Reporter Gene Constructs ppcA-PRFt-DRa(+)Ft, ppcA-PRFt-DRb(+)Ft, and ppcA-PRFt-DRc(+)Ft in Transgenic F. bidentis.
(A) Structures of the ppcA1/GUS chimerical genes.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values are shown. Mu, 4-methylumbelliferone.
(C) to (E) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with the ppcA-PRFt-DRa(+)Ft (C), ppcA-PRFt-DRb(+)Ft (D), or ppcA-PRFt-DRc(+)Ft (E). Incubation times were 6 h (C), 18 h (D), and 48 h (E).
The ppcA-PRFt-DRaFt and ppcA-PRFt-DRbFt promoters directed a clear and reproducible GUS expression in the mesophyll cells (Figures 6C and 6D), although their activities were reduced by ∼5 to 10 times when compared with the activity of the ppcA-PRFt-DR(+)Ft reference promoter (cf Figures 6B and 3B). This indicates that both the ppcA-PRFt-DRaFt and ppcA-PRFt-DRbFt promoters harbor cis-regulatory elements that are sufficient for mesophyll-specific transcription. By contrast, the ppcA-PRFt-DRcFt promotor produced a minute amount of GUS activity that is below the level of histochemical detection (Figure 6E) but that is higher than the activity of the ppcA-PRFt construct (Figure 6B). This suggested that this segment of the C4-DR might contain some weak transcriptional enhancing element(s). These elements are not essential for mesophyll-specific gene expression, but they may interact with the cis-regulatory elements of the a and b parts, thereby increasing their mesophyll enhancer activity.
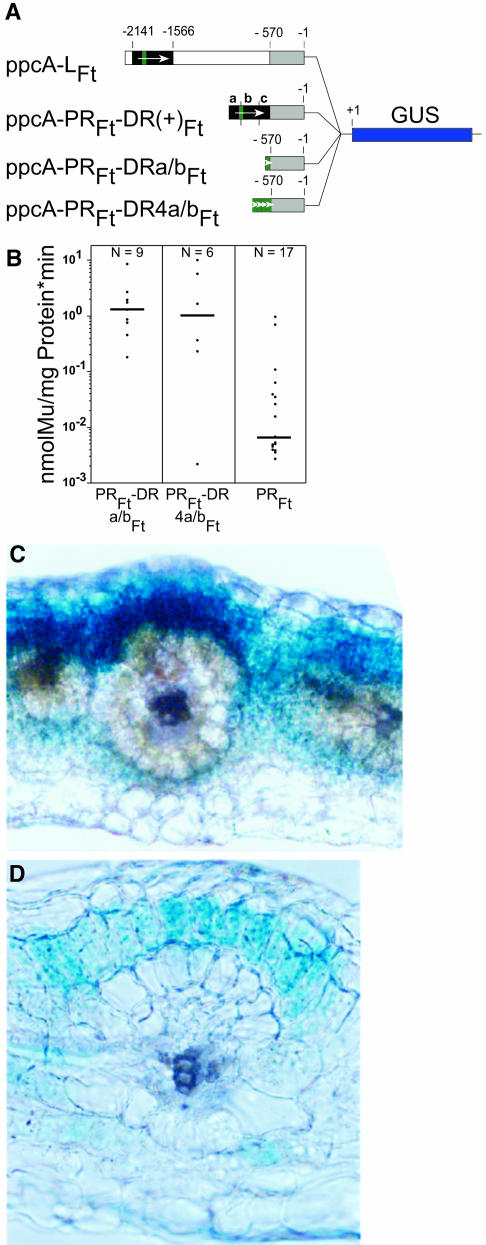
Subfragments a and b of the C4-DR were able to direct a mesophyll-specific expression. This implies that either the two segments contain distinct and different mesophyll specificity elements or that the overlapping stretch of 41 bp harbors a cis-regulatory element for mesophyll expression. To test these possibilities, one and four tandemly oriented copies of the a/b-overlapping fragment were fused in direct orientation with the PR segment of the C4 ppcA1 promoter. The resulting constructs, ppcA-PRFt-DRa/bFt and ppcA-PRFt-DR4a/bFt (Figure 7A), were analyzed in transgenic F. bidentis.
Figure 7.
Analysis of the ppcA1 GUS Reporter Gene Constructs ppcA-PRFt-DRa/b(+)Ft and ppcA-PRFt-DR4a/b(+)Ft in Transgenic F. bidentis.
(A) Structures of the ppcA1/GUS chimerical genes.
(B) GUS activities in leaves of transgenic F. bidentis plants. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values are shown.
(C) and (D) Histochemical localization of GUS activity in leaf sections of transgenic F. bidentis plants transformed with ppcA-PRFt-DRa/b(+)Ft (C) and ppcA-PRFt-DR4a/b(+)Ft (D). Incubation times were 12 h (C) and 6 h (D).
Both constructs exhibited similar expression levels (Figure 7B) and directed a mesophyll-specific expression of the GUS reporter gene (Figures 7C and 7D). It follows that the a/b overlapping C4-DR fragment contains determinants for mesophyll-specific gene expression, and the segment was designated as Mem1.
The Tetranucleotide CACT Is Essential for Mem1 Function
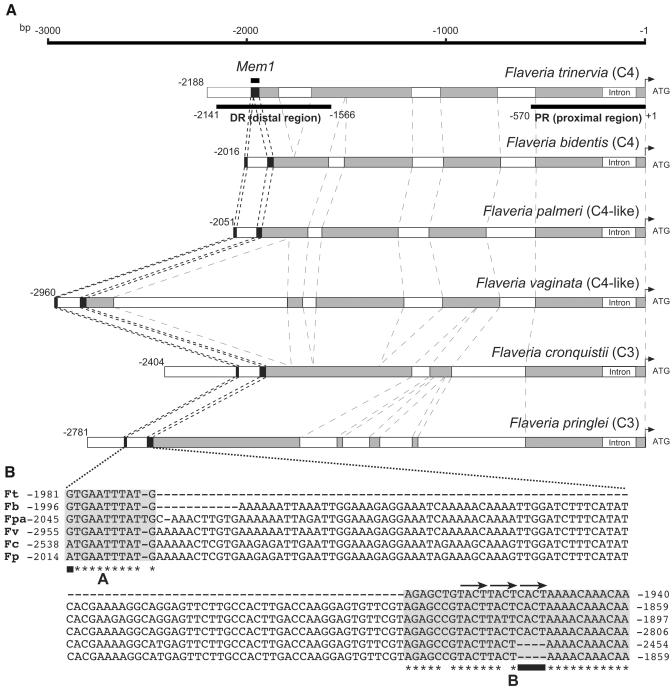
To identify the putative location of mesophyll-specific determinants within the 41-bp Mem1, its sequence was compared with the available ppcA1 promoter sequences of F. pringlei. The search resulted in the unambiguous identification of a C4-DR counterpart in the F. pringlei promoter at its very 5′ end. Sequences matching the 30 bp of the 3′ part of Mem1 (named part B, Figure 8B) were detected in the F. pringlei promoter sequence. However, the 11 bp of the 5′ terminal sequences (named part A, Figure 8B) were lacking in the F. pringlei sequence. This suggests that the C3 ppcA1 promoter of F. pringlei contains only the homolog of part B of Mem1 or, alternatively, that homologous sequences of part A are present in the promoter but have not been detected yet because they are located further upstream. To clarify this ambiguity, the available 5′ flanking sequences of the ppcA1 gene of F. pringlei were extended by vectorette PCR (Siebert et al., 1995). Part A–type sequences were indeed shown to be separated from part B by 108 bp of intervening sequences (Figure 8).
Figure 8.
The Structures of ppcA1 Promoters from C4, C4-like, and C3 Flaveria Species and the Nucleotide Composition of Mem1 and Its Homologs.
(A) Schematic comparisons of the 5′ flanking sequences of the ppcA1 genes of the C4 plants F. trinervia and F. bidentis, the the C4-like species F. palmeri and F. vaginata, and the C3 plants F. pringlei and F. cronquistii. The numbers of nucleotides refer to the translation initiation codons. Regions with high similarity between all promoters (60% or more identical nucleotides) are indicated by gray boxes. The position of Mem1 is indicated by black boxes.
(B) Sequence comparison of Mem1 and its homologs. The A and B segments are shaded. Asterisks label identical nucleotides in the A or B segment of all promoters. Black bars indicate the single nucleotide difference in A and the CACT tetranucleotide in B. The tandemly duplicated T/CACT repeats are labeled by arrows.
The comparison of Mem1 and its homolog in F. pringlei shows two remarkable features. The A part differs only in one single nucleotide at the very 5′ end (labeled in Figure 8). Mem1 of F. trinervia holds a guanine in this position, and there is an adenine in the Mem1 homolog of F. pringlei. More prominent is the difference in part B. A tetranucleotide (CACT) is present in the Mem1 of F. trinervia but is absent in the F. pringlei sequence. The remainder of part B sequences is virtually identical in both promoters.
To elucidate which of the observed differences between the Mem1 of F. trinervia and its homolog in F. pringlei are candidates for mesophyll expression determinants, we pursued a comparative approach. The 5′ flanking sequences of ppcA1-type genes were isolated by vectorette PCR (Siebert et al., 1995) from another C4 species of Flaveria (i.e., F. bidentis), from two C4-like plants, F. palmeri and F. vaginata, and from an additional C3 species (F. cronquistii).
A comparison of the 5′ flanking regions identified in each case Mem1 homologous sequences where the A and B parts were, as in F. pringlei, separated by 97 to 108 bp (Figure 8). The A parts of all C4 and C4-like species showed a guanine at their first nucleotide position. An adenine was present in the A homologs of the two C3 species. A more striking C4-to-C3 associated difference is found for the tetranucleotide CACT. This assemblage is present in the B parts of all C4 and C4-like species but lacking in both C3 promoters. This suggested that the CACT tetranucleotide is critical for Mem1 function.
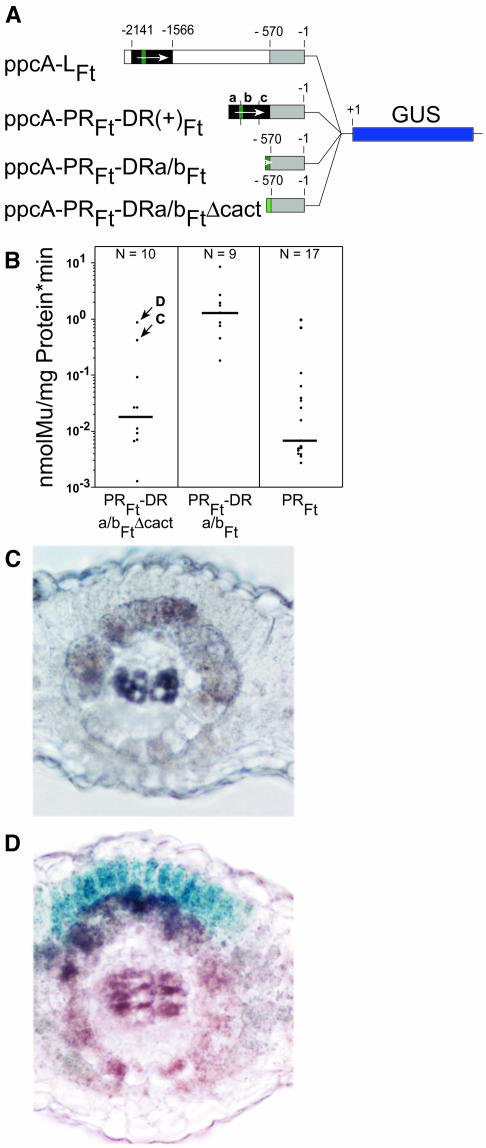
To challenge this assumption, the CACT tetranucleotide was deleted in Mem1 and the modified Mem1, (construct ppcA-PRFt-DRa/bFt-ΔCACT; Figure 9A) was tested for its expression profile in transgenic F. bidentis. Deletion of the CACT tetranucleotide resulted in a 50-fold drop in promoter activity. The resulting activity of the ppcA-PRFt-DRa/bFt-ΔCACT construct was statistically indistinguishable from that of ppcA-PRFt (Figure 9B). It follows that the CACT tetranucleotide is essential for the quantity of mesophyll expression. Histochemical analysis of the 10 transgenic plants obtained showed that nine plants did not reveal any GUS activity in the mesophyll cells (Figure 9C; plant C is shown here as an example because this plant shows the second highest GUS activity). Only one plant, with the highest promoter activity of all 10 plants (labeled D in Figure 9B), directed a mesophyll-specific expression of the GUS reporter gene. Whether the transgene of this plant has been integrated into the genome close to a mesophyll-specific enhancer and the resulting mesophyll expression of the GUS reporter gene represents an artifact is not known. At this stage of investigation, we have to conclude that the CACT-containing cis-regulatory element is necessary for mesophyll expression but may not be sufficient.
Figure 9.
Analysis of the ppcA1 GUS Reporter Gene Construct ppcA-PRFt-DRa/bFt-ΔCACT in Transgenic F. bidentis.
(A) Structure of the ppcA1/GUS chimerical gene.
(B) GUS activities in leaves of transgenic F. bidentis plants. The GUS activity of the ppcA-PRFt-DRa/bFt construct was taken from Figure 7B. The numbers of independent transgenic plants investigated (N) are indicated at the top of each column. Median values are shown. Mu, 4-methylumbelliferone.
(C) and (D) Histochemical analysis of the GUS activity of the ppcA-PRFt-DRa/bFt-ΔCACT driven reporter gene construct in leaf sections of transgenic F. bidentis. In (C), the GUS expression pattern of plant C from (B) is shown, and in (D), that of plant D is shown. Incubation time for (C) was 48 h; incubation time for (D) was 24 h.
DISCUSSION
The C4 cycle genes are largely regulated by transcription (Sheen, 1999). This type of gene regulation involves sets of cis-regulatory modules and their corresponding trans-regulatory factors that interact and thereby control the specific expression of the C4 genes in either mesophyll or bundle-sheath cells. Plants with the C4 photosynthetic pathway are of polyphyletic origin (Kellogg, 1999; Sage et al., 1999), and the networks that regulate cell type–specific gene expression are also likely to have evolved several times independently. To date, it is not known which types of cis- and trans-regulatory elements constitute mesophyll or bundle-sheath cell expression modules at the molecular level and how regulatory networks for mesophyll or bundle-sheath cell-specific gene expression have evolved. Here, a scenario is presented that indicates how the mesophyll cell-specific expression of the C4 PEPC gene in the genus Flaveria may have evolved.
Analysis of chimerical promoter/reporter genes in transgenic F. bidentis identified two segments in the 5′ flanking region of the ppcA1 gene of the C4 plant F. trinervia that are necessary and sufficient for the mesophyll-specific expression of this gene. The DR exhibits enhancer-like properties and, combined with its corresponding PR, confers high levels of mesophyll expression to the reporter gene (Figure 3). The orthologous ppcA1 promoter of the C3 plant F. pringlei directs neither a high nor a mesophyll-specific expression (Stockhaus et al., 1997). One has to conclude therefore that both the distal and proximal promoter regions of the ppcA1 gene have been sites of evolutionary actions. All available evidence in this and previous studies (Stockhaus et al., 1997; Windhövel et al., 2001) supports this point of view.
When the DR of the ppcA1 promoter of F. trinervia (C4 ppcA1 promoter) is fused to the ppcA1 promoter of F. pringlei (C3 ppcA1 promoter), a mesophyll expression component is added to that promoter, but the overall promoter strength does not alter substantially (Figure 4). This may be explained by assuming that the DR of the C4 ppcA1 promoter provides mesophyll specificity, while the PR is responsible for quantitative expression. The quantity elements are not present in the C3 ppcA1 promoter, and, therefore, the chimerical ppcA-MFp-DRFt promoter (Figure 4) does not direct high levels of mesophyll expression. Alternatively, the DR of the C4 ppcA1 promoter may contain transcription repressing sequences that reduce ppcA1 expression in the bundle-sheath cells and the vascular bundle and that thereby relatively increase mesophyll expression. However, the activity of all constructs containing the C4 DR or its subfragments is clearly higher than the activity of the PR of the C4 ppcA1 promoter alone (Figures 3, 6, and 7). This demonstrates that the C4 DR contains mesophyll transcriptional enhancer sequences. Whether there are, in addition, bundle-sheath repressing sequences remains an open question.
The proposed attributes of the DRs and PRs of the C4 ppcA1 promoter may not be easily identified by experiments. The PR of the C4 ppcA1 promoter (C4-PR) alone shows only a very basic level of expression (Figure 3). This demonstrates that the DR of the C4 promoter (C4-DR) is absolutely essential for the C4-typical high expression potential of the corresponding PR. On the other hand, the C4-DR does not result in any mesophyll expression when it is fused to the −46 fragment of the 35S promoter of the Cauliflower mosaic virus (Burscheidt, 1998). This indicates that the C4-DR exhibits its mesophyll expression potential only when it is combined with a PR from either the C4 or the C3 ppcA1 promoter. To achieve a high mesophyll-specific expression, the C4-DR has to be combined with its cognate PR. It has to be concluded, therefore, that the distal and proximal promoter regions do not function as separate modules and act additively but, rather, as a synergistic transcriptional controlling system that evolved together.
Are the C4-DR and C4-PR segments the only parts of the 5′ flanking region of the C4 ppcA1 gene that are involved in controlling the transcription of that gene? When the ppcA-PRFt-DRFt construct and its derivatives are compared with that of the full C4 ppcA1 promoter (Figures 3 and 6), there is clearly a significant loss in expression quantity. This indicates that the smaller promoter constructs lack quantitative cis-regulatory elements that are present in the full promoter. Alternatively, the reduced expression levels of the ppcA-PRFt-DRFt construct and its derivatives may also be attibutable to the changed distance between the DR and PR segments in these promoters and exhibit topological constraints (Rippe et al., 1995). The difference in expression levels between the ppcA-PRFt-DR(+)Ft and ppcA-PRFt-DR(−)Ft constructs (Figure 3) supports this possibility.
Whether the C4 ppcA1 promoter sequences between −570 and −1049 encompasses further C4-relevant cis-regulatory elements remains unclear. The C4 ppcA1 promoter deletion experiments with the heterologous C3 plant tobacco suggest that the intermediate region may contain repressing sequences (cf. the expression levels of the ppcA-1,5Ft and ppcA-1,0Ft constructs with the ppcA-PRFt promoter; Figure 2). However, the ppcA-1,5Ft and ppcA-1,0Ft constructs have not been analyzed in the homologous C4 system, and the biological meaning of the tobacco data therefore remains questionable. Even though we cannot exclude that the segment between the DR and PR region contains cis-regulatory elements, we conclude that they are most probably only of minor importance. The DR and PR segments are the major and essential cis-regulatory modules for the high and mesophyll-specific expression of the C4 ppcA1 gene.
To date, the cis-regulatory elements of the PR have not been mapped precisely. Using the yeast one-hybrid system, it was found that the PR of the C4 ppcA1 promoter interacts with homeobox transcription factors of the zinc finger subclass (Windhövel et al., 2001), whereas the PR of the C3 ppcA1 promoter does not contain detectable binding sites for these zinc finger homeobox proteins. At least one binding site is located in the first intron, which is inserted in the 5′ leader region of the C4 ppcA1 gene (Windhövel et al., 2001). The in planta significance of the zinc finger homeobox proteins and their exact target sequences need to be investigated.
A cis-regulatory module for mesophyll-specific gene expression named Mem1 has been identified in the DR of the C4 ppcA1 promoter. The module is mapped at 41-bp resolution and overlaps with the a and b parts of the C4-DR segment. Fusing the 41-bp segment to the PR of the C4 ppcA1 promoter is sufficient to confer mesophyll-specific expression to the GUS reporter gene. Mem1, therefore, has to carry cis-regulatory elements for mesophyll-specific gene expression. Whether there are other mesophyll expression elements in the a or b part of the C4-DR segment is unknown. If these elements exist, they are probably redundant to Mem1. Whether Mem1 harbors also a bundle-sheath repressing element remains an open question and should be investigated in the future.
A comparative analysis with ppcA1 promoter sequences from other C4, C4-like, and C3 Flaveria species identified Mem1 homologous sequences in all examined plants. Their comparison hinted at elements for mesophyll-specific ppcA1 gene expression. The most notable C4-to-C3 difference detected between Mem1 of the C4/C4-like plants and its counterpart in the C3 species is a CACT tetranucleotide (Figure 8). The motif is present in all Mem1 sequences of the C4/C4-like plants but lacking in the Mem1 homologs of the C3 species. The CACT tetranucleotide is found in a sequence segment, the B region of Mem1, which is fully conserved in the C4 and C3 ppcA1 promoters. This finding suggested that the CACT motif is essential for mesophyll-specific gene expression. Functional analyses with transgenic plants confirmed this assumption. Deletion of the CACT tetranucleotide from Mem1 abolished the mesophyll expression of the GUS reporter gene. We conclude that the addition of the CACT tetranucleotide to the C3 promoter during C3-to-C4 evolution created a new cis-regulatory element that was necessary for confering mesophyll expression to the promoter.
Although the deletion of the CACT tetranucleotide reduced promoter activity almost completely, one single transgenic plant out of 10 (i.e., that with the highest activity) expressed the transgene in the mesophyll cells. We do not know whether the expression pattern of this transgene reflects an artifact because of the nearby presence of a mesophyll enhancer within the genome. Therefore, we have to conclude that the CACT-containing cis-regulatory element may not be the only cis-regulatory element in Mem1. The CACT-containing cis-element is necessary but may not be sufficient for mesophyll expression.
The CACT tetranucleotide is embedded in a sequence context (TTACTCACTAA) that can form an imperfect palindrome. The palindrome resembles a binding site for a GCN4-like basic leucine zipper transcription factor (Arndt and Fink, 1986; Oñate et al., 1999; Matys et al., 2003). A DNA protein interaction screen with the yeast one-hybrid system (Li and Herskowitz, 1993) supports this notion. Using Mem1 as a bait, a basic leucine zipper protein was isolated that interacts with Mem1 of F. trinervia but not with the Mem1 homolog of F. pringlei (M. Akyildiz and P. Westhoff, unpublished data). Taken together, all available evidence suggests that the tetranucleotide CACT is part of a cis-regulatory element that may be targeted by a basic leucine zipper transcription factor.
How did this novel cis-regulatory element evolve? Adjacent to the CACT motif in the 5′ direction, two tandem TACT repeats are observed in all C4/C4-like ppcA1 promoters but also in the C3 promoters (Figure 8). Short direct repeats are known to be an important source of genetic change in all organisms because replication misalignment may lead to the deletion or addition of repeat units (Bzymek and Lovett, 2001; Li et al., 2002). We propose that such a mechanism was responsible for adding the CACT motif to the C3 promoter. The addition of this third imperfect repeat unit resulted in the formation of a novel cis-regulatory element. This element could be targeted by transcription factors available already in the mesophyll cells, and thereby a new expression pattern was created.
A C4-to-C3 associated nucleotide difference was also observed in the A segment of Mem1 (Figure 8). Whereas all C4 and C4-like species have a guanine at the outermost 5′ position of Mem1, it is an adenine in the C3 species. No putative transcription factor binding site is detectable in this Mem1 segment; thus, it is not clear whether this nucleotide difference is of functional importance.
The A segment is contiguous with the B segment only in Mem1 of F. trinervia, but in all other C4 and C3 ppcA1 promoters, the two segments are separated from each other by ∼100 bp. This suggests that the contiguous arrangement of the A and B parts is not of functional importance. If the A segment contains a cis-regulatory element, then the A and B parts should form separate cis-regulatory units.
Which scenario can be envisaged for the evolution of the C4 ppcA promoter in the genus Flaveria? The C3 reference promoter from F. pringlei is weak and does not show any cell specificity. The activity of this promoter is even higher in the bundle-sheath cells and the vascular bundle than in the mesophyll cells. By contrast, the C4 promoter is strong and is active only in the mesophyll cells. Evolution toward C4 could therefore have started by increasing promoter strength. This is supported by ppcA1 mRNA quantification in C3-C4 intermediate Flaveria species. Even C3-C4 intermediates with a low degree of C4 trait expression (i.e., F. chloraefolia; Edwards and Ku, 1987) show already elevated ppcA mRNA amounts (Engelmann et al., 2003). It is reasonable to assume that the increase in ppcA expression was restricted to the mesophyll cells. This implies the evolution of mesophyll expression elements, for instance, by modifying rudimentary progenitor elements that were already present in the C3 promoter (i.e., Mem1). The isolation and functional analysis of ppcA promoters from C3-C4 intermediate species of Flaveria has been initiated and should clarify this point. With a delay or maybe even in parallel the cis-regulatory modules for expression in bundle-sheath cells and the vascular bundle had to be inactivated. This could have been achieved by direct mutational modification of these modules and/or by the addition of bundle-sheath repressor elements. Which of these strategies nature has pursued is unknown.
In his review on biochemical evolution, A.C. Wilson (Wilson et al., 1977) pointed out that “quantitative mutations affecting enzyme levels may have had a major role in the adaptative metabolic evolution of multicellular organisms” and that “these quantitative effects can result from point mutations in control genes.” In the meantime, evolutionary biologists have collected convincing evidence that supports the view that changes in the spatiotemporal expression patterns of genes are the principal mechanism for the evolution of novelty, both in morphological and biochemical traits (Doebley and Lukens, 1998; Carroll, 2000).
Our investigations on the molecular evolution of C4 PEPC in the genus Flaveria are in line with this concept. The studies show that at the onset of the transition from C3 to C4 photosynthesis, the enzyme is still rather C3-like with respect to its kinetic and regulatory properties; it becomes C4-like only much later (reviewed in Svensson et al., 2003). By contrast, the expression pattern of the ppcA1 gene was modified very early in evolution from C3 to C4 (Engelmann et al., 2003). The data presented here indicate that comparatively small changes in the nucleotide sequence should be responsible for these changes that give rise to a novel mode of expression. It will be interesting to see whether the evolution of mesophyll- or bundle-sheath cell-specific gene expression in Flaveria always relied upon the same set of cis- and trans-regulatory elements. The analysis of another mesophyll specifically expressed gene, for instance, carbonic anhydrase (Badger, 2003), would therefore be highly desirable, and the study of bundle-sheath specific gene expression should be initiated. Because the C4 photosynthetic pathway evolved several times independently (Sage, 2004), it will be even more interesting to investigate whether the various C4 species pursued similar or different strategies to achieve the same goal, a differential expression of their C4-photosynthesis associated genes.
METHODS
Construction of Chimerical Promoters
DNA manipulations and cloning were performed according to Sambrook and Russell (2001). All promoter GUS reporter gene fusions used in this work are based on the constructs ppcA-PRFt, ppcA-LFt, and ppcA-LFp, which were formerly designated ppcA-S-Ft, ppcA-L-Ft, and ppcA-L-Fp. Their construction has been described in detail (Stockhaus et al., 1994). In all constructs, the 3′ border of the ppcA1 5′ flanking sequences of Flaveria trinervia and F. pringlei (Figure 1) is located just upstream of the AUG initiation codon. For cloning purposes, a SmaI site was added to the 3′ border of each fragment by PCR amplification with an appropriately designed oligonucleotide. The 5′ borders of ppcA-LFt and ppcA-LFp are defined by HindIII (ppcA-LFt) or XbaI sites (ppcA-LFp), which occur naturally in these promoter regions (Figure 1). The 5′ border of the ppcA-PRFt promoter fragment of F. trinervia corresponds to nucleotide position −570. A XbaI site was added to this border by PCR amplification. All promoter fragments were assembled in pBluescribe M13− (Stratagene Cloning Systems, La Jolla, CA) and confirmed by sequencing. They were excised by HindIII/SmaI digestion and transferred to HindIII/SmaI-restricted pBI121 (Clontech Laboratories, Palo Alto, CA) in front of the GUS reporter gene (Stockhaus et al., 1994). The ppcA1 promoter reporter gene constructs prepared in the course of this study were cloned as described below. All DNA fragments generated by PCR were confirmed by DNA sequencing.
Construction of ppcA-1,5Ft and ppcA1,0Ft
The ppcA-LFt promoter plasmid (pBluescribe M13−) was digested with AccI (−1566) or Asp718 (−1049) (Figure 1). Blunt ends were generated by fill-in synthesis with the Klenow fragment of Escherichia coli DNA polymerase I followed by ligation of HindIII linkers. The AccI and Asp718 restricted plasmids were digested with HindIII, the 5′ located ppcA-LFt promoter fragments were removed by agarose gel electrophoresis, and the HindIII ends of the remaining plasmids were religated. After an intermediate cloning step in E. coli, the resulting ppcA-1,5Ft and ppcA1,0Ft promoter regions were excised by HindIII/SmaI restriction and inserted into pBI121 in front of the GUS reporter gene.
Construction of ppcA-PRFt-DR(+)Ft and ppcA-PRFt-DR(−)Ft
The ppcA-LFt promoter plasmid (pBluescribe M13−) was digested with AccI (−1566). Blunt ends were generated by fill-in synthesis, and XbaI linkers were ligated. The DNA was restricted with XbaI (−2141), and the released 575-bp XbaI fragment (named DR) was isolated. The DR fragment was cloned into XbaI-digested ppcA-PRFt pBI121 and led to ppcA-PRFt-DR(+)Ft (DR inserted in correct orientation) or ppcA-PRFt-DR(−)Ft (DR in opposite orientation).
Construction of ppcA-MFp
The ppcA-LFp promoter plasmid (pBluescribe M13−) was digested with BclI (−1854) and XbaI (−2584) (Figure 1). The released 685-bp BclI/XbaI fragment was removed by agarose gel electrophoresis, and the remaining ppcA1 promoter plasmid DNA was recovered. Blunt ends were generated by treatment with Klenow polymerase followed by religation of the promoter plasmid. The resulting ppcA-MFp promoter fragment was excised by HindIII/SmaI and inserted into pBI121.
Construction of ppcA-MFp-DRFt
The ppcA-LFp promoter plasmid (pBluescribe M13−) was digested with BclI (−1854), and blunt ends were generated by treatment with Klenow polymerase. The DNA was restricted with XbaI (−2584) (Figure 1). The released 685-bp BclI/XbaI fragment was removed by agarose gel electrophoresis, and the 5′ deleted ppcA1 promoter plasmid DNA was recovered. The DR of the ppcA1 promoter of F. trinervia was isolated by digesting ppcA-LFt with AccI, creating blunts by treatment with Klenow polymerase, and releasing the DR by restriction with XbaI. The DR was ligated with the 5′ deleted ppcA1 promoter of F. pringlei. The resulting ppcA-MFp-DRFt promoter was excised by HindIII/SmaI and inserted into pBI121.
Construction of ppcA-PRFt-DRaFt, ppcA-PRFt-DRbFt, and ppcA-PRFt-DRcFt
The DR of the ppcA1 promoter of F. trinervia was divided into the three overlapping segments a (−2141 to −1940), b (−1981 to −1668), and c (−1713 to −1566). These were amplified by PCR with the ppcA-PRFt-DR(+)Ft promoter plasmid as template. Each 3′ oligonucleotide primer carried a XbaI, and each 5′ oligonucleotide primer carried a HindIII site (Table 1). After digestion with HindIII and XbaI, the resulting PCR products were used to replace the DR fragment in the ppcA-PRFt-DR(+)Ft construct.
Table 1.
Oligonucleotide Primers Used for the Construction of Chimerical Promoters
| FtDEa5′ | 5′-GGGAAGCTTAGAACATGAAAAAAGGACTCACCAGG-3′ |
| FtDEa3′ | 5′-GGGTCTAGATTGTTTGTTTTAGTGAGTAAG-3′ |
| FtDEb5′ | 5′-GGGAAGCTTGTGAATTTATGAGAGCTGTAC-3′ |
| FtDEb3′ | 5′-GGGTCTAGAGTACTTAATTTCCATTTCTC-3′ |
| FtDEc5′ | 5′-GGGAAGCTTTGTGTGTGTGAATATGTTGC-3′ |
| FtDEC3′ | 5′-GGGTCTAGATACATACTTTCCAAATTCATAG-3′ |
| FtDEa3′-Xho | 5′-GGGCTCGAGTTGTTTGTTTTAGTGAGTAAG-3′ |
| FtDEb5′-Sal | 5′-GGGGTCGACGTGAATTTATGAGAGCTGTAC-3′ |
| FtDEa/bΔ5′ | 5′-AGCTTGTGAATTTATGAGAGCTGTACTTACTAAAACAAACAAT-3′ |
| FtDEa/bΔ3′ | 5′-CTAGATTGTTTGTTTTAGTAAGTACAGCTCTCATAAATTCACA-3′ |
Construction of ppcA-PRFt-DRa/bFt and ppcA-PRFt-DR4a/bFt
The a/b-overlapping region (−1981 to −1940) was amplified by PCR using the FtDEb5′ and FtDEa3′ primers. After digestion with HindIII and XbaI, the a/b-fragment was inserted into ppcA-PRFt-DR(+)Ft to replace the DR fragment. The resulting promoter was named ppcA-PRFt-DRa/bFt.
Tandem repeats of the a/b-overlapping region were generated as described by de Pater et al. (1993) using the primers FtDEa3′-Xho and FtDEb5′-Sal (Table 1), which contain XhoI and SalI sites instead of XbaI and HindIII sites. The resulting multimeric DNAs were used as a template for PCR amplification with the FtDEb5′ and FtDEa3′ primers (Table 1). The fragment that contains four tandem repeats of the a/b-overlapping region was isolated by agarose gel electrophoresis, digested with HindIII and XbaI, and inserted into ppcA-PRFt-DR(+)Ft to replace the DR fragment. The resulting promoter was named ppcA-PRFt-DR4a/bFt.
Construction of ppcA-PRFt-DRa/bFt-ΔCACT
The a/b-overlapping region (−1981 to −1940) without the CACT tetranucleotide was generated by annealing the two oligonucleotides FtDEa/bΔ5′ and FtDEa/b3′. The annealed oligonucleotides were inserted into ppcA-PRFt-DR(+)Ft to replace the DR fragment. The resulting promoter was named ppcA-PRFt-DRa/bFt-ΔCACT.
Plant Transformation
The promoter/GUS constructs were introduced by electroporation into the Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991). Tobacco (Nicotiana tabacum) plants were transformed as described (Horsch et al., 1985; Stockhaus et al., 1994). The transformation of F. bidentis plants was performed according to Chitty et al. (1994). Integration of the chimerical genes into the F. bidentis genome was examined by DNA gel blot analysis or by PCR (Stockhaus et al., 1997). In all tested transgenic plants, the hybridizing fragment or the PCR product had the expected size, indicating that the promoter fragment and the GUS gene were linked in the genomic DNA and that each transgenic plant contains at least one copy of the respective chimerical gene.
Measurement of GUS Activity and Histochemical Analysis
Regenerated plants or T1 plants were used for the analysis of the GUS activity. Tobacco plants grown from tissue culture were used for the measurements of GUS activity. For the histochemical analysis, tobacco plants were transferred to soil and grown in a greenhouse. F. bidentis were greenhouse plants, 40 to 50 cm tall and before flower initiation. GUS activities were measured quantitatively (Jefferson et al., 1987; Kosugi et al., 1990) or in situ (Stockhaus et al., 1997). The average values of the data are expressed by medians, and the Mann-Whitney U test statistics as implemented in the software package Kaleidagraph 3.6 for Mac OS X (Synergy Software, Reading, PA, www.synergy.com) were used to test whether two data series differ from each other.
DNA Isolation
Nucleic acids were isolated from leaf tissue (Westhoff et al., 1991). DNA was recovered from the 2 M LiCl soluble nucleic acid fraction by isopropanol precipitation. The DNA was dissolved in double-distilled water, and residual RNA was digested by RNase A treatment. After phenol/chloroform extraction, the DNA was precipitated with isopropanol, dissolved in double-distilled water, and stored at −20°C until use.
Isolation of 5′ Flanking Sequences from the ppcA1 Genes of F. bidentis, F. vaginata, F. palmeri, and F. cronquistii
The 5′ flanking regions of ppcA1 genes of F. bidentis, F. vaginata, F. palmeri, and F. cronquistii were isolated from total DNA by vectorette PCR (Siebert et al., 1995) with the Universal Genome Walker Kit (Clontech Laboratories) as recommended by the manufacturer. For each plant species, DraI, EcoRV, PvuII, and StuI DNA libraries were constructed. The gene-specific primers for the primary and secondary PCR reactions (GSP1 and GSP2; Table 2) of the first walking step were designed to hybridize to the very 5′ part of the coding region of the ppcA1 genes of both F. trinervia and F. pringlei. The primers were expected to hybridize to the 5′ coding regions of the other ppcA1 genes as well. In the second walking step, gene-specific primers were designed according to the sequence of the promoter fragments isolated in the first walking step (Table 2). The resulting PCR fragments were cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced.
Table 2.
Oligonucleotide Primers Used for Genome Walking
| GSP1 | 5′-CGAATCGATGTAATTTCTCCACATTCCGG-3′ |
| GSP2 | 5′-TCATACTCAACAAGCTTATCATCCTCAGAA-3′ |
| GSP3Fv | 5′-TAAGTCARTCTATGACTCGCGCGTTGTG-3′ |
| GSP4Fv | 5′-CGCGTCGACGTAAAAACATTGAAGCCACAY-3′ |
| GSP3Fc | 5′-CACGCTTAGCTAAATGGGTAAGTGTAGAG-3′ |
| GSP4Fc | 5′-ATGATGTGTTCATGAGTTCATCTGGTTA-3′ |
| GSP3Fpa | 5′-CGTTGTGACGGGGCCATCAAATGGA-3′ |
| GSP4Fpa | 5′-ATGCGCACGTTGCCGCGTGTAAACTCGT-3′ |
| GSP1Fp | 5′-CGCCTCTATGTACAGAGAATACCTTTGTTC-3′ |
| GSP2Fp | 5′-GGCTCTACGAACACTCCTTGGTCAAG-3′ |
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the accession numbers X64143 (F. trinervia), AY297087 (F. bidentis), AY297088 (F. palmeri), AY297090 (F. vaginata), AY297089 (F. cronquistii), and X64144 (F. pringlei).
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft initially within the Graduiertenkolleg Molekulare Physiologie and later by SFB 590 Inhärente und adaptive Differenzierungsprozesse at the Heinrich Heine University of Düsseldorf. Additional support from the Fonds der Chemischen Industrie is gratefully acknowledged. We are indebted to Uwe Santore for carefully reading the manuscript.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter Westhoff (west@uni-duesseldorf.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019729.
References
- Arndt, K., and Fink, G.R. (1986). GCN4 protein, a positive transcription factor in yeast, binds general promoters at all 5′ TGACTC 3′ sequences. Proc. Natl. Acad. Sci. USA 83, 8516–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger, M. (2003). The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosyn. Res. 77, 83–94. [DOI] [PubMed] [Google Scholar]
- Blackwood, E.M., and Kadonaga, J.T. (1998). Going the distance: A current view of enhancer action. Science 281, 60–63. [DOI] [PubMed] [Google Scholar]
- Bläsing, O.E., Westhoff, P., and Svensson, P. (2000). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275, 27917–27923. [DOI] [PubMed] [Google Scholar]
- Burscheidt, J. (1998). Cis-Regulatorische Determinanten für Mesophyll- und Bündelscheidenspezifische Genexpression in C4-Spezies der Gattung Flaveria—Die Promotoren der Phosphoenolpyruvat-Carboxylase- und der Glycin-Decarboxylasegene. Master's thesis (Düsseldorf, Germany: Heinrich-Heine-Universität).
- Bzymek, M., and Lovett, S.T. (2001). Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98, 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S.B. (2000). Endless forms: The evolution of gene regulation and morphological diversity. Cell 101, 577–580. [DOI] [PubMed] [Google Scholar]
- Chitty, J.A., Furbank, R.T., Marshall, J.S., Chen, Z., and Taylor, W.C. (1994). Genetic transformation of the C4 plant, Flaveria bidentis. Plant J. 6, 949–956. [Google Scholar]
- Crétin, C., Santi, S., Keryer, E., Lepiniec, L., Tagu, D., Vidal, J., and Gadal, P. (1991). The phosphoenolpyruvate carboxylase gene family of Sorghum: Promoter structures, amino acid sequences and expression of genes. Gene 99, 87–94. [DOI] [PubMed] [Google Scholar]
- de Pater, S., Pham, K., Chua, N.H., Memelink, J., and Kijne, J. (1993). A 22-bp fragment of the pea lectin promoter containing essential TGAC-like motifs confers seed-specific gene expression. Plant Cell 5, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, G.E., and Ku, M.S.B. (1987). Biochemistry of C3-C4 intermediates. In The Biochemistry of Plants, Vol. 10, M.D. Hatch and N.K. Boardman, eds (New York: Academic Press), pp. 275–325.
- Engelmann, S., Bläsing, O.E., Gowik, U., Svensson, P., and Westhoff, P. (2003). Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria—A gradual increase from C3 to C4 characteristics. Planta 217, 717–725. [DOI] [PubMed] [Google Scholar]
- Ernst, K., and Westhoff, P. (1996). The phosphoenolpyruvate carboxylase (ppc) gene family of Flaveria trinervia (C4) and F. pringlei (C3): Molecular characterization and expression analysis of the ppcB and ppcC genes. Plant Mol. Biol. 34, 427–443. [DOI] [PubMed] [Google Scholar]
- Hatch, M.D. (1987). C4 photosynthesis: A unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106. [Google Scholar]
- Hermans, J., and Westhoff, P. (1990). Analysis of expression and evolutionary relationships of phosphoenolpyruvate carboxylase genes in Flaveria trinervia (C4) and F. pringlei (C3). Mol. Gen. Genet. 224, 459–468. [DOI] [PubMed] [Google Scholar]
- Hermans, J., and Westhoff, P. (1992). Homologous genes for the C4 isoform of phosphoenolpyruvate carboxylase in a C3- and a C4-Flaveria species. Mol. Gen. Genet. 234, 275–284. [DOI] [PubMed] [Google Scholar]
- Höfer, M.U., Santore, U.J., and Westhoff, P. (1992). Differential accumulation of the 10-, 16- and 23-kDa peripheral components of the water-splitting complex of photosystem II in mesophyll and bundle-sheath chloroplasts of the dicotyledonous C4 plant Flaveria trinervia (Spreng.) C. Mohr. Planta 186, 304–312. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, F.E., Hoffmann, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (1999). Phylogenetic aspects of the evolution of C4 photosynthesis. In C4 Plant Biology, R.F. Sage and R.K. Monson, eds (San Diego, CA: Academic Press), pp. 411–444.
- Kosugi, S., Ohashi, Y., Nakajima, K., and Arai, Y. (1990). An improved assay for β-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci. 70, 133–140. [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. [DOI] [PubMed] [Google Scholar]
- Li, J.J., and Herskowitz, I. (1993). Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262, 1870–1874. [DOI] [PubMed] [Google Scholar]
- Li, Y.-C., Korol, A.B., Fahima, T., Beiles, A., and Nevo, E. (2002). Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 11, 2453–2465. [DOI] [PubMed] [Google Scholar]
- Matys, V., et al. (2003). TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson, R.K. (1999). The origins of C4 genes and evolutionary pattern in the C4 metabolic phenotype. In C4 Plant Biology, R.F. Sage and R.K. Monson, eds (San Diego, CA: Academic Press), pp. 377–410.
- Monson, R.K., and Moore, B.D. (1989). On the significance of C3-C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant Cell Environ. 12, 689–699. [Google Scholar]
- Oñate, L., Vicente-Carbajosa, J., Lara, P., Díaz, I., and Carbonero, P. (1999). Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 274, 9175–9182. [DOI] [PubMed] [Google Scholar]
- Powell, A.M. (1978). Systematics of Flaveria (Flaveriinae-Asteraceae). Ann. Mo. Bot. Gard. 65, 590–636. [Google Scholar]
- Rippe, K., von Hippel, P.H., and Langowski, J. (1995). Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci. 20, 500–506. [DOI] [PubMed] [Google Scholar]
- Sage, R.F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage, R.F., Li, M., and Monson, R.K. (1999). The taxonomic distribution of C4 photosynthesis. In C4 Plant Biology, R.F. Sage and R.K. Monson, eds (San Diego, CA: Academic Press), pp. 551–584.
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sheen, J. (1999). C4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 187–217. [DOI] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhaus, J., Poetsch, W., Steinmüller, K., and Westhoff, P. (1994). Evolution of the C4 phosphoenolpyruvate carboxylase promoter of the C4 dicot Flaveria trinervia: An expression analysis in the C3 plant tobacco. Mol. Gen. Genet. 245, 286–293. [DOI] [PubMed] [Google Scholar]
- Stockhaus, J., Schlue, U., Koczor, M., Chitty, J.A., Taylor, W.C., and Westhoff, P. (1997). The promoter of the gene encoding the C4 form of phosphoenolpyruvate carboxylase directs mesophyll specific expression in transgenic C4 Flaveria spp. Plant Cell 9, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, P., Bläsing, O., and Westhoff, P. (1997). Evolution of the enzymatic characteristics of C4 phosphoenolpyruvate carboxylase: A comparison of the orthologous ppcA phosphoenolpyruvate carboxylases of Flaveria trinervia (C4) and F. pringlei (C3). Eur. J. Biochem. 246, 452–460. [DOI] [PubMed] [Google Scholar]
- Svensson, P., Bläsing, O.E., and Westhoff, P. (2003). Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 414, 180–188. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., and Gowik, U. (2004). Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: A case study with the genus Flaveria. Ann. Bot. 93, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff, P., Offermann-Steinhard, K., Höfer, M., Eskins, K., Oswald, A., and Streubel, M. (1991). Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta 184, 377–388. [DOI] [PubMed] [Google Scholar]
- Wilson, A.C., Carlson, S.S., and White, T.J. (1977). Biochemical evolution. Annu. Rev. Biochem. 46, 573–639. [DOI] [PubMed] [Google Scholar]
- Windhövel, A., Hein, I., Dabrowa, R., and Stockhaus, J. (2001). Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 45, 201–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.