Abstract
Highly organized interphase cortical microtubule (MT) arrays are essential for anisotropic growth of plant cells, yet little is known about the molecular mechanisms that establish and maintain the order of these arrays. The Arabidopsis thaliana spiral1 (spr1) mutant shows right-handed helical growth in roots and etiolated hypocotyls. Characterization of the mutant phenotypes suggested that SPR1 may control anisotropic cell expansion through MT-dependent processes. SPR1 was identified by map-based cloning and found to encode a small protein with unknown function. Proteins homologous to SPR1 occur specifically and ubiquitously in plants. Genetic complementation with green fluorescent protein fusion proteins indicated that the SPR1 protein colocalizes with cortical MTs and that both MT localization and cell expansion control are conferred by the conserved N- and C-terminal regions. Strong SPR1 expression was found in tissues undergoing rapid cell elongation. Plants overexpressing SPR1 showed enhanced resistance to an MT drug and increased hypocotyl elongation. These observations suggest that SPR1 is a plant-specific MT-localized protein required for the maintenance of growth anisotropy in rapidly elongating cells.
INTRODUCTION
Directional cell expansion is fundamental to plant morphogenesis. In cells undergoing diffuse growth, directional cell elongation (anisotropic cell expansion) requires both turnover and reorganization of the cell wall–constituting polysaccharides, such as cellulose, hemicelluloses, and pectins (Brett and Waldron, 1996). Among them, bundles of cellulose polymers (cellulose microfibrils) appear to act as the major load-bearing polymer that specifies growth direction by preventing turgor pressure–driven wall yielding parallel to their alignment while permitting it in the opposite direction (Baskin, 2001).
It has been proposed that oriented deposition of cellulose microfibrils is controlled by cortical microtubules (MTs). Cortical MTs and cellulose microfibrils are often found to run in parallel (Baskin, 2001 and the references therein). In Arabidopsis thaliana roots, this similar orientation only occurs during the early phase of cell elongation (Sugimoto et al., 2000). Such parallelism has been explained by a putative mechanism in which the cellulose-synthesizing protein complex may track along cortical MTs beneath the plasma membrane (monorail model), or the movement of the complex may be restricted by the cortical MT deposition (guard rail model) (reviewed in Foster et al., 2003). By the use of sophisticated histological techniques and genetic methods, however, recent publications suggest that cortical MTs and cellulose microfibrils control anisotropic cell expansion in at least partly independent pathways (Sugimoto et al., 2003 and the references therein).
Cortical MTs comprise a specialized cytoskeletal array found particularly in walled plant cells. Over the past decade, time-lapse imaging and photobleaching experiments of cortical MTs in living plant cells has increased our understanding of their dynamic properties (Wasteneys et al., 1993; Hush et al., 1994; Yuan et al., 1994; Shaw et al., 2003). Earlier work demonstrated that cortical MTs are nucleated at the cell cortex (Wasteneys et al., 1989). Most recently, green fluorescent protein (GFP)-tubulin expression in epidermal cells of Arabidopsis has shown that some plasma membrane–associated MTs can translocate by a differential treadmilling at the polymer ends (Shaw et al., 2003). As can be predicted from animal studies, regulated treadmilling and membrane attachment of plant cortical MTs should require concerted operation of specific MT-associated proteins (MAPs). Indeed, several MAPs have been identified in plants and shown to localize to cortical MTs (for reviews, see Azimzadeh et al., 2001; Hussey et al., 2002; Wasteneys, 2002), though all plant MAPs so far identified at the molecular level have related proteins in animals (Hashimoto, 2003).
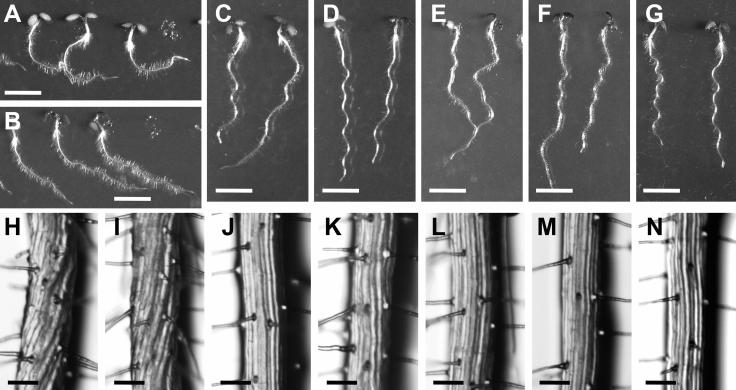
The Arabidopsis spiral1 (spr1) mutant was isolated because of its characteristic helical root growth (Furutani et al., 2000). Epidermal cell files of spr1 roots are twisted to form right-handed helices (Figures 1H and 1I). On vertically oriented hard agar plates, spr1 roots grow to the right when viewed from above the agar plates (Figures 1A and 1B). This skewed root growth is driven by the friction between agar surface and helical epidermal cell files. The spr1 mutant phenotype is enhanced under the conditions that accelerate cell elongation. Under such conditions, epidermal cells of spr1 mutants undergo isotropic cell expansion, resulting in spherically shaped cells protruding from the organ surface (Furutani et al., 2000).
Figure 1.
Phenotypes of spr1 Mutants and Transgenic Plants.
Top panels show 4-d-old seedlings grown on agar plates in the light. Bottom panels show magnification of the root epidermal cell files. Bars in (A) to (G) = 5 mm; bars in (H) to (N) = 100 μm.
(A) and (H) spr1-1 mutants (in ecotype Ler).
(B) and (I) spr1-2 mutants (in ecotype Col).
(C) and (J) Wild-type Ler.
(D) and (K) Wild-type Col.
(E) and (L) spr1-1 plants transformed with a 2.1-kb genomic DNA fragment spanning the At2g03680 gene sequence.
(F) and (M) PSPR1:SPR1:GFP transgenic plants made in the spr1-2 mutant background.
(G) and (N) P35S:SPR1 transgenic plants made in the Col background.
Cortical MT orientation in spr1 plants is abnormal. In root epidermal cells, cortical MTs are oriented obliquely to form left-handed helices, whereas in the ground tissue of etiolated hypocotyls, a mixture of transverse, oblique, and longitudinal arrays are observed (Furutani et al., 2000). The helical growth phenotype is suppressed by low concentrations of MT drugs, oryzalin, propyzamide, and taxol. When used at higher concentrations, these drugs were shown to depolymerize (oryzalin and propyzaminde) or bundle (taxol) MTs, thereby inducing radial expansion of plant cells. The spr1 phenotype is also enhanced at low temperatures, which are known to destabilize MT polymers in some plant cell types. From these observations, we hypothesized that the SPR1 protein is required for the functional integrity of cortical MTs and therefore is essential for anisotropic cell expansion (Furutani et al., 2000).
In this study, we identified the SPR1 gene by a map-based cloning strategy. We determined that SPR1 encodes a small protein with unknown biochemical functions. Proteins homologous to SPR1 are found to occur ubiquitously in plants, but no related sequence was found outside the plant kingdom. Consistent with our hypothesis on the SPR1 functions, SPR1 protein was shown to associate with cortical MTs. Expression analysis and overexpression experiments suggested that SPR1 is required to maintain growth anisotropy in cells undergoing rapid expansion.
RESULTS
Map-Based Cloning of SPR1
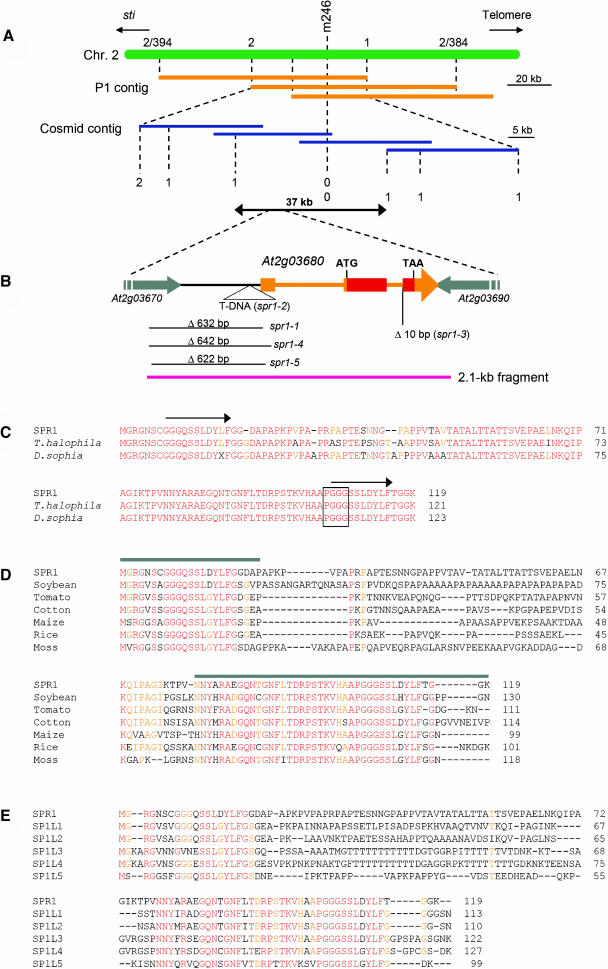
Using the spr1-1 mutant allele, SPR1 was initially mapped close to the cleaved amplified polymorphisms (CAPS) marker m246 on chromosome 2 (Figure 2A). Because sequence information was not available at the time, P1 and cosmid contigs were constructed for the mapped locus. New CAPS markers were created based on the partial DNA sequences obtained from the clones in the contig and used to restrict the SPR1 gene to a 37-kb region (Figure 2A). Because spr1-1 was derived from a seed pool irradiated with fast neutrons, we searched for a deletion or a rearrangement in the 37-kb region of the spr1-1 genome. Restriction fragment length polymorphism analysis and PCR amplification of the spr1-1 and Landsberg erecta (Ler) genomic DNA within the target region revealed a 0.6-kb deletion in a 5.5-kb HindIII fragment. Transformation of spr1-1 mutants with the full-length Ler fragment rescued the helical growth defects (data not shown), indicating that the SPR1 gene lay in this region. Sequence analysis revealed a 632-bp deletion in the spr1-1 allele that disrupted two genes, At2g03670 and At2g03680 (Figure 2B), both of which were predicted to be expressed based on the presence of ESTs.
Figure 2.
Cloning Procedure and Protein Sequence Alignments of SPR1.
(A) Fine mapping of the SPR1 locus. The region flanking the CAPS marker m246 was covered by P1 phage clones (orange bars). CAPS markers were created from the P1 sequences and used to locate the SPR1 locus to a 60-kb region. This region was covered by cosmid clones (blue bars), and new CAPS markers were generated from the cosmid sequences. Mapping with these markers finally confined the SPR1 locus to a 37-kb region. Numbers of recombination events are shown.
(B) Genomic region surrounding the SPR1 gene (At2g03680). Transcribed region of SPR1 is colored orange or red. Orange boxes represent exons, and thin orange bars represent introns. The SPR1-coding region is red. Parts of the two flanking genes are shown with gray arrows. Molecular lesions found in five spr1 mutant alleles are shown. The pink bar represents the 2.1-kb genomic fragment shown to complement spr1-1 mutants.
(C) Protein sequence alignment of SPR1 and two Brassicaceae family homologs. Residues identical in all three proteins are shown in red, and those identical in two proteins are shown in orange. Arrows above the SPR1 sequence indicate peptide segments with sequence identity. The PGGG motif found in some mammalian MAPs is boxed.
(D) Protein sequence alignment of SPR1 and six non-Brassicaceae family homologs. Residues identical in all seven proteins are shown in red, and those identical in five or six proteins are orange. Gray bars above the SPR1 sequence indicate the region with high sequence identity among all SPR1 homologs. These regions were used for the GFP fusion experiments shown in Figure 5.
(E) Protein sequence alignment of SPR1 and five Arabidopsis homologs. Residues identical in all six proteins are shown in red, and those identical in five are orange.
Accession numbers for the sequences used in the alignments are listed in Methods.
To identify which of the two genes was responsible for the spr1 phenotype, four more mutant alleles (spr1-2 through spr1-5; all proved to be allelic by noncomplementation) were sequenced. spr1-4 and spr1-5 contained deletions similar to that in the spr1-1 (Figure 2B). spr1-2 harbored a T-DNA insertion immediately downstream of the putative TATA box element of At2g03680 (Figure 2B). spr1-3 had a 10-bp deletion at an intron/exon junction of At2g03680 (Figure 2B). Furthermore, a 2.1-kb genomic fragment that spanned the entire coding region and the 0.4-kb 5′ region of At2g03680, but only a small C-terminal region of At2g03670 (Figure 2B), rescued the spr1-1 mutant (Figures 1E and 1L). Taken together, we concluded that At2g03680 encodes the SPR1 gene.
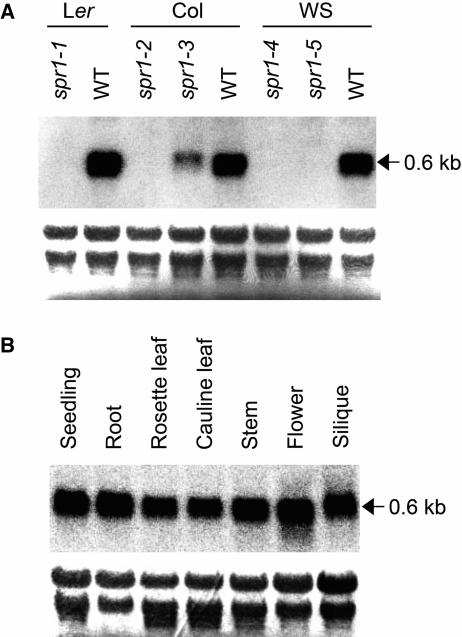
Consistent with large deletions or an insertion in the 5′ region, spr1-1, spr1-2, spr1-4, and spr1-5 mutants did not accumulate SPR1 transcripts (Figure 3A). By contrast, spr1-3 had reduced SPR1 transcript level but slightly larger transcript size than that generated in the wild-type plants (Figure 3A). Sequencing of RT-PCR products revealed that the second intron of SPR1 was not spliced in the spr1-3 mutant. This is consistent with the 10-bp deletion in the spr1-3 allele destroying the consensus acceptor sequence of the SPR1 second intron (Figure 2B).
Figure 3.
RNA Gel Blot Analysis of SPR1.
(A) An RNA gel blot from 9-d-old wild-type and spr1 mutant seedlings.
(B) An RNA gel blot of various organs from wild-type Col plants.
Each lane was loaded with 5 μg of total RNA. After transfer to nylon membranes, rRNA was stained with methylene blue to confirm equal loading (bottom panels). Membranes were then hybridized to detect SPR1 transcripts (top panels).
SPR1 Encodes a Novel Plant-Specific Protein
The predicted SPR1 polypeptide consists of 119 amino acids (Figure 2C). A TBLASTN search of the GenBank nucleotide database revealed no homologous gene with known biological or biochemical functions. Numerous EST sequences, however, were found to encode polypeptides homologous to SPR1. These SPR1-related genes appear to exist ubiquitously in plants: homologs were found in all 18 plant species for which >10,000 EST sequences have been deposited (http://www.plantgdb.org). SPR1-related sequences also were found in a moss EST database (http://moss.nibb.ac.jp/). Alignments of the protein sequences deduced from some of the EST clones are shown in Figures 2C and 2D. Only the proteins from two Brassicaceous species, Thellungiella halophila and Descurainia sophia, show amino acid identity with SPR1 along the entire polypeptide length (Figure 2C). By contrast, the closest homologs from each of the non-Brassicaceous species show sequence identity only in N- and C-terminal regions, whereas the internal regions are highly variable (Figure 2D). The Arabidopsis genome contains five SPR1 homologs that we refer to as SPIRAL1-LIKE1 (SP1L1) through SP1L5. Again, these SP1L proteins share high sequence identity only in the N- and C-terminal regions (Figure 2E). The N- and C-terminal regions show limited sequence similarity to each other (Figure 2C, arrows). Based on the EST sequences deposited in the database, all SP1L genes, except for SP1L1, are predicted to be transcribed.
SPR1 Protein Is Colocalized with Cortical MTs
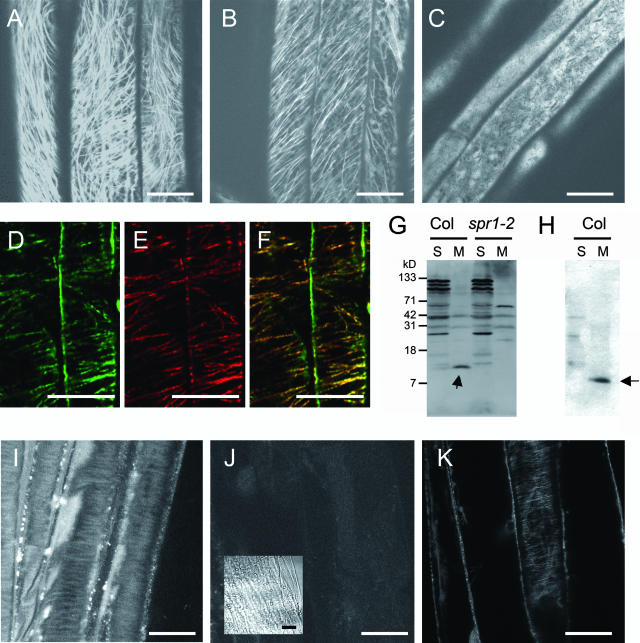
Because SPR1 protein sequence analysis did not provide information on likely functions, we sought to determine where the SPR1 protein is localized in Arabidopsis cells. To this end, we created transgenic plants that expressed a SPR1:GFP fusion protein under the control of the 0.4-kb 5′ region and the first intron of SPR1 (for convenience, we refer to this genomic fragment as the SPR1 promoter or PSPR1). Among the 33 transgenic lines generated in the spr1-2 mutant background (PSPR1:SPR1:GFP plants), 30 lines complemented the helical growth phenotype (Figures 1F and 1M), indicating functional integrity of the SPR1:GFP fusion protein. Confocal laser scanning microscopy (CLSM) observation of these transgenic plants clearly showed fibrous structures in the cortical regions of hypocotyl and root epidermal cells (Figures 4A and 4B). Similar structures were observed in other organs, including cotyledons, petioles, rosette leaves, and inflorescences (data not shown). When PSPR1:SPR1:GFP plants were pretreated with the MT depolymerizing drug oryzalin, the fibrous structures disappeared, indicating that the observed structures were MTs (Figure 4C). Localization of SPR1:GFP to cortical MTs was further confirmed by double immunostaining for GFP and tubulins (Figures 4D to 4F).
Figure 4.
SPR1 Protein Localization.
(A) to (C) Confocal images of PSPR1:SPR1:GFP transgenic plants.
(A) Upper hypocotyl of 5-d-old dark-grown seedlings.
(B) Root elongation zone of 5-d-old dark-grown seedlings.
(C) Upper hypocotyl of 5-d-old dark-grown seedlings treated with 200 nM oryzalin for 30 min before observation.
(D) to (F) Confocal images of a PSPR1:SPR1:GFP hypocotyl stained simultaneously with anti-GFP and anti-tubulin antibodies.
(D) Immunofluorescence with anti-GFP antibodies.
(E) Immunofluorescence with anti-tubulin antibodies.
(F) An overlaid image of (D) and (E) showing colocalization of SPR1:GFP and MTs (yellow).
(G) and (H) Protein gel blot analyses of SPR1 in plant protein extracts.
(G) Detection by a crude antiserum. SPR1 (arrow) was recovered mainly in a microsomal fraction (M) of wild-type Col extracts but was absent in extracts of the null spr1-2 mutant. Several nonspecific bands were present, especially in soluble fractions (S) of both wild-type and spr1-2 extracts.
(H) Detection by a purified antibody preparation. In wild-type Col extracts, the affinity-purified SPR1 antibody detected SPR1 in the microsomal fraction. Nonspecific bands in soluble and microsomal fractions were highly reduced.
(I) and (J) Confocal images of the dark-grown seedlings stained with antibodies for SPR1. Cortical cell surface in upper hypocotyl regions of Col (I) and spr1-2 (J) seedlings. Inset in (J), bright-field image.
(K) Confocal image of a dark-grown GFP:α-tubulin transgenic seedling.
Bars = 20 μm.
To confirm the MT localization of SPR1 in wild-type plants, an antiserum was produced with recombinant SPR1 proteins. On a protein gel blot, this antiserum recognized a protein band at expected size for SPR1 (12 kD) mainly in a microsomal fraction of wild-type Columbia (Col) but not in spr1-2 (Figure 4G, arrow). Because the antiserum gave many nonspecific bands in both Col and spr1-2 extracts, affinity purification was performed to give a single major band on a protein gel blot (Figure 4H, arrow). This antibody preparation was then used for immunofluoresent staining on dark-grown hypocotyls of wild-type Col seedlings. A CLSM observation of cortical cell surface revealed fibrous structures characteristic of cortical MTs (Figure 4I). In a negative control experiment, spr1-2 mutant seedlings showed only negligible fluorescence (Figures 4J). Similar fibrous structures were visualized by a CSLM observation of GFP:α-tubulin transgenic plants (Figure 4K) (Ueda et al., 1999), indicating that the SPR1 antibody stained the MTs. The observed colocalization of SPR1 with cortical MTs is consistent with the proposed role of SPR1 in MT-dependent processes (Furutani et al., 2000).
N- and C-Terminal Regions of SPR1 Are Sufficient for MT Localization
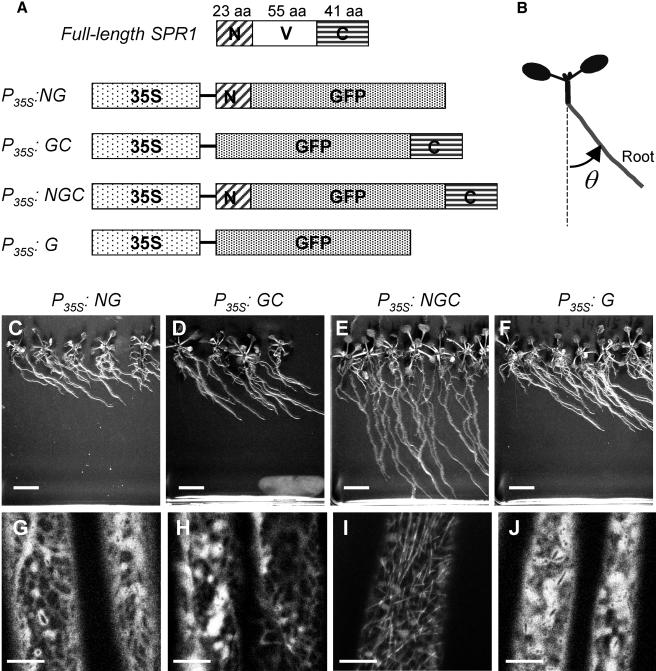
As described above, SPR1 and its homologs share sequence identity only in the N- and C-terminal regions, whereas the internal sequence is highly variable. This raised a question as to whether the internal region is required for the function(s) specific to each protein or primarily serves as a structural linker between the N- and C-terminal regions. Furthermore, we wanted to know if the conserved regions alone are sufficient for targeting SPR1 to cortical MTs. To address these questions, we created chimeric DNA constructs that express GFP proteins fused with N- and/or C-terminal regions of SPR1 under the 35S promoter (P35S) of Cauliflower mosaic virus (Figure 5A). These constructs were introduced into spr1-3 mutants.
Figure 5.
Functional Analysis of Conserved SPR1 Domains.
(A) Schematic pictures of full-length SPR1 coding sequence (top) and four chimeric DNA constructs. 35S, 35S promoter of Cauliflower mosaic virus; GFP, GFP-coding sequence; N, coding sequence for the 23–amino acid N-terminal region of SPR1; C, coding sequence for the 41–amino acid C-terminal region of SPR1; V, coding sequence for the 55–amino acid internal variable region of SPR1; aa, amino acids. See Figure 2D for exact excision points.
(B) Definition of the root skewing angle (θ) listed in Table 1.
(C) to (F) Independent T1 transgenic plants growing on hard agar plates.
(G) to (J) Confocal images of upper hypocotyl regions of dark-grown seedlings.
(C) and (G) P35S:NG; (D) and (H) P35S:GC; (E) and (I) P35S:NGC; (F) and (J) P35S:G.
Bars in (C) to (F) = 1 cm; bars in (G) to (J) = 10 μm.
As shown in Figure 5E, replacing the internal sequence of SPR1 with GFP (P35S:NGC; N, G, and C stand for SPR1 N-region, GFP, and SPR1 C-region, respectively) partially rescued the root skewing phenotype of spr1-3. Microscopic analysis revealed that the helical growth of root epidermal cell files was also partially rescued (data not shown). By contrast, GFP fused with either the N- or C-terminal region alone (P35S:NG or P35S:GC) or free GFP (P35S:G) failed to rescue the spr1-3 defects (Table 1, Figures 5C, 5D, and 5F).
Table 1.
Root Skewing Angles of GFP Transgenic Plants, Wild-Type Col, and spr1-3 Mutants
| Plant | Root Skewing Angle (θ)a | n |
|---|---|---|
| P35S:NG | 52.0 ± 8.7 | 138 b |
| P35S:GC | 50.8 ± 8.6 | 86 b |
| P35S:NGC | 24.2 ± 8.5 | 145 b |
| P35S:G | 53.4 ± 5.4 | 50 b |
| Col | −4.2 ± 3.4 | 120 c |
| spr1-3 | 54.7 ± 5.9 | 120 c |
See Figure 5B for the definition of θ.
Number of independent T1 plants used to score θ.
Number of individuals used to score θ.
Transgenic plants were then examined by CLSM to observe the distribution of the GFP fusion proteins. Only the NGC fusion protein was localized to cortical MTs (Figure 5I), whereas the NG, GC, and G proteins were all distributed throughout the peripheral cytoplasm (Figures 5G, 5H, and 5J). These results indicate that the conserved N- and C-terminal regions act together in targeting SPR1 proteins to cortical MTs and that the internal region may play minor roles as compared with the conserved N- and C-terminal regions. The coincidence between phenotypic rescue and MT localization further suggests that the primary function of SPR1 is related to its association with cortical MTs.
Expression Patterns of SPR1
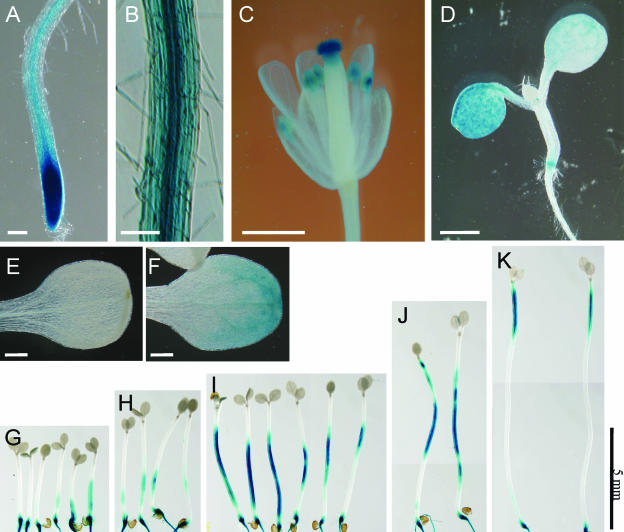
An RNA gel blot analysis using total RNAs from various organs of wild-type Arabidopsis plants indicated ubiquitous expression of SPR1 (Figure 3B). To analyze SPR1 expression at the tissue level, we made a construct in which the β-glucuronidase (GUS) reporter gene was placed under the 1.0-kb SPR1 promoter described above and the 0.3-kb 3′ region of SPR1 (TSPR1). This construct (PSPR1:GUS:TSPR1) was introduced into wild-type Col plants by Agrobacterium tumefaciens–mediated transformation. Seedlings from 28 independent transgenic lines were stained for GUS activity. All lines showed a very similar staining pattern, except for two that showed no staining. Five lines harboring homozygous T-DNA insertion at a single locus were established and used for detailed expression analysis described below. Strong GUS staining was found at the root tip, including early elongation zone (Figure 6A). In mature roots, GUS stain was present in all cell layers (Figure 6B). In flowers, strong GUS activity was found in anthers, stigmas (Figure 6C), and pollen (data not shown in detail).
Figure 6.
Histochemical GUS Staining of PSPR1:GUS:TSPR1 Transgenic Plants.
(A) Root apical region.
(B) Root basal region.
(C) Flower.
(D) Four-day-old seedlings grown under continuous light.
(E) A cotyledon of a 6-d-old seedling grown in the dark.
(F) A cotyledon of a seedling grown for 5 d in the dark and then exposed to continuous light for 1 d.
(G) to (K) Seedlings grown in the dark for 3 d (G), 3.5 d (H), 4 d (I), 5 d (J), and 6 d (K).
Bars in (A) and (B) = 100 μm; bars in (C) and (D) = 1 mm; bars in (E) and (F) = 200 μm; bar in (G) to (K) = 5 mm.
In young seedlings, the expression of SPR1 appeared to be dependent on light and developmental stage. When seedlings were grown under continuous illumination of white light, hypocotyls showed negligible staining, whereas cotyledons started to express GUS after 4 d of growth (Figure 6D). By contrast, when seedlings were grown in the dark, hypocotyls started to express GUS after 3 to 4 d of growth, whereas cotyledons remained unstained (Figures 6G to 6K). The light-dependent induction of GUS expression in the cotyledons was confirmed by exposing dark-grown seedlings to continuous light. One day after the transfer, cotyledons of the light-exposed seedlings started to express GUS, whereas those of control seedlings left in the darkness remained unstained (cf. Figures 6E and 6F). GUS staining in the root was constitutive; no difference was observed between light- and dark-grown seedlings (data not shown).
Interestingly, the region of strong GUS stain moved acropetally along the hypocotyls of dark-grown seedlings. Three- or 3.5-day-old seedlings (corresponding to 24 to 36 h after germination) showed GUS stain preferentially at the basal part of hypocotyls (Figures 6G and 6H). Four- or five-day-old seedlings were stained in the middle region of hypocotyls (Figures 6I and 6J). In 6-d-old seedlings, GUS stain revealed expression only in the apical region of fully elongated hypocotyls (Figure 6K). A similar expression pattern was detected by immunostaining of dark-grown wild-type hypocotyls (see supplemental data online).
In summary, high SPR1 expression was associated with tissues undergoing rapid cell expansion, including the root elongation zone, hypocotyls of dark grown-seedlings, and cotyledons of light-grown seedlings (Schiefelbein and Benfey, 1991; Neff and van Volkenburgh, 1994; Gendreau et al., 1997).
Overexpression of SPR1
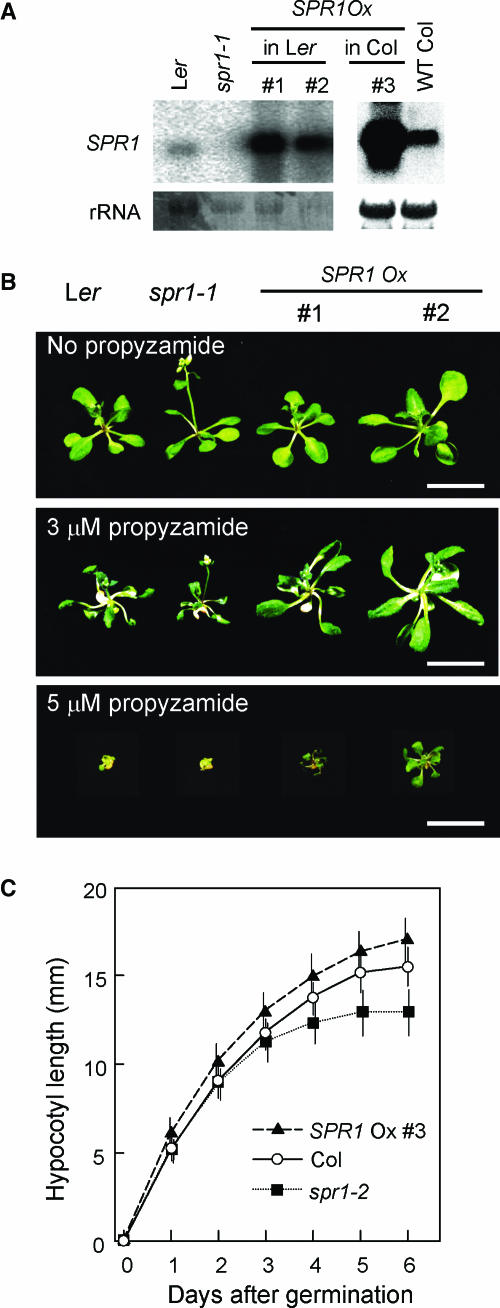
To gain further insight into the SPR1 function, we generated transgenic plants that overexpressed SPR1 under the 35S promoter of Cauliflower mosaic virus (P35S:SPR1 plants) in the wild-type Col and Ler backgrounds. RNA gel blot analysis revealed that P35S:SPR1 plants accumulated 20 to 40 times more SPR1 transcript than corresponding nontransformants (Figure 7A). The P35S:SPR1 plants were apparently normal (Figure 1G). It is notable that the root epidermal appearance of the P35S:SPR1 roots was indistinguishable from those of the wild type (Figure 1N): increased SPR1 level did not cause left-handed twisting of root epidermal cell files. This suggests that SPR1 does not simply act in biasing the cell elongation toward a certain direction. Rather, it suggests that SPR1 is necessary to maintain straight cell elongation.
Figure 7.
Overexpression of SPR1.
(A) RNA gel blots showing overexpression of SPR1 in P35S:SPR1 transgenic plants. Procedure for the RNA gel blot analysis is given in the legend for Figure 3. Top panels, hybridization with SPR1 probe; bottom panels, methylene blue staining.
(B) P35S:SPR1 transgenic plants show enhanced resistance to propyzamide. Plants of Ler, spr1-1, and two lines of P35S:SPR1 in Ler background were grown on nutrient agar media either without (top panel) or with 3 μM (middle panel) or 5 μM propyzamide (bottom panel) for 18 d. Bars = 1 cm.
(C) Hypocotyl elongation kinetics of Col (open circles), spr1-2 (filled squares), and a transgenic line of P35S:SPR1 in Col background (filled triangles). Plotted data is the average (symbols) and standard errors (vertical bars) from the measurements of 73, 87, and 65 individuals of Col, spr1-2, and P35S:SPR1, respectively.
The P35S:SPR1 plants were found to be moderately more resistant to long-term treatment with the MT-depolymerizing drug propyzamide. When grown on a medium containing 5 μM propyzamide, plants from two independent P35S:SPR1 lines (1 and 2) showed marginal survival, whereas both Ler and spr1-1 plants died (Figure 7B).
To investigate the role of SPR1 in cell elongation, we compared the cell elongation kinetics of P35S:SPR1, wild-type Col, and spr1-2 plants using dark-grown hypocotyls, the extension of which relies solely on cell expansion but not on cell division (Gendreau et al., 1997). Seedlings of wild-type Col, spr1-2, and a P35S:SPR1 line made in the Col background (line 3) were grown in the dark. At 24-h intervals, positions of the shoot apical meristem were marked on the surface of Petri dishes under a dissecting microscope illuminated with dim green light. At all time points, hypocotyls of the P35S:SPR1 plants were longer than those of wild-type plants (Figure 7C). The differences were statistically significant at all time points (P < 10−7, Student's t test). We repeated the same experiment and obtained a similar result. This observation suggests that enhancement of SPR1 expression levels can increase cell elongation. By contrast, hypocotyls of spr1-2 mutants elongated at a rate similar to that of wild-type plants up to 2 d after germination. At this time point, spr1-2 hypocotyls did not show the helical growth (data not shown). Three days after germination, elongation of spr1-2 hypocotyls started to decline. It was at this time point that the upper part of the spr1-2 hypocotyls developed the helical growth phenotype (Furutani et al., 2000).
DISCUSSION
SPR1 Encodes a Plant-Specific MT-Localizing Protein
Our map-based cloning lead to the identification of SPR1 as a novel protein composed of 119 amino acid residues. In the nucleotide database, the SPR1 cDNA sequence is annotated as a nitrilase-associated protein, based on an interaction in the yeast two-hybrid system (GenBank accession number Z96936; D. Bartling, personal communication). Our yeast two-hybrid assay, however, could not reproduce this interaction (R. Prieto and T. Hashimoto, unpublished results). The deposited sequence has a single base pair deletion close to the SPR1 stop codon. If this is not a simple sequencing error but has in fact been incorporated accidentally during the cloning procedure, the translated protein would have 27 unrelated amino acids translated from the 3′ noncoding region. Such an artificial sequence might account for the reported interaction with nitrilase in yeast. Under these circumstances, we will solely use SPR1 as the gene symbol.
The SPR1 amino acid sequence did not show homology to known proteins with defined functions, nor did it contain a structural motif indicative of its activity. Nevertheless, a database search revealed that SPR1-related proteins occur ubiquitously in plants; many monocot and dicot species as well as a moss species possess multiple SPR1-related genes. The deduced protein sequences from those ESTs and SPR1 are similar only in N- and C-terminal regions, except for two from other species in the Brassiceae, and therefore are closely related to Arabidopsis. The internal regions are highly variable but are similar in length. This suggests that amino acid residues important for the SPR1 function(s) reside mainly in the N- and C-terminal regions, whereas the internal region functions as a spacer that holds the two conserved polypeptides in a defined distance. The results of our GFP fusion protein expression study shown in Figure 5 support this hypothesis. Only GFP fused with the SPR1 polypeptides at both N and C termini could partially rescue the spr1 defect. The incomplete rescue may be attributable to either nonoptimal spacing by the GFP polypeptide, imperfect excision of the SPR1 N- and C-terminal domains, conformational constraint at the fusion points, or a combination of these problems. Use of the 35S promoter should not be a problem because intact SPR1 proteins expressed under P35S could completely rescue the spr1 defects (data not shown). Taken together, these findings suggest that SPR1 and its homologs have a similar function, even though their internal regions are highly divergent.
Observation of PSPR1:SPR1:GFP transgenic plants and immunostaining revealed that SPR1 colocalizes with cortical MTs in plant cells. Previously, we hypothesized that SPR1 is involved in MT-dependent processes based on the observation of cortical MTs and growth phenotype under the conditions that either stabilize or destabilize MTs (Furutani et al., 2000). Therefore, the observed colocalization of SPR1 with cortical MTs is consistent with our hypothesis. The results shown in Figure 5 further support the function of SPR1 on MTs; GFP fused with both N- and C-terminal regions of SPR1 could partially rescue the spr1 defects and concomitantly localize to cortical MTs. By contrast, free GFP or GFP fused with either the N- or C-terminal region of SPR1 alone did not rescue the spr1 defects and did not localize to cortical MTs. The SPR1 C-terminal region contains a PGGG sequence that is conserved in the MT binding repeats of several mammalian MAPs (Figure 2C) (Hyams and Lloyd, 1994). Moreover, transgenic plants overexpressing SPR1 showed slightly increased resistance to propyzamide, a drug that promotes MT depolymerization (Figure 7B), further correlating the SPR1 function to MT organization.
Highly ordered cortical MT arrays are a plant-specific cytoskeletal structure that play essential roles in directed expansion of plant cells (Burk et al., 2001; Whittington et al., 2001; Sugimoto et al., 2003). In this sense, it is noteworthy that no SPR1 homologs were found outside the plant kingdom. Possibly, SPR1 is required only for plant-specific MT-dependent processes but is dispensable for the MT functions common to animals and plants. Consistent with this view, no cell division–related defects were detected in the spr1 mutants (Furutani et al., 2000), though SPR1:GFP was also localized to noncortical MTs, such as preprophase band, spindles, and phragmoplasts (data not shown).
Role of SPR1 in Cell Elongation
Based on phenotypic characterization, we have previously hypothesized that the spr1 mutant is primarily impaired in directional cell expansion (Furutani et al., 2000). Present molecular analysis further supports the role of SPR1 in the cell elongation processes. First, as discussed above, SPR1 protein is localized to MTs. Several reports have shown that treatment of plant tissues with MT drugs results in isotropic cell expansion (Morejohn, 1991; Baskin et al., 1994; Hasenstein et al., 1999; Sugimoto et al., 2003). Recent molecular genetic analysis of Arabidopsis mutants also suggests that the functional integrity of cortical MTs is required for anisotropic cell expansion. Mutation in the MICROTUBULE ORGANIZATION1 (MOR1) gene that encodes a plant homolog of the TOGp/XMAP215 family of MAPs causes left-handed twisting and isotropic cell expansion in all plant organs (Whittington et al., 2001; Sugimoto et al., 2003). Similarly, cells in the fra2/bot1/lue1/erh3 mutants, which are impaired in the p60 subunit of the MT-severing enzyme katanin, are radially swollen (Burk et al., 2001; Bichet et al., 2001; Webb et al., 2002; Bouquin et al., 2003). The lefty mutants, which have mutations in α-tubulin genes, show constitutive left-handed helical growth in various organs (Thitamadee et al., 2002). lefty mutants were originally isolated as suppressors of spr1-1, indicating genetic interaction between SPR1 and α-tubulins.
Second, SPR1 is expressed preferentially in tissues with rapid cell elongation. In light-grown seedlings, SPR1 is expressed in cotyledons but not in hypocotyls. By contrast, in dark-grown seedlings, SPR1 is expressed in hypocotyls but not in cotyledons. The opposite effect of light on the expression of SPR1 in hypocotyls and cotyledons suggests that light is not a simple switch for the SPR1 expression. Rather, SPR1 expression seems to be controlled as a part of a light-dependent developmental program. In the light, plants undergo photomorphogenesis in which cotyledons rapidly expand, whereas hypocotyls show only limited elongation. In the dark, plants adopt skotomorphogenesis in which hypocotyls rapidly elongate, whereas cotyledons do not change their size (Neff and Van Volkenburgh, 1994; Fankhauser and Chory, 1997; Gendreau et al., 1997). Furthermore, the site of high SPR1 expression moved acropetally along dark-grown hypocotyls (Figures 6G to 6K). Gendreau et al. reported that the cell elongation of Arabidopsis etiolated hypocotyls is initiated at the basal cells and moves up toward the apical cells. The temporal and spatial distribution of SPR1-expressing cells in hypocotyls generally matches the zone of rapid cell elongation (Gendreau et al., 1997).
Third, transgenic plants overexpressing SPR1 showed a small but reproducible increase in the elongation of dark-grown hypocotyls. The relatively small increase suggests that the wild-type level of SPR1 is already close to the amount required for maximum elongation or that SPR1 can accelerate ongoing cell elongation but cannot trigger new cell elongation. The same experiment also showed reduced hypocotyl length of spr1 mutants. The measurements of spr1 hypocotyls, however, should be interpreted with caution. Hypocotyl elongation of spr1 started to deviate from that of wild-type plants only 3 d after germination. This is exactly the time when upper hypocotyls of spr1 mutant started to show helical growth, thereby reducing the overall length of hypocotyls. Therefore, the role of SPR1 in hypocotyl elongation should be deduced only from gain-of-function experiments such as overexpression.
Origin of the Helical Growth Phenotype
Several observations suggest that helical growth results from partial loss of anisotropic cell expansion. First, when the spr1 phenotype was enhanced either by growth at low temperature or by crossing with spr2 mutant that acts in cell expansion in a separate pathway, mutant epidermis underwent isotropic expansion to the extent of producing spherical epidermal cells (Furutani et al., 2000). Second, the temperature-sensitive mor1 mutant shows radial cell expansion at 31°C, whereas at a less restrictive temperature (29°C), left-handed helical growth is observed before the emergence of spherical cells (Whittington et al., 2001). Third, treatment of plant roots with high concentrations of MT drugs results in radial cell expansion (Morejohn, 1991; Baskin et al., 1994; Hasenstein et al., 1999; Sugimoto et al., 2003), whereas at lower concentrations, these drugs predominantly cause left-handed helical growth of the Arabidopsis roots. Forth, the lefty mutants that show left-handed helical growth are thought to have compromised MT status because of the incorporation of defective α-tubulins into their MT polymers (Thitamadee et al., 2002).
If helical growth is a manifestation of partially impaired anisotropic cell expansion, then why do null spr1 alleles show helical growth rather than isotropic cell expansion? A trivial possibility would be that SPR1 does not actively participate in maintaining anisotropic cell expansion. However, given the fact that SPR1 functions through conserved N- and C-terminal regions, it is more plausible that SPR1 and SP1Ls share partially redundant functions and that the null spr1 mutation represents partial loss of the SPR1/SP1L gene family. Our preliminary characterization of double mutants of SPR1 and some SP1Ls suggests that this is the case (K. Nakajima, T. Kawamura, and T. Hashimoto, unpublished results).
We have previously proposed a model in which the helical growth phenotype of spr1 arises from differential effects of the spr1 mutation on epidermis and ground tissue (i.e., relatively strong radial expansion of inner ground tissue causes the outer and inner cells to have different longitudinal length). To compensate for the different cell length, epidermal cells become inevitably skewed (Furutani et al., 2000). Spatial expression profile of SPR1 in this study did not provide experimental data for this model. SPR1 is expressed in all root tissues, including epidermis and ground tissue. Our preliminary experiments of cell layer–specific complementation of SPR1 suggest that a complex non-cell-autonomous mechanism is behind the helical growth phenotype (K. Nakajima and T. Hashimoto, unpublished results). Extensive analysis of SPR1 and SP1Ls expression patterns, as well as their interaction with cellular components affecting growth anisotropy (e.g., cortical MTs, cellulose microfibrils, and other wall constituents), is necessary to elucidate the complex mechanism that underlies the helical growth phenotype.
METHODS
Plant Materials and Growth Conditions
Isolation of spr1-1 (Ler), spr1-2 (Col), spr1-3 (Col), and spr1-4 (Wassilewskija ecotype) mutant alleles have been described (Furutani et al., 2000). spr1-5 (Wassilewskija ecotype) was isolated from the Institut National de la Recherche Agronomique T-DNA population (http://flagdb-genoplante-info.infobiogen.fr/projects/fst/DocsIntro/introCollection.html). Plants were grown as described previously (Furutani et al., 2000), unless noted otherwise.
Map-Based Cloning
F2 progenies from a cross between spr1-1 (in Ler) and wild-type Col were used as a mapping population. The completely recessive spr1-1 allele allowed simple linkage analysis between the SPR1 locus and the Ler chromosome. The SPR1 locus was found to be closely linked to the m246 CAPS marker that was located at the top end of chromosome 2 (no recombinants in 282 chromosomes). A P1 genomic library (generated at Mitsui Gyosai Bio) was screened using the m246 PCR fragment as a probe. Three clones were obtained and aligned into a contig based on the BamHI restriction map. New CAPS markers were generated from the insert-end sequences of the P1 clones. Use of these markers confined the SPR1 locus in a 60-kb segment. A cosmid minilibrary was constructed from a pool of partially digested P1 DNAs and the pOCA18 vector (Olszewski et al., 1988). Four cosmid clones that cover the 60-kb genomic region were digested with HindIII and the fragments subcloned into pBluescript KS+. DNA sequences derived from these subclones were used to generate a new set of CAPS markers and used for 394 F2 chromosomes. The SPR1 locus was finally restricted to a 37-kb interval.
Genomic DNAs from spr1-1 and Ler were digested with HindIII and subjected to a genomic DNA gel blot using each of the two cosmid clones spanning the 37-kb segment as a probe. A restriction fragment length polymorphism was found in a 5.4-kb Ler fragment, which in spr1-1 was 4.7 kb. A series of PCR amplification steps were performed to scan the 37-kb region and successfully detected 0.6-kb deletion within the 5.4-kb HindIII fragment. This fragment was introduced into the spr1-1 mutant using pBIN19 binary vector and shown to rescue the spr1 mutant defects. Subsequent sequencing of five spr1 alleles predicted that the gene At2g03680 encodes SPR1. This was further confirmed by genetic complementation of spr1-1 with a 2.1-kb SpeI-SnaBI genomic fragment that spanned the 0.4-kb upstream region relative to the putative TATA box, all exons and introns, and the 0.6-kb downstream region of At2g03680.
RNA Gel Blot Analysis
Total RNA was extracted from Arabidopsis thaliana plants with the RNAeasy plant mini kit (Qiagen, Valencia, CA). RNA was separated by a formaldehyde gel and transferred to Hybond-N+ membrane (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions. After UV cross-linking, rRNAs were visualized by 0.02% (w/v) methylene blue in 0.5 M sodium acetate, pH 5.2. Membranes were preincubated with the hybridization buffer consisting of 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 1% (w/v) SDS, 10% (w/v) dextran sulfate, 100 μg/mL of denatured herring sperm DNA, and 50% (v/v) deionized formamide at 42°C. The full-length SPR1-coding region was 32P-labeled and incubated with the membranes overnight at 42°C. Membranes were washed with a solution consisting of 0.1% (w/v) SDS and 0.1× SSC at 55°C. Radioactive signal was scanned by the BAS2500 image analyzer (Fuji Film, Tokyo, Japan).
Plasmid Construction, Plant Transformation, and Protein Production in Escherichia coli
Plasmids for plant transformation and recombinant protein production were constructed by a standard molecular biology technique (Sambrook et al., 1989). The SPR1 genomic or cDNA fragments used were produced by PCR from cloned DNA or genomic DNA. All PCR-derived clones were sequenced along their entire length to exclude the possibility of PCR-induced errors. GFP-coding sequences used in this study were derived from pAVA319 (von Arnim et al., 1998). The 35S promoter of Cauliflower mosaic virus and the GUS-coding region were derived from pBI221 (Clontech, Palo Alto, CA). For recombinant protein expression, full-length SPR1-coding region was inserted into the vectors pMALc2x (New England Biolabs, Beverly, MA) and pET32b (Novagen, Madison, WI) to produce the fusion proteins with maltose binding protein (MBP:SPR1) and thioredoxin (TRX:SPR1), respectively. Detailed construction procedures are available upon request.
For plant transformation, each DNA construct was transferred to either the pBIN19 (Frisch et al., 1995) or the pBIB vector that harbored a hygromycin-resistant marker (Becker, 1990) and then introduced into the Agrobacterium tumefaciens strain PMP90. Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998).
Recombinant protein was produced in the E. coli strain BL21. All proteins were expressed as a soluble form at 37°C. Proteins were extracted by sonication and affinity purified using amylose resin (New England Biolabs) for MBP:SPR1 or nickel-nitrilotriacetic acid agarose (Qiagen) for TRX:SPR1 according to the manufacturers' instructions. The eluted proteins were further purified by anion-exchange chromatography using either a Mono Q fast-protein liquid chromatography column or Q-Sepharose fast flow resin (Amersham Biosciences).
Immunological Technique
Polyclonal rabbit antisera were produced using TRX:SPR1 as an antigen. For protein gel blotting, antiserum was directly added at 1/2000 dilution. Signal was detected with the ECL-plus kit (Amersham Biosciences). For histochemical staining, antiserum was purified in three steps to reduce nonspecific binding. The first step used protein A-Sepharose (Amersham Biosciences) to purify whole IgG fraction. The second step was to remove antibodies specific for the TRX tag. This was accomplished by the use of recombinant TRX immobilized on an NHS-activated Sepharose column (Amersham Biosciences). The third step involved affinity purification using purified MBP:SPR1 protein immobilized on NHS-activated Sepharose. All antibody purification procedures were performed according to the described methods (Harlow and Lane, 1999).
For plant protein extraction, Arabidopsis seedlings were sliced into small pieces by razor blades and ground in ice-cold extraction buffer (50 mM Hepes-KOH, pH 7.5, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 0.25 M sucrose, 20 μg/mL of pepstatin A, and 20 μg/mL of leupeptin) containing 0.1 mg/mL of butylated hydroxytoluene and 10% (v/v) polyclar AT. After filtration through two layers of Miracloth (Calbiochem, San Diego, CA), the insoluble debris was removed by centrifugation at 7000g for 15 min at 4°C. Microsomal proteins then were separated from the supernatant by centrifuging the protein extracts twice at 50,000g for 30 min at 4°C. Immunoblot analysis was performed with Hybond-P membrane and the ECL-plus kit (Amersham Biosciences) according to the manufacturer's instructions.
Whole mount immunostaining was done by the method described for MT labeling (Sugimoto et al., 2000) with some modifications. Seedlings were fixed for 40 min in a solution containing 4% (w/v) paraformaldehyde (TAAB, Berkshire, UK) made in PEMT buffer [50 mM piperazine-1,4-bis(2-ethanesulfonic acid), pH 7.2, 2 mM EGTA, 2 mM MgSO4, and 0.05% (w/v) Triton X-100]. After NaBH4 treatment, seedlings were incubated in blocking buffer (1% [w/v] BSA and 0.05% Tween 20 in PBS) overnight. Seedlings were then incubated with affinity-purified anti-SPR1 antibody in the blocking buffer for 3 h at room temperature. Alexa Fluor 488–conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR) was used as a secondary antibody.
Microscopy
CSLM observation was performed with a Leica LSM510 confocal microscope (Wetzlar, Germany). The 488-nm argon laser beam was used for the excitation of GFP and Alexa Fluor 488. Some images were processed by the Photoshop program (Adobe Systems, San Jose, CA) to enhance image contrast.
Histochemical GUS Staining
GUS staining was done in a solution containing 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 50 mM potassium phosphate, pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 0.2% (v/v) Tween 20, and 20% (v/v) methanol. After staining, plants were incubated in 70% (v/v) ethanol at 37°C overnight, then transferred to a series of decreasing concentrations of ethanol (50, 30, and 0% [v/v]).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: Arabidopsis SPR1, AY464947; Arabidopsis SPR1 homologs, SP1L1, NM_102400 (At1g26360); SP1L2, NM_105590 (At1g69230); SP1L3, NM_111085 (At3g02180); SP1L4, NM_121564 (At5g15600); SP1L5, NM_118480 (At4g23496). SPR1 homologs from other species are T. halophila, BM985496; D. sophia, BG321626; Soybean, BI787806; Tomato, AW219464; Cotton, BE052696; Maize, BI431085; and Rice, CA758698. The moss SPR1 homolog sequence (contig 465; Physcomitrella patens) was retrieved from the Physcobase database (http://moss.nibb.ac.jp). Nucleotide sequence for SP1L1 is very likely to be misannotated in the database, where coding regions for SP1L1 and the preceding hydrolase-like gene are fused together. In this study, we separated the deposited nucleotide sequence into two genes, and the latter half was designated SP1L1.
Supplementary Material
Acknowledgments
We thank J. Schiefelbein for the spr1-2 mutant, K. Nakamura and A. Morikami for the spr1-3 mutant, A. Arnim for GFP plasmids, and Tomomi Kawamura for technical assistance. We are grateful to G.O. Wasteneys for valuable comments on the manuscript and T. Fujita for introducing the moss database. P1 and cosmid genomic libraries were provided by Mitsui Gyosai Bio. Some seeds were obtained through ABRC. This work was supported in part by grants from the Ministry of Education, Science, Sports, and Culture and from Japan Atomic Energy Research Institute to T.H.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Takashi Hashimoto (hasimoto@bs.naist.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017830.
References
- Azimzadeh, J., Traas, J., and Pastuglia, M. (2001). Molecular aspects of microtubule dynamics in plants. Curr. Opin. Plant Biol. 4, 513–519. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Wilson, J.E., Cork, A., and Williamson, R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35, 935–942. [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Hofte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Mattsson, O., Naested, H., Foster, R., and Mundy, J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801. [DOI] [PubMed] [Google Scholar]
- Brett, C., and Waldron, K. (1996). Physiology and Biochemistry of Plant Cell Walls, 2nd ed. (London: Chapman & Hall).
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- Foster, R., Mattsson, O., and Mundy, J. (2003). Plants flex their skeletons. Trends Plant Sci. 8, 202–204. [DOI] [PubMed] [Google Scholar]
- Frisch, D.A., Harris-Haller, L.W., Yokubaitis, N.T., Thomas, T.L., Hardin, S.H., and Hall, T.C. (1995). Complete sequence of the binary vector Bin 19. Plant Mol. Biol. 27, 405–409. [DOI] [PubMed] [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1999). Using Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hasenstein, K.H., Blancaflor, E.B., and Lee, J.S. (1999). The microtubule cytoskeleton does not integrate auxin transport and gravitropism in maize roots. Physiol. Plant 105, 729–738. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T. (2003). Dynamics and regulation of plant interphase microtubules: A comparative view. Curr. Opin. Plant Biol. 6, 568–576. [DOI] [PubMed] [Google Scholar]
- Hush, J.M., Wadsworth, P., Callaham, D.A., and Hepler, P.K. (1994). Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107, 775–784. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., Hawkins, T.J., Igarashi, H., Kaloriti, D., and Smertenko, A. (2002). The plant cytoskeleton: Recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50, 915–924. [DOI] [PubMed] [Google Scholar]
- Hyams, J.S., and Lloyd, C.W. (1994). Microtubules. (New York: John Wiley & Sons).
- Morejohn, L.C. (1991). The molecular pharmacology of plant tubulins and microtubules. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (London: Academic Press), pp. 29–43.
- Neff, M.M., and Van Volkenburgh, E. (1994). Light-stimulated cotyledon expansion in Arabidopsis seedlings (the role of phytochrome B). Plant Physiol. 104, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N.E., Martin, F.B., and Ausubel, F.M. (1988). Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 16, 10765–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiefelbein, J.W., and Benfey, P.N. (1991). The development of plant roots: New approach to underground problems. Plant Cell 3, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., Kamyar, R., and Ehrhardt, D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Himmelspach, R., Williamson, R.E., and Wasteneys, G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15, 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee, S., Tuchihara, K., and Hashimoto, T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196. [DOI] [PubMed] [Google Scholar]
- Ueda, K., Matsuyama, T., and Hashimoto, T. (1999). Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206, 201–206. [Google Scholar]
- von Arnim, A.G., Deng, X.W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., Gunning, B.E.S., and Hepler, P.K. (1993). Microinjection of fluorescent brain tubulin reveals dynamic properties of cortical microtubules in living plant cells. Cell Motil. Cytoskeleton 24, 205–213. [Google Scholar]
- Wasteneys, G.O., Jablonsky, P.P., and Williamson, R.E. (1989). Assembly of purified brain tubulin at cortical and endoplasmic sites in perfused internodal cells of the alga Nitella tasmanica. Cell Biol. Int. Rep. 13, 513–528. [Google Scholar]
- Webb, M., Jouannic, S., Foreman, J., Linstead, P., and Dolan, L. (2002). Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3—A katanin-p60 protein. Development 129, 123–131. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Yuan, M., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl. Acad. Sci. USA 91, 6050–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.