Abstract
To date, a large number of sequences of protein kinases that belong to the sucrose nonfermenting1–related protein kinase2 (SnRK2) family are found in databases. However, only limited numbers of the family members have been characterized and implicated in abscisic acid (ABA) and hyperosmotic stress signaling. We identified 10 SnRK2 protein kinases encoded by the rice (Oryza sativa) genome. Each of the 10 members was expressed in cultured cell protoplasts, and its regulation was analyzed. Here, we demonstrate that all family members are activated by hyperosmotic stress and that three of them are also activated by ABA. Surprisingly, there were no members that were activated only by ABA. The activation was found to be regulated via phosphorylation. In addition to the functional distinction with respect to ABA regulation, dependence of activation on the hyperosmotic strength was different among the members. We show that the relatively diverged C-terminal domain is mainly responsible for this functional distinction, although the kinase domain also contributes to these differences. The results indicated that the SnRK2 protein kinase family has evolved specifically for hyperosmotic stress signaling and that individual members have acquired distinct regulatory properties, including ABA responsiveness by modifying the C-terminal domain.
INTRODUCTION
Plants have developed various mechanisms to cope with abiotic stresses, such as drought, high salinity, and cold. Under these adverse conditions, plants suffer from water stress, which imposes osmotic stress to the cells, and respond by inducing genes and modifying protein functions to protect cellular activities and to maintain whole plant integrities (Bray, 1997; Xiong and Zhu, 2002; Zhu, 2002). Stress-induced gene expression leads to synthesis of proteins with various functions, including proteins that prevent denaturation and oxidative damage of cellular components and the enzymes for the synthesis of osmolytes, such as sugars and amino acids (Delauney and Verma, 1993; Shinozaki and Yamaguchi-Shinozaki, 1996, 2000; Oono et al., 2003). Parts of these responses, such as the regulation of stomata openings, are mediated by the stress hormone abscisic acid (ABA), the synthesis of which is induced by water stress (Zeevaart, 1999).
A large number of studies have been conducted on water deficit or osmotic stress responses, and their signaling transduction mechanisms have been intensively studied via biochemical, physiological, molecular biological, and genetic approaches (Bray, 1997; Knight, 2000; Xiong and Zhu, 2002; Xiong et al., 2002; Zhu, 2002). These studies have indicated the involvement of signaling factors or second messengers such as Ca2+, phospholipids and phosphoinositides and enzymes that generate them, protein kinases, and phosphatases (Munnik et al., 1998, 1999, 2000; Pical et al., 1999; Wang, 1999; DeWald et al., 2001; Xiong et al., 2002). However, many questions have remained unanswered: whether these components act in the same signaling pathway; whether they are core components of the main pathway or those that function in the modification of signaling events, such as cross talk with other signals and feedback or feed-forward controls; and what molecular changes are brought about to each component by the immediate upstream factor to relay the signal.
Protein kinases appear to play key roles in osmotic signaling as in many other signaling cascades. Osmotic and other abiotic and biotic stresses are known to cause increases in cytosolic Ca2+ concentration (Knight, 2000). It has been shown that Ca2+-dependent protein kinases (CDPKs) are induced by water deficit (Urao et al., 1994) and that overexpression of OsCDPK7 results in increased cold and osmotic stress tolerance in rice (Oryza sativa; Saijo et al., 2000). Furthermore, expression of a constitutively active form of AtCDPK1 has been shown to activate an osmotic stress–related promoter (Sheen, 1996).
Another group of Ca2+-regulated protein kinases of potential importance in stress signaling are the members of the CIPK (calcineurin B–like protein [CBL]–interacting protein kinase)/PKS (SOS2-like protein kinase)/SnRK3 (sucrose nonfermenting1–related protein kinase3) family (Halford and Hardie, 1998; Liu et al., 2000; Guo et al., 2001; Luan et al., 2002; Hrabak et al., 2003). They do not bind Ca2+ by themselves, but instead each interacts with a specific member(s) of the CBL/SCaBP (SOS2-like Ca2+-binding protein) family Ca2+ sensors (Liu and Zhu, 1998; Shi et al., 1999; Kim et al., 2000; Albrecht et al., 2001; Guo et al., 2001; Luan et al., 2002). Notably, the interacting pair of SOS2 (CIPK/PKS) and SOS3 (CBL/SCaBP) CIPK/SPK-CBL/SCaBP regulates the activity of an Na+/H+ antiporter, which functions to maintain ionic homeostasis upon high salinity stress (Qiu et al., 2002). The PSK3-SCaBP5 pair is implicated in ABA signaling (Guo et al., 2002; Kim et al., 2003). Similarly, CBL1 and the interacting CIPKs appear to play a role in decoding Ca2+ signals generated by osmotic stress (Cheong et al., 2003).
In yeast (Saccharomyces cerevisiae), hyperosmotic signals are perceived by a sensor His kinase, transmitted to a mitogen-activated protein kinase (MAPK) cascade, via a His-Asp phosphorelay and result in protective responses (Maeda et al., 1994). In Arabidopsis thaliana, a drought/osmotic stress–inducible sensor-like His kinase, AtHK1, has been identified and suggested to have a role in osmotic stress signaling (Urao et al., 1999). Pairs of MAPK and MAPK kinase that may be involved in osmotic stress signaling have been identified in alfalfa (Medicago sativa; SIMKK-SIMK; Kiegerl et al., 2000) and in tobacco (Nicotiana tabacum; NtMEK2-SIPK/WIPK; Yang et al., 2001; Zhang and Klessig, 2001). Arabidopsis MAPKs, AtMPK4 and AtMPK6, have also been reported to be activated by hyperosmotic stress (Ichimura et al., 2000). However, these MAPKs are activated not only by osmotic stress but also by other stresses such as cold and wounding, as well as by signal molecules such as salicylic acid, H2O2, and elicitors (Zhang and Klessig, 1998; Hoyos and Zhang, 2000; Mikolajczyk et al., 2000; Nuhse et al., 2000; Desikan et al., 2001; Yuasa et al., 2001). Expression of a constitutively active form of a MAPK kinase kinase, ANP1/NPK1, has been shown to activate MAPKs that are activated by hyperosmotic stress and result in the activation of oxidative stress–inducible promoters and resistance to a broad range of stresses (Kovtun et al., 2000). These observations suggest that those MAPKs may function in a stress response pathway that involves reactive oxygen species and/or are related to protection against oxidative stresses.
In several studies, in-gel protein kinase assays, using myelin basic protein (MBP) as a substrate, detect Ca2+-independent protein kinase activities with molecular masses at ∼40 kD, which are rapidly activated by hyperosmotic stress together with the above-mentioned MAPKs in tobacco cultured cells (Droillard et al., 2000; Hoyos and Zhang, 2000; Mikolajczyk et al., 2000; Monks et al., 2001) and leaf tissues (Monks et al., 2001), as well as in alfalfa (Munnik et al., 1999) and Arabidopsis (Droillard et al., 2002) cultured cells. A hyperosmotic stress–activated 42-kD protein kinase has been purified from tobacco cells (Mikolajczyk et al., 2000). Partial amino acid sequencing of this protein kinase suggested that it is a homolog of Arabidopsis ASK1, which belongs to the SnRK2 family (Halford and Hardie, 1998; Hrabak et al., 2003). The ∼40-kD hyperosmotic stress–activated non-MAPK protein kinases detected in the studies mentioned above are presumably members of the SnRK2 family, based on the similarities to the tobacco ASK1 homolog in apparent molecular mass, rapid activation kinetics, and substrate specificities. Two members of the soybean (Glycine max) SnRK2 family, SPK1 and SPK2, are activated by hyperosmotic stress in yeast cells, although the activation in plant cells has not been demonstrated (Monks et al., 2001).
Vicia faba AAPK and Arabidopsis OST1/SRK2E, which are also included in the SnRK2 family, have been shown to be activated by ABA and involved in the ABA regulation of stomata closing and ABA-regulated gene expression (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). A wheat (Triticum aestivum) SnRK2 member, PKABA1, has been reported to be induced by ABA and hyperosmotic stress and to repress the activities of gibberellic acid (GA)–inducible promoters when transiently overexpressed in barley (Hordeum vulgare) aleurone layers, thus suggesting involvement in the ABA signaling that leads to GA signal suppression (Gomez-Cadenas et al., 1999, 2001; Shen et al., 2001; Johnson et al., 2002).
In this article, all members of the SnRK2 family encoded by the rice genome were analyzed with respect to their activity regulation and expression. We found that all 10 members are activated by hyperosmotic stress and that three of them are also activated by ABA. In addition to the responsiveness to ABA, distinctions were found among the members regarding the dependence of activation on the strength of the osmotic stress. Furthermore, the regulatory roles of the relatively divergent C-terminal domains of these kinases were investigated for the observed functional distinctions among the members. The results suggested that this family of protein kinases has evolved specifically for the hyperosmotic stress signaling and that individual members have acquired distinct regulatory properties, including ABA responsiveness by modifying the C-terminal as well as the kinase domain.
RESULTS
The Rice Genome Encodes 10 SnRK2 Protein Kinase Family Members
By searching genome sequence databases, 10 genes were identified that encode SnRK2 family protein kinases in the rice genome. These protein kinases were designated SAPK1 through SAPK10, which stand for osmotic stress/ABA–activated protein kinase1 through 10 based on our observations described below. The same number of SnKR2 family proteins has been reported to be encoded in the Arabidopsis genome (Mustilli et al., 2002). SAPK3 is identical to the previously reported rice Ser/Thr protein kinase REK (Hotta et al., 1998). The amino acid sequences of the SnRK2 protein can be divided into two parts, the N-terminal highly conserved kinase domain and the divergent C-terminal domain containing regions rich in acidic amino acids (Halford and Hardie, 1998). A phylogenetic tree was constructed with the amino acid sequences of the kinase domains of all the members of the rice and Arabidopsis SnRK2 family and of some members functionally characterized in other species (Figure 1A). The tree suggests that these protein kinases can be divided into three subclasses, denoted here subclasses I, II, and III, which contain SAPK3 through SAPK7, SAPK1 and SAPK2, and SAPK8 through SAPK10, respectively (Figure 1A). The tree based on the sequences of the C-terminal domain (data not shown) gave essentially the same result except for the position of SAPK3, which appeared to be isolated both from subclasses I and II. Therefore, classification for SAPK3 is tentative. Because each subclass contains Arabidopsis members, the distinction of the subclasses is believed to have been established before the divergence of dicot and monocot phyla. V. faba AAPK and Arabidopsis SnRK2.6 (OST1/SRK2E), both of which have been reported to be activated by ABA and involved in the ABA regulation of stomatal closing, are included in subclass III. Halford and Hardie (1998) have divided the family into SnRK2a (corresponding to subclass I) and SnRK2b (corresponding to subclass II) in a comparison that does not include the subclass III members. More structural similarities were found between subclasses II and III than other pairs: (1) the C-terminal sequence blocks are conserved (Figure 1B); (2) the number of amino acids is fewer compared with subclass I; and (3) the acidic patches are rich in Asp, whereas those of subclass I are abundant in Glu (Figure 1B). The last difference was previously pointed out as a difference between SnRK2a and SnRK2b (Halford and Hardie, 1998), indicating that subclass III can be included in SnRK2b.
Figure 1.
Rice and Arabidopsis SnRK2 Protein Kinase.
(A) A phylogenetic tree constructed with the amino acid sequences of the kinase domains (amino acid residues 1 to 268 of SAPK1 and corresponding residues of other sequences) of all SnRK2 family members of rice and Arabidopsis and some selected members from other species. The rice and Arabidopsis members are highlighted with black and white backgrounds, respectively. Nomenclature of the Arabidopsis members followed that of the PlantsP database (http://plantsp.sdsc.edu/; Hrabak et al., 2003). OST1/SRK2E are synonymous to SnRK2.6. Alignment of the full-length sequences is in the supplemental data online. The bootstrap values are in percent.
(B) Alignment of the amino acid sequences of the C-terminal domains. Identical amino acid residues are boxed, and similar residues are shaded in gray. Dashes indicate gaps in the sequences to allow maximal alignment.
Both SAPK1 and SAPK2 are highly homologous to wheat PKABA1 and barley HvPKABA1. The amino acid sequences of PKABA1 and HvPKABA1 show the highest identities to SAPK1 among the rice family members. Searching the EST databases led to the identification of wheat and barley SnRK2 members (TaSAPK1 and HvSPAK2, respectively) that are more homologous to SAPK2 than to SAPK1 (Figure 1A). Thus, SAPK1 and SAPK2 are considered orthologs of PKABA1/HvPKABA1 and TaSAPK2/HvSAPK2, respectively.
Expression of Some Members of the Rice SnRK2 Family Is Regulated by Hyperosmotic Stress and ABA
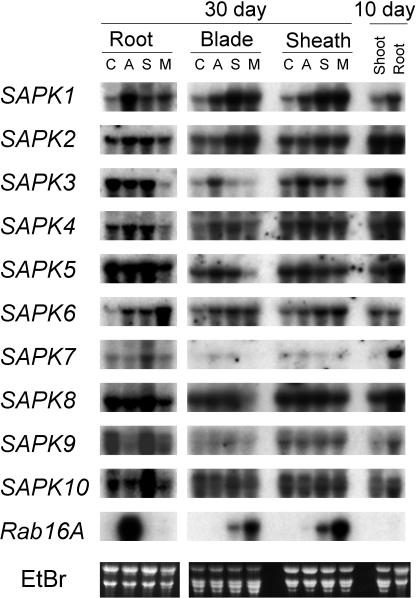
Expression of the SnRK2 members was analyzed by RNA gel blot hybridization (Figure 2). The expression of all the members of the SnRK2 family was detected in the leaf blades and the sheath as well as in the roots. The following effects were observed upon treatment with ABA (50 μM), NaCl (150 mM), or mannitol (600 mM) on the expression of each gene. SAPK1 was upregulated by all three treatments in both root and the above-ground organs, although the effect of ABA was weaker than those of the other two treatments. SAPK2 was upregulated by NaCl or mannitol treatment significantly in blades and weakly in sheaths but not in other organs. SAPK3 was upregulated by ABA but not by other treatments in the blades and sheaths and downregulated by mannitol treatment in the roots. SAPK4 was upregulated by ABA or NaCl treatment in the blades. SAPK5 was downregulated by mannitol treatment strongly in the blades and weakly in the roots. SAPK6 was weakly upregulated by all treatments in the blades and the sheaths, and weakly by ABA or NaCl and strongly by mannitol treatment in the roots. SAPK7 was upregulated to low levels by ABA or NaCl treatment in the blades but not in the sheaths and to a higher level by NaCl in the roots. SAPK8 was downregulated only by mannitol treatment strongly in the blades and weakly in the sheaths and the roots. SAPK9 was downregulated by ABA or mannitol in the roots but not in other organs. SAPK10 was similar to SAPK9. The transcript levels were significantly higher in roots than shoots of 10-d-old plants, except for SAPK2 and SAPK6. This tendency was also observed for the 30-d-old plants (data not shown).
Figure 2.
The Effects of Various Stresses on the Expression of SAPK1 through SAPK10.
Ten micrograms of total RNA isolated from leaf blades, sheaths, and roots of 30-d-old rice plants, which had been subjected to control (C) or 50 μM ABA (A), 150 mM NaCl (S), or 600 mM mannitol (M) for 12 h, or shoots and roots of untreated young plants (10 d old) were analyzed by gel blot hybridization with the indicated probe. Because the blots with the root RNA from 30-d-old plants were separately hybridized from those with the other RNAs, signal intensities cannot be directly compared between them. A Rab16A probe was also hybridized to the blot to monitor the treatments. EtBr, ethidium bromide staining image.
Expression of REK (SAPK3) had not been detected in roots in the previous report (Hotta et al., 1998). However, we detected a significant level of the SAPK3 transcript in roots. Whereas they used field-grown plants for RNA preparation, we analyzed the expression with plants grown hydroponically in a greenhouse. Such differences in the growth condition may have resulted in the difference in the expression in roots.
All the Members of the SnRK2 Protein Kinase Family Are Activated in Response to Hyperosmotic Stress
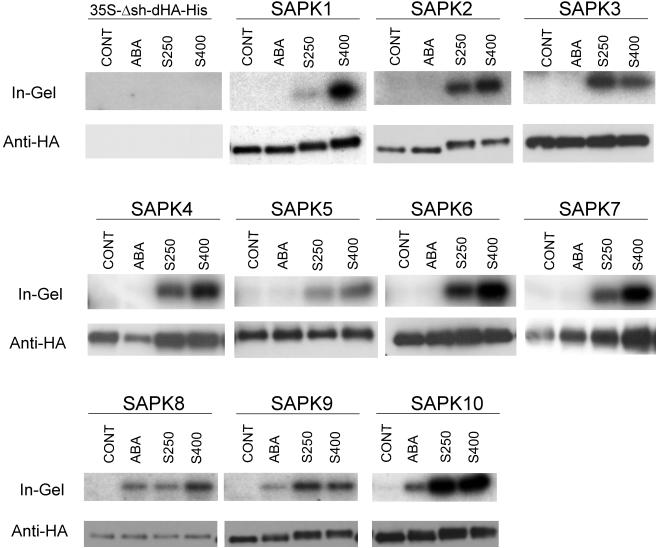
Whereas V. faba AAPK and Arabidopsis SRK2E are activated by ABA (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002), the tobacco 42-kD ASK1-like protein kinase has been shown to be activated by hyperosmotic stress (Mikolajczyk et al., 2000). Thus, we tested whether each of the rice SnRK2 members was activated by hyperosmotic stress and ABA. To do this, cultured cell protoplasts were transiently transformed with a vector that could express each of the rice SnRK2 members as a fusion protein with double haemagglutinin epitope (dHA) and polyhistidine affinity (His) tags. Transformed cells were treated with 250 mM or 400 mM NaCl or 50 μM ABA. Then, the expressed protein was recovered with Ni-agarose affinity resin and subjected to the in-gel kinase assay using MBP as a substrate and to immunoblot analysis using anti-HA antibody.
MBP kinase activities were detected in the affinity-recovered protein fractions from the cells expressing any of the SnRK2 members when they were treated with either concentration of NaCl, whereas essentially no specific kinase activities were detected when the cells were subjected to control treatment (Figure 3). The cells transformed with an empty vector gave no specific kinase activities and HA-reactive bands in the in-gel kinase assay and immunoblot analysis, respectively, regardless of treatment (Figure 3). Therefore, the detected kinase activities were judged to represent those of the expressed SnRK2 kinases. These results indicated that all of the rice SnRK2 members are activated in response to NaCl treatment of the cells. Based on further detailed analyses on SAPK1 and SAPK2 described below, the observed effects of NaCl treatment were considered responses to hyperosmotic stress.
Figure 3.
Hyperosmotic Stress Activates All the Members of the SnRK2 Protein Kinase Family, among Which SAPK8, SAPK9, and SAPK10 Are Also Activated by ABA.
Protoplasts transformed with an empty vector (35S-Dsh-dHA-His) or the expression vector for each of the dHA-His–tagged SAPK1 through SAPK10 were treated with 50 μM ABA, or 250 (S250) or 400 mM (S400) NaCl as indicated for 30 min. Control treatment (CONT) was performed by adding R2P medium alone. After treatment, the expressed protein was affinity recovered and subjected to the in-gel kinase assay with MBP as a substrate (top; In-Gel) and immunoblot analysis with anti-HA antibody (bottom; Anti-HA). In the immunoblot analysis, sometimes two bands were detected. Although slower migrating bands are likely to be authentic judging from size, faster bands always behaved in parallel with the upper bands and had kinase activities. They could be N-terminally truncated forms produced by proteolysis or products translated from a second Met.
Interestingly, dependence of the relative activation level on the NaCl concentration differed between family members. SAPK1 was activated to a much higher level by treatment with 400 mM NaCl than with 250 mM NaCl. Although SAPK2, SAPK4, SAPK5, SAPK6, SAPK7, and SAPK8 were also activated to higher levels by 400 mM NaCl than by 250 mM NaCl, the differences between the two concentrations were not as great compared with that observed for SAPK1. SAPK10 was activated to the same level by the two NaCl concentrations. The activation levels tended to be lower by 400 mM NaCl than by 250 mM for SAPK3 and SAPK9.
Only Limited Members of the SnRK2 Protein Kinase Family Are Activated in Response to ABA
Whereas all the members of the rice SnRK2 family were activated by NaCl treatment, only SAPK8, SAPK9, and SAPK10 were activated by ABA (Figure 3). To our surprise, the results mean that there are no SnRK2 members that are activated only by ABA. As mentioned above, these three kinases are members of subgroup III, which includes AAPK and OST1/SRK2E, the two previously characterized ABA-activated protein kinases. The structures specific to the kinases of this subgroup are suggested to be responsible for the regulation of activities by ABA signals.
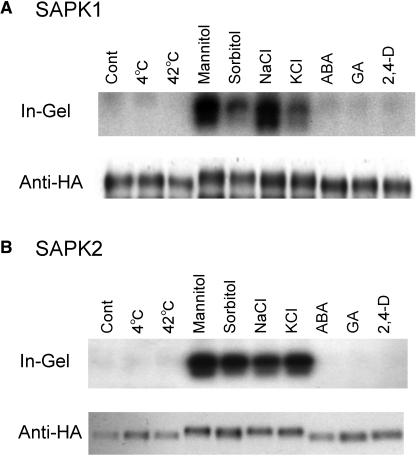
SAPK1 and SAPK2 Are Rapidly Activated Specifically in Response to Hyperosmotic Stress
To further characterize the rice SnRK2 family protein kinases, we focused on two highly homologous members, SAPK1 and SAPK2. When the cells were treated with 600 mM mannitol or sorbitol or 400 mM KCl, these kinases were also activated (Figure 4). Therefore, the activation effect of NaCl treatment on SAPK1 and SAPK2, as well as the other members of SnRK2, was considered a response to hyperosmotic stress. For unknown reason, sorbitol and KCl appeared to be not as effective as mannitol and NaCl, respectively, at the same concentrations. The difference was highly pronounced for SAPK1 because it requires higher levels of the stress for activation. Treatment with 2,4-D or GA3, or high or low temperature stress, did not result in the activation of SAPK1 and SAPK2 (Figure 4).
Figure 4.
SAPK1 and SAPK2 Are Activated Specifically by Hyperosmotic Stress.
Protoplasts expressing dHA-His–tagged SAPK1 (A) or SAPK 2 (B) were treated with NaCl (400 mM), KCl (400 mM), mannitol (600 mM), sorbitol (600 mM), ABA (50 μM), GA (5 μM), or 2,4-D (20 μM) or subjected to a temperature shift (4°C or 42°C) for 30 min. Control treatment (Cont) was performed by adding R2P medium alone. After treatment, the expressed protein was affinity recovered and analyzed as in Figure 3.
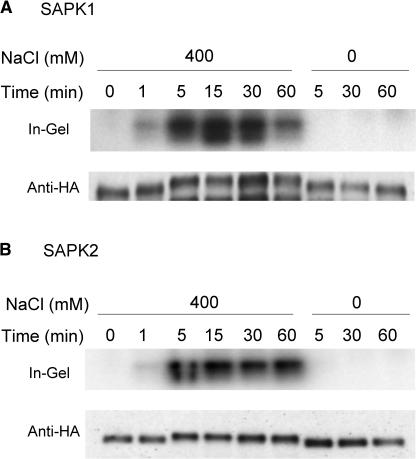
Time-course analysis revealed that activation of SAPK1 and SAPK2 was already detectable after 1 min and reached near maximal levels after 5 min of NaCl treatment (Figure 5). The activation level of SAPK1 but not of SAPK2 declined after 60 min (Figure 5). Similar rapid activation in response to hyperosmotic stress has been observed for other SnRK2 (or SnRK2-like) protein kinases, such as the tobacco ASK1-like 42-kD protein kinase, as well as other hyperosmotically activated ∼40-kD protein kinases (Mikolajczyk et al., 2000). Therefore, it is likely that the family members, in general, function in early steps of hyperosmotic signaling.
Figure 5.
Activation of SAPK1 and SAPK2 by Hyperosmotic Stress Is Rapid.
Protoplasts expressing dHA-His–tagged SAPK1 (A) or SAPK 2 (B) were treated with NaCl (0 or 400 mM) for the indicated time. After treatment, the expressed protein was affinity recovered and analyzed as in Figure 3.
Differential Responses to Osmotic Strengths between SAPK1 and SAPK2 Are Determined by the C-Terminal Domain
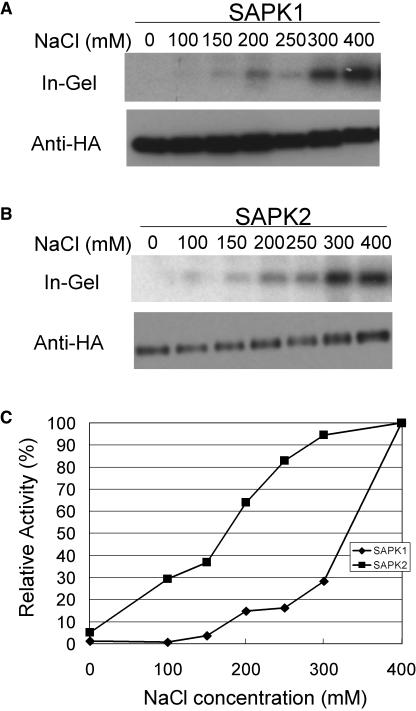
As mentioned above, differential responses to osmotic strengths were observed even between SAPK1 and SAPK2, the two highly homologous SnRK2 members; a higher level of relative activation at a lower NaCl concentration was observed for SAPK2 compared with SAPK1 (Figure 3). To further explore such differential regulation of these two SnRK2 members, a dose–response analysis was performed in a more quantitative manner (Figure 6). Whereas essentially no activation was observed for SAPK1 by 100 mM NaCl, SAPK2 was activated to 30% of the level obtained by 400 mM NaCl. SAPK1 activation was observed only at NaCl concentrations higher than 200 mM, and a sharp rise in the activation level was observed above 300 mM.
Figure 6.
SAPK1 and SAPK2 Exhibit Distinct Osmotic Strength Dependence for Activation.
(A) and (B) Protoplasts expressing dHA-His–tagged SAPK1 (A) or SAPK2 (B) were treated with NaCl at the indicated concentration for 30 min and analyzed as in Figure 3.
(C) Kinase activities obtained in (A) and (B) were normalized with the amount of SAPK protein by quantifying radioactivity in the in-gel kinase assay and the chemiluminescent signals in immunoblot analysis using a phosphor imager and a CCD camera, respectively, and plotted against the NaCl concentration. Relative kinase activities are expressed as a percent of the maximum activity obtained at 400 mM. Diamonds, SAPK1; squares, SAPK2.
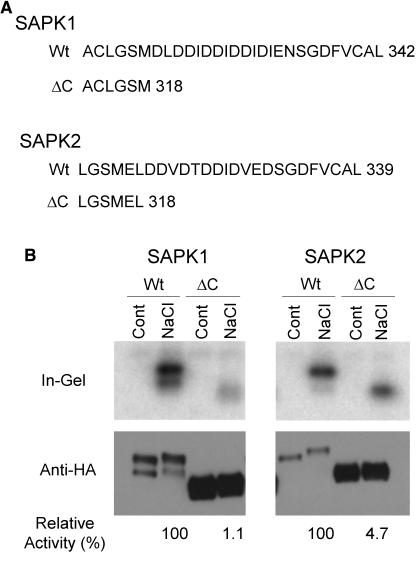
Because the kinase domains of SAPK1 and SAPK2 are highly conserved (92% amino acid identity), we speculated if the difference in the regulatory property might reside in the more divergent C-terminal domain. Before testing this hypothesis, we examined the role of the C-terminal domain for the kinase activity by deleting the domain. When a portion of the domain, which included the acidic amino acid patch, was deleted, specific activities normalized with the SAPK protein were dramatically decreased, although a very low level of activation by hyperosmotic stress was still observed, indicating the importance of the C-terminal domain for full activity (Figure 7). It should be noted that levels of the expressed proteins greatly increased by the deletions both for SAPK1 and SAPK2. This suggested that the C-terminal domain may also play a role in determining protein stability.
Figure 7.
Deletion of the C-Terminal Acidic Region Dramatically Reduces the Activities of SAPK1 and SAPK2.
(A) The C-terminal amino acid sequences of the wild-type (Wt) SAPK1 and SAPK2 or their C-terminally deleted derivatives (ΔC). The C-terminal amino acid residue number is indicated at right of each sequence.
(B) Protoplasts expressing dHA-His–tagged wild-type (Wt) SAPK1 or SAPK2 or their C-terminal deletion derivative (ΔC) were treated with 400 mM NaCl for 30 min, analyzed, and quantified as in Figures 3 and 6B, respectively. At the bottom of the immunoblot images, relative normalized kinase activities of the hyperosmotically activated enzyme, determined as in Figure 6, are indicated.
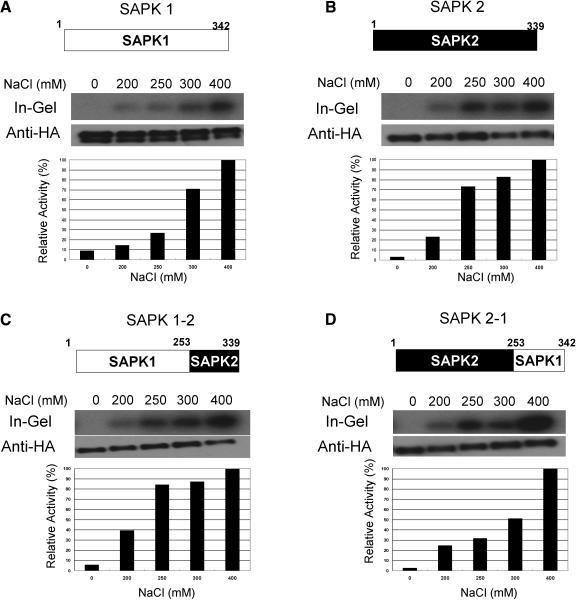
To test whether the C-terminal domain has a regulatory function in the differential response to osmotic strengths between SAPK1 and SAPK2, domain exchange experiments were performed (Figure 8). When the SAPK1 C-terminal domain was substituted with that of SAPK2, the chimeric kinase responded to increasing concentrations of NaCl similarly to that of SAPK2 (Figure 8C). Conversely, the dose response of the chimeric kinase with the SAPK2 kinase domain and the SAPK1 C-terminal domain was similar to that of SAPK1 (Figure 8D). These results indicated that the C-terminal domain of these kinases is responsible for the differential activation response to osmotic strengths.
Figure 8.
Differential Responses to Osmotic Strengths between SAPK1 and SAPK2 Are Determined by the C-Terminal Domain.
Protoplasts expressing dHA-His–tagged SAPK1 (A) or SAPK2 (B) or a chimeric kinase consisting of the SAPK1 kinase domain and the SAPK2 C-terminal domain (C) or consisting of the SAPK2 kinase domain and the SAPK1 C-terminal domain (D) were treated with NaCl at the indicated concentration for 30 min, analyzed, and quantified as in Figures 3 and 6B. On the top of each panel, the structure of the wild type or chimeric kinase, including the amino acid residues at the borders of the kinase and the C-terminal domains, is illustrated.
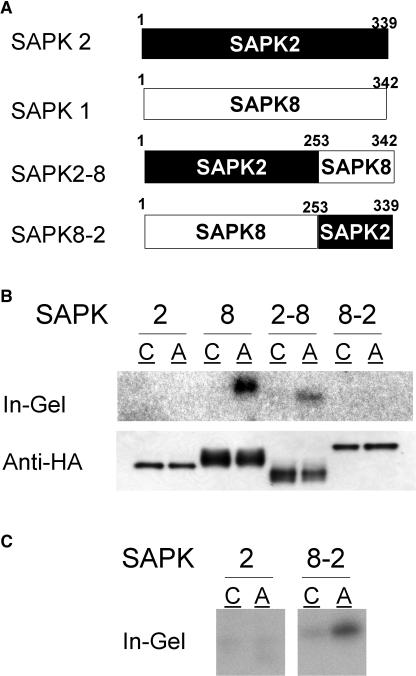
The C-Terminal Domain of an ABA-Activated SnRK2 Member Can Confer ABA Responsiveness to a Member That Is Not Activated by ABA
As described above, despite the high sequence conservation in the kinase domain among all members, only limited SnRK2 members were found to be activated by ABA (Figure 3). Therefore, it is likely that ABA activation is also specified by the divergent C-terminal domain. To determine whether the C-terminal domain possesses such a regulatory role, the C-terminal domains were exchanged between SAPK2 and SAPK8 (Figure 9A). When the chimeric kinase SAPK2-8 having the SAPK2 kinase domain and the SAPK8 C-terminal domain was expressed in protoplasts, it was clearly activated by ABA (Figure 9B). This result indicated that the SAPK8 C-terminal domain plays a central role in the activation by ABA signal. Conversely, the chimeric kinase SAPK8-2 having the SAPK8 kinase domain and SAPK2 C-terminal domain failed to exhibit detectable ABA activation when comparable amounts of SAPK protein were used (Figure 9B). However, residual ABA activation of SAPK8-2 was detected when a 20 times larger amount of the kinase protein was loaded (Figure 9C). Therefore, the kinase domain itself can also receive ABA signals, though at a low efficiency.
Figure 9.
The C-Terminal Domain of SAPK8 Can Confer ABA Responsiveness to the SAPK2 Kinase Domain.
(A) Schematic illustrations of chimeric kinases consisting of the SAPK2 kinase domain and the SAPK8 C-terminal domain (SAPK2-8) and consisting of the SAPK8 kinase domain and the SAPK2 C-terminal domain (SAPK8-2), including the amino acid residues at the borders of the kinase and C-terminal domains
(B) Protoplasts expressing dHA-His–tagged SAPK2 or SAPK8 or the chimeric kinase SAPK2-8 or SAPK8-2 were treated with 50 μM ABA (lane A) or control solution (lane C) for 30 min and analyzed as in Figure 3.
(C) Twenty times larger amounts of SAPK2 and SAPK8-2 proteins used in (B) were analyzed by the in-gel kinase assay. Under these conditions, basal or contaminating activities were detected for control-treated samples.
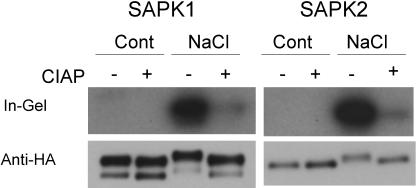
SAPK1 and SAPK2 Are Activated by Phosphorylation in Response to Hyperosmotic Stress
Mobility decreases in response to NaCl or ABA treatment were observed for some of the SnRK2 members, including SAPK1 and SAPK2, in the immunoblot analyses in Figures 3 to 9. Such mobility shifts always paralleled enzyme activation. Activation of kinase activities observed in the in-gel kinase assay, which involves a denaturation process, is likely to represent covalent modifications of the enzyme, such as phosphorylation. Therefore, we tested whether the activation of SAPK1 and SAPK2 in response to hyperosmotic stress requires phosphorylation (Figure 10). SAPK1 and SAPK2 proteins expressed in protoplasts and activated by hyperosmotic stress were recovered and treated with calf intestine alkaline phosphatase (CIAP). CIAP treatment of SAPK1 and SAPK2 resulted in the loss of kinase activities, which was concomitant with the decreased mobility, indistinguishable from those of their inactive forms (Figure 10). These results indicated that the activation of SAPK1 and SAPK2 in response to hyperosmotic stress was mediated by phosphorylation, which was detected as mobility shifts. In the time course and dose–response analyses (Figures 5 and 6), a gradual decrease in the motilities occurred along with increasing activity levels. This may suggest that phosphorylation occurs at multiple sites, the degree of which correlates to activation levels.
Figure 10.
SAPK1 and SAPK2 Are Activated by Phosphorylation.
Unactivated or activated dHA-His–tagged SAPK1 or SAPK2 obtained from transfected control (Cont) or NaCl (400 mM)-treated protoplasts, respectively, was incubated with (+) or without (−) CIAP as described in Methods and analyzed as in Figure 3.
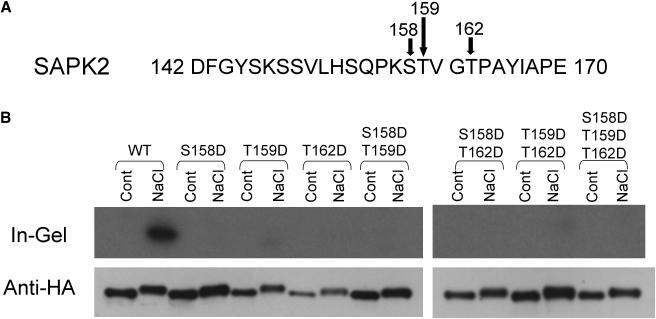
Ser and Thr Residues in the Activation Loop Are Essential for the Activity and/or Activation of SAPK1 in Response to Hyperosmotic Stress
In an attempt to create constitutive active forms of SAPK1, mutant forms of SAPK1 were expressed, which had substitution(s) for Ser and/or Thr to Asp within the regions corresponding to the activation loop because similar mutations have been reported to create constitutively active forms in several protein kinases, including a member of the SnRK1 protein kinase family (Gong et al., 2002). Based on some examples of such constitutively active mutants (Asai et al., 2002; Gong et al., 2002), we created several Ser and/or Thr to Asp mutant forms of SAPK1. However, none of these mutations [Ser-158 to Asp (S158D), Thr-159 to Asp (T159D), Thr-162 to Asp (T162D), or their combinations] resulted in constitutively active forms (Figure 11). Instead, all of them almost completely lost kinase activity, even when expressing cells were subjected to hyperosmotic stress. The results indicated that Ser-158, Thr-159, and Thr-162 are essential for activity or activation. Nevertheless, hyperosmotic stress–induced mobility shifts similar to that of the wild-type protein were observed. In other words, the SAPK proteins were phosphorylated without their kinase activities, suggesting that the observed phosphorylation was not caused by intermolecular autophosphorylation. Hotta et al. (1998) reported that REK (SAPK3) undergoes Ca2+-dependent autophosphorylation. It needs to be tested whether such an activity is related to the phosphorylation of SAPK proteins required for enzyme activation.
Figure 11.
Ser and Thr Residues in the Activation Loop Is Essential for the Activity and/or Activation of SAPK1 in Response to Hyperosmotic Stress.
(A) The amino acid residues for the Ser or Thr that were converted to the Asp residue are indicated by arrows in the SAPK2 partial amino acid sequence.
(B) Protoplasts expressing the wild-type (WT) dHA-His–tagged SAPK2 or its mutant form (S158D, T159D, T162D, S158D/T159D, T159D/T162D, or S158D/T159D/T162D) were treated with 400 mM NaCl or subjected to control (Cont) treatment and analyzed as in Figure 3.
DISCUSSION
Partial amino acid sequences of a hyperosmotic stress–activated 42-kD protein kinase, which was biochemically purified from tobacco cultured cells, were shown to have high similarities to cloned SnRK2 sequences (Mikolajczyk et al., 2000). However, cloning of the cDNA or genomic sequence for the 42-kD tobacco protein kinase has not been reported. Although the observation that soybean SnRK2 members SPK1 and SPK2 were activated by hyperosmotic stress in yeast cells is also suggestive of their involvement in hyperosmotic stress signaling, the activation in plant cells was not demonstrated (Monks et al., 2001). Thus, our knowledge of SnRK2 protein kinases in relation to hyperosmotic stress has been only fragmentary. On the other hand, two cloned SnRK2 protein kinases, V. faba AAPK and Arabidopsis OST1/SRK2E, have been shown to be activated by ABA and to function in the major pathway of ABA signaling, at least in guard cells (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). A suggestion from these observations is that each member or subgroup of SnRK2 is activated by a distinct signal and involved in a specific transduction pathway. Contrary to such an implication, however, we have demonstrated here that all members of the SnRK2 family protein kinases encoded by the rice genome are activated by hyperosmotic stress and that some of them are activated by ABA as well. Thus, there are no members that are activated only by ABA. These results indicated that the entire family has specifically evolved as a part of the signal transduction machinery for hyperosmotic stress response and that some of the members have additionally acquired regulation by ABA. Although we have shown that low or high temperature stress does not activate SAPK1 and SAPK2, it remains to be determined whether the SnRK2 members are activated by other signals, such as elicitors and H2O2 as observed for the hyperosmotic stress–activated MAPKs.
Our genome-wide functional analysis of the rice SnRK2 family combined with the phylogenetic comparison, including the full members of the family in rice and Arabidopsis, the representative monocot and dicot genomes, respectively, is of significance in knowing the functional properties of the uncharacterized members of Arabidopsis as well as of other plant species. The ABA-activated SnRK2 members, rice SAPK8 through SAPK10, V. faba AAPK, and Arabidopsis OST1/SRK2E are evolutionarily more related to one another than to other rice members and classified into subclass III. Therefore, the ancestral SnRK2 protein kinase common to the subclass III kinases may have evolved by gaining the capacity to be activated by ABA. From the phylogenetic relationships, other Arabidopsis members included in subclass III (SnRK2.2 and SnRK2.3) are predicted to be activated by ABA. It is also highly likely that AAPK and OST1/SRK2E are also activated by hyperosmotic stress.
A wheat SnRK2 protein kinase, PKABA1, has been implicated in the negative regulation of GA signaling by ABA in aleurone tissue. This implication is based on two types of observations (Gomez-Cadenas et al., 1999, 2001; Shen et al., 2001; Johnson et al., 2002). First, PKABA1 expression is induced by ABA. Second, transient overexpression of this kinase in aleurone tissue suppresses the GA-induced activity of several GA-regulated promoters. SAPK1 is considered to be orthologous to PKABA1 based on sequence homology. Expression of SAPK1 was induced by ABA like PKABA1. However, our results indicated that SAPK1 is inactive or has only barely detectable activity without hyperosmotic signal and is not activated by ABA. Thus, it remains to be determined whether SAPK1 functions in the negative regulation by ABA of GA signaling in the aleurone tissue. We demonstrated that SAPK1 and SAPK2 are activated by hyperosmotic stress via phosphorylation. It has been shown that a phosphorylated state of the enzyme is essential for AAPK activity and the tobacco ASK1-like 42-kD protein kinase (Li et al., 2000; Mikolajczyk et al., 2000). However, our study has shown that the phosphorylation of SnRK2 members is actually induced by hyperosmotic stress and required for activation, by correlating the stress-induced mobility shift with phosphorylation, and activation with phosphorylation. The phosphorylation-mediated activation mechanism of SAPK1 and SAPK2 can be extrapolated to other members because most members exhibited mobility shifts that are induced by hyperosmotic stress, as seen for SAPK1 and SAPK2. In general, phosphorylation of a protein does not always accompany a mobility shift on SDS-PAGE. Therefore, those SnRK2 members that did not display a clear mobility shift upon activation could also be activated via phosphorylation, considering the high sequence similarities among the members.
The activation of the subclass III members by ABA is also suggested to be mediated by phosphorylation. A careful examination of the SAPK8 immunoblot bands revealed a slight mobility shift that was confirmed upon activation by ABA to a similar extent to that observed for the activation by hyperosmotic stress (Figure 3). For SAPK9 and SAPK10, only small portions of the immunoblot signals were observed as shifted bands. This appeared to be reflected by the lower levels of the relative activation by ABA compared with those by hyperosmotic stress. By contrast, there was not a large difference in the levels of SAPK8 activation by ABA and by hyperosmotic stress, as in the case with SAPK9 and SAPK10. Such differences are likely to be derived from the difference in the amounts of protein expressed per cell, which were considerably lower for SAPK8 than for SAPK9 and SAPK10. Therefore, the relatively low level of activation by ABA is likely as a result of the limitation of the activation capacity by ABA signal but not to the difference in the activity per molecule in the fully activated state.
Although it is suggested that the activation of the subclass III members by both hyperosmotic and ABA signals is mediated by phosphorylation, it remains to be determined whether the same phosphorylation sites are used for the two types of activation. However, in either case, hyperosmotic and ABA signals are assumed to be separately relayed to the subclass III members if a common upstream mechanism (activating kinases) is operating for hyperosmotic stress activation of the SnRK2 members. Otherwise, the members of other subclasses should be activated by ABA. Alternatively, members of subclasses I and II may use the upstream kinases regulated only by hyperosmotic signal, whereas subclass III members are activated by distinct upstream kinases, which are regulated by both hyperosmotic and ABA signals. However, the high sequence conservation among the members of the three subclasses favors the former possibility.
Despite the high sequence similarities and the common regulation by hyperosmotic stress, the rice SnRK2 members exhibited functional distinctions in two aspects. First, only certain members are activated by ABA signals as discussed above. Second, differences were found among the members in the osmotic strength dependence of activation, even between the two closest members, SAPK1 and SAPK2. Domain exchange experiments revealed that the divergent C-terminal domains are responsible for both distinctions. One likely possibility for the biochemical function for the C-terminal domain is, therefore, that it provides a binding site for the upstream activating kinase. SAPK1 and SAPK2 may respond to different levels of osmotic stress by binding to the same activating kinase via the C-terminal domain with a lower and higher affinity, respectively, or to distinct activating kinases that are activated by higher and lower osmotic strengths, respectively.
As discussed above, the putative kinases that activate SnRK2 members in response to ABA and hyperosmotic signals are likely to be distinct. Therefore, the C-terminal domain of subclass III members may bind both types of activating kinases, one regulated by ABA and the other regulated by hyperosmotic stress, whereas the C-terminal domain of other subclasses would bind only to the latter type. The putative activating kinase appeared to be able to recognize and phosphorylate the target SnRK2 protein kinases without the aid of the C-terminal domain, though at a low efficiency because partial deletion of the domain resulted in dramatic loss of activities, but still responded at low levels to hyperosmotic stress. In other words, a specific recognition could be achieved by the interaction via the structure around the target phosphorylation site to some extent and facilitated by the specific interaction with the C-terminal domain. This hypothesis will explain the observation that the chimeric kinase, consisting of the SAPK8 kinase domain and the SAPK2 C-terminal domain, was activated by ABA, though at a low level.
The biological significance of the differential osmotic responses of the SnRK2 members is currently unknown. We have not investigated the spatial expression patterns of each family member; they may be differentially expressed depending on tissue/cell types having different sensitivities to the stress. Phosphorylation of a soybean phosphatidylinositol transfer protein, Ssh1p, is rapidly induced by NaCl at concentrations higher than 500 mM in leaf tissues but by NaCl concentrations of 200 mM in root tissues. The identity of Ssh1p kinase has been proposed to be that of SnRK2 family protein kinases because soybean SPK1 (classified as a subclass I SnRK2 member; Figure 1) can phosphorylate Ssh1p in yeast cells and features Ssh1p kinase activities detected by in-gel kinase assays using cell extracts that resemble those of the tobacco ASK1-like 42-kD kinase (Monks et al., 2001). The activation of these kinase activities requires a NaCl concentration higher than 500 mM in leaf tissues, whereas it is activated by 250 mM NaCl or less in cultured cells (Monks et al., 2001). These observations appear to be related to the differential response of the SnRK2 members to osmotic strengths described in this study.
The ABA signal itself is produced as a consequence of hyperosmotic stress. In a simplified model, hyperosmotic or drought response pathways are divided into two types, ABA-dependent and -independent ones (Shinozaki and Yamaguchi-Shinozaki, 2000). From this model, our finding that the subclass III SnRK2 members are activated by both ABA and hyperosmotic stress is quite puzzling; the dual regulation of these kinases indicates the mergence of the two signals and would predict the production of common responses. One possibility is that the subclass III protein kinases may not be involved in the pathway for many of the known ABA responses. However, this possibility is less likely because not only the stomatal response but also the ABA-inducible expression of rd22 and rd29B are suppressed in srk2e/ost1 mutants (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). One of the most critical criteria for whether a certain response is ABA dependent or independent is whether it is abolished in ABA-deficient mutants. However, drought/hyperosmotic stress–inducible genes that are also induced by applied ABA-inducible genes tend to be considered as those mediated via the ABA-dependent pathway without testing using the mutant. In addition, at least some drought/hyperosmotic stress–inducible genes have both ABA-responsive elements (ABREs) and dehydration-responsive elements, which act as cis elements for ABA signal and ABA-independent drought signal, respectively. Thus, it appears not easy to discriminate between ABA-dependent and -independent responses. Taking this into consideration, our results may suggest that hyperosmotic signal activates the downstream pathway common to the ABA-response pathway without increasing the ABA level. Such a cross-regulation is reminiscent of the synergistic activation of the wheat Em gene by ABA and osmotic stress (Bostock and Quatrano, 1992).
At least one of the rice class III members is expected to function in the ABA regulation of stomatal closure as AAPK and OST1/SRK2E do. Conversely, these two kinases from V. faba and Arabidopsis are expected to be activated not only by ABA but also by hyperosmotic stress, just as SAPK8, SAPK9, and SAPK10 are activated. This may suggest that hyperosmotic stress will also be able to induce stomatal closure without being mediated by ABA. Low humidity–induced stomatal closure has been shown to occur in an ABA-independent manner (Assmann et al., 2000). Although how low relative humidity is sensed by the leaf is poorly understood, the activation of subclass III members by hyperosmotic stress may be related to such ABA-independent stomatal regulation.
Recently, an interaction between PKABA1 and an ABRE-binding factor, TaABF, has been demonstrated by a yeast two-hybrid assay (Johnson et al., 2002). If the ABRE-binding factor is an in vivo substrate for PKABA1, expression of ABA-responsive genes would be induced or modified by hyperosmotic stress because PKABA1 is suggested to be activated by hyperosmotic stress but not by ABA based on the orthologous relationship between SAPK1 and PKABA1. The synergistic effect of hyperosmotic stress on the ABA-induced expression of the wheat Em gene (Bostock and Quatrano, 1992) may also be one of the consequences of such an interaction of the two signals.
Although the hyperosmotic stress activation of all the family members revealed in this study strongly suggests their important roles in the hyperosmotic signal transduction, the downstream components and the outputs of the signaling pathway are yet to be elucidated. Overexpression of these kinases in transgenic plants could result in the activation of downstream responses because very low but significant basal activities of SAPKs were detectable without applying hyperosmotic stress (data not shown). It will also be possible to trace upstream of the pathway, first by identifying the immediate upstream kinase, which will greatly contribute to the overall understanding of hyperosmotic stress as well as ABA signal transduction mechanisms. Such studies are currently under way.
METHODS
Plasmid Construction
cDNA fragments of SAPK1 through SAPK10 were obtained by PCR using EST clones obtained from the National Agrobiological Institute (Tsukuba, Japan; GenBank accession numbers C22414 [SAPK1]; C22640 [SAPK5]; AU056506 [SAPK6]; AU075635 [SAPK7]) or cDNAs reverse transcribed from total RNA prepared from rice (Oryza sativa) seedlings (14 d old), developing seeds (1 to 12 d after flowering) or suspension cultured cells (Oc line). The 5′ and 3′ primers contained EcoRI or BamHI and SalI or XhoI sites, respectively, for subcloning. The amplified cDNA was cut with EcoRI and SalI and cloned into the corresponding site of p35S-shΔ-dHA-His (Kagaya et al., 2002), which allows the expression of C-terminally dHA-His–tagged SAPK protein in cultured cell protoplasts. Domain-exchanged or amino acid substituted derivatives of SAPK cDNA fragments were produced by multiplex PCR and similarly cloned into p35S-shΔ-dHA-His. Sequences of the primers used for cloning are provided in the supplemental data online.
Expression and Affinity Recovery of SAPKs in Cultured Cell Protoplasts
The expression plasmids (10 μg) described above were transfected into protoplasts prepared from rice cultured cells (Oc line) by electroporation as described previously (Kagaya et al., 2002). Electroporated cells were concentrated by natural sedimentation and cultured in 1.2 mL of R2P medium (Kyozuka and Shimamoto, 1991) at a cell density of ∼4 × 106 cells/mL) for 15 h. One-fourth volume of R2P medium containing NaCl, KCl, sorbitol, mannitol, ABA, GA3, or 2,4-D at a 5× concentration of treatment was added to a 1-mL aliquot of the protoplast culture. After the treatment for the desired time, the protoplast suspension was mixed with 255 μL of a concentrated His-binding buffer to give the following final concentrations of the components in 1.7 mL: 20 mM sodium phosphate buffer, pH 7.4, 10 mM imidazol, 1% Tween 20, 50 mM β-glycerophosphate, 100 μM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and Complete EDTA-free (protease inhibitor cocktail tablets; Roche, Penzberg, Germany), frozen in liquid N2. After thawing, a 10 M NaCl solution and distilled water were added to the cell suspension to give the final concentration of 500 mM and to make the final volume of 1.7 mL, respectively. Cells were further disrupted by sonication, and debris was removed by centrifugation. The obtained cell extract was mixed with 50 μL (as a packed volume) Ni-NTA agarose (Qiagen, Valencia, CA) and incubated at 4°C for 30 min with rotation to allow the expressed protein to bind. Then, the resin was washed six times with a washing buffer (20 mM sodium phosphate buffer, pH 7.4, 20 mM imidazol, 500 mM NaCl, 50 mM β-glycerophosphate, 100 μM Na3VO4, 0.5 mM PMSF, and Complete EDTA-free), and the bound protein was eluted with 40 μL of an elution buffer (20 mM sodium phosphate buffer, pH 7.4, 250 mM imidazol, 150 mM NaCl, 0.5 mM EDTA, 50 mM β-glycerophosphate, 100 μM Na3VO4, 0.5 mM PMSF, and Complete EDTA-free).
Immunoblot Analysis
Protein samples prepared as described above were separated by electrophoresis on a 7.5% SDS-polyacrylamide gel until a 25-kD prestained maker (Bio-Rad, Hercules, CA) ran out from the gel. Proteins on the gel were electroblotted onto nitrocellulose membrane (Trans-BlotN transfer medium; Bio-Rad) in blotting buffer (4.8 mM Tris, 3.9 mM Gly, 20% methanol, and 0.13 mM SDS). Immunodetection of the HA-tagged proteins was performed as described previously (Kagaya et al., 2002). For quantification, immunoblot signals were captured by a cooled CCD camera (Imagemaster VDS-CL; Amersham Pharmacia Biotech, Buckinghamshire, UK).
In-Gel Kinase Assay
Protein samples were separated by electrophoresis on a 10% polyacrylamide gel embedded with 0.2 mg/mL of MBP (Upstate, Charlottesville, VA) and subjected to denaturation/renaturation treatment as described previously (Mikolajczyk et al., 2000). The in-gel kinase reaction solution contained 10 mM Tris, pH 7.5, 2 mM DTT, 0.1 mM EGTA, 15 mM MgCl2, and 20 μM ATP and 185 kBq/mL [γ-32P]ATP (110 TBq/mmol; Amersham). The reaction was terminated by transferring the gels into a bath containing 5% trichloroacetic acid and 1% sodium phosphate. The unincorporated [γ-32P]ATP was removed from the gels by repeated washing in the bath solution. Finally, the gels were exposed to BioMax MS autoradiography film (Eastman Kodak, Rochester, NY).
Phosphatase Treatments
dHA-His–tagged SAPK1 or SAPK2 protein in the protoplast extracts was bound to Ni-NTA agarose resin and washed three times as described above. The resin was further washed three times by CIAP reaction buffer (50 mM Tris-HCl, pH 7.5, and Complete EDTA-free), then incubated with or without CIAP (Takara-Shuzo, Kyoto, Japan) at 37°C for 24 h. After treatment, the resin was washed by the reaction buffer three times, and the proteins were eluted with 40 μL of an elution buffer (50 mM Tris-HCl, pH 7.5, 250 mM imidazol, 50 mM EDTA, and Complete EDTA-free) and subjected to the in-gel kinase assay and immunoblot analysis.
Phylogenetic Analysis
Multiple sequence alignments were conducted using ClustalX (version 1.8.1; Thompson et al., 1997) and presented using BOXSHADE (version 3.21; http://www.ch.embnet.org/software/BOX_form.html). An unrooted neighbor-joining tree (Saitou and Nei, 1987) was also built using ClustalX (version 1.8.1) and presented using TreeView (Page, 1996). Support for the tree obtained was assessed using the bootstrap method (Felsenstein, 1985). The number of bootstrap replicates was 1000.
Plant Treatments
Germinated seeds of rice (O. sativa subsp japonica cv Nipponbare) were sown on a nylon net with a styrofoam float and grown hydroponically with one-fifth strength of MS salt solution under the greenhouse conditions for 30 d or in a growth chamber (continuous light; 28°C) for 10 d. After the growth periods, the plants were transferred to a solution containing 50 μM ABA, 600 mM mannitol, or 150 mM NaCl for treatment. For ABA treatment, the same solution was also sprayed onto above-ground organs.
RNA Isolation and Gel Blot Analysis
Total RNA was isolated by the aurintricarboxylic acid method described by Wadsworth et al. (1988). RNA was fractionated by electrophoresis on a formaldehyde-containing agarose gel and blotted to a nylon membrane (Hybond-XL; Amersham Pharmacia Biotech) by standard methods. The RNA gel blot was hybridized in a hybridization solution (250 mM Na2HPO4, pH 7.4, 7% SDS, and 1 mM EDTA) containing 32P-labeled DNA probe at 65°C overnight, washed four times in washing buffer (40 mM Na2HPO4, pH 7.4, 1% SDS, and 1 mM EDTA), and exposed to x-ray film.
Gene-specific probes for SAPK1 through SAPK10 were prepared by PCR using cDNA clones as templates. The PCR-amplified regions as expressed by nucleotide number relative to the putative translation start site were: SAPK1, 1031 to 1235; SAPK2, 1022 to 1176; SAPK3, 872 to 1149; SAPK4, 751 to 1210; SAPK5, 1029 to 1471; SAPK6, 1038 to 1408; SAPK7, 1037 to 1341; SAPK8, 1082 to 1351; SAPK9, 1060 to 1471; and SAPK10, 799 to 1223.
Sequence data from this article have been deposited with GenBank. Accession numbers for the rice SnRK2 members described here are as follows: SAPK1 (AB125302), SAPK2 (AB125303), SAPK3 (AB125304), SAPK4 (AB125305), SAPK5 (AB125306), SAPK6 (AB125307), SAPK7 (AB125308), SAPK8 (AB125309), SAPK9 (AB125310), and SAPK10 (AB125311). Arabidopsis Genome Initiative identification numbers of the Arabidopsis SnRK2 members are as follows: SnRK2.1 (At5g08590), SnRK2.2 (At3g50500), SnRK2.3 (At5g66880), SnRK2.4 (At1g10940), SnRK2.5 (At5g63650), SnRK2.6/OST1/SRK2E (At4g33950), SnRK2.7 (At4g40010), SnRK2.8 (At1g78290), SnRK2.9 (At2g23030), and SnRK2.10 (At1g60940). GenBank accession numbers of the sequences from other species are as follows: AAPK (AAF27340), PKABA1 (BAB61735), HvPKABA1 (BAB61735), SPK1 (L01453), and SPK2 (L19360). The accession numbers of SAPKs are given in Methods. The sequences of TaSAPK2 and HvSAPK2 are obtained by connecting overlapping EST sequences (accession numbers BJ251848 and BJ317818 for TaSAPK2; AV834998, BJ471725, and BJ463508 for HvSAPK2).
Supplementary Material
Acknowledgments
This work was funded in part by Grants-in-Aid for Scientific Research on Priority Areas (Grant 12138204) and the Center of Excellence from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project IP-5002) and the Japan Society for the Promotion of Sciences (Research for the Future Grant JSPS-ooL1603). We thank Tomiko Chikada for excellent technical assistance.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tsukaho Hattori (hattori@agr.nagoya-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019943.
References
- Albrecht, V., Ritz, O., Linder, S., Harter, K., and Kudla, J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M., Snyder, J.A., and Lee, Y.-R.J. (2000). ABA-deficient (aba1) and ABA-insensitive (abi1–1, abi2–1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant Cell Environ. 23, 387–395. [Google Scholar]
- Bostock, R.M., and Quatrano, R.S. (1992). Regulation of Em gene expression in rice, interaction between osmotic stress and abscisic acid. Plant Physiol. 98, 1356–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Cheong, Y.H., Kim, K.N., Pandey, G.K., Gupta, R., Grant, J.J., and Luan, S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15, 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney, A., and Verma, D.P.S. (1993). Proline biosynthesis and osmoregulation in plants. Plant J. 25, 215–223. [Google Scholar]
- Desikan, R., Hancock, J.T., Ichimura, K., Shinozaki, K., and Neill, S.J. (2001). Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol. 126, 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald, D.B., Torabinejad, J., Jones, C.A., Shope, J.C., Cangelosi, A.R., Thompson, J.E., Prestwich, G.D., and Hama, H. (2001). Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol. 126, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard, M.J., Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2002). Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 527, 43–50. [DOI] [PubMed] [Google Scholar]
- Droillard, M.J., Thibivilliers, S., Cazale, A.C., Barbier-Brygoo, H., and Lauriere, C. (2000). Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: Two crossroad MAP kinases and one osmoregulation-specific protein kinase. FEBS Lett. 474, 217–222. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.H., and Walker-Simmons, M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Zentella, R., Walker-Simmons, M.K., and Ho, T.H. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–679. [PMC free article] [PubMed] [Google Scholar]
- Gong, D., Guo, Y., Jagendorf, A.T., and Zhu, J.K. (2002). Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 130, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Halfter, U., Ishitani, M., and Zhu, J.K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L., Song, C.P., Gong, D., Halfter, U., and Zhu, J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3, 233–244. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Hardie, D.G. (1998). SNF1-related protein kinases: Global regulators of carbon metabolism? Plant Mol. Biol. 37, 735–748. [DOI] [PubMed] [Google Scholar]
- Hotta, H., Aoki, N., Matsuda, T., and Adachi, T. (1998). Molecular analysis of a novel protein kinase in maturing rice seed. Gene 213, 47–54. [DOI] [PubMed] [Google Scholar]
- Hoyos, M.E., and Zhang, S. (2000). Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol. 122, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak, E.M., Chan, C.W.M., Gribskov, M., Harper, J.F., Choi, J.H., Halford, N., Kudla, J., Luan, S., Nimmo, H.G., Sussman, M.R., Thomas, M., Walker-Simmons, K., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Yoshida, R., Yuasa, T., and Shinozaki, K. (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24, 655–665. [DOI] [PubMed] [Google Scholar]
- Johnson, R., Wagner, R., Verhey, S.D., and Walker-Simmons, M.K. (2002). The ABA-responsive kinase PKABA1 interacts with a seed-specific ABA response element binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya, Y., Hobo, T., Murata, M., Ban, A., and Hattori, T. (2002). Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14, 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., Eklof, S., Till, S., Bogre, L., Hirt, H., and Meskiene, I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-N., Cheong, Y.H., Grant, J.J., Pandey, G.K., and Luan, S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-N., Cheong, Y.H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–325. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., and Shimamoto, K. (1991). Transformation and regeneration of rice protoplasts. In Plant Tissue Culture Manual, B1, K. Lindsey, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–16.
- Li, J., Wang, X.Q., Watson, M.B., and Assmann, S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287, 300–303. [DOI] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.S., and Zhu, J.K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Plant Cell 12, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.), S389–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T., Wurgler-Murphy, S.M., and Saito, H. (1994). A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk, M., Olubunmi, S.A., Muszynska, G., Klessig, D.F., and Dobrowolska, G. (2000). Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12, 165–178. [PMC free article] [PubMed] [Google Scholar]
- Monks, D.E., Aghoram, K., Courtney, P.D., DeWald, D.B., and Dewey, R.E. (2001). Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 13, 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik, T., Irvine, R.F., and Musgrave, A. (1998). Phospholipid signaling in plants. Biochim. Biophys. Acta 1389, 222–272. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Ligterink, W., Meskiene, I., Calderini, O., Beyerly, J., Musgrave, A., and Hirt, H. (1999). Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 20, 381–388. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Meijer, H.J.G., ter Riet, B., Frank, W., Bartels, D., and Musgrave, A. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. [DOI] [PubMed] [Google Scholar]
- Mustilli, A.-C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse, T.S., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK6. J. Biol. Chem. 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Oono, Y., Seki, M., Nanjo, T., Narusaka, M., Fujita, M., Satoh, R., Satou, M., Sakurai, T., Ishida, J., Akiyama, K., Iida, K., Maruyama, K., et al. (2003). Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J. 34, 868–887. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Pical, C., Westergren, T., Dove, S.K., Larsson, C., and Sommarin, M. (1999). Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J. Biol. Chem. 274, 38232–38240. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99, 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Gomez-Cadenas, A., Zhang, P., Walker-Simmons, M.K., Sheen, J., and Ho, T.H. (2001). Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol. Biol. 47, 437–448. [DOI] [PubMed] [Google Scholar]
- Shi, J., Kim, K.-N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., Luan, S., and Kudla, J. (1999). Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1996). Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 7, 161–167. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Katagiri, T., Mizoguchi, T., Yamaguchi-Shinozaki, K., Hayashida, N., and Shinozaki, K. (1994). Two genes that encode Ca2+-dependent protein kinases are induced by drought and high salt stresses in Arabidopsis thaliana. Mol. Gen. Genet. 224, 331–340. [DOI] [PubMed] [Google Scholar]
- Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, B., Hirayama, T., and Shinozaki, K. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11, 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth, G.J., Redinbaugh, M.G., and Scandalios, J.G. (1988). A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal. Biochem. 172, 279–283. [DOI] [PubMed] [Google Scholar]
- Wang, X. (1999). The role of phospholipase D in signaling cascades. Plant Physiol. 120, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14S, 165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.-K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. [DOI] [PubMed] [Google Scholar]
- Yang, K.Y., Liu, Y., and Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Alonso, J., Ecker, J.R., and Shinozaki, K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yuasa, T., Ichimura, K., Mizoguchi, T., and Shinozaki, K. (2001). Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 42, 1012–1016. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1999). Abscisic acid metabolism and its regulation. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Amsterdam, The Netherlands: Elsevier Science), pp. 189–207.
- Zhang, S., and Klessig, D.F. (1998). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95, 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.