Abstract
The Arabidopsis thaliana CORONATINE INSENSITIVE1 (COI1) gene encodes an F-box protein to assemble SCFCOI1 complexes essential for response to jasmonates (JAs), which are a family of plant signaling molecules required for many essential functions, including plant defense and reproduction. To better understand the molecular basis of JA action, we screened for suppressors of coi1 and isolated a coi1 suppressor1 (cos1) mutant. The cos1 mutation restores the coi1-related phenotypes, including defects in JA sensitivity, senescence, and plant defense responses. The COS1 gene was cloned through a map-based approach and found to encode lumazine synthase, a key component in the riboflavin pathway that is essential for diverse yet critical cellular processes. We demonstrated a novel function for the riboflavin pathway that acts downstream of COI1 in the JA signaling pathway and is required for suppression of the COI1-mediated root growth, senescence, and plant defense.
INTRODUCTION
Jasmonates (JAs), which include jasmonic acid and its cyclopentane derivatives as well as cyclopentenones (Reymond and Farmer, 1998), are synthesized from the octadecanoid/hexadecanoid pathways and widely distributed throughout the plant kingdom. JAs modulate the expression of numerous genes (Reymond et al., 2000; Schenk et al., 2000; Sasaki et al., 2001; Feng et al., 2003) and mediate responses to stress, wounding, insect attack, pathogen infection, and UV damage (Staswick, 1992; McConn et al., 1997; Wasternack and Parthier, 1997; Reymond and Farmer, 1998; Farmer, 2001). They also play pivotal roles in reproduction (Feys et al., 1994; McConn and Browse, 1996; Xie et al., 1998) and regulate many other plant developmental processes (Creelman and Mullet, 1997). Without JA, Arabidopsis thaliana plants, such as the JA biosynthetic mutants fad (McConn and Browse, 1996), opr3/dd1 (Sanders et al., 2000; Stintzi and Browse, 2000; Stintzi et al., 2001), and aos (Park et al., 2002), are unable to generate viable pollen and therefore fail to complete their life cycle. Without JA signaling or JA adenylation, Arabidopsis plants, including jar1 (Staswick et al., 1992, 1998), coronatine insensitive1 (coi1) (Feys et al., 1994), and jin (Berger et al., 1996), are unresponsive to JA-inhibitory root growth, unable to accumulate JA-inducible genes, and susceptible to pest attack and pathogen infection.
The coi1 mutation defined a key regulator in the JA signaling pathway. The recessive coi1 mutants fail to respond to JA and coronatine, a phytotoxin structurally similar to jasmonic acid (Feys et al., 1994), displaying defects in JA-regulated gene expression, exhibiting male sterility, and showing susceptibility to insect attack and pathogen infection (Feys et al., 1994; McConn et al., 1997; Vijayan et al., 1998; Reymond et al., 2000; Stintzi et al., 2001; Feng et al., 2003). The COI1 gene encodes a 592–amino acid protein containing an F-box motif and 16 leucine-rich repeat sequences (Xie et al., 1998), which interact with Arabidopsis CULLIN1, RBX1, and Skp1-like proteins ASK1 or ASK2 to assemble SCFCOI1 complexes in planta (Xu et al., 2002). SCFCOI1 is assumed to regulate the abundance of the substrate proteins, which may suppress a set of transcription factors and/or affect the expression of appropriate target genes essential for JA responses (Xie et al., 1998; Xu et al., 2002).
Genetic screens for suppressors have been used to further investigate gene functions and to dissect signaling transduction pathways. In Arabidopsis, suppressor screens have been used to identify genes/mutants functional in many pathways, including the auxin (Cernac et al., 1997), gibberellin (Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995), and abscisic acid (Koorneef et al., 1982) pathways as well as the systemic acquired resistance pathway (Li et al., 1999; Zhang et al., 2003).
To better understand the molecular mechanism via which COI1 regulates JA responses, we conducted a screen for suppressors of the coi1 mutant. A coi1 suppressor1 (cos1) recessive mutant was identified and found to regain wild-type–like phenotypes of JA-sensitive root elongation, gene expression, senescence, and defense response in the coi1 background. The COS1 gene was cloned using a map-based approach and found to encode lumazine synthase, a key component in the riboflavin pathway that is essential for diverse yet critical cellular processes. The riboflavin pathway appears to act downstream of COI1 and to be required for suppression of the COI1-mediated root growth, senescence, and plant defense.
RESULTS
Genetic Screening for the coi1 Suppressors
Because null mutant alleles of COI1 are male sterile (Feys et al., 1994; Xie et al., 1998) and unsuitable for suppressor screening, we isolated the leaky allele coi1-2 (Xu et al., 2002), which is resistant to JA but partially fertile and able to produce a small quantity of seeds. The coi1-2 seeds were mutagenized with ethyl methanesulfonate (EMS), and the M2 seeds harvested from ∼20,000 M1 EMS-mutagenized coi1-2 plants were then used in a screen for coi1 suppressors based on the suppression of JA resistance in the elongating root of coi1-2. Approximately 100,000 M2 seeds were germinated on plant growth medium containing 25 μM methyl JA (MeJA) that inhibits root growth in wild-type seedlings but not in coi1-2 seedlings.
Fourteen seedlings were isolated from the mutagenized coi1-2 M2 population because they regained the JA-sensitive phenotype displaying short roots and stunted growth when grown on medium containing MeJA. These fourteen plants were therefore selected as suppressor candidates of coi1. Each suppressor candidate was backcrossed to the coi1-2 plant, and all the F1 plants were found to be resistant to MeJA, demonstrating that they are all recessive mutations. Among the fourteen F2 populations, four F2 progeny clearly showed 3:1 segregation of JA resistance/sensitivity, indicating that the restoration of JA inhibitory root growth in each of these four mutants results from a single recessive mutant locus. Genetic crosses between these four lines demonstrate that they are mutated at different loci because all the F1 progeny show resistance to MeJA. One locus, which is named cos1, was chosen for further studies because of the strong suppression phenotypes (Figure 1A). This cos1 coi1-2 mutant (homozygous for both cos1 and coi1-2 mutations) was backcrossed to coi1-2 four times to segregate away other potential mutations.
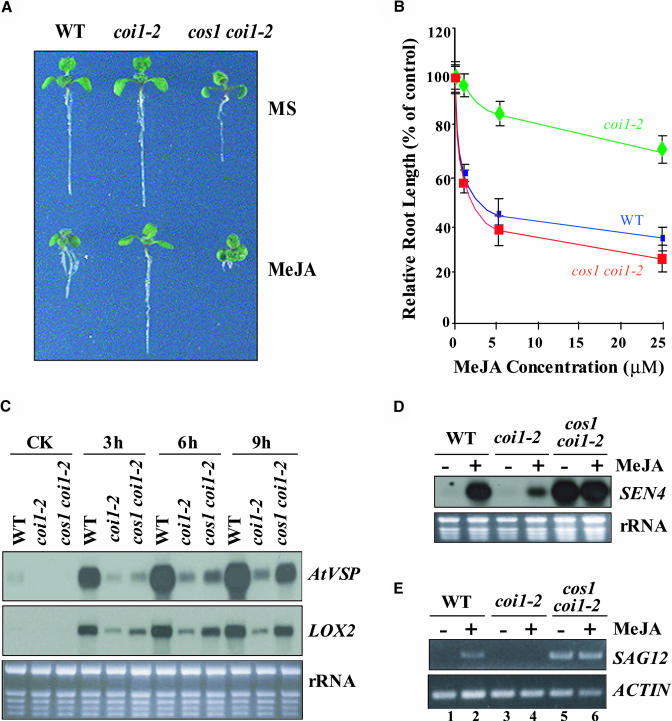
Figure 1.
cos1 Restores the Wild-Type–like JA Sensitivity and JA-Inducible Gene Expression in coi1-2.
(A) Phenotypes of 10-d-old wild type (ecotype Columbia), coi1-2, and cos1 coi1-2 grown on MS medium without (MS) or with 10 μM MeJA.
(B) MeJA dose–response curve of root growth. Root length of the seedlings grown on MS containing 1, 5, or 25 μM MeJA is expressed as a percentage of root length on MS (control). The experiment was repeated five times. Error bars represent se (n > 15).
(C) RNA gel blotting of the JA-inducible gene AtVSP and LOX2 in 20-d-old seedlings untreated (CK) or treated with 100 μM MeJA for 3, 6, and 9 h. Ethidium bromide staining of rRNAs shown at bottom indicates the loading amount of total RNA on the gel.
(D) RNA gel blotting of JA-inducible senescence marker gene SEN4 in plants treated with MeJA (+) or water (−) for 48 h.
(E) The total RNA described in (D) was used in RT-PCR to examine the expression of the senescence-associated gene SAG12. An ACTIN fragment was amplified as a control.
Suppression of cos1 on coi1-Mediated JA Insensitivity
The basis for the initial screen for coi1 suppressors was the suppression of JA insensitivity in the elongating coi1-2 root. To further confirm the effect of the cos1 mutation, we measured the root length of seedlings grown on MS medium supplemented with various concentrations of MeJA (0, 1, 5, and 25 μM). As expected, the coi1-2 is resistant to JA (Figures 1A and 1B). However, root growth of the cos1 coi1-2 seedlings was inhibited by MeJA (Figures 1A and 1B), demonstrating that the cos1 mutation fully suppresses coi1-mediated JA insensitivity. In contrast with JA-sensitive root growth rescued by cos1 in coi1-2, the cos1 mutation was unable to restore the male fertility defect of coi1-2 plants (data not shown).
Restoration of JA-Inducible Gene Expression in the coi1 Mutant by cos1
When assayed for JA-inducible expression of Arabidopsis VEGETATIVE STORAGE PROTEIN (AtVSP) (Benedetti et al., 1995) and LIPOXYGENASE2 (LOX2) (Bell et al., 1995), the cos1 mutation is able to restore defect of the JA-inducible gene expression in coi1-2. Figure 1C shows that the levels of AtVSP and LOX2 transcripts upon JA induction are higher in cos1 coi1-2 than those in coi1-2.
Mutations in COI1 were found to cause a defect in JA-inducible expression of senescence-associated genes (Y. He et al., 2002; S. Xiao and D. Xie, unpublished data). To investigate if the cos1 mutation can restore the defect in JA-inducible expression of senescence marker genes in coi1-2, the rosette leaves excised from 20-d-old plants were treated with MeJA for 48 h and then used for RNA extraction. A senescence marker gene, SEN4 (Nam, 1997), was used as a probe in RNA gel blot hybridization. As shown in Figure 1D, SEN4 transcript is highly accumulated in the wild type upon JA induction (Figure 1D, lane 2); however, the SEN4 expression upon JA induction was severely reduced in the coi1-2 mutant (Figure 1D, lane 4), consistent with previous observations that mutations in COI1 lead to defect of JA-inducible senescence-associated gene expression. Interestingly, cos1 coi1-2 highly accumulated SEN4 transcript even without JA induction (Figure 1D, lanes 5 and 6), suggesting that the cos1 mutation constantly suppresses the defect of JA-inducible expression of senescence-associated genes in the coi1 background. When assayed for expression of other senescence marker genes using RT-PCR, cos1 also was found to constantly suppress the coi1 mutation (Figure 1E; data not shown). For example, the cos1 coi1-2 (Figure 1E, lanes 5 and 6), but not coi1-2 (Figure 1E, lanes 3 and 4), constantly accumulated the senescence-associated gene SAG12 that is believed to be a good marker of senescence (Weaver and Amasino, 2001) (Figure 1E).
Restoration of JA-Dependent Senescence in the coi1 Mutant by cos1
JA was found to induce leaf senescence in wild-type Arabidopsis but not in the JA-insensitive mutant coi1 plant (Y. He et al., 2002). Having shown the suppression of cos1 on the defect of JA-inducible expression of senescence genes, we attempted to investigate whether cos1 can rescue the defect of the age-dependent expression of senescence genes. The soil-grown coi1-2, cos1 coi1-2, and wild-type plants at various developmental stages were harvested for RNA extraction and then used in RNA gel blot analysis with the senescence marker genes as probes. As shown in Figure 2A, the SEN4 transcript was found not to express in the 20-d-old wild type or coi1-2 plants; with respect to the 30-d-old plants, a high level of SEN4 expression was detected in the wild type but not in the coi1-2 mutant, suggesting that mutations in COI1 affect the age-dependent senescence. Interestingly, the cos1 coi1-2 plants constantly accumulated a high level of SEN4 transcript as indicated at the age of 20 and 30 d (Figure 2A), suggesting that the cos1 mutation continuously suppresses the defect of age-dependent expression of senescence genes in coi1-2. Similar conclusion was obtained when other senescence marker genes were examined (data not shown).
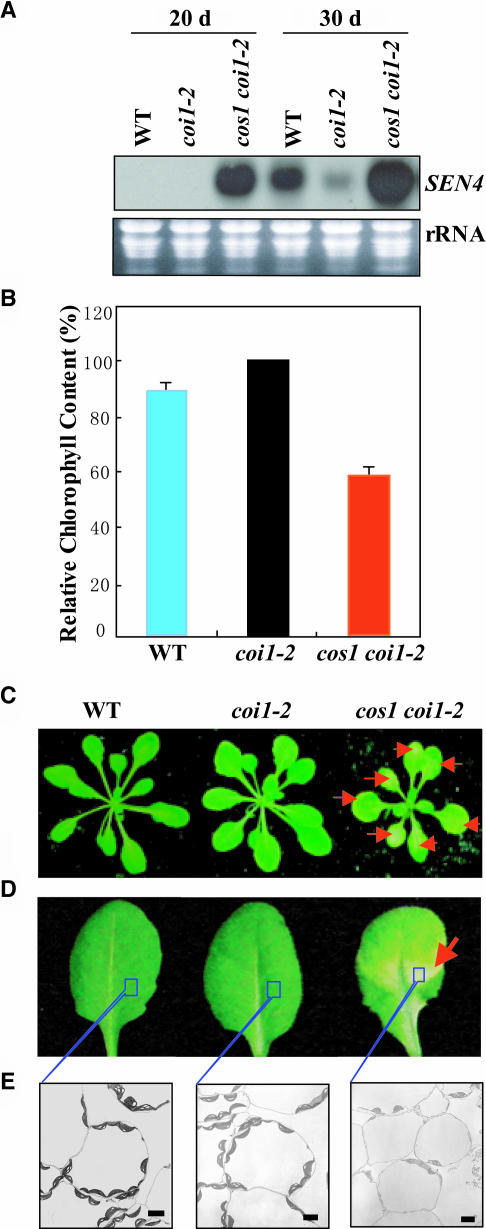
Figure 2.
The cos1 Mutation Suppresses the Defect of Senescence in coi1-2.
(A) The wild-type, coi1-2, and cos1 coi1-2 plants grown in soil for 20 or 30 d were harvested for extraction of total RNAs used in RNA gel blot analysis with the senescence marker gene SEN4 as a probe.
(B) Chlorophyll content of leaves from the plants grown in soil for 20 d. The chlorophyll content in coi1-2 was set to 100%, and the relative chlorophyll content in the wild type and cos1 coi1-2 was calculated accordingly. Experiments were repeated five times. Error bars represent se (n > 10).
(C) and (D) Phenotypes of the wild type, coi1-2, and cos1 coi1-2 grown in soil for 20 d. The red arrows indicate the senescence-associated yellowing phenotypes in resettle leaves.
(E) Electronic microscopic examination on chloroplasts of resettle leaves described in (D). Scale bars = 5 μm.
Senescence also was assessed by measuring chlorophyll content, a typical senescence-associated physiological marker, which is known to decline with the progression of age-dependent senescence (Yoshida et al., 2002). The rosette leaves of 20-d-old plants grown in soil were harvested and used in the chlorophyll content measurement. As shown in Figure 2B, the relative chlorophyll content of the coi1-2 mutant (100%) is higher than that of the wild type (90%). The chlorophyll content in cos1 coi1-2 is severely reduced (60%), suggesting that cos1 severely suppresses the defect of leaf senescence in coi1-2.
The leaves of Arabidopsis wild-type seedlings treated with JA displayed visible yellowing, which is one of the striking precocious symptoms associated with leaf senescence (Y. He et al., 2002). This kind of yellowing is mostly caused by the preferential breakdown of chlorophyll and chloroplasts (Gut et al., 1987). The cos1 coi1-2 plant was found to constantly display the visible yellowing phenotypes in the leaves, stems, and siliques (Figures 2C and 2D; data not shown). We used electron microscopy to investigate if these visible yellowing phenotypes in cos1 coi1-2 are caused by a senescence-related preferential breakdown of chloroplasts. As shown in Figure 2E, the chloroplasts within the yellowing leaves from the 20-d-old cos1 coi1-2 plants were significantly broken down, whereas no preferential breakdown of chloroplasts was observed in the wild type and coi1-2 at similar developmental stages. These results were consistent with the RNA gel blot analysis data that senescence marker genes were expressed in 20-d-old cos1 coi1-2 but not in the wild type or coi1-2. Taking all the data together, we conclude that cos1 may act downstream of COI1 and constantly suppresses the defect of the JA-dependent senescence in coi1-2.
Restoration of the JA-Regulated Defense Response in the coi1 Mutant by cos1
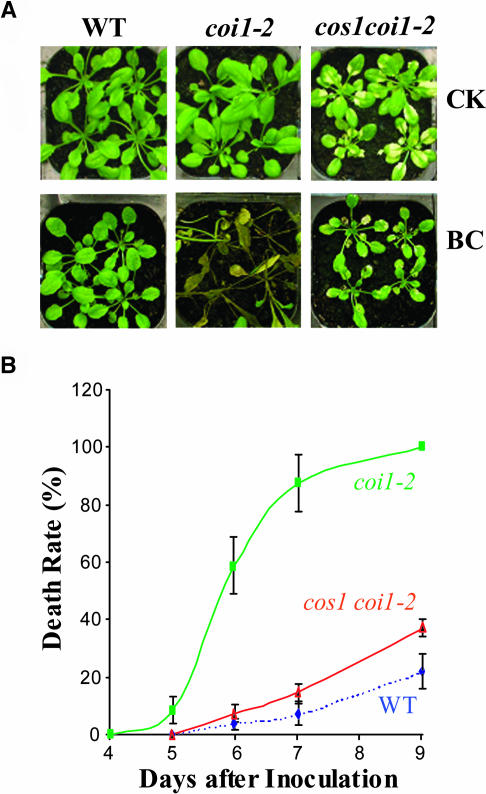
The coi1 mutations previously were found to cause loss of resistance against insects and pathogens in Arabidopsis (McConn et al., 1997; Vijayan et al., 1998; Stintzi et al., 2001). To investigate whether the cos1 mutation restores the defense responses in coi1-2, we inoculated the fungus pathogen (Botrytis cinerea) onto the wild type, coi1-2, and cos1 coi1-2 (Figure 3). By 6 d after inoculation, 60% of coi1-2 plants had died, whereas the cos1 coi1-2 plants showed resistance to fungal infection and <10% of cos1 coi1-2 had died, which was similar to the wild type (7%). By 9 d after inoculation, all of the coi1-2 plants (100%) were dead; however, the cos1 coi1-2 has only ∼35% of death rate similar to that of the wild type (Figures 3A and 3B). These results clearly demonstrate that the cos1 mutation restores the resistance against pathogens in coi1-2.
Figure 3.
The cos1 Mutation Restores the Defect of Disease Resistance in coi1-2.
(A) Fourteen-day-old plants were inoculated with the fungus pathogen B. cinerea (BC) or with water as control (CK) and photographed by 9 d after inoculation.
(B) The death rate for each line (WT, coi1-2, and cos1 coi1-2) was recorded at 4, 5, 6, 7, or 9 d after inoculation. The experiment was repeated three times. Error bars represent se (n > 40).
The Effect of cos1 on transport inhibitor response1
In addition to COI1, the JAR1 gene was the only cloned gene in the signaling pathway defined by the JA-insensitive mutants (Staswick et al., 2002). To examine the effects of the cos1 mutation in the JA-insensitive mutant jar1 (Staswick, 1992; Staswick et al., 1998), we attempted to generate the cos1 jar1 double mutant through sequence verification of the jar1-1 allele among cos1 homozygous plants, which were identified from F2 progeny of a cross between jar1-1 and cos1 coi1-2 based on cos1-conferred lesion-like yellow spots (Figures 2C and 2D). Surprisingly, we were unable to identify the cos1 jar1-1 mutants because none of the cos1 homozygous F2 plants contained the jar1-1 allele, though >100 plants were sequence verified. These data indicate that the cos1 locus has tight genetic linkage with the JAR1 locus, which makes it difficult to generate the cos1 jar1-1 double mutants. Similarly, we also failed to generate cos1 in the wild-type COI1 or coi1-1 null mutant background through genetic crossing (data not shown), suggesting a tight linkage of cos1 with COI1 that is physically close to the JAR1 gene (Arabidopsis genome sequencing project).
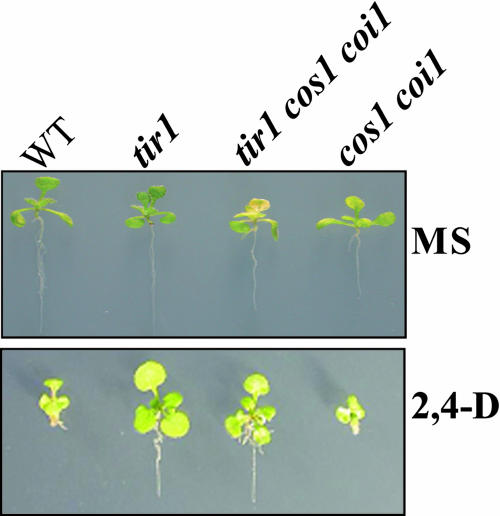
We then attempted to investigate the role for the cos1 mutation in transport inhibitor response1 (tir1), an auxin-insensitive mutant, which defines the F-box protein TIR1 that is most closely related to COI1 (Ruegger et al., 1998; Xie et al., 1998). The tir1-1 cos1 coi1-2 mutant was identified through sequence verification of the tir1-1 allele among the cos1 coi1-2 homozygous plants from F2 progeny of a cross between tir1-1 and cos1 coi1-2. When assayed for auxin inhibitory root elongation, the tir1-1 and tir1-1 coi1-2 mutant seedlings are resistant to the synthetic auxin 2,4-D as expected, and the tir1-1 cos1 coi1-2 mutant seedlings exhibit auxin resistance similar to the tir1-1 or tir1-1 coi1-2 mutant seedlings (Figure 4). These data indicate that the cos1 mutation is unable to alter the auxin responses in the tir1-1 mutant and therefore has no suppression on tir1.
Figure 4.
cos1 Is Not a Suppressor of the tir1 Mutation.
Phenotypes of 10-d-old wild type (ecotype Columbia), tir1-1, tir1-1 cos1 coi1-2, and cos1 coi1-2 grown on MS medium or MS containing 0.1 μM 2,4-D.
Map-Based Cloning of COS1
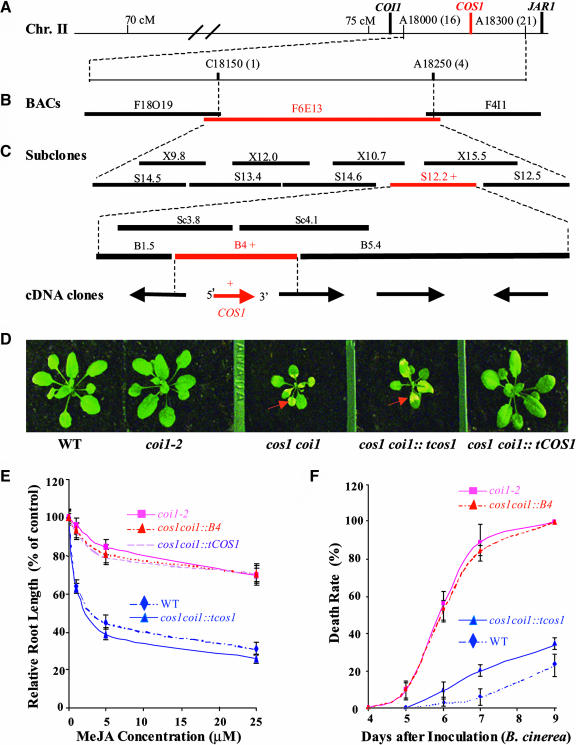
To genetically map the COS1 locus, we identified a Landsberg erecta JA-insensitive mutant allele coi1-12, which contains a single amino acid replacement from Phe 359 to Lys in COI1 (D. Xie, unpublished data) and has a polymorphic genetic background from the Columbia cos1 coi1-2 mutant. We made the genetic crossing between the coi1-12 and cos1 coi1-2 plants and subsequently screened for their F2 progeny (homozygous for both cos1 and coi1) those that are sensitive to JA. Based on the linkage analysis among molecular markers and the cos1-conferred JA-sensitive phenotype using these JA-sensitive F2 progeny, we localized the COS1 locus on chromosome II between two amplified fragment length polymorphism (AFLP) markers, A18000 and A18300. As shown in Figure 5A, the COS1 locus was mapped between the JAR1 and COI1 loci, consistent with our previous observation that cos1 has tight genetic linkage with JAR1 and COI1. Further mapping using a cleaved amplified polymorphic sequence marker, C18150 (one recombinant), and AFLP marker A18250 (four recombinants) placed COS1 on a BAC, F6E13, which contains a 110-kb Arabidopsis genomic insert (Figure 5B).
Figure 5.
Map-Based Cloning of COS1.
(A) to (C) The COS1 locus is located on chromosome II between A18000 (16 heterozygous recombinants) and A18300 (21 recombinants), further mapped onto a single BAC (F6E13), and was finally placed onto an ∼4-kb genomic fragment (B4) corresponding to COS1 by a complementation test with subcloned BAC inserts. Restriction endonucleases used to generate subclones (B, BamHI; Sc, SacI; S, SpeI; X, XbaI) are shown. The fragments marked with red and plus signs can complement cos1. cM, centimorgan.
(D) Phenotype of the 20-d-old plants of the wild type, coi1-2, cos1 coi1-2, and cos1 coi1-2 transgenic for either the mutant cos1 cDNA (cos1 coi1::tcos1) or the wild-type COS1 cDNA (cos1 coi1::tCOS1). The red arrows indicate the senescence-associated yellowing phenotypes.
(E) COS1 complements cos1-conferred JA sensitivity. Root length of the indicated seedlings grown on MS containing MeJA (1, 5, or 25 μM) is expressed as a percentage of root length on MS medium. The experiment was repeated five times. Error bars represent se (n > 15).
(F) COS1 complements cos1-conferred disease resistance in coi1-2. The death rate for each indicated line was recorded at various days after inoculation with B. cinerea. The experiment was repeated three times. Error bars represent se (n > 40).
The COS1 gene was finally localized onto an ∼4-kb BamHI fragment (B4) by functional complementation with the subcloned F6E13 fragments (Figure 5C). The B4 fragment identifies a single 684-bp full-length cDNA (Figure 5C). Thus, the 684-bp cDNA is the COS1 gene. Further analysis of the cos1 coi1-2 plants transgenic for either the B4 fragment or the COS1 cDNA that was 5′ myc-tagged under the control of the 35S promoter of Cauliflower mosaic virus (CaMV 35S) (referred to as tCOS1) demonstrates that both the B4 fragment and the COS1 cDNA can complement the cos1-conferred senescence (Figure 5D; data not shown), JA sensitivity (Figure 5E), and defense response (Figure 5F).
The corresponding sequence from the cos1 mutation deviated from that of the wild type by a single nucleotide change (G238A), G to A, at position + 238 relative to the translation start codon of the COS1 genomic DNA (Figure 6). This single nucleotide, G238, is the first nucleotide of the second exon and therefore is crucial for RNA splicing. The G238A point mutation abolished the splicing recognition sequence (from TA/GG to TA/AG), and the TA/GC sequence located 18 bp downstream of G238A mutation serves as a new RNA splicing recognition site resulting in an 18-nucleotide deletion in the mutant cos1 cDNA (Figure 6), which was confirmed by sequence verification of the RT-PCR–amplified cos1 mutant cDNA.
Figure 6.
Sequence Comparison of the Wild Type and Mutant COS1 Gene.
DNA sequence of exons is indicated with capital letters; the cos1 mutation G238A is indicated by arrows; the 18-bp DNA sequence absent from the cos1 mutant cDNA is underlined; and the six amino acids encoded by the 18-bp DNA is also underlined.
To investigate whether the 18-bp deletion in the mutant cos1 mRNA affects the protein expression of the mutant cos1, we made transgenic plants expressing the mutant cos1 cDNA that was 5′ myc-tagged under the control of the CaMV 35S constitutive promoter (referred to as tcos1). Protein gel blotting with α-Myc antibody shows that the tcos1-encoded mutant protein was accumulated at a level similar to the tCOS1-encoded protein in the transgenic lines (data not shown); however, tCOS1 is able to fully complement the cos1 mutation, whereas tcos1 could not (Figures 5D to 5F; data not shown). The result indicates that the 18-bp deletion in cos1 does not disrupt COS1 protein stability; however, the six amino acids encoded by the deleted 18 nucleotides are crucial for COS1 function.
Sequence analysis reveals that the COS1 gene encodes lumazine synthase, which was previously shown to functionally complement the bacterial lumazine synthase–deficient mutant (Jordan et al., 1999) and shares high identity at amino acid levels with lumazine synthases from bacteria (∼53%), yeast (∼57%), and other plants, including spinach (Spinacia oleracea) and tobacco (Nicotiana tabacum; >70%) (Jordan et al., 1999; Persson et al., 1999).
DISCUSSION
Null mutations in COI1 abolish all the JA responses exhibiting resistance to JA inhibition of plant growth, defect in pollen fertility, reduction in senescence, and susceptibility to pathogen infection and insect attack. The COI1 protein previously was found to assemble into SCFCOI1 complexes that were speculated to recruit substrate proteins, which may function as suppressors to negatively regulate the expression of appropriate downstream genes essential for JA responses (Xie et al., 1998; Xu et al., 2002). To further dissect COI1-mediated JA responses and to gain insight into the mechanism of COI1 function, we have isolated a coi1 suppressor mutant, here referred as to cos1. The cos1 mutation can restore the coi1-related phenotypes, including defects in JA sensitivity, senescence, and plant defense responses in the coi1 background. However, the cos1 mutation failed to restore the male fertility defect of the coi1-2 plants, which further dissects the JA signal transduction pathway and indicates that JA-mediated pollen development is regulated by other component(s) independent of COS1. The cos1 mutation also failed to restore the auxin insensitivity in the tir1-1 mutant, demonstrating that cos1 is not a suppressor of tir1.
The COS1 gene was cloned via a map-based approach and found to encode lumazine synthase. Lumazine synthase catalyzes the penultimate step in biosynthesis of riboflavin (vitamin B2) in plants, fungi, and microorganisms (Persson et al., 1999), which is a key component in the riboflavin pathway essential for diverse yet critical cellular processes. Riboflavin is essential for the basic metabolism and serves as a precursor of coenzymes riboflavin monophosphate and flavin adenine dinucleotide; the riboflavin pathway-derived flavocoenzymes were found to function as indispensable redox cofactors in all cells and also serve a variety of other roles, such as DNA photorepair, light sensing, and bioluminescence (Muller, 1992; Sancar, 1994; Ahmad et al., 1998; Christie et al., 1998; Vande Berg and Sancar, 1998; Briggs and Huala, 1999; Aubert et al., 2000; Briggs and Olney, 2001; Salomon et al., 2001; Q.Y. He et al., 2002). The evidence that cos1 restores the defects in coi1-2 demonstrates a novel function for the riboflavin pathway in JA signaling transduction, suggesting that the wild-type COS1 represents the riboflavin pathway essential for suppression action exerted by unidentified negative regulator(s) (such as SCFCOI1 substrates) of JA responses.
Because the riboflavin pathway is essential for diverse yet critical cellular processes, a complete abolishment of the riboflavin pathway would cause pleiotrophic phenotypes and probably affect the survival of plants. It is likely that the cos1 mutant identified in this work is a leaky allele and that the cos1 mutant protein is partially functional. In the cos1 coi1-2 mutant plants, the 6–amino acid deletion in the cos1 mutant protein appears to attenuate the riboflavin pathway but not to completely disrupt this pathway. Therefore, the cos1 mutation does not affect the plant survival but suppresses the defects in the JA sensitivity, senescence, and plant defense responses in the coi1-2 mutant background via attenuation of the unidentified negative regulator(s) that require the riboflavin pathway to exert their suppression action.
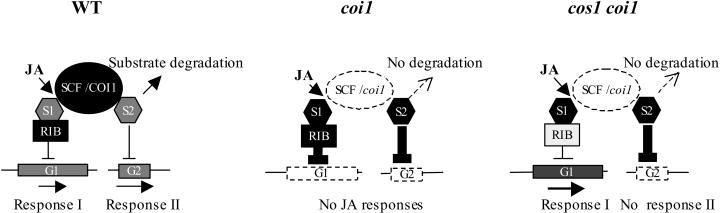
Here, we propose a working model for the function of the riboflavin pathway in the JA signaling transduction chain. As shown in Figure 7, different SCFCOI1 substrates (indicated as groups S1 and S2), which will be degraded through SCFCOI1-26S proteasome in responsive to JA signaling, may negatively regulate different transcription factors and/or appropriate downstream genes (indicated as two sets of genes, G1 and G2) that mediate their corresponding JA responses (indicated as response I and response II). The first group of substrates (S1) may require the riboflavin pathway (indicated as RIB) to exert their suppression action on the downstream G1 target genes, whereas the action of the second group of substrates (S2) may not require the riboflavin pathway. In wild-type plants (Figure 7, left panel), the endogenous JA, and developmental and environmental cues (such as flowering and insect attack) that activate JA signaling, promote the ubiquitylation and degradation of substrate proteins. As a result, the downstream genes are expressed to maintain JA responses. In the coi1 null mutant plants (Figure 7, middle panel), mutation of COI1 disrupts SCFCOI1, resulting in a high level of accumulation of the SCFCOI1 substrates, which constantly exert suppression on the downstream genes, leading to loss of all the JA responses. In the cos1 coi1 double mutants (Figure 7, right panel), the cos1 mutation attenuates the riboflavin pathway and therefore decreases the suppression action exerted by the S1 substrates and finally activates the expression of their downstream genes (G1), leading to restoration of their corresponding JA responses, including JA sensitivity, senescence, and defense (response I). However, cos1 is unable to restore fertility in coi1 because the riboflavin pathway is not required for the suppression exerted by the S2 substrate that suppresses the G2 downstream genes, leading to defect in pollen fertility (response II).
Figure 7.
Model of the Function of the Riboflavin Pathway in the JA Signal Transduction Chain.
S1 and S2 represent two groups of substrates that are activated by the JA signals (JA) for ubiquitylation and degradation via SCFCOI1-26S proteasome (SCF/COI1). G1 and G2 represent different sets of JA response genes that mediate distinct JA responses (I and II). Arrows indicate positive regulation action; blunted lines indicate negative regulation; the weight of each line indicates the predicted regulation level. The shading, which was used to mark the putative components, indicates abundance of these components. RIB, riboflavin pathway.
In the cos1 coi1-2 double mutants, application of exogenous JA was found to cause enhanced inhibition of root growth (response I) and induction of gene expression (Figure 1). This induction of JA response by exogenous JA in the cos1 coi1-2 double mutants may result from activation of substrate (S1) degradation because coi1-2 is a leaky mutant allele. Alternatively, JA-responsive G1 genes may be repressed by substrate S1 but also activated by some unknown JA-regulated positive components, probably including dephosphorylated regulator(s) previously proposed (Rojo et al., 1998); the exogenous JA was therefore able to activate the unknown JA-regulated positive components that modulate the expression of JA-responsive G1 genes in the cos1 coi1-2 double mutants. More experiments will be required to test our working model and explanation. Identification and functional analysis of the SCFCOI1 substrates will be useful to understanding the novel role for the riboflavin pathway in JA signaling and will be essential for illustrating the molecular mechanism via which JA regulates plant defense and development.
METHODS
Plant Growth Conditions
Seeds were surface sterilized, plated on plant growth medium (MS; Sigma, St. Louis, MO), chilled at 4°C for 3 d, and then transferred to a growth chamber under a 16-h-light (22 to 24°C)/8-h-dark (16 to 19°C) photoperiod. For root length measurement experiments, the seedlings were grown on MS medium supplemented with various concentrations of MeJA (Aldrich, Milwaukee, WI) for 9 d before measurement.
Mutant Screening
The coi1-2 leaky mutant was identified previously (Xu et al., 2002). The coi1-2 seeds (∼30,000) were mutagenized with 0.3% EMS following routine procedures. About 70% of the mutagenized seeds (referred as to the M1 population) could grow in soil and generate M2 seeds under the growth conditions of a 16-h-light (21 to 23°C)/8-h-dark (16 to 19°C) photoperiod.
M2 Seeds were routinely plated on MS medium containing 25 μM MeJA to screen for mutants sensitive to MeJA exhibiting phenotypes of short root and stunted growth.
Generation of the tir1-1 cos1 coi1-2 Mutant
The tir1-1 mutant, which harbors a point mutation from guanine 440 to adenine (g440a) leading to an amino acid replacement from G147 to D (G147D) (Ruegger et al., 1998), was crossed to the cos1 coi1-2 mutant plant. The cos1 homozygous plants were identified from the F2 progeny based on cos1-conferred lesion-like yellow spots, and all of the cos1 homozygous plants were also homozygous for the coi1-2 allele because cos1 has close genetic linkage with coi1-2. These cos1 coi1-2 homozygous plants were then used in sequence verification of the tir1-1 g440a mutation; the plants containing the homozygous tir1-1 allele were the tir1-1 cos1 coi1-2 mutants. Seeds harvested from the tir1-1 cos1 coi1-2 mutant plant were used to assay for auxin inhibitory elongation in MS medium containing 0.1 μM 2,4-D.
RNA Gel Blot and RT-PCR Analysis
Specific primers were designed based on their DNA sequences to PCR amplify AtVSP (Benedetti et al., 1995), LOX2 (Bell et al., 1995), and SEN4 (Nam, 1997) and used as RNA gel blot probes. The probe labeling and RNA gel blot hybridization methods were described previously (Xu et al., 2001).
RT-PCR analysis was performed following routine procedures. The senescence marker gene SAG12 was amplified with primers 5′-CAGCTGCGGATGTTGTTG-3′ and 5′-CCACTTTCTCCCCATTTTG-3′, and the ACTIN gene was amplified with primers 5′-CACCGCTTAACCCGAA-3′ and 5′-GTGAGGTCACGACCAG-3′.
Total RNA used in Figure 1 was extracted from 20-d-old plants that were untreated (CK) or treated with 100 μM MeJA in daytime for 3, 6, and 9 h (Figure 1C) or for 48 h (Figures 1D and 1E).
Molecular Markers
The cleaved amplified polymorphic sequence marker C18150 reveals a DNA polymorphism between Columbia and Landsberg erecta when Sau3AI is used to digest a PCR fragment amplified with the primers 5′-CGTTACAAGATCTGATAATT-3′ and 5′-TCTCGCCATTAGCACGTTA-3′. The AFLP markers A18000, A18250, and A18300 reveal a polymorphism (difference in size of 15, 29, and 58 bp, respectively) when the PCR fragment was amplified with their corresponding primers (A18000, 5′-AGAGCTATGTTGTGCCTGATA-3′ and 5′ -ATTCCTCCATCGATGCGGCT-3′; A18250, 5′-TTGGGGATTGATAACGACAA-3′ and 5′-ACCAAACCATTCTAAAGTGA-3′; A18300, 5′-TATGCATATGCATTGAGCGTAA-3′ and 5′-TCTTTTATACAAGAAACCTCAACCTT-3′).
Complementation Test
The F6E13 inserts were digested with the indicated restriction enzymes. Each digested fragment was recovered and cloned into pCambia 1300 vector. The mutant and wild-type COS1 cDNAs were amplified with the primers 5′-ATGAAGTCATTAGCTTCGCC-3′ and 5′-CTATTTCAGGTGGTGCTCAA-3′ using RT-PCR from RNAs isolated from cos1 coi1-2 and the wild type, respectively. The cDNA fragment was in-frame tagged with the myc epitope at 5′ and driven by the CaMV 35S promoter in the pMYC2 vector (Xu et al., 2002). The constructs were mobilized into Agrobacterium tumefaciens by electroporation and then introduced into cos1 coi1-2 by in planta vacuum infiltration.
The cos1 coi1-2 plants transgenic for the mutant cos1 cDNA, the wild-type COS1 cDNA, and an ∼4-kb genomic fragment containing COS1 were identified and referred as to cos1 coi1-2::tcos1, cos1 coi1-2::tCOS1, and cos1 coi1-2::B4.
Pathogen Infection
Fourteen-day-old plants were inoculated with the fungus pathogen Botrytis cinerea (500,000 spores/mL) or with water as the control, placed in a growth chamber at the appropriate temperature (22°C) and high humidity (∼100%) for 2 d to stimulate infection, and then transferred to a growth room under the growth conditions of a 16-h-light (21 to 23°C)/8-h-dark (16 to 19°C) photoperiod. The death rate for each line was recorded at various days after inoculation. In each treatment, at least 40 plants from each line (the wild type, coi1-2, or cos1 coi1-2) were investigated. The experiment was repeated three times.
Measurement of Chlorophyll and Transmission Electron Microscopy
Chlorophyll was extracted from equal volumes of leaf discs by immersion in 1 mL of N,N-dimethylformamide for 48 h in the dark at 4°C. Absorbance was recorded at 664 and 647 nm, and total chlorophyll concentration was calculated according to methods described previously (Yoshida et al., 2002).
Samples examined by transmission electron microscopy were fixed in 2% paraformaldehyde and 4% glutaraldehyde in 100 mM cacodylate buffer for 3 h and then postfixed with 2% osmium tetroxide in 100 mM sodium cacodylate buffer for 1 h at 4°C. Samples were then dehydrated through a series of 30, 50, 70, 90, and 100% ethanol and finally in propylene oxide. Samples were embedded in 100% Spurr's resin (Electron Microscopy Sciences, Fort Washington, PA) and polymerized at 65°C overnight. Ultrathin sections were cut on a Jung Reichert ultramicrotome and examined with a transmission electron microscope (JEM1010; JEOL, Tokyo, Japan) at 100 kV.
Sequence data from this article have been deposited with the GenBank data library under accession number NM_129967.
Acknowledgments
We thank Nottingham Arabidopsis Stock Centre and the ABRC for seeds and BAC clones and Paul Staswick and Mark Estelle for jar1 and tir1 mutants. L.D. was supported by the Science and Technology Department of Hunan Province, China. This work was supported by a grant to D.X. from Singapore Agency of Science, Technology, and Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daoxin Xie (daoxin@imcb.a-star.edu.sg).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020370.
References
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998). Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392, 720–723. [DOI] [PubMed] [Google Scholar]
- Aubert, C., Vos, M.H., Mathis, P., Eker, A.P.M., and Brettel, K. (2000). Intraprotein radical transfer during photoactivation of DNA photolyase. Nature 405, 586–590. [DOI] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, C.E., Xie, D.X., and Turner, J.G. (1995). Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 123–142. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Olney, M.A. (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac, A., Lincoln, C., Lammer, D., and Estelle, M. (1997). The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124, 1583–1591. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. (2001). Surface-to-air signals. Nature 411, 854–856. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCF(COI1) and modulates jasmonate responses. Plant Cell 15, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut, H., Ruts, C., Matile, P., and Thomas, H. (1987). Leaf senescence in a non-yellowing mutant of Festuca pratensis: Degradation of carotenoids. Plant Physiol. 70, 659–663. [Google Scholar]
- He, Q.Y., Cheng, P., Yang, Y.H., Wang, L.X., Gardner, K.H., and Liu, Y. (2002). White collar-1, a DNA binding transcription factor and a light sensor. Science 297, 840–843. [DOI] [PubMed] [Google Scholar]
- He, Y., Fukushige, H., Hildebrand, D.F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, D.B., Bacot, K.O., Carlson, T.J., Kessel, M., and Viitanen, P.V. (1999). Plant riboflavin biosynthesis. Cloning, chloroplast localization, expression, purification, and partial characterization of spinach lumazine synthase. J. Biol. Chem. 274, 22114–22121. [DOI] [PubMed] [Google Scholar]
- Koorneef, M., Jorna, M.L., Brinkhorst-van der Swan,, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (aba) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, F. (1992). Chemistry and Biochemistry of Flavoenzymes. (Boca Raton, FL: CRC Press).
- Nam, H.G. (1997). The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 8, 200–207. [DOI] [PubMed] [Google Scholar]
- Park, J.H., Halitschke, R., Kim, H.B., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Persson, K., Schneider, G., Jordan, D.B., Viitanen, P.V., and Sandalova, T. (1999). Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly. Protein Sci. 8, 2355–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Titarenko, E., Leon, J., Berger, S., Vancanneyt, G., and Sanchez-Serrano, J.J. (1998). Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 13, 153–165. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, M., Eisenreich, W., Durr, H., Schleicher, E., Knieb, E., Massey, V., Rudiger, W., Muller, F., Bacher, A., and Richter, G. (2001). An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. USA 98, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar, A. (1994). Structure and function of DNA photolyase. Biochemistry 33, 2–9. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., Amagai, M., Kuwata, C., Tsugane, T., Masuda, T., Shimada, H., Takamiya, X., et al. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153–161. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. (1992). Jasmonate, genes, and fragrant signals. Plant Physiol. 99, 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W.P., and Howell, S.H. (1992). Methyl jasmonate inhibition of root-growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Yuen, G.Y., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Berg, B.J., and Sancar, G.B. (1998). Evidence for dinucleotide flipping by DNA photolyase. J. Biol. Chem. 273, 20276–20284. [DOI] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., and Parthier, B. (1997). Jasmonate signalled plant gene expression. Trends Plant Sci. 2, 302–307. [Google Scholar]
- Weaver, L.M., and Amasino, R.M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127, 876–886. [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., and Somerville, C.R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Wang, Z., Peng, W., Huang, R., Huang, D., and Xie, D. (2001). An Arabidopsis mutant cex1 exhibits constant accumulation of jasmonate-regulated AtVSP, Thi2.1 and PDF1.2. FEBS Lett. 494, 161–164. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., Ito, M., Nishida, I., and Watanabe, A. (2002). Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 29, 427–437. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]