Abstract
Balance of physiological levels of iron is essential for every organism. In Aspergillus fumigatus and other fungal pathogens, the transcription factor HapX mediates adaptation to iron limitation and consequently virulence by repressing iron consumption and activating iron uptake. Here, we demonstrate that HapX is also essential for iron resistance via activating vacuolar iron storage. We identified HapX protein domains that are essential for HapX functions during either iron starvation or high-iron conditions. The evolutionary conservation of these domains indicates their wide-spread role in iron sensing. We further demonstrate that a HapX homodimer and the CCAAT-binding complex (CBC) cooperatively bind an evolutionary conserved DNA motif in a target promoter. The latter reveals the mode of discrimination between general CBC and specific HapX/CBC target genes. Collectively, our study uncovers a novel regulatory mechanism mediating both iron resistance and adaptation to iron starvation by the same transcription factor complex with activating and repressing functions depending on ambient iron availability.
Keywords: fungi, iron regulation, sensing, siderophores, transcription factor complex
Introduction
The redox-active metal iron is an indispensable cofactor in a variety of essential cellular processes such as oxidative phosphorylation, biosynthesis of numerous metabolites, and detoxification of oxidative stress. Paradoxically, the same redox property makes this metal potentially toxic by causing oxidative stress (Halliwell & Gutteridge, 1984; Lin et al, 2011). Thus, iron homeostasis requires precise regulation of iron uptake and storage to satisfy the cellular needs but to avoid toxic iron excess.
In Aspergillus fumigatus, iron homeostasis is maintained by two central transcription factors, which are interconnected in a negative transcriptional feed-back loop: the GATA-factor SreA and the bZIP-factor HapX (Haas, 2012). During iron sufficiency, SreA represses iron uptake, including reductive iron assimilation and siderophore-mediated iron uptake, to avoid toxic effects (Schrettl et al, 2008). During iron starvation, HapX activates siderophore-mediated iron acquisition and represses iron-consuming pathways, including heme biosynthesis and respiration, to spare iron (Schrettl et al, 2010). As shown in Aspergillus nidulans, HapX functions via physical interaction with the CCAAT-binding complex (CBC) (Hortschansky et al, 2007). The CBC is a heterotrimeric DNA-binding complex, which is conserved in all eukaryotes. In A. nidulans, inactivation of either one of its subunits, phenocopied HapX inactivation with respect to defects in adaptation to iron starvation (Hortschansky et al, 2007). However, the CBC has HapX-independent functions in Aspergillus spp. (Kato, 2005; Thon et al, 2010). Humans lack HapX and genome-wide identification resulted in 5,000–15,000 CBC-binding sites depending on the type of human cell analyzed (Fleming et al, 2013).
Deficiency in HapX, but not SreA, attenuates virulence of A. fumigatus in murine models of aspergillosis (Schrettl et al, 2008, 2010), which emphasizes the crucial role of adaptation to iron limitation in pathogenicity. With the exception of Saccharomyces cerevisiae and closely related Saccharomycotina species, most fungal species possess orthologs to SreA and HapX (Haas et al, 2008; Kaplan & Kaplan, 2009). The important role of HapX orthologs in virulence is conserved in Candida albicans, Cryptococcus neoformans, and Fusarium oxysporum (Jung et al, 2010; Chen et al, 2011; Hsu et al, 2011; Lopez-Berges et al, 2012). Both SreA and HapX appear to be regulated post-translationally by iron, blocking HapX function and activating SreA function (Haas et al, 1999; Hortschansky et al, 2007). In Schizosaccharomyces pombe, post-translational iron sensing by the HapX and SreA orthologs involves the monothiol glutaredoxin Grx4 (Labbe et al, 2013).
Recently, iron resistance of A. fumigatus was shown to involve SreA-mediated repression of iron uptake and vacuolar iron storage mediated by the vacuolar iron importer CccA (Gsaller et al, 2012). In A. nidulans and A. fumigatus, inactivation of both HapX and SreA is synthetically lethal underlining the critical role of iron homeostasis in cellular survival (Hortschansky et al, 2007; Schrettl et al, 2010). In agreement with their expression pattern and characterized mode of action, the detrimental effects of SreA or HapX inactivation identified so far were confined to growth during iron sufficiency or starvation, respectively, which does not explain the synthetic lethality of their inactivation. Here, we provide an explanation for this synthetic lethality by demonstrating that HapX mediates both repression of vacuolar iron storage during iron starvation and activation of vacuolar iron storage during iron excess, i.e. HapX displays inverse activities depending on the ambient iron availability. In line, we identified protein domains that are essential for mediating adaptation to iron starvation or iron excess, exclusively. Moreover, we demonstrate for the first time that HapX not only acts via protein–protein interaction with the CBC but also directly recognizes an evolutionary conserved motif in the cccA promoter. As the CBC has HapX/iron-independent targets, the latter data reveal the mechanism for discrimination of general CBC and specific HapX/CBC target genes.
Results and Discussion
HapX mediates iron resistance by activating CccA-mediated vacuolar iron storage
HapX functions were analyzed in A. fumigatus ATCC 46645 (Schrettl et al, 2010) and, to facilitate the studies, in its ΔakuA-derivative AfS77, which lacks non-homologous recombination (Hartmann et al, 2010). We did not observe any phenotypic differences between respective ATCC46645- and AfS77-derivative strains (data not shown). For clarity, however, the genetic background used is given for all experiments.
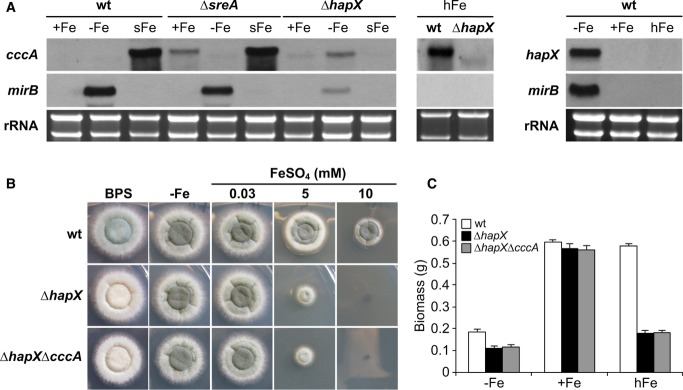
Previously, genome-wide transcriptional profiling revealed that during iron starvation HapX activates genes involved in iron acquisition (including siderophore transporter-encoding mirB) and represses the vacuolar iron transporter-encoding cccA as well as numerous genes involved in iron-consuming processes (see below) (Schrettl et al, 2010). CccA-mediated vacuolar iron storage was recently shown to mediate iron resistance (Gsaller et al, 2012). Consistently, the cccA transcript level is upregulated by iron and particularly by SreA-deficiency (Gsaller et al, 2012). The latter is consistent with SreA-deficiency increasing the cellular iron content (Schrettl et al, 2008) but also shows that transcriptional activation of cccA is mediated by an SreA-independent regulatory mechanism.
Northern analysis demonstrated that HapX-deficiency (strain ΔhapX) impairs not only repression of cccA during iron starvation but also induction of cccA during a 1-h shift from iron starvation to iron sufficiency as well as during growth in high-iron medium (Fig1A). As shown previously (Schrettl et al, 2010), HapX-deficiency caused downregulation of mirB during iron starvation, but did not affect repression of mirB by iron (Fig1A).
Figure 1. HapX is important for adaptation to both iron limitation and iron excess.
- HapX represses cccA during iron starvation and activates cccA during iron excess. Northern analysis was performed with liquid cultures under conditions of iron starvation (−Fe), iron sufficiency (+Fe, 0.03 mM FeSO4), and high-iron availability (hFe, 3 mM FeSO4) at 37°C for 24 h or from mycelia shifted for 1 h from −Fe to +Fe (sFe).
- On agar plates, HapX-deficiency impairs sporulation on BPS-plates, and growth during iron excess. Growth pattern of wild-type (wt), ΔhapX and ΔhapXΔcccA on solid minimal media containing the indicated iron concentration is shown after 48 h at 37°C. The greenish color of the fungal colonies originates from the spore pigment, and its decrease indicates reduced sporulation. The original size of fungal colony photographs is 2.3 × 2.3 cm in all figures.
- HapX-deficiency impairs submerged growth during both iron starvation and iron excess. Liquid biomass production was monitored after 24 h of growth at 37°C under the indicated iron availability. The data represent the mean ± standard deviation (SD) of biological triplicates. The difference between mutant and wild-type strains was statistically significant during −Fe and hFe but not +Fe (two-tailed, unpaired t-test; P < 0.05).
Data information: The iron-sensitive phenotype of ΔcccA was previously analyzed in Gsaller et al (2012) and was further characterized in Fig2. Moreover, the response of hapX transcript levels to a 1-h shift from iron starvation to sufficiency (sFe) was analyzed in Fig4. Strains are derivatives of A. fumigatus AfS77.
The role of HapX in transcriptional control of cccA during iron excess implicated a role of HapX in iron detoxification. In agreement, HapX-deficiency not only decreased sporulation on agar plates in the presence of the iron starvation-inducing, iron-specific chelator bathophenanthroline disulfonate (BPS) and decreased biomass production in liquid cultures during iron starvation, as shown previously (Schrettl et al, 2010), but also dramatically decreased growth on solid and in liquid high-iron media (Fig1B and C). As reported previously (Schrettl et al, 2010), HapX-deficiency did affect neither growth rate nor sporulation under iron-replete conditions.
A mutant strain lacking both HapX and CccA, ΔhapXΔcccA, displayed the same growth pattern as ΔhapX on solid and in liquid media (Fig1B and C). The epistasis of HapX- to CccA-deficiency strongly suggests that lack of cccA expression is responsible for the ΔhapX growth defect during iron excess.
Taken together, HapX acts as a Janus-type transcription factor mediating both repression and activation of cccA and consequently vacuolar iron storage depending on the ambient iron availability.
HapX additionally controls CccA-independent mechanisms involved in iron detoxification
Notably, HapX-deficiency rendered A. fumigatus more susceptible to iron toxicity than CccA-deficiency on solid (Fig2A) and in liquid (Supplementary Table S1) high-iron media. These data indicate that HapX is also required for the activity of iron detoxification mechanisms other than CccA-mediated iron storage. In support, conditional expression of cccA using the xylose-inducible xylP promoter (Zadra et al, 2000; Gsaller et al, 2012) increased iron resistance of ΔhapX under inducing but not repressing conditions (Fig2A; compare strains ΔhapX and ΔhapXcccAOE). However, the radial growth of this strain did not reach that of the wild-type or the ΔcccAcccAOE strain (a cccA deletion mutant expressing cccA under control of the inducible xylP promoter). Compared to ΔhapX, ΔhapXcccAOE also displayed a significant decrease in growth and sporulation on BPS- and low-iron agar plates, demonstrating that activation of vacuolar iron storage is particularly detrimental in a HapX-deficient background.
Figure 2. HapX-deficiency renders A. fumigatus more susceptible to iron toxicity than CccA-deficiency and impairs induction of genes involved in iron-consuming processes.
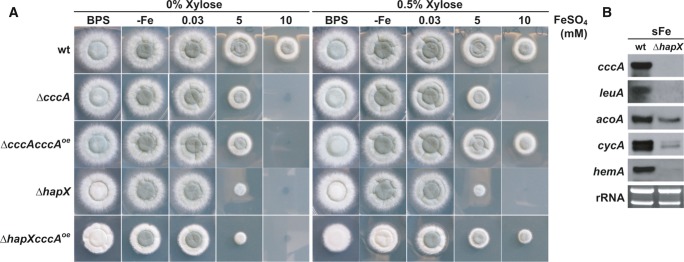
- Strains were grown on solid minimal medium with the given iron availability under xylP-driven cccAOE non-inducible (0% xylose) and inducible (0.5% xylose) conditions for 48 h at 37°C.
- Northern blot analysis was performed after liquid growth for 24 h at 37°C under iron limitation and a subsequent 1-h shift to iron sufficiency (sFe). rRNA is shown as a control for RNA quantity and quality.
Data information: Strains are derivatives of A. fumigatus ATCC 46645.
As previously indicated by genome-wide transcriptional profiling (Schrettl et al, 2010), apart from cccA numerous other genes involved in iron-consuming processes are repressed by HapX during iron starvation. Northern analysis revealed that in a 1-h shift from iron-limited to iron-replete conditions, which reflects short-term iron excess, HapX-deficiency impairs the transcriptional activation not only of cccA but also of genes encoding iron-consuming functions (Fig2B). These proteins include the iron–sulfur cluster-containing LeuA (α-isopropylmalate isomerase) and AcoA (aconitase), involved in leucine biosynthesis and the TCA cycle, respectively, the heme-containing CycA (cytochrome c) involved in respiration, and the heme biosynthesis protein HemA (δ-aminolevulinic acid synthase). These data indicate that HapX might help to detoxify iron excess via general upregulation of iron-dependent proteins and processes. In agreement with the iron-detoxifying activity of iron-dependent proteins, overexpression of iron–sulfur cluster enzymes has been shown to attenuate iron toxicity in S. cerevisiae (Li et al, 2011).
HapX levels are significantly higher during iron starvation compared to sufficiency or excess of iron
In contrast to iron sufficiency, A. fumigatus hapX and its orthologs in A. nidulans, F. oxysporum, C. albicans, S. pombe, and C. neoformans, are transcriptionally upregulated by iron starvation (Mercier et al, 2006; Hortschansky et al, 2007; Jung et al, 2010; Schrettl et al, 2010; Hsu et al, 2011; Lopez-Berges et al, 2012). Remarkably, Northern analysis did not detect hapX transcripts during iron excess despite the HapX requirement under this condition (Fig1A). To increase the sensitivity of transcript detection, hapX transcript levels were quantified by qRT-PCR and compared to that of sreA (Fig3A). This analysis confirmed highest hapX expression during iron starvation, i.e. 33-fold higher compared to iron sufficiency, and 17-fold downregulation after a 1-h shift from iron starvation to iron sufficiency. As reported previously (Schrettl et al, 2008), sreA expression was increased (about threefold) during iron sufficiency compared to iron starvation and highly upregulated (about 29-fold) during a 1-h shift from iron starvation to iron sufficiency. During iron excess, a condition in which SreA was previously found to be important for iron resistance (Schrettl et al, 2008; Gsaller et al, 2012), the sreA transcript level was about threefold increased compared to that of hapX (data not shown), i.e. hapX was clearly expressed, although below the Northern sensitivity level.
Figure 3. HapX production decreases during ambient and high-iron availability.
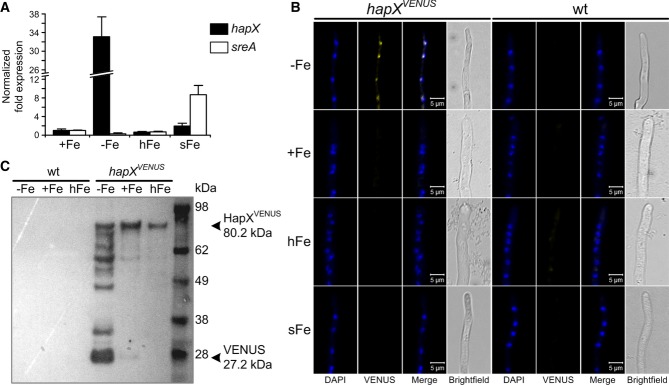
- qRT-PCR revealing iron-dependent sreA and hapX transcript abundance. Transcript levels of hapX and sreA were determined during iron starvation (−Fe), iron sufficiency (+Fe, 0.03 mM), iron excess (hFe, 3 mM) and after a 1-h shift from iron starvation to iron sufficiency (sFe) and normalized to that of γ-actin (AFUA_6G04740) using the
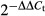 method. Data represent the mean ± SD of two biological and three PCR technical replicates and are presented relative to the transcript levels during iron sufficiency. All differences found are statistically significant with exception of the hapX transcript level during iron sufficiency compared to high-iron conditions (two-tailed, unpaired t-test; P < 0.05).
method. Data represent the mean ± SD of two biological and three PCR technical replicates and are presented relative to the transcript levels during iron sufficiency. All differences found are statistically significant with exception of the hapX transcript level during iron sufficiency compared to high-iron conditions (two-tailed, unpaired t-test; P < 0.05). - In epifluorescence microscopy, HapXVENUS is detectable in the nuclei only during iron starvation. 104 spores of the respective strain were grown in 24-well plates in liquid media at 37°C for 18 h. DAPI was used for staining of nuclei.
- Western blot analysis after GFP-trap enrichment revealing significantly increased HapXVENUS production during iron starvation. The molecular mass of HapXVENUS is 80.2 kDa (27.2 kDa Venus + 53 kDa HapX).
Data information: Strains are derivatives of A. fumigatus AfS77.
To analyze the expression and localization of A. fumigatus HapX at the protein level, we generated an A. fumigatus strain expressing HapX N-terminally tagged with the Venus fluorescent protein (a derivative of yellow fluorescent protein) (Nagai et al, 2002), under the control of the endogenous hapX promoter in single copy at the hapX locus in ΔhapX (strain hapXVENUS). This cured all mutant phenotypes on solid and in liquid media, indicating that the HapXVENUS protein is fully functional (Supplementary Table S1). In agreement with the transcriptional data, in epifluorescence microscopy HapXVENUS was detectable during iron starvation but not during iron sufficiency, iron excess or a 1-h shift from iron starvation to iron sufficiency (Fig3B). As previously observed in A. nidulans and S. pombe (Hortschansky et al, 2007; Mercier & Labbe, 2009), A. fumigatus HapXVENUS localized to the nucleus during iron starvation. These data indicate that lower protein levels of HapX are required for its functions during iron excess compared to iron starvation. Consistently, S-tagged HapX (strain hapXR) was detectable only during iron starvation (Fig4E) but not during iron excess (data not shown) in Western blot analyses. These data also demonstrate that expression pattern-based prediction of gene functions can be misleading. In order to increase the sensitivity of HapX protein detection, we enriched HapXVENUS by GFP-trap, a commercially available GFP pull-down (Rothbauer et al, 2008), before Western blot analysis with a GFP-directed antiserum was applied. This way, the HapXVENUS protein was detected in mycelia grown under iron starvation, iron-replete as well as high-iron conditions with the lowest amount present under high-iron conditions (Fig3C). Under iron starvation, significant HapX proteolyses was found. Most likely, the Venus-HapX degradation was caused during the non-denaturating GFP enrichment procedure. The highly increased degradation during iron starvation conditions can be explained by the strong induction of protease activity during iron starvation conditions (data not shown and Supplementary Table S2). However, it cannot be ruled out that this proteolysis reflects a higher HapX turnover during iron starvation, which might be related to the increased transcript level under this condition. The reduced HapX protein content during iron excess compared to iron starvation might be explained by the reduced number of target genes expressed under this condition.
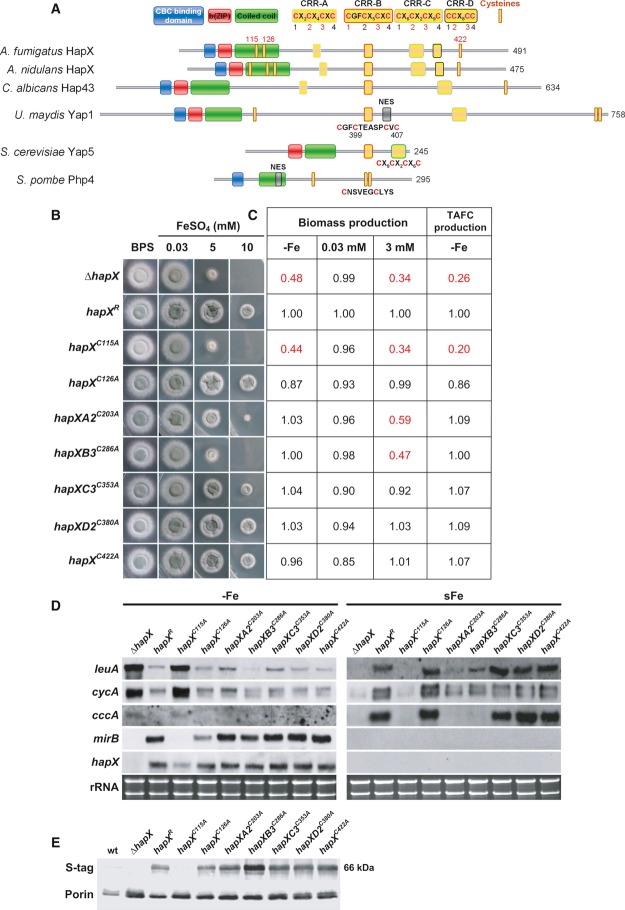
Figure 4. CRR-B and, to a lesser degree, CRR-A are crucial for iron detoxification but not adaptation to iron starvation.
- Schematic view of the HapX Cys and domain organization including comparisons of HapX orthologs from A. fumigatus, A. nidulans, C. albicans, S. pombe, Ustilago maydis Yap1, and S. cerevisiae Yap5.
- Strains were grown for 48 h at 37°C on agar plates with the given iron concentration.
- Production of biomass during iron starvation (−Fe), iron sufficiency (0.03 mM, +Fe), and iron excess (3 mM, hFe), as well as production of siderophores under iron starvation was monitored after liquid growth for 24 h at 37°C. The data represent the mean ± SD of biological triplicates; the values were normalized to that of strain hapXR carrying a non-mutated S-tagged hapX. Statistically significant differences compared to hapXR are shown in red (two-tailed, unpaired t-test; P < 0.05). Original data with standard deviations are given in Supplementary Table S1.
- Northern blot analyses were performed after liquid growth for 24 h at 37°C under iron limitation (−Fe) or after a subsequent 1-h shift into iron sufficiency (sFe). rRNA is shown as a control for RNA quantity and quality.
- Western blot analyses were performed after liquid growth for 24 h at 37°C under iron limitation using antisera recognizing the S-tag for detection of HapX, or porin as control for loading. We were unable to detect S-tagged HapX during iron sufficiency or high-iron conditions with this method (data not shown).
Data information: For simplicity, only one mutant per CRR is shown, the respective, phenotypically identical second mutant is shown in Supplementary Fig S3. Strains are derivatives of A. fumigatus AfS77.
Two cysteine-rich regions (CRR), CRR-A and CRR-B, are crucial for HapX-mediated iron resistance
Aspergillus fumigatus HapX, 491 amino acid residues in length, contains the following domains: a “b(ZIP)” basic and a “coiled-coil” domain, which together mediate DNA-binding in bZIP-type transcription factors, and an N-terminal CBC-binding domain that is essential for HapX function due to its requirement for interaction with the CBC subunit HapE (Hortschansky et al, 2007). Moreover, HapX harbors the enormous number of 19 cysteine residues (Cys), whereby 16 are organized in 4 clusters, termed CRR-A, CRR-B, CRR-C, and CRR-D containing four Cys each (Fig4). Two single Cys (Cys115 and Cys126) are localized in the coiled-coil region, and another one (Cys422) is localized in the C-terminus. The importance of these Cys is supported by their evolutionary conservation, for example all Cys are conserved in seven Aspergillus species (Supplementary Fig S1); CRR-A, CRR-B, CRR-C as well as the C-terminal Cys are conserved even in distantly related fungal species such as C. albicans (Fig4A and Supplementary Fig S2).
Due to the potential role of Cys in iron sensing (Lill et al, 2012), we studied the impact of 11 of these Cys on HapX functions by site-directed mutagenesis replacing Cys by alanine residues (Fig 4 and Supplementary Fig S3). This analysis included all three single as well as two Cys from each CRR. For simplicity, only one mutant per CRR is shown in Fig 4, the respective, phenotypically identical second mutant is shown in Supplementary Fig S3. All analyzed hapX versions, including the non-mutated (strain hapXR), were expressed under the control of the endogenous promoter, contained a C-terminal S-tag (Terpe, 2003) and were integrated at the hapX locus in the ΔhapX strain.
Mutations in CRR-B (strains hapXB1C277A and hapXB3C286A) dramatically decreased adaptation to iron excess, similar to HapX-deficiency (ΔhapX), reflected by decreased radial growth under high-iron conditions, decreased biomass production in high-iron media as well as impaired transcriptional induction of cccA and leuA during a shift from iron starvation to iron sufficiency (Fig 4 and Supplementary Fig S3). Compared to CRR-B mutations, CRR-A mutations (strains hapXA2C203A and hapXA3C208A) similarly impaired the transcriptional response of cccA and leuA to iron, but the growth of the mutant strain was significantly increased on solid as well as in liquid high-iron media (Fig 4 and Supplementary Fig S3). Mutations in CRR-C (strains hapXC2C350A and hapXC3C353A) led to a slightly decreased growth on solid and in liquid high-iron media, but did not affect the transcription pattern of cccA and leuA (Fig 4 and Supplementary Fig S3). Notably, mutation of two different Cys in the same CRR impaired iron resistance to the same degree in CRR-A, CRR-B, and CRR-C (Fig 4 and Supplementary Fig S3) suggesting that the Cys in the same CRR act as a domain rather than individually. Remarkably, neither of the mutations in CRR-A, CRR-B and CRR-C did affect the growth during iron sufficiency or limitation (Fig4). Consistently, siderophore production and transcript levels of HapX-repressed genes (cccA, leuA, cycA) as well as HapX-activated mirB were wild-type-like (strain hapXR) in all these mutants under iron limitation (Fig4). Taken together, these data demonstrate that mainly CRR-B, but to a lower degree also CRR-A and even less CRR-C are required for the HapX functions in iron detoxification, while these CRR are dispensable for the HapX functions in adaptation to iron starvation.
Mutation of Cys126 (strain hapXC126A) slightly decreased liquid biomass production and TAFC production during iron starvation but did not impact HapX functions in any other assays performed (Fig4). Mutation of Cys422 (strain hapXC422A) slightly decreased liquid biomass production under both iron starvation and excess but did not cause any other alterations (Fig4). Mutations in CRR-D (strains hapXD2C380A and hapXD3C389A) were phenotypically inconspicuous in all analyses performed (Fig4).
In contrast, mutation of the Cys115 (strain hapXC115A) phenocopied HapX-deficiency during both iron limitation and iron excess (Fig4). The most likely explanation is that this mutation results in the loss of the HapX protein as seen in Fig4E, possibly due to improper folding of this HapX derivative. Noteworthy, this mutation results in a decrease of hapX transcript level during iron starvation suggesting positive transcriptional autoregulation of HapX.
The HapX C-terminus is essential for the adaption to iron starvation
To further characterize HapX domains, we generated A. fumigatus strains expressing different C-terminal hapX truncations (strains hapX464 – hapX158), here untagged, under the control of the endogenous promoter and integrated at the hapX locus in ΔhapX (Fig5D).
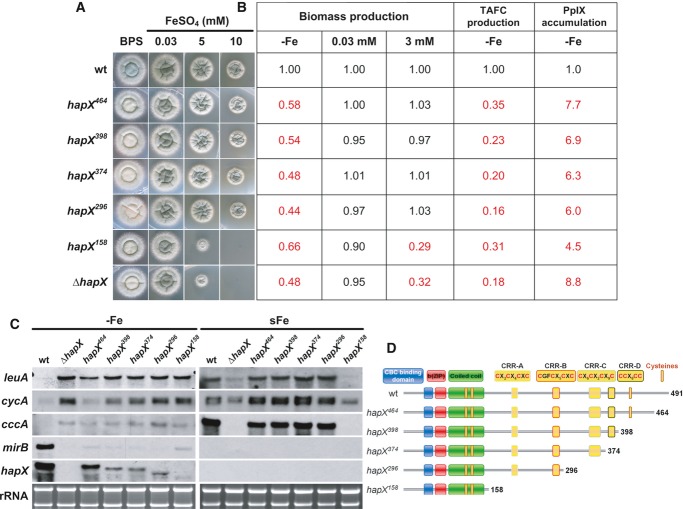
Figure 5. The HapX C-terminus is crucial for adaption to iron starvation but not iron detoxification.
- Strains were grown for 48 h at 37°C on agar plates with the given iron concentration.
- Production of biomass during iron starvation (−Fe), iron sufficiency (0.03 mM, +Fe), and iron excess (3 mM, hFe), as well as production of siderophores and PpIX under iron starvation was monitored after liquid growth for 24 h at 37°C. The data represent the mean ± SD of biological triplicates; the values were normalized to the wild-type. Statistically significant differences compared to the wild-type are shown in red (two-tailed, unpaired t-test; P < 0.05). Original data ± SD are found in Supplementary Table S1.
- Northern blot analyses were performed after liquid growth for 24 h at 37°C under iron limitation (−Fe) or after a subsequent 1-h shift to iron sufficiency (sFe). rRNA is shown as a control for RNA quantity and quality.
- Schematic view of the HapX truncations analyzed.
Data information: Strains are derivatives of A. fumigatus AfS77.
Truncation of the C-terminal 27 amino acid residues (strain hapX464) decreased sporulation on BPS agar plates (Fig5A). In iron-limited liquid media, this truncation decreased biomass production, TAFC production, mirB transcript levels but increased protoporphyrine IX (PpIX) accumulation and transcript levels of cccA and leuA (Fig5). Taken together, this truncation was similar to a ΔhapX phenotype during iron starvation, however, less severe (with regard to biomass and TAFC production as well as the cycA transcript level), but did not affect growth and transcription of iron-related genes under iron excess, i.e. cccA and leuA (Fig5).
Truncation of the C-terminal 93, 117, and 195 amino acid residues (strains hapX389, hapX374 and hapX296, respectively) perfectly phenocopied ΔhapX under iron limitation but did not affect iron detoxification (Fig5). The latter is consistent with the presence of CRR-A and CRR-B that are crucial for iron resistance in the respective HapX versions (see above). The functionality of the C-terminus is supported by the high evolutionary conservation not only in Aspergillus species (Supplementary Fig S1) but also for example in C. albicans with 41% identical amino acids in the C-terminal 66 amino acid residues (data not shown).
Truncation of the C-terminal 333 amino acid residues (strain hapX158) impaired iron detoxification to the same extent as HapX-deficiency, which is in agreement with the lack of CRR-A and CRR-B (Fig5). This truncation also reduced adaptation to iron starvation, but not to the same extent as seen in hapX464 – hapX296 or ΔhapX, i.e. during iron limitation liquid biomass and TAFC production as well as the mirB transcript level were higher, while the PpIX accumulation was lower. These data indicate that this HapX version, comprised of only the CBC-binding domain and the bZIP region, still executes residual functions in activation of siderophore biosynthesis and repression of iron consumption. These functions appear to be masked in the HapX versions encoded by hapX464 – hapX296.
Notably, hapX398 – hapX158 displayed not only decreased transcriptional activation of mirB but also decreased hapX transcript levels as seen in the HapX loss of function hapXC115A mutant (see above). These data indicate positive transcriptional autoregulation of HapX.
Taken together, both the cysteine-to-alanine mutations and the C-terminal truncations indicate that CRR-A and CRR-B are required for HapX-mediated iron detoxification, while the C-terminal 93 amino acid residues are crucial for both activation as well as repression functions that are required for adaptation to iron starvation.
An evolutionary conserved cccA promoter element is recognized by a protein complex consisting of the CBC and a HapX homodimer involving direct DNA binding by both the CBC and HapX
To identify putative, evolutionary conserved, regulatory motifs in the cccA promoter, the 1-kb 5′-upstream regions of cccA homologs from 28 fungal species including A. fumigatus, A. nidulans and F. oxysporum were subject to MEME analysis (Bailey & Elkan, 1994). The identified sites and their positions in the promoters of the different species are shown in Supplementary Fig S4. The highest scoring sequence (e-value of 3.4 × 10−115), present in all 28 species, was a bipartite motif separated by a spacer region with low conservation (Fig6A). Consistent with the HapX-independent regulation, the highest scoring motif was not found in the promoter of the S. cerevisiae cccA homolog (data not shown). The 5′-conserved submotif matches the CBC consensus DNA-binding motif (CCAAT box), CCAAT(C/T)(A/G) (Huber et al, 2012). This is in perfect agreement with the previous finding that HapX acts via physical interaction with the CBC (Hortschansky et al, 2007). Interestingly, binding of the CCAAT box by CBC would cover the entire spacer region as identified in the CBC/DNA binary complex crystal structure (Huber et al, 2012), which indicates that the 3′-submotif is the first accessible region for binding of another DNA-binding protein. The 3′-conserved non-palindromic submotif does not match any known transcription factor consensus binding sequence. This is intriguing, because bZIP proteins usually bind short palindromic or pseudo-palindromic target sequences. Furthermore, based on the amino acid signature sequence of its basic region NXXAQXXFR (Supplementary Fig S1), HapX belongs to the Pap1/Yap1 subfamily of bZIP transcription factors that are known to recognize TTACGTAA and TTAGTAA consensus motifs (Fujii et al, 2000).
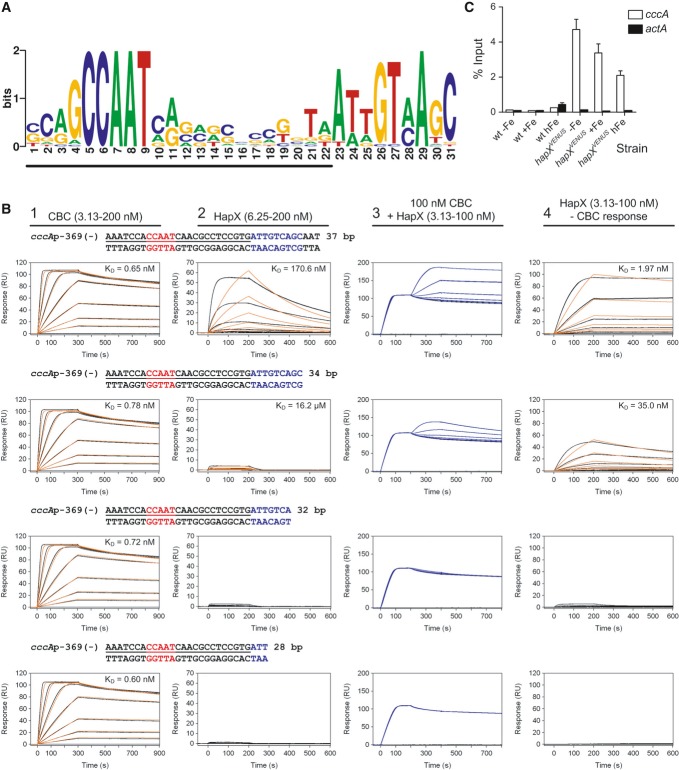
Figure 6. HapX binds in vitro and in vivo to an evolutionary conserved motif identified in promoters of cccA homologs.
- An evolutionary conserved, bipartite motif in promoters of cccA homologs identified by MEME analysis. The underlined nucleotides would be covered upon CBC-binding as can be predicted based on the identified CBC/DNA binary complex crystal structure (Huber et al, 2012).
- Real-time SPR characterization of in vitro formation of the CBC/DNA-HapX ternary complex on the conserved cccA promoter motif from A. fumigatus. SPR analyses included binding of the CBC to DNA (panel 1), HapX to DNA (panel 2) and HapX to preformed CBC/DNA complexes (panel 3). The SPR sensorgrams are shown from sensor-immobilized 37 base pair duplexes covering the full as well as 3′-truncated duplexes. Nucleotides marked in blue represent the HapX consensus binding site in fungal cccA promoters identified by MEME analysis. Binding responses of the indicated CBC or HapX concentrations injected in duplicate (black lines) are shown overlaid with the best fit derived from a 1:1 interaction model including a mass transport term (red lines). Binding responses of CBC/DNA-HapX ternary complex formation (panel 3, blue lines) were obtained by concentration-dependent co-injection of HapX on preformed binary CBC/DNA complexes after 200 s within the steady-state phase. Sensorgrams in panel 4 depict the association/dissociation responses of HapX on preformed CBC/DNA and were generated by CBC response (co-injection of buffer instead of HapX) subtraction from HapX co-injection responses. Dissociation constants (KD) are plotted inside the graphs.
- ChIP analysis demonstrating in vivo binding of HapX to the conserved cccA promoter motif from A. fumigatus. ChIP qPCR was performed on wild-type or the strain containing Venus-tagged HapX (hapXVENUS) grown for 18 h, then shifted to fresh media with no iron (−Fe), 0.03 mM iron (+Fe), or 3 mM iron (hFe) for 8 h. DNA was immunoprecipitated with either a control IgG antibody, or anti-GFP polyclonal antibody that recognizes the Venus protein. Binding of HapXVENUS to the DNA region was assessed by qPCR. HapX binding is represented as percent enrichment of input control samples ± SD from triplicates. The actA (actin) promoter served as a negative control.
Nevertheless, we postulated that the 3′-submotif might be recognized by HapX. To address this hypothesis, we overexpressed and purified the A. fumigatus CBC (comprising the conserved domains of subunits HapB, HapC, and HapE) as well as a peptide corresponding to residues 24–158 of HapX, which includes the CBC-binding domain, basic region, and coiled-coil domain (Supplementary Fig S5A). Light scattering analysis of purified HapX (24–158) revealed a molar mass of 31.38 kDa, demonstrating that this domain is dimeric in solution (theoretical mass of 31.36 kDa), as expected for a bZIP protein (Supplementary Fig S5B). To examine the putative in vitro interaction of the CBC, HapX and the identified common promoter element of cccA homologs, we applied surface plasmon resonance (SPR) analyses (Fig6B). These measurements indicated high-affinity (KD = 0.7 nM) recognition of the CCAAT box by the A. fumigatus CBC independent of the presence of the 3′-submotif, i.e. its binding affinity was not affected by truncation of the 3′-submotif (compare first column in Fig6B). This affinity is similar to that found for the interaction of the A. nidulans CBC with a CCAAT motif from the sreA promoter (Thon et al, 2010). HapX binds the 3′-submotif with low affinity (KD = 170.6 nM) as its binding was abolished by truncation of the 3′-submotif (compare second column in Fig6B). However, on CBC-coated DNA, the binding affinity of HapX was 87-fold increased (KD ≈ 1.97 nM), whereby the binding again strictly depended on the presence of the 3′-submotif (compare fourth column in Fig6B). Furthermore, by taking advantage of the fact that SPR responses correspond to bound masses, we were able to unravel the stoichiometry of the CBC/DNA-HapX ternary complex. Saturating CBC responses on the 37-bp DNA duplex reached a value of ≈ 100 RU (upper left sensorgram in Fig6B) and nearly the same responses were observed by co-injection of HapX on preformed CBC/DNA complexes (upper right sensorgram in Fig6B) due to the similar molecular masses of the CBC (33.44 kDa) and the HapX dimer (31.36 kDa). Therefore, we conclude that one binary CBC/DNA complex is bound by one HapX dimer. Taken together, our data demonstrate that HapX interacts in vitro not only with the CBC but also with DNA with 2:1:1 stoichiometry by recognizing the 3′-submotif located adjacent to the CCAAT box in the evolutionary conserved cccA promoter element.
Chromatin immunoprecipitation (ChIP) analysis confirmed that HapX also binds to this promoter element in vivo, notably independent of the ambient iron concentration (Fig6C). The constitutive presence of HapX at the cccA promoter suggests that transcription of cccA is primarily mediated by post-translational modification of HapX, i.e. iron sensing, rather than by transcriptional regulation of hapX. In agreement, the potential HapX iron-sensing CRR-B motif, CGFCX5CXC, is essential for the transcriptional activation of cccA in response to high-iron stress (see above).
In summary, we show that HapX not only physically interacts with the CBC but also directly recognizes a distinct DNA motif. As the CBC has numerous HapX/iron-independent functions (Kato, 2005; Fleming et al, 2013), these data reveal for the first time the mechanism for discrimination of general CBC and specific HapX/CBC target genes.
Both functions, adaptation to iron limitation and excess, are evolutionary conserved in HapX orthologs
Similar to A. fumigatus, HapX orthologs repress iron-dependent pathways and the cccA orthologs during iron starvation in A. nidulans, F. oxysporum, S. pombe, C. neoformans, and C. albicans (Mercier et al, 2006; Hortschansky et al, 2007; Jung et al, 2010; Schrettl et al, 2010; Hsu et al, 2011; Lopez-Berges et al, 2012). Here, we found that HapX-deficiency also impairs growth of A. nidulans and F. oxysporum on high-iron media (Fig7) demonstrating that the function of HapX in iron detoxification is evolutionary conserved. Inspection of genome-wide transcriptional profiling data indicated that the cccA ortholog FOXG_04047 in F. oxysporum is repressed similar to A. fumigatus during iron starvation and upregulated during iron sufficiency in a HapX-dependent manner (Lopez-Berges et al, 2012). These data suggest that in F. oxysporum the decreased iron resistance caused by HapX-deficiency also results from impaired vacuolar iron storage.
Figure 7. HapX-mediated iron detoxification is evolutionary conserved.
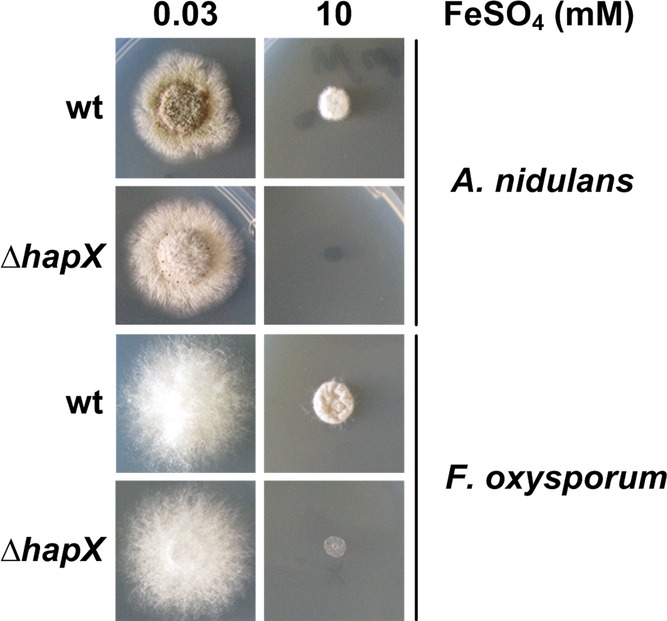
A. nidulans and F. oxysporum deletion mutants were grown for 48 h at 37°C on agar plates with the given iron concentration.
In agreement with the evolutionary conserved function in iron resistance, CRR-A and CRR-B, which were identified in this study to be crucial for transcriptional activation of cccA and consequently iron detoxification in A. fumigatus, are conserved in most HapX orthologs (Fig4A and Supplementary Fig S2).
In S. cerevisiae, the cccA ortholog, ccc1, is post-transcriptionally repressed during iron starvation by Cth1/2-mediated mRNA decay (Puig et al, 2005), which is transcriptionally induced by Aft1. Transcriptional activation during iron excess is mediated by the bZIP-type transcription factor Yap5. S. cerevisiae lacks HapX and SreA orthologs and, vice versa, A. fumigatus lacks orthologs of Aft1, Cth1/2, and Yap5 (Haas et al, 2008). Like A. fumigatus HapX, S. cerevisiae Yap5 comprises a bZIP domain. However, Yap5 lacks a CBC-binding domain, does not function via interaction with the CBC, and is involved in adaptation to iron excess but not iron starvation (Li et al, 2008). Thus, transcriptional activation of vacuolar iron storage is mediated by different regulatory mechanisms in S. cerevisiae and A. fumigatus. Nevertheless, it is particularly interesting that CRR-B, which is mainly important for the function of HapX in iron resistance, shows significant similarity to a CRR that is likewise essential for the iron resistance function of S. cerevisiae Yap5 (Fig4A).
The common CRR-B motif, CGFCX5CXC, which is essential for the function in iron resistance of HapX and Yap5, is reminiscent of the CGFS motif found in monothiol glutaredoxins. Here, this motif is essential for the formation of [2Fe-2S]-bridged homodimers (Picciocchi et al, 2007; Bandyopadhyay et al, 2008; Iwema et al, 2009) that participate in mitochondrial and cytosolic iron–sulfur protein biogenesis (Rouhier et al, 2010), delivery and transfer of iron–sulfur clusters into proteins and subcellular compartments (Muhlenhoff et al, 2010), and the relay of the cellular iron status to iron-responsive transcription factors (Rutherford et al, 2005; Ojeda et al, 2006; Pujol-Carrion et al, 2006; Kaplan & Kaplan, 2009). Consistent with a role of CRR-B in sensing the cellular iron status via iron–sulfur clusters in S. cerevisiae Yap5, and potentially A. fumigatus HapX, inactivation of iron–sulfur cluster biosynthesis blocks Yap5 activation in yeast (Li et al, 2012). Moreover, the second CRR present in Yap5 shows similarity to CRR-C of HapX (Fig4A).
Php4, which transcriptionally represses iron-dependent pathways in S. pombe (Vachon et al, 2012), is an atypical HapX ortholog comprising a CBC-binding domain and a coiled-coil domain, but lacking the basic DNA-binding region of the bZIP domain. Only three Cys are present with two being clustered in the sequence CNSVEGCLYS (Fig4A). In a two-hybrid approach, the first of these Cys, Cys172, was found to be essential for iron-dependent association of Php4 with the monothiol glutaredoxin Grx4, indicating a role in iron sensing via iron–sulfur cluster availability. However, the function of this Cys has not been analyzed directly in Php4.
Recently, sulfur starvation caused by deficiency in the sulfur regulator MetR was found to increase the cellular iron content, iron sensitivity and transcript levels of genes involved in iron uptake but to decreased the cccA transcript level (Amich et al, 2013). These data demonstrate that sulfur homeostasis is required for proper iron regulation, most likely via iron-sulfur cluster and/or glutathione biosynthesis as shown for S. cerevisiae and S. pombe (Li & Outten, 2012). Together with the results presented here, these data furthermore indicate that sulfur homeostasis is required not only for SreA-mediated regulation of iron uptake but also HapX-mediated regulation of vacuolar iron detoxification.
Taken together, CRR-A and -B, but not the C-terminus, are required for sensing of high-iron conditions by HapX to mediate activation of vacuolar iron detoxification via a conformational change of and/or interaction with different protein partners compared to iron starvation conditions. In contrast, the C-terminal 93 amino acid residues, but not CRR-A and CRR-B, are crucial for both the activating (e.g. mirB) as well as repressing (e.g. cccA) functions of HapX during iron starvation. The intriguing question why some promoter interactions result in repression and some in activation has still to be resolved; again this might be dependent on interaction with different partners present in different promoter sets.
Strikingly, certain domains of HapX and Yap5 are also found in the Ustilago maydis peroxide sensor, named Yap1 (Molina & Kahmann, 2007). This protein was identified as an ortholog of Yap1 from S. cerevisiae and regulates the oxidative stress response of the plant pathogen. A closer inspection of the Yap1 amino acid sequence revealed the presence of common HapX domains, i.e. the N-terminal CBC-binding and bZIP domains, a central HapX/Yap5 CRR-B type motif and the C-terminal CRR-C domain. Two additional Cys (CPC-motif) are located close to the C-terminus, whereas HapX-like domains CRR-A and CRR-D are lacking. Within the CRR-B domain, Cys399, and Cys407 were found to be crucial for nuclear localization and function of U. maydis Yap1, probably due to oxidative masking of a putative nuclear export sequence located between CRR-B and CRR-C (Fig4A and Supplementary Fig S2) (Delaunay et al, 2000). Taken together, it seems that different fungi use a toolbox-like set of domains for sensing distinct environmental stimuli. Furthermore, these data indicate a possible role of Yap1 as a Yap1/HapX chimera in regulation of U. maydis' iron homeostasis.
Conclusion
Collectively, our study uncovered a novel regulatory mechanism in A. fumigatus and other filamentous fungi mediating both iron resistance and adaptation to iron starvation by the same transcription factor complex, comprising the CBC and a HapX homodimer, with activating and repressing functions depending on ambient iron availability. Comparison of HapX target promoter regions coupled with protein-DNA interaction analysis identified an evolutionary conserved CBC/HapX binding motif and revealed the discriminatory mechanism for CBC and CBC/HapX targets. Moreover, mutational analysis identified HapX protein domains that are essential for adaptation to either limitation or excess of iron, which will be instrumental for the further characterization of iron sensing.
Most fungal species encode HapX orthologs indicating wide evolutionary conservation of this iron regulatory mechanism. In contrast, the role model S. cerevisiae lacks a HapX ortholog and employs iron regulatory transcription factors that are not found in most other fungal species (Haas et al, 2008; Kaplan & Kaplan, 2009). In addition to the high-iron-sensing Yap5, iron regulation in S. cerevisiae involves the low-iron-sensing paralogous transcription factors Aft1 and Aft2. Aft1 and Aft2 transcriptionally activate iron acquisition as well as Cth1 and Cth2. These two paralogous mRNA-binding proteins mediate post-transcriptional repression of iron consumption by promoting respective mRNA degradation, including the homolog of A. fumigatus cccA (Martinez-Pastor et al, 2013). These data demonstrate complete regulatory rewiring of vacuolar iron storage in this yeast compared to A. fumigatus, with different regulators being required for activation and repression. Nevertheless, the mechanistically different iron regulatory systems of A. fumigatus and S. cerevisiae seem to employ common protein motifs for mediation of iron regulation. As an example of the modular toolbox using a common protein motif for transmitting different signals, the N-terminal CBC-binding domain of HapX is also present in S. cerevisiae Hap4. Hap4 was recently suggested to participate in iron regulation (Ihrig et al, 2010). In contrast to HapX, however, Hap4 lacks DNA binding and CRRs and therefore its mode of action is significantly different from the HapX mechanism (McNabb & Pinto, 2005; Hortschansky et al, 2007). In mammals, iron acquisition (transferrin receptor) and iron detoxification (ferroportin-mediated iron export and ferritin-mediated iron storage) are coupled at the post-transcriptional level by responding inversely to binding of iron regulatory proteins (IRPs) to iron-responsive elements in the untranslated regions of respective mRNA′s (Wang & Pantopoulos, 2011).
Taken together, the comparison of different organisms underlines the essentiality of iron handling with similar readouts mediated by different regulatory mechanisms.
Materials and Methods
Strains, oligonucleotides, and growth conditions
All strains and oligonucleotides used in this study are listed in Supplementary Tables S3 and S4, respectively. Generally, A. fumigatus strains were cultivated at 37°C in Aspergillus minimal medium (AMM) according to Pontecorvo et al (1953) containing 1% (w/v) glucose and 20 mM glutamine as carbon and nitrogen sources, respectively. For iron-depleted conditions, iron was omitted and iron amounts used in this study are given in the figures. To increase iron starvation in solid media, the ferrous iron chelator BPS was used at a final concentration of 0.2 mM in iron-depleted media. Production of conidia was performed on AMM agar plates containing 0.03 mM FeSO4. Depending on the transformed resistance gene, A. fumigatus strains were selected on media containing 0.1 μg/ml pyrithiamine or 0.2 mg/ml hygromycin B.
Generation of a hapX mutant strain with conditional cccA expression
For heterologous expression of cccA, the plasmid pxylPPcccA (Gsaller et al, 2012) was transformed in A. fumigatus ΔhapX (ATCC 46645) (Supplementary Fig S6). The plasmid harbors cccA under control of xylPP, a xylan/xylose-inducible promoter derived from the xylanase xylP of Penicillium chrysogenum (Zadra et al, 2000). pAN7.1 containing a hygromycin resistance cassette (hph) was co-transformed for selection of transformants.
Generation of a hapX deletion mutant in ΔakuA background and a ΔhapXΔcccA double deletion strain
The hapX coding sequence was deleted in AfS77 using the bipartite marker technique (Supplementary Fig S7A) (Nielsen et al, 2006). AfS77 was transformed with two DNA fragments each containing overlapping but incomplete fragments of the pyrithiamine resistance-conferring gene ptrA as described previously (Schrettl et al, 2010) yielding strain ΔhapX (Supplementary Fig S7A). Deletion constructs were amplified with primers oAfhapX-5/oAoPtrA2 (PCR1, 2.6 kb) and oAfhapX-6/oAoPtrA1 (PCR2, 2.2 kb) using genomic DNA of ΔhapX (ATCC 46645 derivative) as template.
Disruption of cccA in ΔhapX background (AfS77) was also carried out using the bipartite marker technique (Supplementary Fig S7B). For this purpose, the cccA 5′-flanking region was amplified from genomic DNA using primers oAfcccA1/oAfcccAr4. For amplification of the 3′-flanking region, primers oAfcccA3/oAfcccAr2 were employed. Generated DNA fragments were digested with StuI (5′-flanking region) and HindIII (3′-flanking region). The hygromycin resistance cassette was released from plasmid pAN7.1 by digestion with StuI and HindIII and ligated with the 5′- and 3′-flanking region, respectively. The transformation construct A (3.3 kb, fusion of the cccA 5′-flanking region and the hph split marker) was amplified from the ligation product using primers oAfcccA5 and ohph14. For amplification of the transformation construct B (2.8 kb, fusion of the cccA 3′-flanking region and the supplementary hph split marker) primers oAfcccAr6 and ohph15 were employed. For transformation of A. fumigatus strains, both constructs A and B were used simultaneously. ΔhapX (AfS77) was transformed with the fragments each containing overlapping but incomplete fragments of the hygromycin resistance gene hph, yielding strain ΔhapXΔcccA.
ΔhapX complementation, site-directed mutagenesis, C-terminal truncation, S-tagging, and venus-tagging of HapX
For these studies, the basic plasmid phapXR-hph was generated employing the fusion PCR-technique (Nielsen et al, 2006). The resulting plasmid contains the hapX coding sequence C-terminally linked with an S-tag under control of its native promoter and terminator region as well as a hygromycin resistance cassette. The different plasmid versions were integrated at the hapX deletion locus in AfS77 ΔhapX (Supplementary Fig S8A).
As a first step, the sequence encoding the S-tag (72 bp), including a 5′-sequence coding for a GA4-linker and a 3′-stop codon (TAA), was PCR amplified from plasmid pAO81 (Yang et al, 2004) using primers oAfhapX-S1 and oAfhapX-S2 (Supplementary Table S4). These primers carry extensions, which are complementary to sequences 30 bp upstream (oAfhapX-S1) and downstream (oAfhapX-S2) of the hapX stop codon (TGA), thereby generating a 132-bp-long PCR product. Primers oAfhapX-1 and oAfhapX-S3 were employed to amplify 2.9 kb of genomic DNA sequence, comprising 5′-flanking region (including the hapX promoter) and the hapX gene lacking the endogenous stop codon (TGA). Primers oAfhapX-S2 and oAfhapX-S4 were used to amplify 1.0 kb of genomic sequence, comprising 3′-flanking region (hapX terminator region). After gel purification, equal molar amounts of the PCR products were applied as template for fusion PCR. By using the (nested) primers oAfhapX-7 and oAfhapX-8 a 3.7 kb long fragment was amplified, which contains the hapX promoter, the hapX coding sequence comprising the S-tag, and the hapX terminator region. Subsequently, 3′ A-overhangs were added and the construct was subcloned into pGEM-T-Easy (Promega) via T/A cloning, yielding phapXR (6.7 kb). Finally, a hygromycin resistance cassette was subcloned in phapXR, yielding plasmid phapXR-hph. Therefore, a DNA fragment containing the resistance cassette (2.4 kb) was amplified with primers ohyg-1/ohyg-2 using pAN7.1 as template. Next, phapXR was opened with SphI and blunt-ended using the Klenow enzyme (NEB). Eventually, the amplified DNA fragment was phosphorylated with polynucleotide kinase (NEB) and ligated into the blunt-ended plasmid backbone.
In order to substitute specific amino acids by site-directed mutagenesis the QuikChange kit (Stratagene) and oligonucleotides listed in Supplementary Table S4 were used. For the introduction of each mutation (Fig4B and Supplementary Fig S3), complementary primers, around 40 bp in length including the desired mutation in its center, were designed. Plasmid phapXR-hph was amplified with respective primers in a PCR with a total volume of 50 μl (18 cycles, TDen = 95°C, TMelt = 56°C, TElong = 72°C). Next, 1 μl DpnI restriction enzyme was added to the solution followed by 3 h of incubation at 37°C. E. coli DH5α cells were transformed with 10 μl of the digested solution. After amplification of plasmid DNA, plasmids containing mutated coding sequences were screened by digestion. Resulting plasmids were named phapXC115A-hph, phapXC126A-hph, phapXA2C203A-hph, phapXA3C208A-hph, phapXB1C277A-hph, phapXB3C286A-hph, phapXC2C350A-hph, phapXC3C353A-hph, phapXD2C380A-hph, phapXD3C389A-hph, and phapXC422A-hph.
For C-terminal truncation of HapX, reverse primers were designed that anneal to the hapX coding sequence at specific sites (Supplementary Table S4). Additionally, reverse primers comprise a STOP codon and a BstBI recognition site. Each PCR was performed with the same forward primer – hapXtrunc-f. Amino acid lengths of truncated HapX proteins (full-length in Af293 = 491 aa) are listed in Supplementary Table S3 and Fig5D. After amplification, PCR fragments were digested with XbaI/BstBI and subcloned into XbaI/BstBI opened phapXR-hph. Plasmids comprising the truncated hapX versions were designated phapX464-hph, phapX398-hph, phapX374-hph, phapX296-hph, and phapX158-hph.
For N-terminal tagging of HapX with Venus fluorescent protein plasmid phapXVENUS-hph was generated (Supplementary Fig S8B). Therefore, hapX promoter DNA was amplified using primers 5′hapX-f/5′hapXvenus-r (PCR1, 1.3 kb; template: phapXR-hph) (Supplementary Table S4). Primers 5′hapXvenus-f/venus-r were employed to amplify the codon optimized venus coding sequence (PCR2, 0.7 kb; template: pMA-Venus). In a third PCR, hapX coding sequence comprising the 3′ untranslated region was amplified using primers venushapX-f/hapX3′-r (PCR3, 1.6 kb; template: phapXR-hph). Subsequently, the hapX promoter region and Venus coding sequence were combined via fusion PCR using primers 5′hapX-f2/venushapX-r (PCR4, 2.1 kb; template: PCR1 & PCR2). In the final PCR, hapX promoter DNA linked to venus coding sequence was fused to hapX coding sequence including the 3′ terminator region with primers hapXtrunc-f/hapX-r (PCR5, 3.5 kb; template PCR3 & PCR4). The resulting DNA fragment was digested with XbaI/BstBI and subcloned into XbaI/BstBI opened phapXR-hph. The 5′hapX promoter region includes an XbaI recognition site and primer hapX-r contains the palindromic recognition sequence for BstBI.
For transformation, 5 μg of the respective plasmid was linearized through digestion with SnaBI, the recognition site of which is located 761 bp downstream of the stop codon.
Fluorescence microscopy, PpIX analysis, siderophore analysis, Northern analysis, and qRT-PCR analysis
For imaging of hyphae, cells were grown on coverslips in 0.3 ml AMM containing 1% (w/v) glucose, 20 mM glutamine and the desired iron concentration. Microscopy images were captured using an Axio Imager. M2 microscope (Carl Zeiss) equipped with a 63× oil immersion objective lens (numerical aperture, 1.40), a HPX 120 V compact light source (Carl Zeiss) and the AxioCam MRm camera (Carl Zeiss). Images were processed using ZEN 2012 imaging software (Carl Zeiss). DAPI was used to stain nuclei. PpIX content, siderophore production, RNA isolation, and Northern analysis were carried out as described previously (Hortschansky et al, 2007). The hybridization probes used in this study were generated by PCR using DIG-labeled nucleotides. For qRT-PCR analysis, RNA was digested with DNase I and column eluted using RNA Clean & Concentrator™-25 kit (ZYMO Research). cDNA was synthesized using GoScript™ Reverse Transcription System (Promega) and random primers. qPCR was performed in a StepONE Plus Instrument (Applied Biosystems) with POWER SYBR® Green PCR Master Mix (Applied Biosystems). Primers for hapX, sreA, and actA are listed in Supplementary Table S4.
Immunoprecipitation of Venus-HapX fusion protein from A. fumigatus cell extracts
For protein extraction from AfS77 (wt) and hapXVENUS mycelia, a modification of a published procedure was used (Liu et al, 2010). Briefly, mycelia were washed, frozen in liquid nitrogen, lyophilized and homogenized by grinding with mortar and pestle. 100 mg cell powder was resuspended in 1 ml lysis buffer (25 mM Tris/HCl, 300 mM NaCl, 0.5% (v/v) NP-40, 5 mM EDTA, 15 mM EGTA, 1 mM AEBSF, 1 mM DTT, 1× cOmplete protease inhibitor (Roche), pH 7.5). Extracts were cleared by two centrifugation steps (30 and 10 min at 53,200 × g at 4°C). Twenty-five microliter of pre-equilibrated GFP-Trap agarose beads (ChromoTek) were added to the supernatant and incubated for 1 h on an end-over-end rotor at 4°C. The supernatant was removed by centrifugation, and the beads were washed four times with wash buffer (25 mM Tris/HCl, 300 mM NaCl, 5 mM EDTA, 1 mM AEBSF, 1× complete protease inhibitor (Roche), pH 7.5) followed by two washing steps with water. Bound proteins were eluted with 100 μl of 10% (v/v) acetonitrile, 5% (v/v) acetic acid. 50 μl of each protein eluate was dried down in a vacuum centrifuge, boiled in 1× SDS-PAGE sample buffer, and loaded onto 4–12% NuPAGE Bis-Tris gels. Proteins were transferred to a PVDF membrane using the iBlot system (Invitrogen). After antibody incubation, the membrane was developed using 1-Step Ultra TMB-Blotting substrate (Thermo). The following antibodies were used: anti-GFP antibody (ab290, Abcam); HRP-conjugated anti-rabbit IgG antibody (GGHL-15P, ICL, Inc.).
Western blot detection of S-tagged HapX
For Western blots, 100 mg dry weight of lyophilized mycelia were rehydrated in 1 ml of TNETG buffer (20 mM Tris, pH 7.4, 2.5 mM EDTA, 150 mM NaCl, 10% (v/v) glycerol, 0.5% (v/v) Triton X-100, 1 mM PMSF). Cells were lysed by 5 bursts of 1 min each in the presence of 1/3 volume of glass beads. Cell debris were removed by centrifugation. For Western blots, 50 μg protein were loaded per lane and separated on a 12.5% SDS-PAGE. Blots were immuno-decorated with a polyclonal anti-S-tag antibody (ICL, Inc.) for detection of S-tagged HapX or a polyclonal antibody directed against S. cerevisiae Porin (Por1).
MEME analysis
Protein blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify the most similar homologs of A. fumigatus CccA (XP_751578.1). The 1-kb 5′-upstream sequences of the top 26 hits from unique species (Supplementary Fig S6) as well as A. fumigatus and F. oxysporum, which is known to require HapX for iron detoxification (see above), were subject to MEME analysis (Bailey & Elkan, 1994) to identify putative common motifs of size 6–35 bp, occurring once or zero times.
Bacterial expression and purification of the CBC and HapX for SPR analysis
The A. fumigatus core CBC was produced and purified as described for the CBC from A. nidulans (Huber et al, 2012). Briefly, synthetic genes coding for HapC(40–137) and HapE(47-164) were cloned in the pnCS vector (Diebold et al, 2011) for expression of a bicistronic transcript. A synthetic gene encoding HapB(230–299) was cloned into pET39b (Novagen) and E. coli BL21(DE3) cells were co-transformed with both plasmids. After overnight autoinduction and cell lysis, the heterotrimeric CBC was purified to homogeneity by subsequent cobalt chelate affinity and size-exclusion chromatography.
A cDNA fragment encoding A. fumigatus HapX(24–158) (covering the CBC-binding domain, basic region, and coiled-coil domain) with an extended N-terminus including a cleavage site for tobacco etch virus (TEV) protease was amplified and subcloned into the pMAL-c2X (New England Biolabs) vector. The resulting plasmid was transformed into E. coli Rosetta 2 (DE3) cells for overnight autoinduction. Crude bacterial lysates were purified by Dextrin Sepharose affinity chromatography (GE Healthcare). The maltose-binding protein HapX(24–158) fusion was cleaved with TEV protease and further purified sequentially using CellufineSulfate (Millipore) affinity chromatography, (NH4)2SO4 precipitation (50% w/v), and Superdex 75 prep grade (GE Healthcare) size exclusion chromatography. The absolute molecular mass of HapX(24–158) was determined by static light scattering experiments on a miniDawn TREOS monitor in series with an Optilab T-rEX differential refractometer (Wyatt). HapX(24–158) was chromatographed on a Superdex 200 10/300 GL column (GE Healthcare), and molar mass was calculated using ASTRA 6 software (Wyatt).
Surface plasmon resonance measurements
Real-time analyses were performed on a Biacore T200 system (GE Healthcare) at 25°C. DNA duplexes containing CCAAT box at position −369 in the 5′-upstream region of the A. fumigatus cccA gene were produced by annealing complementary oligonucleotides using a fivefold molar excess of the non-biotinylated oligonucleotide. The dsDNA was injected on flow cells of a streptavidin (Sigma)-coated CM3 sensor chip at a flow rate of 10 μl/min until the calculated amount of DNA that gives a maximum CBC-binding capacity of 100 RU had been bound. CBC and HapX(24–158) samples containing 10 μg/ml poly(dI-dC) were injected in running buffer (10 mM phosphate pH 7.4, containing 2.7 mM KCl, 137 mM NaCl, 1 mM DTT and 0.005% (v/v) surfactant P20) at a flow rate of 30 μl/min. Co-injection of HapX(24–158) on preformed binary CBC/DNA complexes within the equilibrium phase was performed by using the dual injection command. Each injection was performed at least two times. The chip surface was regenerated with 10 mM Tris/HCl pH 7.5, containing 0.5 M NaCl, 1 mM EDTA and 0.005% (w/v) SDS for 1 min. Refractive index errors due to bulk solvent effects were corrected with responses from DNA-free flow cell 1 as well as subtracting blank injections. Kinetic raw data were processed and globally fitted with Scrubber 2.0c (BioLogic Software) using a 1:1 interaction model including a mass transport term.
ChIP analysis
1 × 106 spores/ml of wild-type (AfS77) or the strain carrying Venus-tagged HapX (hapXVENUS) cultures were grown in liquid AMM media for 18 h at 37°C, then shifted to fresh media with no iron, 0.03 mM, or 3 mM iron for 8 h. ChIP was performed as previously described (Blatzer et al, 2011). Briefly, cells were exposed to 1% (v/v) formaldehyde for crosslinking, DNA was collected by lysis of powdered tissue in ChIP lysis buffer and sheared with sonication. ChIP was performed with 1 μg anti-GFP polyclonal antibody (ab290; Abcam) or IgG control on Dynabeads Protein A magnetic beads (Invitrogen). ChIP'd DNA was treated with RNase A and binding was assessed by qPCR. qPCR was performed in triplicate using 0.5 μl of ChIP'd DNA or input control DNA in a 20 μl reaction with 1× iQ Sybr Green Supermix and 0.2 mM final concentration of each primer. Cycle parameters for qPCR were 40 cycles of 94°C for 30 s and 60°C for 30 s, using BioRad iQ single color real-time PCR detection system. Percent input was calculated according to the Life Technologies ChIP analysis website. Briefly, for each sample, the input controls were adjusted to correct for amount of DNA template using [(mean input control Ct)-log2(100/30)]. Percent enrichment was calculated using 100*(2(Adjusted inputCt-ChIPCt)). As an additional negative control, a region of actA promoter region (nt −980 to −725 relative to the translation start) not predicted to bind HapX was PCR amplified. Oligonucleotides used for ChIP analysis are listed in Supplementary Table S4.
Acknowledgments
This work was supported in part by the Austrian Science Foundation (FWF P21643-B11 to HH), the joint D-A-CH program “Novel molecular mechanisms of iron sensing and homeostasis in filamentous fungi” (FWF I1346-B22 to HH, Deutsche Forschungsgemeinschaft (DFG) BR 1130/14-1 to AAB, and DFG HO 2596/1-1 to PH), DFG-SFB 987 to UM, and the National Institute of Allergy and Infectious Diseases (NIH R01AI081838 to RAC). We are grateful to Dr. Sven Krappmann for providing A. fumigatus AfS77, Dr. Antonio Di Pietro for providing F. oxysporum strains, and Sylke Fricke (HKI) for excellent technical assistance.
Author contributions
FG, HH, PH, AAB, RAC, DC, UM and MK experimental design. FG, PH, VK, SRB, BEL, NR, ERW, AAV and KT experiments. FG, HH, PH and AAB interpretation of generated data. HH, FG, PH and AAB writing the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Amich J, Schafferer L, Haas H, Krappmann S. Regulation of sulphur assimilation is essential for virulence and affects iron homeostasis of the human-pathogenic mould Aspergillus fumigatus. PLoS Pathog. 2013;9:e1003573. doi: 10.1371/journal.ppat.1003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, Cornish EJ, Mazurie A, Grahl N, Haas H, Cramer RA. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 2011;7:e1002374. doi: 10.1371/journal.pgen.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A, Isnard AD, Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold ML, Fribourg S, Koch M, Metzger T, Romier C. Deciphering correct strategies for multiprotein complex assembly by co-expression: application to complexes as large as the histone octamer. J Struct Biol. 2011;175:178–188. doi: 10.1016/j.jsb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- Gsaller F, Eisendle M, Lechner BE, Schrettl M, Lindner H, Muller D, Geley S, Haas H. The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus. Metallomics. 2012;4:1262–1270. doi: 10.1039/c2mt20179h. [DOI] [PubMed] [Google Scholar]
- Haas H, Zadra I, Stoffler G, Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem. 1999;274:4613–4619. doi: 10.1074/jbc.274.8.4613. [DOI] [PubMed] [Google Scholar]
- Haas H, Eisendle M, Turgeon GB. Siderophores in fungal physiology and virulence. Ann Rev Phytopathol. 2008;46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- Haas H. Iron – a key nexus in the virulence of Aspergillus fumigatus. Front Microbiol. 2012;3:28. doi: 10.3389/fmicb.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, Dumig M, Jaber BM, Szewczyk E, Olbermann P, Morschhauser J, Krappmann S. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl Environ Microbiol. 2010;76:6313–6317. doi: 10.1128/AEM.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thon M, Kniemeyer O, Abt B, Seeber B, Werner ER, Kato M, Brakhage AA, Haas H. Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron. EMBO J. 2007;26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell. 2011;10:207–225. doi: 10.1128/EC.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber EM, Scharf DH, Hortschansky P, Groll M, Brakhage AA. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure. 2012;20:1757–1768. doi: 10.1016/j.str.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Ihrig J, Hausmann A, Hain A, Richter N, Hamza I, Lill R, Mühlenhoff U. Iron regulation through the back door: iron-dependent metabolite levels contributeto transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2010;9:460–471. doi: 10.1128/EC.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D'Souza C, White R, Kronstad JW. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Kaplan J. Iron acquisition and transcriptional regulation. Chem Rev. 2009;109:4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- Kato M. An overview of the CCAAT-box binding factor in filamentous fungi: assembly, nuclear translocation, and transcriptional enhancement. Biosci Biotechnol Biochem. 2005;69:663–672. doi: 10.1271/bbb.69.663. [DOI] [PubMed] [Google Scholar]
- Labbe S, Khan MG, Jacques JF. Iron uptake and regulation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2013;16:669–676. doi: 10.1016/j.mib.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Li L, Bagley D, Ward DM, Kaplan J. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol. 2008;28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jia X, Ward DM, Kaplan J. Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J Biol Chem. 2011;286:38488–38497. doi: 10.1074/jbc.M111.286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miao R, Bertram S, Jia X, Ward DM, Kaplan J. A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J Biol Chem. 2012;287:35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Lin H, Li L, Jia X, Ward DM, Kaplan J. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem. 2011;286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Osmani AH, Ukil L, Son S, Markossian S, Shen KF, Govindaraghavan M, Varadaraj A, Hashmi SB, De Souza CP, Osmani SA. Single-step affinity purification for fungal proteomics. Eukaryot Cell. 2010;9:831–833. doi: 10.1128/EC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Berges MS, Capilla J, Turra D, Schafferer L, Matthijs S, Jochl C, Cornelis P, Guarro J, Haas H, Di Pietro A. HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell. 2012;24:3805–3822. doi: 10.1105/tpc.112.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, de Llanos R, Romero AM, Puig S. Post-transcriptional regulation of iron homeostasis in Saccharomyces cerevisiae. Int J Mol Sci. 2013;14:15785–15809. doi: 10.3390/ijms140815785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Pinto I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1829–1839. doi: 10.1128/EC.4.11.1829-1839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A, Pelletier B, Labbe S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A, Labbe S. Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem. 2009;284:20249–20262. doi: 10.1074/jbc.M109.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L, Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell. 2007;19:2293–2309. doi: 10.1105/tpc.107.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol. 2006;43:54–64. doi: 10.1016/j.fgb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the Cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, Heinekamp T, Jacobsen I, Werner ER, Brakhage AA, Haas H. SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol. 2008;70:27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, Werner ER, Niermann WC, Brakhage AA, Haas H. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6:e1001124. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, Brakhage AA. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010;38:1098–1113. doi: 10.1093/nar/gkp1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon P, Mercier A, Jbel M, Labbe S. The monothiol glutaredoxin Grx4 exerts an iron-dependent inhibitory effect on Php4 function. Eukaryot Cell. 2012;11:806–819. doi: 10.1128/EC.00060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra I, Abt B, Parson W, Haas H. xylP promoter-based expression system and its use for antisense downregulation of the penicillium chrysogenum nitrogen regulator. Appl Environ Microbiol. 2000;66:4810–4816. doi: 10.1128/aem.66.11.4810-4816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.