Abstract
Glioblastoma is one of the most devastating cancers, in which tumor cell infiltration into surrounding normal brain tissue confounds clinical management. This review describes basic and translational research into glioma proliferation and invasion, in particular the phenotypic switch underlying a stochastic “go or grow” model of tumor cell behavior. We include recent progress in system genomics, cancer stem cell theory, and tumor–microenvironment interaction, from which novel therapeutic strategies may emerge for managing this malignant disease. We suggest that an effective therapeutic strategy should target both adaptive glioblastoma cells and the stroma–tumor interaction.
Keywords: glioblastoma, invasion, proliferation, targeted therapy
Primary glial tumors are classified into 4 histologic grades of increasing aggressiveness. Glioblastoma multiforme (GBM), World Health Organization grade IV, the most common and malignant, has an annual incidence of 5.26 per 100 000 population (17 000 new diagnoses per year).1 Unlike most other cancers, in which metastasis from the site of origin is the leading cause of death, GBM very seldom metastasizes outside the neuraxis2; instead, local invasion/tumor recurrence is the leading cause of death. The short survival time after diagnosis (ie, patients dying before metastasis occurs) may account for infrequent metastasis.3 Another possibility is that the brain's unique vasculature, with a blood–brain barrier and an absence of lymphatic vessels, denies tumor cells access to the systemic circulation. Moreover, under normal conditions, the immune system likely plays an important role in limiting the seeding potential of circulating glioma cells and development of extracranial micrometastases. This is highlighted by occurrence of extracranial GBM in immunosuppressed transplant recipients.4 A well-characterized process describing a phenotypic switch between proliferation and invasion in solid tumor metastasis is the epithelial-mesenchymal transition (EMT).5 Although GBM is of neuroepithelial origin, a number of studies have shown that invasive GBM shares common molecular features with metastatic cancers.6,7
Successful isolation of a glioma stem cell (GSC)–like subpopulation of tumor cells and the methodology of using glioma patient-derived xenograft (PDX) models that contain GSCs greatly aid in understanding mechanisms and adaptation accounting for the invasiveness of GBM.8 Compared with traditionally grown long-term glioma cells propagated in vitro, GSCs and PDX models demonstrate invasive intracranial tumor growth in mice and recapitulate gene expression patterns similar to human GBM; findings using these models support a working hypothesis that tumor stem cells are a primary cause of GBM invasiveness.
Invasion by cancer cells is believed to depend upon crosstalk between various cellular components of the host's normal tissue-remodeling mechanisms, which encourages the study of how the host microenvironment participates in the invasive process. Hypoxia is a prominent microenvironmental feature in brain cancer. It is believed that rapid growth of glioma drives excessive oxygen needs, which, in combination with tumor-related intravascular thrombosis and hemorrhage, leads to tissue hypoxia. The responses of tumor cells to hypoxia may include migration from the hypoxic area, production of angiogenic factors to induce new blood vessel formation, and enhancement of anaerobic glycolysis. Consequently, the local accumulation of lactate generates a low pH environment that further stimulates cells to invade.9,10
This review highlights aspects of GBM with a focus on ways that these malignant cells survive, invade, and recur in the brain. Specific attention is given to genomics and epigenomics, to the shifting phenotype between proliferation and invasion, especially how cancer stem cell behavior is impacted, and to the role of tumor interaction with the microenvironment, which contributes to the challenges of medical management of GBM. We conclude with comments on emerging opportunities and pressing challenges to shifting the survival for patients with this aggressive cancer.
Genetic and Epigenetic Modification of GBM Proliferation and Invasion
Infiltrative growth of single malignant cells invading normal brain parenchyma is a hallmark of GBM. Identifying the key molecules associated with invasion is critical to devising therapies to control this disease. At an experimental level, laboratory studies of established GBM cell lines reveal that the cultures are heterogeneous, with some subpopulations prone to proliferation and others prone to invasion; these distinct subpopulations can be isolated.11,12 U87MG is a proliferative GBM line that forms fast-growing tumors when inoculated orthotopically into a mouse. However, after harvesting individual invasive cells from the mouse tumor–brain boundary and reculturing them, such cells showed an enhanced invasive phenotype in subsequent intracranial inoculation. Comparative gene expression between invasive and proliferative subpopulations of U87MG cells identified the p75 neurotrophin receptor as a central regulator of GBM invasion.11 Similarly, using laser capture microdissection, cells from a glioma's invasive border and proliferative core were captured from hematoxylin and eosin–stained GBM sections, and protein extracts from the invasive and proliferative subpopulations were compared. The invasive GBM cells protruding into the brain parenchyma expressed higher levels of the guanine nucleotide exchange factors ELMO1 and Dock180 compared with cells from the tumor core.12 The targeting of these 2 molecules may affect invasive growth.
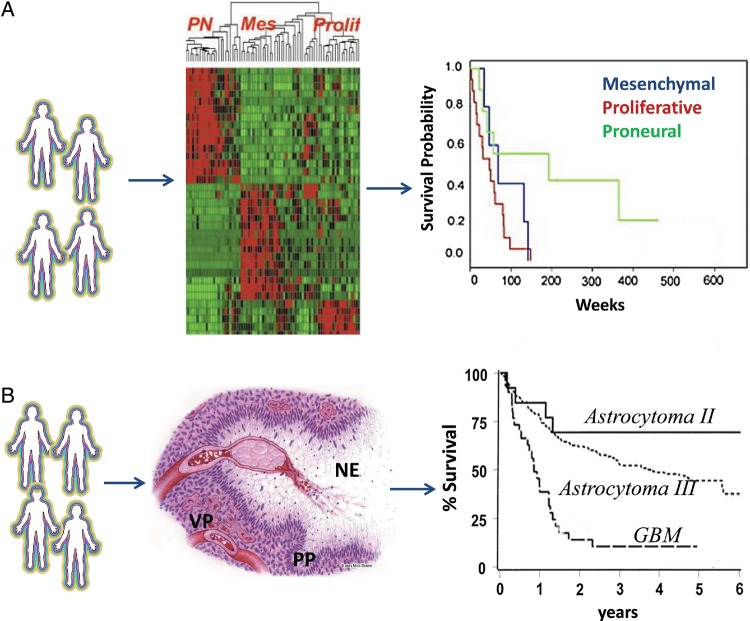
The proliferative and invasive phenotypes of GBM have been characterized at the genomic level. Using in silico analysis, Phillips et al13 classified GBM into “mesenchymal,” “proliferative,” and “proneural” subtypes and showed that both the mesenchymal and proliferative types correlated with shorter survival time relative to the proneural type (Fig. 1A). In parallel, The Cancer Genome Atlas (TCGA) was established to generate a comprehensive catalogue of somatic genomic changes in cancer. With nearly 500 primary GBM tumors being processed using multiple genomic approaches to explore core signaling pathways and key genomic and epigenomic alterations associated with GBM progression,13–15 Verhaak et al14 further classified GBM into 4 molecular subtypes with slightly different terms. Tumors whose molecular profile fits a “classical” signature represent a more proliferative phenotype and those with a “mesenchymal” signature a more invasive one; both are associated with worse prognosis. In contrast, the “proneural” signature represents a GBM subtype associated with better prognosis. The study also describes a “neural” signature associated with normal brain tissue in which tumor cells express neuronal markers. Although this study provides additional evidence contrasting the proliferative and invasive phenotypes in GBM, guarded interpretation is warranted, since clinical observations show that virtually all subtypes show infiltrative growth in brain. Thus, how these molecular subtypes may directly inform proliferation versus invasion requires further investigation. Broad-based genomic characterization of GBM provoked the idea to use molecular pathology to complement or even replace the traditional histopathology (Fig. 1B). Relative to histology, in which diagnoses are based on the morphological changes in tumor tissue, molecular pathology identifies detailed genetic alterations in individual samples and is anticipated to be more relevant to the development of precision medicine. In fact, the most recent work by Brennan et al15 strongly demonstrated that systematic genomic analyses with detailed clinical information, such as treatment and survival outcomes, can be used to discover genomic-based predictive and therapeutic biomarkers. Compared with previous studies, this study further included the data sets of whole genomes, coding exomes, transcriptome sequencing, and microRNA (miRNA) expression profiles. The authors confirmed a survival advantage in GBM patients whose tumor was of the proneural subtype; such tumors are associated with a cytosine–phosphate–guanine island (CPG) methylator phenotype, and MGMT DNA methylation. These serve as predictive biomarkers for treatment response, but only in classical-subtype GBM.
Fig. 1.
Molecular and histolopathological features of GBM. (A) Molecular pathology uses a genomic signature in association with clinical outcomes for diagnosis (adapted from Phillips et al13 with permission). Initial in silico analysis classifies glioblastoma into 3 molecular subsets. A mesenchymal signature indicates a more invasive phenotype. Patients with mesenchymal or proliferative signatures are associated with poor clinical outcome (shorter survival). A proneuronal signature correlates with better prognosis (longer survival). (B) Histopathology relies on morphological changes in association with clinical outcomes for diagnosis. The histological hallmarks of glioblastoma include necrosis (NE), which is often surrounded by pseudopalisading (PP) tumor cells that are migrating away. Vascular proliferation (VP) indicates the rebuilding of the vasculature (adapted from Brat and Van Meir56 with permission). These classic morphological features of glioblastoma correlate with shorter survival (adapted from Prados et al92 with permission).
Studies based on TCGA data have also reported genetic and epigenetic determinants of GBM phenotypes. Mutations were increased in members of receptor tyrosine kinase (RTK)/Ras/phosphatidylinositol-3 kinase, p53, and retinoblastoma 1 signaling, the leading aberrant pathways in GBM.16 MET and CD44 overexpression and nuclear factor-kappaB (NFкB) signaling activation were associated with the mesenchymal phenotype. Genetically, individual gene amplifications or mutations were associated with specific disease progression. For example, epidermal growth factor receptor (EGFR) amplification was frequently found in samples having the classical signature; isocitrate dehydrogenase 1 (IDH1) mutation and platelet-derived growth factor receptor alpha amplification were often associated with the proneural signature; and neurofibromatosis type 1 loss or mutation and phosphatase and tensin homolog loss frequently occurred in mesenchymal GBM (Fig. 1).13,16,17 Integrated analysis of gene expression profiles and array comparative genomic hybridization revealed no correlation between mean expression and the DNA copy number of genes in proneural, mesenchymal, and proliferative tumors. Surprisingly, it seems that transcriptional networks regulate mesenchymal transformation of malignant glioma cells. In human glioma, signal transducer and activator of transcription 3 (STAT3) and CCAAT/enhancer-binding protein β (C/EBPβ) are transcription factors that reside upstream of networks that correlate with a mesenchymal phenotype and predict poor clinical outcome.18
The Principle of “Go or Grow”
Clinically, GBM often shows highly infiltrative growth patterns that disperse tumor cells throughout the brain.19 Recent findings in lower-grade (diffuse) astrocytoma (World Health Organization grades II and III) uncovered widespread infiltration of tumor cells that stained positive for the specific IDH1 R132H mutation,20 confirming that invasion occurs early in gliomagenesis, because IDH1 mutations are early events in the progression toward certain subtypes of malignant glioma.21 These tumors present with a bulky, proliferative, angiogenic tumor core, but they also include invasive tumor cells penetrating into the surrounding normal brain parenchyma, which manifests as an infiltrative border and contributes to incomplete surgical removal and inevitable tumor recurrence primarily along the resection margin. After surgical debulking, residual invasive GBM cells adopt, or revert to, a proliferative phenotype, growing into a more aggressive, locally recurrent tumor. Sequential switching between proliferation and invasion characterizes tumor progression. At the microscopic level, proliferation and migration appear to be temporally, mutually exclusive phenotypes, as indicated by recent in vivo imaging data that glioma cells migrate in a salutatory fashion. While proliferating, cells pause for as short as an hour to divide before the daughter cells move again.22 Gao et al23 experimentally demonstrated that the 2 phenotypes can switch under certain conditions. By growing the same subsets of GBM cells in soft agar, proliferative cells can be selected for and grown into subclones. Further growth of proliferative clones in Matrigel, a gelatinous basement membrane extract that mimics the complex extracellular environment, changes their morphology from spherical to branching, consistent with a switch from proliferation to invasion. When the 2 subpopulations of cells were compared in terms of response to hepatocyte growth factor (HGF) stimulation, the proliferative cells showed a faster growing but less invasive phenotype and activation of the Myc signaling pathway. In contrast, the invasive cells showed enhanced ability to penetrate Matrigel, lower proliferation, and elevated Ras signaling.23 Recently, Dhruv et al24 affirmed the role of c-Myc activation at the crowded, proliferative center of GBM tumors and of an increase in NFкB activation in those same GBM cells that were radially dispersed in a highly invasive pattern. Such studies provide a solid rationale for targeting these pathways to interfere with GBM proliferation and invasion.
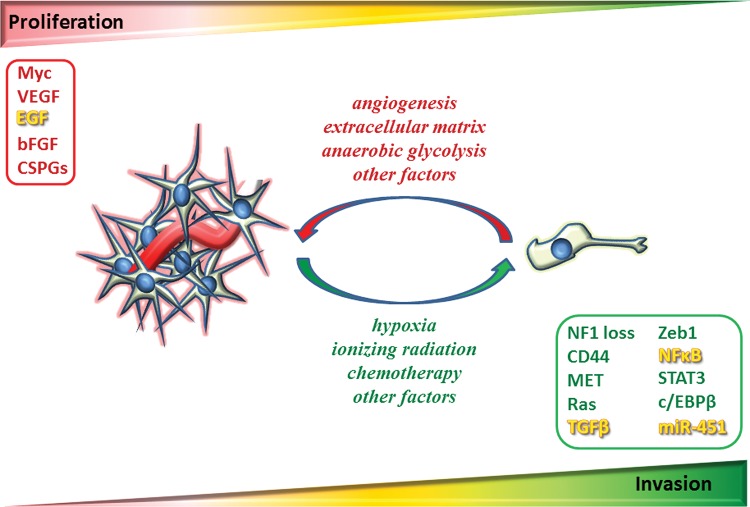
Although both experimental observations and mathematical modeling25 support proliferation and migration as mutually exclusive states of glioma cells, from the standpoint of clinical relevance, one could question how these data may actually be applied to the real tumors that are developed within a heterogeneous, multifaceted living environment that includes extracellular matrix (ECM), fiber tracts, vasculature, and a host of soluble factors. In particular, many molecular players in the pathways affecting proliferation and invasion actually overlap (eg, epidermal growth factor [EGF],8,26,27 transforming growth factor beta [TGFβ],28,29 NFкB24,30). From an evolutionary perspective, there is diversity within a polyclonal cell population, in which switching between phenotypes can be regulated either through intrinsic (spontaneous stochastic switching) or extrinsic (responsive switching) factors or a combination of both.31 By assuming that glioma cells are a mixture of proliferative and invasive subpopulations, Mansury et al32 showed that individual cells' phenotypic behavior depends on both the genotype of the cells they interact with and the microenvironment to which they are exposed. The dynamics of tumor invasion are therefore determined by the optimal payoff in which tumor growth is ensured.32 Experimentally, a recent study demonstrated that miRNA-451 regulates the adaption to proliferation and invasion in glioma cells based on the metabolic stress through the same signaling pathway of liver kinase B1/adenosine monophosphate–activated protein kinase.33 As such, a dogmatic view that glioma cells exist in a mutually exclusive “go or grow” state indeed oversimplifies the complexity of malignant gliomas. A more likely scenario is that glioma cells can effectively adapt from proliferating and invasive phenotypes based on the balance of cues from various regulating factors in the local environment (Fig. 2).
Fig. 2.
Diagram of proliferation and invasion and the key pathways. Progression of glioblastoma is determined by 2 key factors: cell proliferation rate and speed of cell migration. In theory, proliferation (illustrated in red) and invasion (illustrated in green) are temporally, mutually exclusive phenotypes. As such, the highly invasive glioma cells have a lower proliferative rate; in turn, the highly proliferative cells are less invasive. At a macroscopic level, cell-cell interactions and microenvironmental factors both play important roles in regulating the phenotypic switching. While ionizing radiation, chemotherapy, and hypoxia can force tumor cells (eg, GSCs) into a “go” phenotype, angiogenesis, tumor ECM, and anaerobic glycolysis promote repopulation of tumor cells and formation of recurrent tumors. The dynamic phenotypic switching between proliferative and migratory states of individual cells determines the overall phenotype of tumor growth. The key pathways associated with proliferation and invasion are outlined. Note that there are pathways regulating both proliferation and invasion (illustrated in yellow). bFGF, basic fibroblast growth factor; CSPGs, chondroitin sulfate proteoglycans; NF1, neurofibromatosis type 1.
In human solid tumors of epithelial origin such as breast and prostate cancers, EMT seems to be an iterative, adaptive process. The successful switch between the 2 states requires programmed changes in gene expression and functional behaviors that have marked consequences on the cellular phenotype.34 EMT has been found to play an important role in cancer progression, controlling the switch between cancer proliferation and metastasis. Proliferative tumors often show an “epithelial” morphology, with tight cell junctions and overexpression of E-cadherin at the cell membrane as a marker. Following EMT, the tumor cells become elongated and fibroblast-like, the tight cell junctions disappear, E-cadherin expression is lost, and β-catenin appears in the nucleus. Cells degrade the ECM, producing a more invasive and migratory (mesenchymal) phenotype.5 The newly programmed mesenchymal cells migrate into the blood or lymphatic circulation, where they reside until they encounter suitable extravasation sites. At such sites, the mesenchymal-epithelial transition occurs, in which the mesenchymal cells switch to the epithelial phenotype, becoming less invasive and more proliferative. Expression of β-catenin declines, E-cadherin reappears in the cell membrane, and tight cell-cell junctions reemerge. With this switch, the metastatic cells grow into a secondary tumor, completing the metastasis process.
Because gliomas usually originate from either immature astrocytes, oligodendrocytes, a mixture of the 2,35 or potentially neural stem or progenitor cells,36 and because they seldom metastasize, the EMT paradigm has not been entertained as relevant to this disease. Increasingly, however, evidence has shown that the model of “go or grow” has molecular mechanisms in common with EMT. Hepatocyte growth factor and TGFβ are essential factors regulating EMT, and both are strong stimulators of GBM invasion.29 Elevated MET and CD44 expression, along with an activated NFкB signaling pathway, is found in both metastatic cancer and mesenchymal GBM.34,37–39 The transcription factor Zeb1 is an essential regulator of EMT and is also a driver of GBM invasion.40 Recently, a comprehensive comparison summarizing the commonality between an EMT signature from mammary epithelial cells and those from all 4 molecular subtypes of GBM showed that POSTN is one of the 3 most upregulated genes in both mesenchymal and classical GBM, as well as EMT in epithelial cells.7 Functionally, POSTN plays an important role in breast cancer stem cell–initiated metastasis41 and is tightly associated with the biological processes of cell invasion in GBM. It is also a biomarker for progression of histologic grade in gliomas and indicates a poor prognosis for patients with high-grade gliomas.42 All these results suggest that mesenchymal GBM and metastatic epithelial cancers share common mechanisms that drive the phenotypic switch between proliferation and invasion.
Glioma Stem Cells: The Primary Source of Invasion
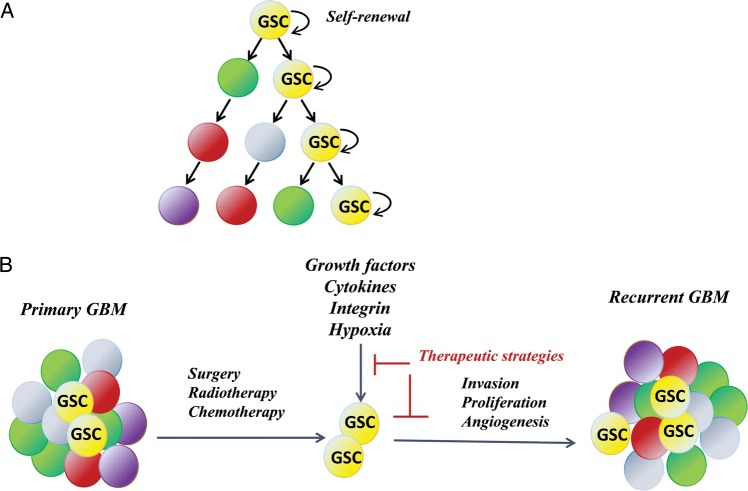
Pioneering work on the cancer stem cell theory began with leukemia patients43 and has extended to breast cancer44 and GBM.45 As stated during a 2006 workshop, “In the cancer stem cell model of tumors, there is a small subset of cancer cells, the cancer stem cells, which constitute a reservoir of self-sustaining cells with the exclusive ability to self-renew and maintain the tumor.”46 In the field of GBM research, the theory includes “a highly tumorigenic subpopulation of cancer cells that display relative resistance to radiation and chemotherapy.”47 In this context, GSCs are such a subpopulation of undifferentiated transformed cells (Fig. 3A). When most differentiated tumor cells die from treatment, it is believed that the GSCs are triggered and repopulate to form recurrent tumors (Fig. 3B). GSCs have a potent plasticity and can differentiate into astrocytes, oligodendrocytes, and neuronal lineages,48 thus providing an unpredictable outcome for the tumor and enabling tumor recurrence.
Fig. 3.
Strategies of targeting GSCs. (A) The GSC theory (adapted from Reya et al93 with permission). Tumors are heterogeneous, and only the subpopulation of GSCs is capable of self-renewal and repopulation into tumors. (B) GSC-targeted therapy. GSCs are resistant to standard therapy and are the cause of tumor recurrence. Therapies should be targeted toward eradication of GSCs rather than the entire tumor cell population. Targeting of the microenvironment should also increase therapeutic efficacy.
A milestone in glioma research has been the success of isolating invasive GSCs from heterogeneous primary tumors.8 Although GBM is highly invasive, established malignant glioma cell lines seldom generate invasive tumors when inoculated into mouse brain. It is believed that long-term monolayer growth in medium with serum induces the differentiation of tumor-initiating cells and drives the cells away from their usual biology (ie, to changes that include the expansion of tumor cells beyond the stem cell stage, and the diminution of inherent invasive capabilities). A major advance in the isolation of GSCs was achieved with a novel “neurosphere” culture system.8 Following surgical removal, a primary GBM tumor can be enzymatically dissociated into single cells. Instead of culturing in the traditional medium containing serum, cells are cultured in serum-free neurobasal medium (NBE) supplemented with basic fibroblast growth factor and EGF. Compared with traditional medium in which cells often adhere to the dishes, tumor cells in NBE grow as nonadherent, multicellular neurospheres in uncoated plates and as an adherent monolayer in polylysine/laminin-coated plates, showing a pattern similar to that of normal neural stem cells. These cells retain the capability to differentiate into multiple lineages under appropriate stimulation and show genetic profiles close to those of human GBM. More importantly, the tumor stem cells are strongly invasive when injected into mouse brain.8,49,50 Therefore, GSCs have been considered the primary cause of GBM invasiveness, and this concept has important implications for both basic biological studies and the development of novel therapeutic strategies.
Bao et al50 published studies showing that by using CD133 as a marker, GSCs from primary tumors can be enriched by flow sorting; such cells are more resistant to radiation and chemotherapy. The CD133+ cells demonstrate remarkable neurosphere formation and induce highly invasive and more angiogenic tumors in mice than do CD133– cells.50 These tumors are also highly resistant to radiotherapy. Fractional focal radiation is an effective therapy for GBM, but on recurrence, the tumors often grow with a more aggressive phenotype. When ionizing radiation produces DNA damage, CD133+ GSCs react with greater repair efficiency, including increased activation of cell cycle checkpoint proteins such as Chk1, Chk2,50 and Rad17, a crucial regulator of the DNA damage checkpoint.51 CD133 mRNA expression is also higher in recurrent tumors than in primary tumors,47 and CD133+ GSCs are reported to be the major cause of resistance to radiation therapy.47,50 CD133+ cells are also responsible for chemoresistance; as such, relative to CD133– cells, CD133+ cells are significantly more resistant to common chemotherapeutic agents, including temozolomide (TMZ), carboplatin, paclitaxel (Taxol), and etoposide (VP16). This is probably because of the higher transcription levels of the drug transporter gene BCRP1 and the DNA repair-associated gene MGMT, as well as of some anti-apoptotic proteins.47
Genomic studies also support targeting GSCs as a therapeutic strategy. Studies have shown that activated STAT3 signaling is essential to GSC survival and development52 and that targeting the STAT3 pathway can suppress stem cell survival and tumor growth.53 A recent study using TCGA data showed that the STAT3 and C/EBPβ transcriptional networks are keys to regulating mesenchymal GBM, the most invasive subtype associated with poorest outcomes. Ectopic coexpression of C/EBPβ and STAT3 can induce neural stem cells to undergo a transition that includes the expression of mesenchymal markers, changes toward a fibroblast-like morphology, and enhanced migration and invasion. The elimination of C/EBPβ and STAT3 can block glioma cell invasion in vitro and reduce tumor aggressiveness in vivo.18 Following surgery, patients with GBM are treated with adjuvant radiation and chemotherapy. While this treatment reduces the number of proliferative tumor cells, the cellular stress also switches the residual infiltrated GSCs into a “go” phenotype to find the proper “soil” and eventually lead to predictable tumor recurrence. In fact, the GSC theory has led to the concept that efforts in developing targeted therapy should be more specifically addressed to the subpopulation of tumor cells (ie, GSCs) that are more adaptive to the standard therapy, as well as to the microenvironmental stimuli that regulate tumor invasion, proliferation, and angiogenesis (Fig. 3B).
Microenvironmental Regulation: Crosstalk Between Tumor and Host
In cancer cell invasion, the dynamic crosstalk between the tumor and the normal host tissue determines the progress of proliferation and invasion. Besides irradiation and chemotherapy, which influence the “go or grow” phenotypes in GSCs, recent work also suggests that a group of tumor ECM factors, chondroitin sulfate proteoglycans, can serve as regulators of the phenotypic switch from “go” to “grow.”54 In addition, matrix metalloproteinases, such as MMP-14, have been recognized to promote angiogenesis (eg, through upregulation of vascular endothelial growth factor [VEGF]) via activation of MMP-2 and MMP-9, which are also key factors in tumor angiogenesis.55 Hypoxia-induced angiogenesis is well accepted as the primary microenvironmental factor regulating GBM proliferation and invasion. Rapid growth of a tumor can directly lead to deficient blood supply and thus a lack of oxygen and nutrients at the tumor core. Also, tumors growing into adjacent brain tissue can cause intravascular thrombosis and hemorrhage. Both situations are common causes of hypoxia. In response, tumor cells migrate away from the hypoxic center and form a hypercellular zone called pseudopalisades. The tumor cells that do not migrate become hypoxic and undergo apoptosis or necrosis, producing an enlarged central necrotic zone.56,57 While moving away from the hypoxic center, tumor cells produce pro-angiogenic factors that attract endothelial cells to rebuild blood vessels in order to provide adequate blood supply for further tumor growth. However, the structure of that vasculature can often simply accrue an aggregation of endothelial cells, which is described as vascular proliferation. Necrosis, pseudopalisades, and vascular proliferation are all classic histopathological hallmarks of GBM (see Fig. 1B). This explanation is mechanistically linked to GSCs, which not only are invasive, but also have a potent capability to induce angiogenesis. GSCs express elevated levels of VEGF and stromal-derived factor 1 (SDF-1, CXCL12),49,58 which are both considered to be pro-angiogenic factors inducing endothelial cell proliferation and tubule organization. In fact, many of the angiogenic factors such as VEGF and SDF-1,49,58 basic fibroblast growth factor and EGF,8 are also essential to GSC maintenance. Compared with nonstem tumor cells, GSCs express more hypoxia-induced factors (HIFs), especially HIF2, which is required for GSC growth and survival. HIF2α knockdown reduces VEGF expression, prevents GSC-induced angiogenesis, and therefore is considered a promising target for anti-GBM therapeutics.59 As the vasculature is rebuilt to restore the blood supply, tumor cells migrate toward these new blood vessels, thereby promoting expansion of the tumor margin. A high degree of vascularization correlates with glioma aggressiveness and is an indicator of poor prognosis.
Hypoxia directly influences a cancer cell's metabolism. Normal differentiated cells use oxidative phosphorylation as the source of ∼90% of their ATP, and they switch to anaerobic metabolism in case of hypoxia. Proliferating tumors, however, use anaerobic glycolysis regardless of oxygen availability, a phenomenon termed the Warburg effect, which consumes less oxygen but produces large amounts of lactate and alanine.60 GBM cells in culture convert as much as 90% of glucose and 60% of glutamine into lactate or alanine through anaerobic glycolysis,61 and hypoxia can further promote the process. Through upregulation of HIF1α, expression of glucose transporters is enhanced,62 while genes that are associated with mitochondrial oxidative phosphorylation are downregulated.63,64 Although anaerobic glycolysis is an inefficient way of generating ATP, it is believed to facilitate the cells' ability to take up and transform nutrients. The excess lactate can be used to produce sufficient NADPH for fatty acid synthesis, and the uptake of glucose and glutamine can provide enough carbon for nucleotide, amino acid, and lipid synthesis,60,61,64 thus offering a replicative advantage for rapidly proliferating cells. Anaerobic glycolysis plays a dual role in regulating tumor proliferation and invasion. From one perspective, it is a malignant characteristic of aggressive GBM rather than a simple adaption to hypoxia, in that tumor cells need anaerobic glycolysis to maintain their proliferative needs.10 Anaerobic glycolysis also positions cells in a specific microenvironmental niche that promotes GBM invasion.9,10 Specifically, the excessive lactate produced by fast-growing tumors can directly lead to local acidification, which results in death of normal neuronal and glial cell populations but favors the growth of neoplastic cells resistant to acidic conditions due to p53 mutations or other molecular mechanisms. The increased H+ concentration induces degradation of the ECM by matrix metalloproteinases and angiogenesis through release of interleukin-8 and VEGF, which serve as chemoattracters to induce tumor cell migration toward the new vasculature.
Targeting Adaptive Glioblastoma
Despite our increasing knowledge of GBM progression, effective therapeutics have not advanced significantly. Altogether there are about 160 FDA-approved oncology drugs, but only 3 are routinely used to treat GBM: biodegradable carmustine (BCNU) wafers (Gliadel; FDA approval in 1995), TMZ (Temodar; FDA approval in 1997), and bevacizumab (Avastin; FDA approval in 2009 for recurrent GBM). The current standard of care for patients harboring GBM consists of multimodal treatment including maximal safe tumor resection followed by the Stupp protocol (radiotherapy plus concurrent daily TMZ followed by adjuvant TMZ).1 TMZ is a DNA-alkylating agent that methylates DNA. Although methylation is at the N7 position of guanine, the O3 position of adenine may contribute to antitumor activity, the most important methylation site being the O6 position of guanine.65 This is supported by the results from clinical trials that MGMT (O6-methylguanidine DNA methyltransferase) gene silencing is associated with longer overall survival (OS) in GBM patients treated with TMZ in addition to radiotherapy.66 MGMT encodes a DNA repair protein that removes alkyl groups at the O6 position of guanine and therefore inhibits DNA repair after TMZ treatment. Because patients whose tumors harbor MGMT promoter methylation clearly benefit the most from TMZ treatment, MGMT promoter methylation is becoming both a predictive factor for TMZ sensitivity and a prognostic factor for survival. However, although standard aggressive therapies generally prolong the median OS time of GBM patients, local tumor recurrence is seen in nearly all cases. Thus, there is a great need to understand the mechanism of TMZ resistance and to develop combination strategies that could improve therapeutic efficacy.
In GBM, an abundant vasculature indicates a poor prognosis, and hypoxia enhances angiogenesis and drives the tumor toward a more aggressive phenotype. Both factors provide a good rationale for targeting angiogenesis in malignant gliomas. Bevacizumab, a humanized monoclonal antibody binding to VEGF-α, was developed to inhibit the VEGF signaling pathway. Promising results from 3 phase II clinical trials indicated that bevacizumab, either alone or in combination with irinotecan, a topoisomerase 1 inhibitor, can prolong progression-free survival (PFS) in recurrent GBM patients.67,68 Thus, this drug received accelerated approval from the FDA for the treatment of recurrent or progressive GBM. However, until now, there has been no evidence that the median OS of patients can be prolonged. More recently, phase II clinical trials tested bevacizumab in combination with the traditional radiochemotherapy for treating newly diagnosed GBM patients.69 Again, studies showed prolonged PFS in these patients, but OS was not significantly changed. Likewise, 2 recent phase III trials (AVAglio and RTOG 0825) showed improved PFS but failed to demonstrate improved OS in patients with newly diagnosed GBM treated with upfront bevacizumab.70 Tumors resistant to and recurring after bevacizumab treatment often show a more aggressive phenotype.71 Moreover, blocking VEGF stimulates the production of other angiogenic factors, such as SDF-1, fibroblast growth factor, and platelet-derived growth factor.72,73 Therefore, the overall benefit versus risk in using bevacizumab to treat newly diagnosed GBM requires serious consideration. Additional studies are clearly needed to determine whether specific GBM subsets may respond more effectively to anti-angiogenic therapy. Identification of biomarkers to determine the patients more likely to respond to bevacizumab should further augment its positive effects on quality of life, neurocognitive functioning, and ability to decrease corticosteroid dependence in a statistically meaningful manner. Despite the uncertainty created by the recent clinical trials, anti-angiogenic treatment using bevacizumab remains a vital tool in the management of GBM patients with progressive disease.74 Further work is needed to optimize existing therapies, determine whether bevacizumab is best used alone or in combination with other agents, and establish biomarkers to improve patient stratification.
A recent study by Lu et al75 explained the molecular mechanism of resistance to bevacizumab treatment in GBM.76 Human GBM tumor specimens that are resistant to bevacizumab appear to have higher MET activity, indicating that long-term treatment with bevacizumab may cause MET pathway activation as a rescue for tumors to recur more aggressively. In a VEGF knockout mouse model that develops aggressive GBM, inhibiting MET can significantly block tumor invasion. Thus, secondary MET pathway activation may be the cause of resistance to VEGF pathway inhibition. Consequently, a combination of VEGF and MET inhibition may be effective. XL184, a small molecule targeting both the MET and VEGF pathways, does show promising effects in clinical trials against GBM.77 A phase II clinical trial for recurrent GBM using the MET antibody onartuzumab (MetMAb) in combination with bevacizumab (relative to each drug alone) is currently recruiting patients (NCT01632228). We anticipate that the combination therapy may eventually prolong OS.
Based upon TCGA data, EGFR amplification occurs in about 45% of GBM patients, and EGFR variant III mutation is found with high frequency. Although tumors with EGFR overexpression and amplification showed good response to EGFR inhibitors in preclinical models, none of the EGFR inhibitors has been effective in clinical trials.78,79 In GBM, inhibiting EGFR signaling is known to result in activation of other RTKs as rescue pathways; for example, MET activation has been frequently observed. Thus, a strategy of using a combination of these 2 inhibitors was developed.80,81 In fact, EGFR crosstalk with the MET pathway turns out to be common. In non–small cell lung cancer patients, EGFRT790M predicts sensitivity to erlotinib, and MET amplification is the major cause of acquired resistance after erlotinib treatment. Importantly, the tumors resistant to erlotinib can be effectively treated by MET inhibitors.82,83
In addition to being a target in recurrent gliomas, the MET pathway by itself is a promising target in cancer therapy.84 Overexpression of MET and its ligand, HGF, are found in most GBM and are associated with poorer prognosis and shorter survival.85 The fact that MET overexpression is directly associated with the mesenchymal subtype provides further rationale for targeting the MET pathway in GBM. Although TCGA data show that MET amplification occurs in 4% of GBM patients, there have been no preclinical GBM models with such a molecular feature for testing sensitivity to MET inhibitors. Studies have shown that tumors with high levels of HGF expression often show autocrine activation, which is a predictive marker of sensitivity to MET inhibitors.86 MET inhibitors are in clinical trials against several types of cancers, including GBM.87 However, due to the inability to identify sensitive tumors, patient stratification has not been included as a component of current clinical trials, which is a major challenge in evaluating drug response where the most vulnerable tumors are in a minority of patients. Furthermore, as with other RTK inhibitors, MET inhibitor therapy alone will also be rapidly challenged by pathway bypass and acquired resistance. As such, developing effective strategies combining various targeted therapies is necessary.
Future Opportunities and Challenges
In recent years, the rapid success in molecular profiling and genomic characterization has greatly accelerated our understanding of the interdependence of GBM proliferation and invasion, which has opened avenues to discoveries that may aid in the differential diagnosis and novel treatment strategies of this devastating disease.
However, although systematic approaches are being established to identify “driver” genes that may directly lead to glioma proliferation and invasion, there is a lack of functional validation of how these driver genes are working alone and in concert with other genes or networks. In malignant GBM, mutations or amplifications of multiple driver genes often occur in the same tumor,88 complicating the problem of finding “the right drug for the right patient.” Such comprehensive genomic landscape of glioblastoma also challenges the classic “go or grow” model, as the dichotomy of proliferative and invasive phenotypes become less significant due to overlap of key molecular pathways. While the concept of GSCs theoretically established a solid therapeutic strategy of targeting the subpopulation of tumor cells more resistant to chemo- and radiotherapy, effective drugs for clinical application are not available; they are, however, accruing for preclinical development. Signals from the microenvironment can also modify the drug response of individual tumors: studies have shown that hypoxia,50 growth factors,89,90 and integrins91 can all increase resistance to certain therapeutics. Hence, continued work is required to understand the molecular mechanisms that drive the invasive behavior of GBM and the phenotypic switch between proliferation and invasion. We foresee significant improvements in therapeutic outcomes in the coming years.
Funding
This work was supported by the American Brain Tumor Association Discovery grant no. 2013; the Steven M. Coffman Charitable Fund; the Jay and Betty Van Andel Foundation (to Q.X.); NIH/NCI R01 CA123451; the Fund for Medical Research and Education, Wayne State University School of Medicine; a Strategic Research Initiative grant, Karmanos Cancer Institute (to S.M.); NIH/NCI U01CA168397; and the Ben and Catherine Ivy Foundation (to M.E.B.).
Acknowledgments
We thank Dr. George Vande Woude for his review and helpful advice on this manuscript. We also thank David Nadziejka for carefully editing the manuscript.
Conflict of interest statement. The authors declare no conflict of interest.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Lun M, Lok E, Gautam S, et al. The natural history of extracranial metastasis from glioblastoma multiforme. J Neurooncol. 2011;105(2):261–273. doi: 10.1007/s11060-011-0575-8. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton JD, Rapp M, Marcel Schneiderhan T, et al. Glioblastoma multiforme metastasis outside the CNS: three case reports and possible mechanisms of escape. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.48.7546. doi:10.1200/JCO.2013.48.7546. [DOI] [PubMed] [Google Scholar]
- 4.Jimsheleishvili S, Alshareef AT, Papadimitriou K, et al. Extracranial glioblastoma in transplant recipients. J Cancer Res Clin Oncol. 2014;140(5):801–807. doi: 10.1007/s00432-014-1625-3. [DOI] [PubMed] [Google Scholar]
- 5.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5(8):711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 6.Xie Q, Thompson R, Hardy K, et al. A highly invasive human glioblastoma pre-clinical model for testing therapeutics. J Transl Med. 2008;6(77):1–13. doi: 10.1186/1479-5876-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarkoob H, Taube JH, Singh SK, et al. Investigating the link between molecular subtypes of glioblastoma, epithelial-mesenchymal transition, and CD133 cell surface protein. PLoS One. 2013;8((5):):e64169. doi: 10.1371/journal.pone.0064169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Gatenby RA, Gawlinski ET, Gmitro AF, et al. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Johnston AL, Lun X, Rahn JJ, et al. The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol. 2007;5(8):e212. doi: 10.1371/journal.pbio.0050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarzynka MJ, Hu B, Hui KM, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67(15):7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giese A, Bjerkvig R, Berens ME, et al. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21(8):1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 20.Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20(1):245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farin A, Suzuki SO, Weiker M, et al. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia. 2006;53(8):799–808. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- 23.Gao CF, Xie Q, Su YL, et al. Proliferation and invasion: plasticity in tumor cells. Proc Natl Acad Sci U S A. 2005;102(30):10528–10533. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhruv HD, McDonough Winslow WS, Amstrong B, et al. Reciprocal activation of transcription factors underlies the dichotomy between proliferation and invasion of glioma cells. PLoS One. 2013;8(8):e72134. doi: 10.1371/journal.pone.0072134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tektonidis M, Hatzikirou H, Chauviere A, et al. Identification of intrinsic in vitro cellular mechanisms for glioma invasion. J Theor Biol. 2011;287:131–147. doi: 10.1016/j.jtbi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Yang W, Aldape K, et al. Epidermal growth factor (EGF)-enhanced vascular cell adhesion molecule-1 (VCAM-1) expression promotes macrophage and glioblastoma cell interaction and tumor cell invasion. J Biol Chem. 2013;288(44):31488–31495. doi: 10.1074/jbc.M113.499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly JJ, Stechishin O, Chojnacki A, et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27(8):1722–1733. doi: 10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- 28.Bruna A, Darken RS, Rojo F, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 30.Meng Q, Zhi T, Chao Y, et al. Bex2 controls proliferation of human glioblastoma cells through NF-kappaB signaling pathway. J Mol Neurosci. 2014;53(2):262–270. doi: 10.1007/s12031-013-0215-1. [DOI] [PubMed] [Google Scholar]
- 31.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309(5743):2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 32.Mansury Y, Diggory M, Deisboeck TS. Evolutionary game theory in an agent-based brain tumor model: exploring the ‘genotype-phenotype’ link. J Theor Biol. 2006;238(1):146–156. doi: 10.1016/j.jtbi.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Godlewski J, Nowicki MO, Bronisz A, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37(5):620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roussos ET, Keckesova Z, Haley JD, et al. AACR special conference on epithelial-mesenchymal transition and cancer progression and treatment. Cancer Res. 2010;70(19):7360–7364. doi: 10.1158/0008-5472.CAN-10-1208. [DOI] [PubMed] [Google Scholar]
- 35.Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2(2):120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 36.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 37.Knudsen BS, Vande Woude G. Showering c-MET-dependent cancers with drugs. Curr Opin Genet Dev. 2008;18(1):87–96. doi: 10.1016/j.gde.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Raychaudhuri B, Han Y, Lu T, et al. Aberrant constitutive activation of nuclear factor kappaB in glioblastoma multiforme drives invasive phenotype. J Neurooncol. 2007;85(1):39–47. doi: 10.1007/s11060-007-9390-7. [DOI] [PubMed] [Google Scholar]
- 39.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siebzehnrubl FA, Silver DJ, Tugertimur B, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malanchi I, Santamaria-Martinez A, Susanto E, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481(7379):85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Wang Y, Jiang C. Stromal protein periostin identified as a progression associated and prognostic biomarker in glioma via inducing an invasive and proliferative phenotype. Int J Oncol. 2013;42(5):1716–1724. doi: 10.3892/ijo.2013.1847. [DOI] [PubMed] [Google Scholar]
- 43.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 44.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 46.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 47.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5(67):1–12. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem Pharmacol. 2010;80(5):654–665. doi: 10.1016/j.bcp.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66(16):7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 50.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 51.Bao S, Tibbetts RS, Brumbaugh KM, et al. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411(6840):969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- 52.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y, Lathia JD, Eyler CE, et al. Erythropoietin receptor signaling through STAT3 is required for glioma stem cell maintenance. Genes Cancer. 2010;1(1):50–61. doi: 10.1177/1947601909356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silver DJ, Siebzehnrubl FA, Schildts MJ, et al. Chondroitin sulfate proteoglycans potently inhibit invasion and serve as a central organizer of the brain tumor microenvironment. J Neurosci. 2013;33(39):15603–15617. doi: 10.1523/JNEUROSCI.3004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulasov I, Yi R, Guo D, et al. The emerging role of MMP14 in brain tumorigenesis and future therapeutics. Biochim Biophys Acta. 2014;1846(1):113–120. doi: 10.1016/j.bbcan.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Brat DJ, Van Meir EG. Vaso-occlusive and prothrombotic mechanisms associated with tumor hypoxia, necrosis, and accelerated growth in glioblastoma. Lab Invest. 2004;84(4):397–405. doi: 10.1038/labinvest.3700070. [DOI] [PubMed] [Google Scholar]
- 57.Brat DJ, Castellano-Sanchez AA, Hunter SB, et al. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64(3):920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- 58.Folkins C, Shaked Y, Man S, et al. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009;69(18):7243–7251. doi: 10.1158/0008-5472.CAN-09-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Pore N, Behrooz A, et al. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276(12):9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 63.Papandreou I, Cairns RA, Fontana L, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 64.Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1(7):552–562. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6(7):2585–2597. [PubMed] [Google Scholar]
- 66.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 67.Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 68.Kreisl TN, Kim L, Moore K, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009;27(5):740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai A, Tran A, Nghiemphu PL, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol. 2011;29(2):142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weller M, Yung WK. Angiogenesis inhibition for glioblastoma at the edge: beyond AVAGlio and RTOG 0825. Neuro Oncol. 2013;15(8):971. doi: 10.1093/neuonc/not106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Groot JF, Fuller G, Kumar AJ, et al. Tumor invasion after treatment of glioblastoma with bevacizumab: radiographic and pathologic correlation in humans and mice. Neuro Oncol. 2010;12(3):233–242. doi: 10.1093/neuonc/nop027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8(8):592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebos JM, Lee CR, Kerbel RS. Tumor and host-mediated pathways of resistance and disease progression in response to antiangiogenic therapy. Clin Cancer Res. 2009;15(16):5020–5025. doi: 10.1158/1078-0432.CCR-09-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Groot JF, Mandel JJ. Update on anti-angiogenic treatment for malignant gliomas. Curr Oncol Rep. 2014;16(4):380. doi: 10.1007/s11912-014-0380-6. [DOI] [PubMed] [Google Scholar]
- 75.Lu KV, Chang JP, Parachoniak CA, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22(1):21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie Q, Vande Woude GF, Berens ME. RTK inhibition: looking for the right pathways toward a miracle. Future Oncol. 2012;8(11):1397–1400. doi: 10.2217/fon.12.130. [DOI] [PubMed] [Google Scholar]
- 77.Wen PY. American Society of Clinical Oncology 2010: report of selected studies from the CNS tumors section. Expert Rev Anticancer Ther. 2012;10(9):1367–1369. doi: 10.1586/era.10.117. [DOI] [PubMed] [Google Scholar]
- 78.Neyns B, Sadones J, Joosens E, et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20(9):1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 79.Raizer JJ, Abrey LE, Lassman AB, et al. A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 2010;12(1):95–103. doi: 10.1093/neuonc/nop015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104(31):12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lal B, Goodwin CR, Sang Y, et al. EGFRvIII and c-Met pathway inhibitors synergize against PTEN-null/EGFRvIII+ glioblastoma xenografts. Mol Cancer Ther. 2009;8(7):1751–1760. doi: 10.1158/1535-7163.MCT-09-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17(1):77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12(2):89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 85.Koochekpour S, Jeffers M, Rulong S, et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57(23):5391–5398. [PubMed] [Google Scholar]
- 86.Xie Q, Bradley R, Kang L, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci U S A. 2012;109(2):570–575. doi: 10.1073/pnas.1119059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eder JP, Vande Woude GF, Boerner SA, et al. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009;15(7):2207–2214. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- 88.Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2012;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 89.Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mitra AK, Sawada K, Tiwari P, et al. Ligand-independent activation of c-Met by fibronectin and alpha(5)beta(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30(13):1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prados MD, Seiferheld W, Sandler HM, et al. Phase III randomized study of radiotherapy plus procarbazine, lomustine, and vincristine with or without BUdR for treatment of anaplastic astrocytoma: final report of RTOG 9404. Int J Radiat Oncol Biol Phys. 2004;58(4):1147–1152. doi: 10.1016/j.ijrobp.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 93.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]