Abstract
AGL15 (for AGAMOUS-Like 15) is a member of the MADS domain family of DNA binding transcriptional regulators that accumulates to its highest amounts during embryo development. To better understand how AGL15 functions, a chromatin immunoprecipitation approach was used to identify directly regulated genes. One DNA fragment that coprecipitated with AGL15 corresponded to a portion of the regulatory region of a gene named DTA1 (for Downstream Target of AGL15-1). The expression of DTA1 was positively correlated with AGL15 abundance during embryogenesis. In this report, a cis element for response to AGL15 was identified, and the activity of DTA1 as a gibberellin (GA) 2-oxidase was confirmed. DTA1 corresponds to AtGA2ox6 and was renamed to indicate this identity. Further experiments related the function of AtGA2ox6 to regulation by AGL15. Constitutive expression of AGL15 and of AtGA2ox6 altered endogenous GA amounts and caused GA-deficient phenotypes in Arabidopsis thaliana that could be at least partially rescued by application of biologically active GA. The phenotype of plants with decreased expression of AtGA2ox6 was the converse of plants overexpressing AtGA2ox6 in terms of seed germination attributes and effects on somatic embryo production.
INTRODUCTION
DNA binding transcriptional regulators are central players in the control of genetic programs. In plants, members of the MADS domain family of regulatory proteins have key roles throughout the plant life cycle, with the best-characterized members involved in inflorescence and floral development (reviewed in Riechmann and Meyerowitz, 1997). MADS domain proteins bind to DNA elements called CArG motifs (for C-[A/T]rich-G) via the conserved 55 to 60 amino acid residue MADS domain and control developmental programs by activating and/or repressing the expression of specific target genes (reviewed in Riechmann and Meyerowitz, 1997; Theissen, 2001). Identification of the genes directly controlled by DNA binding proteins is of critical importance to understanding how transcriptional regulators operate. In addition, determination of the function of the regulated gene products is crucial for defining the events that lead from gene regulation to developmental consequence. However, identification of direct regulatory targets of a transcription factor has been difficult to address by genetic means because of redundancy of gene functions, essential roles for downstream gene products, and the interaction of different regulated gene products to generate a final phenotype (Andrew and Scott, 1992; Graba et al., 1997). Deconvoluting gene regulatory networks is further complicated because a given gene may be regulated by different trans-acting factors, depending on the temporal or spatial context (Arnone and Davidson, 1997; Graba et al., 1997; Fickett and Wasserman, 2000). Other approaches such as differential display or microarray analysis can identify direct targets that respond to a transcriptional regulator but may also isolate indirect targets. Chromatin immunoprecipitation (ChIP) is becoming a widely used approach to obtain DNA fragments directly bound in vivo by a transcription factor and, therefore, potentially directly regulated (reviewed in Orlando et al., 1997; Orlando, 2000; Shannon and Rao, 2002). However, there are cases of in vivo binding of a transcriptional regulator to a cis element, without a corresponding change in gene regulation in the absence of other factors or signals, making assessment of the consequence of binding important (Wyrick and Young, 2002).
ChIP has been used to isolate several direct downstream targets of AGAMOUS-Like 15 (AGL15), a member of the MADS domain family (Wang et al., 2002). AGL15 is currently the only identified member of the Arabidopsis thaliana MIKC subgroup of the MADS box family that is preferentially expressed during embryogenesis (Heck et al., 1995; Rounsley et al., 1995). Other members of the Arabidopsis MIKC subgroup are also expressed in developing embryos but are expressed at similar or higher levels in other tissues and at other stages of development (e.g., Flanagan and Ma, 1994; Burgeff et al., 2002). AGL15 is expressed after germination but generally at much lower levels than during embryo development (Fernandez et al., 2000). In all circumstances studied to date, AGL15 and putative orthologs in other species accumulate in nuclei of angiosperm tissue developing in an embryonic mode, suggesting that AGL15 may be involved in promotion of the embryo phase of the plant life cycle (Heck et al., 1995; Perry et al., 1996, 1999). Furthermore, constitutive expression of AGL15 via the 35S promoter of Cauliflower mosaic virus (CaMV) supports the maintenance of development in an embryonic mode for extended periods of time in culture and enhances the production of somatic embryo formation from the shoot apical meristems of seedlings in a liquid culture system (Harding et al., 2003).
DTA1 (for Downstream Target of AGL15-1) was isolated by ChIP, is expressed in response to AGL15 (Wang et al., 2002), and encodes a protein with similarity to gibberellin (GA) 2-oxidases. GA 2-oxidases are a small family of enzymes in Arabidopsis that catalyze conversion of some biologically active GAs to inactive forms. Does DTA1 function in this manner and, if so, how does this function relate to AGL15? Experimental results presented in this article verify that DTA1 is a direct downstream target of AGL15, identify a DNA binding site for AGL15 in the regulatory regions of DTA1, and demonstrate that DTA1 functions as a GA 2-oxidase with effects on seed germination and somatic embryogenesis.
RESULTS
DTA1 Encodes a Protein That Functions as a GA 2-Oxidase
ChIP was described previously as an approach to isolate DNA fragments, to which AGL15 binds in vivo in embryonic culture tissue constitutively expressing AGL15 (Wang et al., 2002; Harding et al., 2003). One such isolated fragment corresponded to the 5′ regulatory region of a gene encoding an unknown protein that was named DTA1. DTA1 cDNA coding region and 5′ and 3′ untranslated regions were obtained as described previously (Wang et al., 2002). Sequence analysis confirmed the correct prediction of the gene in the Arabidopsis database (gene locus At1g02400, protein identification number AAG00891.1). Database searches indicated that the predicted gene product of DTA1 has homology to GA 2-oxidases, including key conserved residues that would predict function as a GA 2-oxidase. The active site core motif of 2-ketoglutarate–dependent dioxygenases involved in gibberellin metabolism was present, including His-203, Asp-205, and His-260 that bind the active site Fe2+, and Arg-271 and Ser-273 that interact with 2-ketoglutarate (Hedden and Phillips, 2000). DTA1 was more similar to the predicted products of AtGA2ox1 through AtGA2ox3 (similarity from 40.4% to 67.5%) than to AtGA2ox7 (17.3%) and AtGA2ox8 (21.5%).
To test whether DTA1 encodes a gene product with GA 2-oxidase activity, the full-length coding region was expressed in Escherichia coli as a C-terminal fusion to maltose binding protein. The recombinant fusion protein was present in the soluble portion of the induced cell lysate and was used for enzyme assays. As shown in Table 1, 17,17-2H2-GA4 was converted to 2H2-GA34 as identified by gas chromatography–mass spectrometry (GC-MS). Also, 2H2-GA1 and 2H2-GA20 were converted to 2H2-GA8 and 2H2-GA29, respectively (Table 1; data not shown). Thus, DTA1 encodes a functional GA 2-oxidase. Given the sequence similarity, chromosomal location (at the top of chromosome 1), and the fact that DTA1 can catalyze 2β-hydroxylation of gibberellins, DTA1 corresponds to a gene with a product predicted to function as a GA 2-oxidase, AtGA2ox6 (Hedden et al., 2002, and unpublished data therein), and has been renamed to indicate this identity. This gene also corresponds to AtGA2ox4 in Ogawa et al. (2003).
Table 1.
GA 2-Oxidase Enzyme Activity Assay of the Recombinant AtGA2ox6 Protein
| Substrate | Product | Mass Spectra of Products, Mass-to-Charge Ratio (% Relative Abundance) | |
|---|---|---|---|
| 17,17-2H2-GA1 | 2H2-GA8 | 596(100), 581(6), 567(17), 537 (20), 450(24), 377 (10), 240(6) | |
| 17,17-2H2-GA4 | 2H2-GA34 | 508(100), 418(16), 315(10), 290(6), 231(13), 225(16) | |
| 2H2-GA8 standard | 596(100), 581(5), 567(12), 537(13), 450(52), 377 (14), 240(12) | ||
| 2H2-GA34 standard | 508(100), 418(22), 315(12), 290(11), 231(14), 225(34) | ||
Products were identified by GC-MS after incubation of the substrate with recombinant MAL-AtGAox6 protein and conversion to Me-TMsi derivatives.
AtGA2ox6 Is a Direct Downstream Target of AGL15
To confirm that the DNA fragment corresponding to the 5′ regulatory region of AtGA2ox6 was specifically selected in the ChIP experiments, enrichment tests were performed on independently generated ChIP populations (Wang et al., 2002). If a DNA fragment is truly associated with AGL15 in vivo, it should be represented in greater abundance after selection by ChIP than an unbound fragment, for example, from the coding region of ubiquitin extension protein (UBQ6; Callis et al., 1990). Multiplex PCR using primers that amplify the target and control fragments, on input (total) and on ChIP-selected DNA, revealed that the ratio of AtGA2ox6 fragment to control fragment was higher in the ChIP population compared with the input DNA (Figure 1A). After two sequential immunoprecipitations using AGL15-specific antiserum, the AtGA2ox6 fragment was represented in even greater abundance compared with the control fragment (Figure 1A, ChIP2). Chromatin immunoprecipitation populations generated using preimmune serum did not show specific selection of the AtGA2ox6 fragment (data not shown), nor did a series of controls to verify that the immune serum–AGL15-DNA fragment association was specific (Wang et al., 2002).
Figure 1.
AGL15 Interacts in a Sequence-Specific Manner with a Noncanonical CArG Motif Present in the Regulatory Region of AtGA2ox6.
(A) Enrichment of the AtGA2ox6 fragment in a ChIP population selected using AGL15 antiserum. Oligonucleotide primer pairs to amplify the region of AtGA2ox6 cloned in the ChIP population and to amplify control regions not expected to be bound by AGL15 (UBQ6) were used in multiplex PCR on total (Input) DNA diluted 125- and 625-fold, and on DNA recovered by immune precipitation in ChIP after one (ChIP1) and two (ChIP2) sequential immunoprecipitations. PCR products were resolved on an 8% polyacrylamide gel. Size of the PCR products is indicated at right. Numbers refer to the relative enrichment calculated as in Noma et al. (2001).
(B) Autoradiography of EMSAs to assess the interaction of AGL15 with AtGA2ox6 regulatory regions. A 47-bp fragment containing the noncanonical CArG motif CCAATTTAATGG was used in EMSAs. Lanes 1 to 4 contained increasing amounts of AGL15 with a C-terminal T7 tag (0, 40, 80, and 160 ng) expressed and purified from E. coli and incubated with the radiolabeled DNA fragment. Lane 5 added 0.5 μg of anti-T7 antibody to the reaction in lane 4. Lanes 6 and 7 contained incubation of 0 and 160 ng, respectively, of purified AGL15-T7 incubated with a modified radiolabeled DNA fragment, in which the CC within the CArG motif was changed to a TT.
(C) Autoradiography of EMSAs to assess the sequence-specific interaction of AGL15 with the DNA fragment derived from AtGA2ox6. Competitive binding assays with AGL15-T7 (160 ng), 32P-labeled DNA fragment from the AtGA2ox6 promoter with the native CArG of CCAATTTAATGG, and nonradiolabeled competitor corresponding to either the native CArG or a mutated form in which the CC was changed to a TT were analyzed on polyacrylamide gels. Unlabeled competitor was present at 50-, 200-, and 800-fold compared with radiolabeled fragment (lanes 1 to 3, native competitor; lanes 4 to 6, modified competitor).
(D) Quantitative analysis of native AtGA2ox6:GUS and mutated mAtGA2ox6:GUS in wild-type and AGL15 ectopically expressing backgrounds. MUG assays were performed to quantitate the amount of GUS activity to determine expression from the native AtGA2ox6 regulatory region and from a modified form of the AtGA2ox6 promoter, in which the CC in the putative AGL15 binding site was changed to a TT. Expression of the reporter gene was measured in sibling seedlings that contained or lacked a 35S:AGL15 transgene (n = 16 to 21, except for AtGA2ox6:GUS, line 2, where n = 6 to 9; standard error of the mean is shown). Different letters indicate significant difference as assessed by Tukey's mean separation test (SAS, 1999) when wild-type and 35S:AGL15 siblings were compared within a reporter line.
(E) Enrichment of a DNA fragment from a putative Brassica ortholog of AtGA2ox6 in a ChIP population selected using AGL15 antiserum. Oligonucleotide primer pairs were designed to amplify the regulatory region of a putative BnGA2ox that is similar to AtGA2ox6 and to amplify control regions not expected to be bound by AGL15 (BnTUB). Multiplex PCR was performed on total (Input) DNA diluted 125- and 625-fold, and on DNA recovered by immune precipitation (ChIP) or preimmune precipitation (PI-ChIP) from developing Brassica seeds. PCR products were resolved on a 2% agarose gel. Numbers refer to the relative enrichment calculated as in Noma et al. (2001).
(F) Response of expression of AtGA2ox6 to AGL15 levels. RT-PCR products using oligonucleotide primers to assess the expression of AtGA2ox6 in wild-type and agl15 mutant 5- to 8-d developing seeds were analyzed on a 1.5% agarose gel. Tubulin served as a control. A representative gel image is shown; numbers refer to the mean from quantitation of three biological replicates, normalized using tubulin and compared with the wild type.
The selected fragment did not contain any canonical CArG motifs of the form CC[A/T]6GG (reviewed in Riechmann and Meyerowitz, 1997), and no canonical sequences were found nearby. However, in vitro binding site selection studies revealed that AGL15 preferentially binds to sequences with a longer A/T-rich region, of the form [A/T]3C[A/T]8G[A/T]3 (Tang and Perry, 2003). A potential binding site for AGL15 with a longer A/T-rich region was present in the AtGA2ox6 fragment (CC[A/T]8GG). To test whether AGL15 could associate with this sequence in vitro, electrophoretic mobility shift assays (EMSAs) were performed with a 32P-radiolabeled DNA fragment containing this site. AGL15 with a C-terminal T7 epitope was expressed in E. coli, and the protein purified via the T7 epitope. As shown in Figure 1B, purified AGL15-T7 was able to retard the mobility of the probe with the noncanonical CArG motif (Figure 1B, lanes 2 to 4; 40, 80, and 160 ng of AGL15-T7, respectively; lane 1, no AGL15-T7). Addition of a T7-specific antibody caused a decrease in the intensity of the shifted band and appearance of supershifted complexes (Figure 1B, lane 5), indicating that AGL15-T7 was responsible for retardation of the probe. Binding of AGL15 to the DNA fragment was sequence specific. When the CC within the CArG motif was changed to TT, no binding was observed in EMSAs (Figure 1B, lane 7, with 160 ng AGL15-T7; lane 6, no AGL15-T7). Additionally, a 50- to 800-fold excess of unlabeled DNA fragment with the native binding site was able to compete efficiently with the radiolabeled native site for binding to AGL15 (Figure 1C, lanes 1 to 3; 50-, 200-, and 800-fold unlabeled competitor, respectively), but the same amounts of mutated unlabeled fragment had little effect (Figure 1C, lanes 4 to 6; 50-, 200-, and 800-fold unlabeled mutated competitor).
Although AGL15 was able to specifically bind in vitro to a probe containing the CC[A/T]8GG motif, in vitro binding does not necessarily indicate in vivo interaction or function. Enrichment tests indicated that AGL15 interacted in vivo with the AtGA2ox6 fragment that included this site (Figure 1A). But does AGL15 truly contribute to regulation of AtGA2ox6 through the CC[A/T]8GG element? To address this question, quantitative analysis of reporter constructs in response to AGL15 accumulation were performed. Previous work using RT-PCR and reporter constructs demonstrated that expression of AtGA2ox6 is upregulated in response to increased accumulation of AGL15 as found in 35S:AGL15 transgenic plants. The reporter construct (AtGA2ox6:GUS [β-glucuronidase], previously named DTA1:GUS in Wang et al., 2002) used in these studies consisted of ∼2.3-kb sequence 5′ of the translation initiation codon fused to the reporter gene GUS and includes the AtGA2ox6 fragment isolated by ChIP. To quantitate the response to AGL15, transgenic lines carrying the reporter construct were crossed to 35S:AGL15 plants, subsequent progeny separated visually for the presence or absence of the 35S:AGL15 transgene (Fernandez et al., 2000; Wang et al., 2002), and GUS activity in individual seedlings measured using 4-methylumbelliferyl-β-d-glucuronide (MUG) assays. Seedlings with the 35S:AGL15 and reporter transgenes had significantly greater GUS activity than did siblings with only the reporter transgene and no ectopic or increased AGL15 accumulation (Figure 1D). These results were confirmed with two independent reporter lines, each carrying one reporter insert.
To test whether the CC[A/T]8GG motif within the AtGA2ox6 regulatory region was important for the increased expression in response to AGL15, a second reporter construct was generated that was identical to the AtGA2ox6:GUS transgene except that the potential binding site for AGL15 was mutated. Specifically, the CC in the CC[A/T]8GG motif was changed to TT, to which AGL15 cannot bind in vitro (Figure 1B, lane 7). The mutated transgenic lines (mAtGA2ox6:GUS) were crossed to 35S:AGL15 plants, and GUS activities measured in subsequent generations in seedlings with and without the 35S:AGL15 transgene. In contrast with the reporter with the native binding site, the mutated site showed little to no response to AGL15 amounts (Figure 1D). Two independent mutated lines with one insert each were assessed this way. Therefore, the noncanonical CArG motif was a relevant cis element for the response to AGL15 in vivo.
Protein gel blots and immunohistochemical localization studies indicate that the embryonic culture tissue used for ChIP accumulates similar amounts of AGL15 compared with Brassica napus and Arabidopsis zygotic embryos (Wang et al., 2002). However, it is still possible that somewhat higher levels of AGL15 occupy sites that would not be occupied in nontransgenic tissue. It is not reasonable to collect enough Arabidopsis embryos or seeds of early enough developmental stage for ChIP experiments. B. napus is closely related to Arabidopsis, and it is feasible to obtain sufficient tissue for ChIP. The nucleotide sequence corresponding to the coding region of AtGA2ox6 was used to search the Brassica DNA database. A sequence from B. oleracea was 87% identical over 401 nucleotides (accession number BZ033271). B. napus is believed to be an amphidiploid from hybridization of B. oleracea and B. rapa (Parkin et al., 1995), so it is reasonable to expect a closely related sequence in B. napus. The predicted protein product of the identified gene fragment was 87% identical and 93% similar to the N-terminal 133 amino acid residues of the Arabidopsis protein. The first intron–exon boundary was in the same position in both species, but no contiguous Brassica DNA fragments 3′ to this sequence could be identified in the database. However, searches to obtain sequence 5′ to the coding region were successful and allowed assembly of ∼620 bp 5′ to the start codon (accession numbers BH741917 and BZ081538). Overlap between database entries were at least 260 nucleotides in length and 100% identical. The region ∼430 nucleotides 5′ to the start codon is ∼70% identical to that found in Arabidopsis. Interestingly, the 5′ region of the Brassica sequence contains three potential binding sites for MADS domain proteins in similar positions as found in Arabidopsis, including a C-8-G type site of the form GC[A/T]8GG. Oligonucleotide primers were designed corresponding to this region of the Brassica sequence and used to test for enrichment of this fragment in a ChIP population derived from young B. napus seeds containing heart to young bent-cotyledon stage embryos. As shown in Figure 1E, this DNA fragment was enriched in the immune-precipitated sample (ChIP) but not in the preimmune-precipitated sample compared with a DNA fragment from the coding region of a β-tubulin gene (accession number AF258790). The enriched fragment was sequenced to verify identity (data not shown).
To further confirm that regulation of AtGA2ox6 by AGL15 occurred in the context of zygotic embryogenesis, where AGL15 was expressed only from the native promoter, accumulation of AtGA2ox6 mRNA in staged wild-type and agl15 mutant seeds (5 to 8 d postanthesis) was compared by RT-PCR. agl15 contains a T-DNA insertion in the first intron, and no correctly spliced full-length transcript can be detected by RT-PCR (M.D. Lehti-Shiu and D.E. Fernandez, personal communication). As shown in Figure 1F, decreased amounts of AtGA2ox6 mRNA accumulated when no AGL15 was present. This agrees with previous results using staged developing siliques (Wang et al., 2002).
Expression of AtGA2ox6 and Relation to AGL15
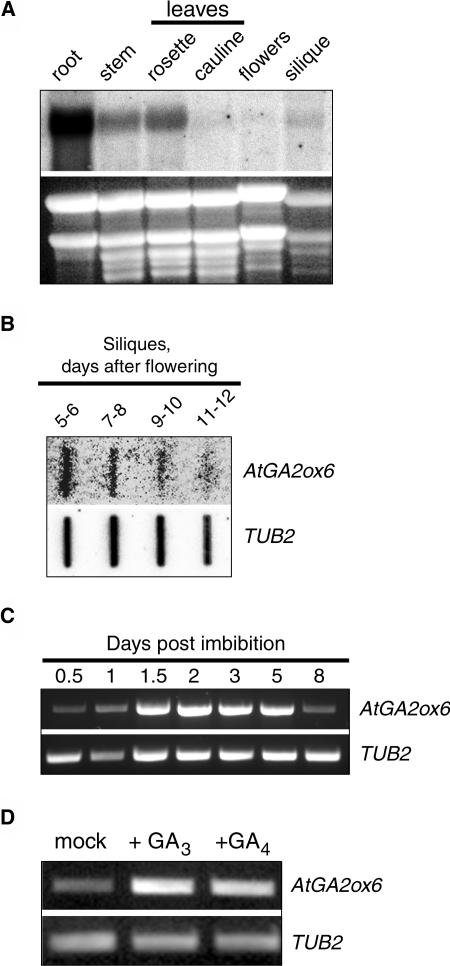
To gain insight into the developmental role of AtGA2ox6, the expression pattern of AtGA2ox6 was analyzed by RNA gel blot using various developmental stages and tissue types from Arabidopsis (Figure 2A). AtGA2ox6 transcript amounts were very high in roots and relatively high in rosette leaves and stems. Signals were also clearly detected from bulk siliques but barely detectable in cauline leaves and flowers. As shown in Figure 2B, expression of AtGA2ox6 in siliques was highest during the early stages of development (5 to 6 d after fertilization [daf]), and mRNA accumulation from this gene declined afterward. During and after germination, mRNA could be detected by RT-PCR (Figure 2C). Increased accumulation of AtGA2ox6 transcript was induced by GA4 or GA3 treatment of plants, indicating a role for feed-forward regulation (Figure 2D).
Figure 2.
Developmental Expression of AtGA2ox6.
(A) RNA gel blot probed to assess AtGA2ox6 expression pattern. Total RNA (20 μg per lane) was electrophoresed through an agarose gel, transferred to membrane, and incubated with 32P-AtGA2ox6 specific probe. The ethidium bromide–stained gel showing rRNA served as a loading control.
(B) RNA slot blot probed to assess AtGA2ox6 expression during silique development. Poly(A+) RNA (1 μg) extracted from different stages of silique development was applied to membrane using a vacuum manifold and incubated with 32P-AtGA2ox6 specific probe. The same blot was stripped and assessed for equal loading with a 32P-2-β-Tubulin (TUB2) specific probe.
(C) RT-PCR products using AtGA2ox6 or 2-β-Tubulin (TUB2) specific oligonucleotide primers and RNA from germinating seeds and establishing seedlings were analyzed on a 1.5% agarose gel.
(D) Feed-forward regulation of AtGA2ox6. RT-PCR analysis to assess AtGA2ox6 expression in rosette leaves from plants treated with 100 μM GA3, GA4, or water (mock treated). 2-β-Tubulin (TUB2) was used as a control.
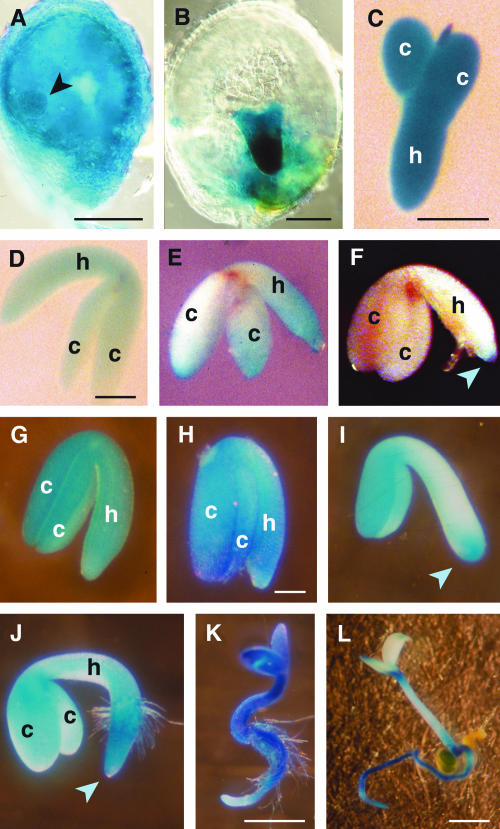
To further investigate the expression of AtGA2ox6 during seed development, the activity of the regulatory regions of this gene was assessed using a GUS reporter construct (AtGA2ox6:GUS). Transgenic plants containing the reporter construct showed a pattern of GUS activity consistent with RT-PCR studies (Wang et al., 2002; Figure 2C) and RNA gel blot analysis (Figures 2A and 2B). Developing seeds or embryos of different stages were stained for GUS activity and demonstrated that the 5′ regulatory region of AtGA2ox6 was active during early embryogenesis, from at least globular stage (∼3 daf) through torpedo stage (∼6 to 8 daf; Figures 3A to 3C). During early stages, GUS activity was also detected in other tissues of the seed (Figure 3A). Decreased GUS activity was observed in bent cotyledon embryos and was nearly undetectable in the late maturing embryo (Figures 3D to 3F). The regulatory regions of AtGA2ox6 were reactivated in imbibed seeds (by 8 h after imbibition; Figure 3G), with high GUS activity persisting throughout the seedling until 5 d after imbibition, when expression became more localized to the root and the shoot apex (Figures 3H to 3L).
Figure 3.
AtGA2ox6:GUS Expression during Embryogenesis, Germination, and Seedling Establishment.
Different stages of development from plants carrying a reporter transgene were stained for GUS activity.
(A) Globular stage embryo (arrowhead) within a developing seed.
(B) Late heart/early torpedo stage embryo within a developing seed.
(C) Isolated torpedo stage embryo.
(D) Bent-cotyledon stage embryo.
(E) and (F) Maturation stage embryos.
(G) to (I) Isolated embryos from seed at 8, 24, and 48 h postimbibition, respectively.
(J) to (L) Seedlings at 3 d ([J] and [K]) and 5 d (L) postimbibition.
Blue arrowheads in (F), (I), and (J) indicate the root apical meristem. h, hypocotyl; c, cotyledons. Bars in (A) to (D) and (H) = 100 μm; in (K) and (L) = 0.5 mm.
Plants that express AtGA2ox6 ectopically and at higher levels and plants that have decreased expression of AtGA2ox6 were obtained to assess in vivo function. The AtGA2ox6 cDNA or the AtGA2ox6 genomic region including introns were introduced into Arabidopsis plants under the control of the CaMV 35S promoter. A greater percentage of transformants containing the genomic version of the gene showed a dwarf phenotype compared with transformants containing the cDNA version of the transgene (∼70%, 19 out of 27 plants show an obvious phenotype for the genomic version of the construct, as shown in Figures 4A and 4B, lines 2 and 6, compared with ∼8%, 3 out of 37 plants, for the cDNA version of the construct). In accordance with the more severe phenotype, the genomic-version transgenic plants were found to accumulate higher amounts of AtGA2ox6 mRNA (Figure 4C; data not shown). Two independent lines were selected that carried one copy of the genomic version of the transgene and produced different degrees of expression for further experiments as described below.
Figure 4.
Phenotypic Analysis of Arabidopsis with Altered Expression of AtGA2ox6 or AGL15.
(A) and (B) Phenotype of two 35S:AtGA2ox6 transgenic lines (lines 2 and 6), a 35S:AGL15 transgenic line (AGL15), and a ga2ox6 knockdown mutant (ga2ox6-1) compared with wild-type plants, at 5 (A) and at 7 to 9 weeks (B).
(C) RNA gel blot to assess the amount of ectopic AtGA2ox6 expression in 35S transgenic plants (lines 2 and 6) compared with wild-type and ga2ox6-1 plants. Total RNAs (7 μg) isolated from 3-week-old plants were analyzed by RNA gel blot with AtGA2ox6 specific radiolabeled probe. rRNA is shown to assess equal loading.
(D) RT-PCR analysis of ga2ox6-1. Oligonucleotide primers corresponding to AtGA2ox6 were used to assess the nature of the insertional mutation in AtGA2ox6. Oligonucleotides that amplify 2-β-Tubulin (TUB2) were used as a control.
(E) Application of biologically active gibberellin rescued the 35S:AtGA2ox6 plants. GA3 was applied to the severe overexpressing line 6, just before or at the onset of bolting. Phenotypes were compared with wild-type and mock-treated line 6 plants 10 d after application of GA.
(F) Application of biologically active gibberellin (GA3) partially rescued the phenotype of 35S:AGL15 plants. GA3 was applied when plants were 3.5 weeks old and were photographed at 5 weeks.
(G) Altered GA profiles in transgenic plants constitutively expressing AGL15 or AtGA2ox6 compared with the wild type. Endogenous GA concentrations in rosette leaves were obtained using GC-MS and internal standards. Means from three replicates and standard error of the mean are shown. Different letters indicate significant difference as assessed by Tukey's mean separation test (SAS, 1999). Uppercase and lowercase letters indicate different comparisons.
In addition to generation of gain-of-function plants, a mutant containing a T-DNA insertion in the first intron of AtGA2ox6 was isolated from the University of Wisconsin–Madison Arabidopsis Knockout Facility α-T-DNA population. To confirm whether the mutant truly represents a knockout allele, RT-PCR was performed after obtaining a plant that was homozygous for a single T-DNA insertion within AtGA2ox6. A very weak DNA band was clearly visible in the mutant after 40 cycles of PCR amplification. Although correctly spliced mRNA was detected, the amount was much less than found in wild-type plants (Figure 4D). Therefore, this allele, named ga2ox6-1, did not represent a complete knockout of the gene but rather was a knockdown mutation (Krysan et al., 1999).
As shown in Figures 4A, 4B, and 4E, the AtGA2ox6 overexpressing plants showed a GA-deficient phenotype (reviewed in Richards et al., 2001). The plants were dwarf, dark green, flowered later than the wild type, showed decreased apical dominance, and remained green longer. Line 6 showed a severe phenotype (Figures 4A, 4B, and 4E) and accumulated more AtGA2ox6 mRNA (Figure 4C) than did line 2, which had a weaker phenotype (Figures 4A to 4C). As summarized in Table 2, plants with the 35S:AtGA2ox6 transgene produced an increased number of rosette leaves, more inflorescence branches, shorter internodes, and smaller fruit size compared with the wild type. The number of seeds per silique was less than in the wild type, and the seeds were more compactly arranged within the silique. Many aspects of the 35S:AtGA2ox6 phenotype were similar to that found when AGL15 is expressed via the CaMV 35S promoter (Fernandez et al., 2000; Figure 4A; Table 2). However, the delay in senescence and abscission of the floral organs reported for the 35S:AGL15 transgenic plants (Fernandez et al., 2000) was not observed in the 35S:AtGA2ox6 plants. All of the aspects of the 35S:AtGA2ox6 phenotype could be rescued by spraying the plants with either GA4 or GA3 (Figure 4E; data not shown). Some aspects of the 35S:AGL15 phenotype could be alleviated by spraying with biologically active GAs, in particular the darker green color, late flowering, and decreased stature (Figure 4F). The delay in floral organ senescence and abscission observed in the 35S:AGL15 plants was not rescued by spraying with GA3 or GA4 (data not shown). The ga2ox6-1 plants exhibited only very mild phenotypes that were not significantly different from the wild type (Figures 4A and 4B; Table 2).
Table 2.
Phenotypic Comparison of the Wild Type to Overexpression Lines of AtGA2ox6 and AGL15 and to ga2ox6-1
| Phenotypic Parameter | Wild Type | 35S:GA2ox6, Line 2 | 35S:GA2ox6, Line 6 | ga2ox6-1 | 35S:AGL15 |
|---|---|---|---|---|---|
| Height (cm) | 40.2 ± 2.6aA | 16.3 ± 3.3 C | 9.7 ± 1.15 D | 37.2 ± 3.1 AB | 32.6 ± 2.2 B |
| Number of rosette leaves at floweringb | 9.0 ± 1.0 A | 16.6 ± 1.1 C | 18.6 ± 1.8 C | 8.0 ± 0.7 A | 13.4 ± 0.9 B |
| Number of inflorescence branchesb | 15.8 ± 1.6 A | 43.6 ± 4.2 C | 53.6 ± 5.2 D | 11.0 ± 2.0 A | 34.0 ± 5.1 B |
| Internode length (mm)b | 8.5 ± 0.9 A | 4.1 ± 0.7 B | 2.1 ± 0.1 C | 9.2 ± 1.1 A | 4.0 ± 0.4 B |
| Fruit length (mm)c | 18.8 ± 0.8 A | 11.4 ± 0.4 C | 8.0 ± 0.6 D | 18.8 ± 0.8 A | 16.4 ± 0.8 B |
| Seed number per siliquec | 59.0 ± 2.0 B | 47.8 ± 1.9 C | 33 ± 2.2 D | 59.2 ± 1.9 B | 63.4 ± 1.3 A |
Mean ± sd.
Methods used for measurement are following the description of Schomberg et al. (2003).
The siliques used for measurement were fully developed and nearly mature.
Comparisons across a row with different letters (in bold) are significantly different as assessed by Tukey's mean separation test (SAS, 1999).
Because the phenotypes of both the AtGA2ox6 and AGL15 overexpressors suggested deficiencies in biologically active GAs, the amounts of select GAs in AtGA2ox6 and AGL15 overexpressors and wild-type Arabidopsis rosette leaves were determined using GC-MS. Compared with the wild-type control, the major biologically active GA in Arabidopsis, GA4 (Hedden, 2002; Ogawa et al., 2003), was significantly decreased in both the AtGA2ox6 and AGL15 overexpressor plants compared with the wild type (Figure 4G; n = 3). GA34 is the GA 2-oxidase metabolite of GA4 (reviewed in Hedden and Phillips, 2000) and was present at increased amounts in both AtGA2ox6 and AGL15 overexpressors, compared with the wild type (Figure 4G).
The Role of AtGA2ox6 during Seed Development and Germination
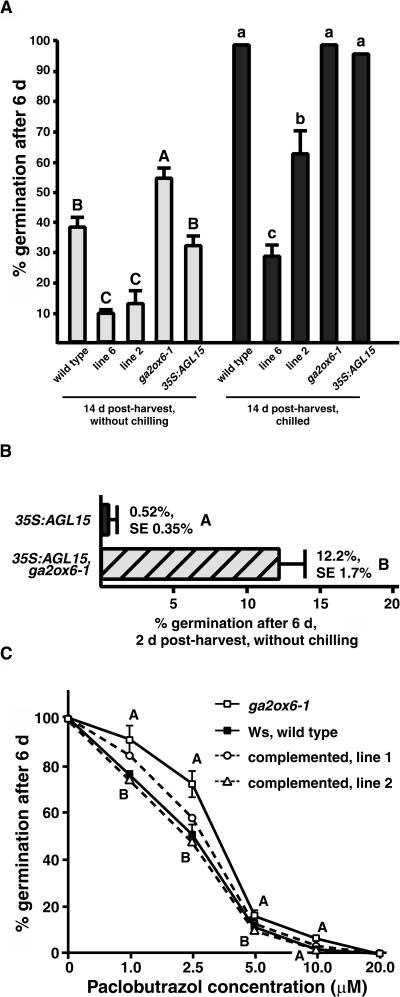
Because of the intriguing expression of AtGA2ox6 during early stages of silique development (Figure 2B) that was subsequently found to be localized predominantly in the developing embryo (Figures 3A to 3F) and the second phase of expression during and after germination (Figures 2C and 3G to 3L), experiments to determine the developmental consequences of AtGA2ox6 expression on seeds were performed. Germination assays were used to test whether AtGA2ox6 expression may influence seed dormancy. Seeds (14 d postharvest) from plants grown at the same time and under identical conditions were sown on moist germination paper and incubated at 25°C under continuous light without any chilling treatment. As shown in Figure 5A, the percentage germination was negatively correlated with the amount of AtGA2ox6 expression. Within 6 d, ∼38% of the seed from wild-type plants completed germination (Figure 5A). However, a higher percentage of seed from plants homozygous for the knockdown mutation ga2ox6-1 completed germination within the same time frame (∼54%; Figure 5A). Seed from plants with higher than normal expression of AtGA2ox6 completed germination to a lower percentage within this time period (only 9% to 13%), and the germination percentage was related to the amount of expression of the transgene. Prechilling for 3 d increased the percentage germination of 35S:AtGA2ox6 seeds and could completely or nearly completely break the dormancy of wild-type, 35S:AGL15, and ga2ox6-1 seeds (Figure 5A). Introduction of the ga2ox6-1 knockdown mutation into 35S:AGL15 plants significantly increased the percentage of seeds capable of completing germination within 6 d, above that observed for 35S:AGL15 seed grown and harvested under identical conditions (Figure 5B).
Figure 5.
Germination Phenotype of Seeds from Gain-of-Function and Loss-of-Function AtGA2ox6 Plants Compared with 35S:AGL15 and Wild-Type Plants.
(A) The amount of expression of AtGA2ox6 was correlated with the degree of seed dormancy. Seeds were stored at room temperature for 14 d after harvest before performing the germination tests.
(B) The ga2ox6-1 knockdown mutant increased the percentage of 35S:AGL15 seed that could complete germination within 6 d. Seeds were stored at room temperature for 2 d after harvest before performing the germination tests.
(C) The ga2ox6-1 knockdown mutant seed was more resistant to the GA biosynthesis inhibitor paclobutrazol. Presence of a complementation transgene in ga2ox6-1 partially or completely rescued the resistance to paclobutrazol. Seeds were stored at room temperature for >1 year before performing germination tests.
The percentage of seeds that completed germination was scored after 6 d at 25°C under continuous light. Standard deviation ([A] and [C]) or standard error of the mean (B) is shown. Different letters indicate significant difference as assessed by Tukey's mean separation test (SAS, 1999). In (A), upper and lower case letters indicate different comparisons. In (C), comparisons are between Wassilewskija (Ws) and ga2ox6-1 at each paclobutrazol concentration.
To test whether the observed seed germination results were as a result of different degrees of dormancy imposed by different endogenous GA amounts, seed germination in response to biologically active GA4+7 (reviewed in Hedden and Phillips, 2000) or to a GA-biosynthesis inhibitor paclobutrazol (reviewed in Yamaguchi and Kamiya, 2002) was investigated. The low percentage germination of the 35S:AtGA2ox6 seed could be completely rescued by the addition of 100 μM GA4+7 (data not shown). The ga2ox6-1 mutant seed was more resistant to paclobutrazol than was wild-type seed (Figure 5C). Both wild-type and ga2ox6-1 seed germination were completely inhibited when the concentration of paclobutrazol exceeded 20 μM, but the mutant seed consistently displayed a higher percentage of germination than the wild type at lower concentrations of paclobutrazol. Transgenic ga2ox6-1 lines containing a complementation construct, in which AtGA2ox6 was expressed from its own regulatory regions, partially or completely rescued the resistance to paclobutrazol (Figure 5C).
AGL15, AtGA2ox6, GAs, and Somatic Embryogenesis
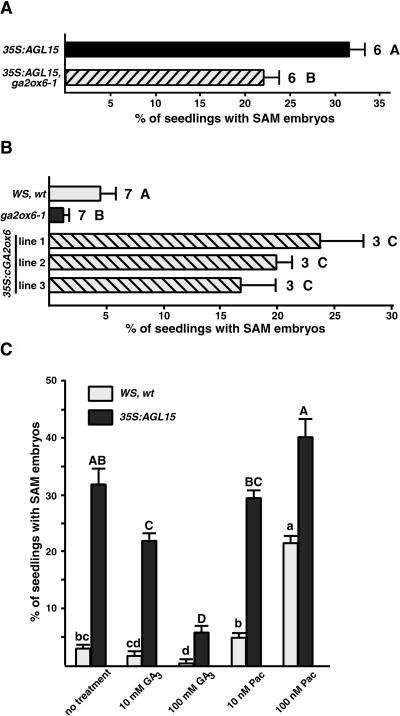
Recently, constitutive expression of AGL15 by the 35S promoter was shown to enhance production of somatic embryos from the shoot apical meristem (SAM) when seeds completed germination in liquid culture containing 2,4-D (Harding et al., 2003). To determine whether expression of AtGA2ox6 was involved in production of SAM embryos, the effect of the knockdown mutant in the 35S:AGL15 transgenic background was investigated. As shown in Figure 6A, inability to express normal amounts of AtGA2ox6 repressed the ability of 35S:AGL15 to promote SAM somatic embryo development. Additionally, ga2ox6-1 in the absence of the 35S:AGL15 transgene produced fewer SAM somatic embryos than did wild-type seedlings (Figure 6B). However, seedlings expressing a cDNA version of AtGA2ox6 via the 35S promoter showed enhanced production of SAM somatic embryos in three independent transgenic lines (Figure 6B).
Figure 6.
Somatic Embryo Production from the Shoot Apex of Seedlings in Culture.
(A) The ga2ox6-1 knockdown mutant decreased the percentage of 35S:AGL15 seedlings that produced somatic embryos from the shoot apex in liquid culture.
(B) Expression of AtGA2ox6 altered frequency of somatic embryo production from the shoot apex. Wild-type seedlings were compared with the ga2ox6-1 mutant and to three independent transgenic lines that constitutively expressed a cDNA version of AtGA2ox6.
(C) Biologically active GAs influenced production of somatic embryos from the shoot apex of wild-type seedlings and seedlings that constitutively expressed AGL15. GA3 or paclobutrazol was added to the culture media.
The percentage of seeds that produced SAM embryos was scored after 21 d. Means and standard error of the mean are shown. The numbers next to the bars in (A) and (B) indicate the number of experiments. All treatments performed in (C) were in triplicate. Different letters indicate significant difference as assessed by Tukey's mean separation test (SAS, 1999). In (B), Tukey's test was performed between Wassilewskija (WS), the wild type, and ga2ox6-1, and between WS, the wild type, and the 35S:GA2ox6 lines separately. Uppercase and lowercase letters in (C) indicate different comparisons.
To further test the effect of biologically active GA on production of SAM somatic embryos, wild-type and 35S:AGL15 seeds were allowed to complete germination in the culture system in the presence of either a biologically active GA, GA3, or in the presence of paclobutrazol. As shown in Figure 6C, a decrease in biologically active GA by addition of paclobutrazol caused an increase in the percentage of seedlings that produced SAM embryos in both wild-type and in 35S:AGL15 seedlings. Conversely, the presence of GA3 decreased production of somatic embryos in both cases.
DISCUSSION
ChIP was described previously as a means to identify nucleotide sequences bound by a transcription factor in vivo as a step toward identification of directly regulated genes (Wang et al., 2002). In this article, further evidence is presented to verify that DTA1 is a direct downstream target of AGL15 and to demonstrate that the protein product of this gene is a functional GA 2-oxidase. DTA1 corresponds to AtGA2ox6 as designated in Hedden et al. (2002) and has been renamed to indicate this identity.
AtGA2ox6 had a somewhat different expression pattern during development than do the previously described GA 2-oxidases (Thomas et al., 1999; Schomburg et al., 2003). Although not expressed exclusively during embryogenesis (Figures 2A and 2C), during this phase of the life cycle, expression of AtGA2ox6 parallels nuclear accumulation of AGL15, with the highest levels of expression and accumulation respectively occurring between globular to early torpedo stage of development (Figures 2B and 3; Heck et al., 1995; Rounsley et al., 1995; Perry et al., 1996). After torpedo stage, expression of AtGA2ox6 and accumulation of AGL15 both decline and are undetectable or almost undetectable in the nearly mature embryo (Figures 2B and 3; Perry et al., 1996).
What developmental role would a GA 2-oxidase have during early embryogenesis? Biologically active GAs have been suggested to have a role in embryogenesis between the torpedo and early cotyledon stages, when cells in the embryo axis elongate (Swain et al., 1997; Hays et al., 2002). Perhaps it is important to keep active GA concentrations low during earlier stages of embryo development, when cell division is more active than cell elongation, and AtGA2ox6 has a role in contributing to control of GA levels during this part of the life cycle. Although viable mature seed was obtained from the ga2ox6-1 mutant, this mutation in the first intron resulted in a decrease in expression rather than a complete loss of function. In addition, other GA 2-oxidases are expressed during silique development (Thomas et al., 1999; Ogawa et al., 2003) that may contribute to functional redundancy. It is also possible that there are more subtle changes early in embryogenesis that are compensated during the later stages of seed development as has been observed in pea (Pisum sativum; Swain et al., 1997), although no obvious differences were noted in morphogenesis in staged seeds (S. Perry, unpublished data). However, the knockdown mutation had a clear effect on seed germination, with the ga2ox6-1 mutant showing a decrease in seed dormancy compared with the wild type (Figure 5A). Furthermore, ga2ox6-1 seeds were more resistant to the effects of a GA biosynthesis inhibitor, paclobutrazol, than were wild-type seeds (Figure 5C). Although mature embryos showed little expression of AtGA2ox6 (Figure 3F), imbibed seeds and establishing seedlings accumulated mRNA from this gene (Figures 2C and 3G to 3L). The germination phenotype may result from the GA status established during seed development and/or GA production and metabolism during germination.
The knockdown mutant partially alleviated the increased dormancy observed in freshly harvested 35S:AGL15 seeds (Figure 5B) and repressed the ability of 35S:AGL15 seedlings to produce somatic embryos from the SAM in liquid culture media containing 2,4-D (Figure 6A). In a nontransgenic background, the knockdown mutant also reduced production of SAM embryos relative to wild-type seedlings (Figure 6B). Conversely, expression of the cDNA corresponding to AtGA2ox6 via the 35S promoter led to enhanced production of SAM embryos in culture (Figure 6B). Furthermore, the production of SAM embryos could be manipulated in wild-type and 35S:AGL15 seedlings by addition of the biologically active GA3 or by the GA biosynthesis inhibitor, paclobutrazol (Figure 6C), indicating that production of SAM embryos is negatively correlated with biologically active GA.
Production of SAM embryos in liquid culture has been correlated with meristem size, with the larger SAMs of amp1, clv1, and clv3 proposed to provide a larger pool of undifferentiated cells capable of following an embryonic default pathway when exposed to 2,4-D (Mordhorst et al., 1998), although mutants lacking a functional SAM are still able to produce somatic embryos from the cotyledonary axils (Mordhorst et al., 2002). Seedlings constitutively expressing AGL15 in wild-type and amp1 mutant backgrounds did not initially have a larger meristem than the corresponding background lacking the transgene (Harding et al., 2003). However, shortly after completion of germination, increased proliferation at the meristem in populations of seedlings showing enhanced SAM embryo production was observed (Harding et al., 2003). Because biologically active GAs have been reported to interfere with SAM function (Hay et al., 2002), one possible explanation for previous and current observations is that reduction of active GAs promotes meristem activity, and other factors allow somatic embryo development from this activated meristem.
Another possible explanation involves transitions between developmental phases. Inhibition of GA biosynthesis enhances the ability of the pickle (pkl) mutant to produce embryos from roots of seedlings, but addition of biologically active GA represses this phenotype (Ogas et al., 1997, 1999). PKL is a CHD3 chromatin-remodeling factor that has been proposed, along with a GA-modulated factor, to repress embryo identity in seedlings (Ogas et al., 1999; Rider et al., 2003). Biologically active GAs are involved in other phase transitions, such as juvenile to adult vegetative development as well as transition to reproductive development (Scott et al., 1999; Sakamoto et al., 2001, and references therein). Therefore, a decrease in biologically active GAs in 35S:AGL15, 35S:AtGA2ox6, or wild-type seedlings treated with paclobutrazol may prevent the transition from embryonic to postembryonic development, enhancing production of somatic embryos from the SAM in culture.
GAs have been reported to have effects on somatic embryo development in contexts other than pkl and outside of the meristem (for example, Hutchinson et al., 1997; Ondrej et al., 2002; Chen and Chang, 2003; Tokuji and Kuriyama, 2003). In carrot (Daucus carota) hypocotyl explants, GA inhibits early stages of somatic embryogenesis, whereas uniconazole, an inhibitor of GA biosynthesis, promotes secondary embryogenesis from primary embryos (Tokuji and Kuriyama, 2003). Interestingly, if GA is added at the time of 2,4-D treatment, instead of after, a slight stimulation of somatic embryo production is observed, suggesting a role for GA during the earliest stages of development. This could explain observations that expression of a genomic version of AtGA2ox6 via the 35S promoter, which accumulates AtGA2ox6 mRNA to higher amounts than the cDNA version of the transgene, nearly completely blocks production of SAM embryos (H. Wang, unpublished data). Also consistent is the observation that the few seeds capable of completing germination in the presence of high levels of paclobutrazol (1 and 10 μM) do not produce any SAM embryos (H. Wang, unpublished data).
Although results presented in this report suggest that part of the developmental consequences of ectopic expression of AGL15 (Fernandez et al., 2000) may be as a result of increased expression of AtGA2ox6 and, therefore, decreased concentrations of biologically active GAs, not all aspects of 35S:AGL15 phenotype were partially or completely rescued by the knock-down mutant. For example, the knockdown mutant does not reduce production of SAM embryos in the 35S:AGL15 background to the level found in the wild-type background. It is important to note that AtGA2ox6 is one of at least five family members that have overlapping patterns of expression. Potential binding sites for MADS domain proteins are present in the 5′ regulatory regions of other family members. If AGL15 is able to upregulate more than one family member, a mutation in ga2ox6 would not be sufficient to completely rescue the phenotypic aspect. Additionally, AGL15 most likely regulates other genes that encode proteins that contribute to particular aspects of observed phenotype. The fact that ectopic expression of AtGA2ox6 was not sufficient to phenocopy all aspects of the 35S:AGL15 phenotype also suggests that other regulated gene products must be involved. Regulatory networks, in general, are likely to be very complicated, with different cohorts of transcriptional regulators orchestrating control of a given gene's expression in different temporal and spatial contexts. This is the likely explanation why AtGA2ox6 was expressed to high abundance in developmental contexts outside of the accumulation pattern of AGL15.
However, AtGA2ox6 has met all of the criteria to demonstrate direct regulation by AGL15 (reviewed in Andrew and Scott, 1992; Carey and Smale, 2000). A direct interaction between AGL15 and regulatory elements of AtGA2ox6 was documented in vitro and in vivo, both in Arabidopsis and Brassica (Figure 1). Binding results in a regulatory consequence: Both increased and decreased amounts of AGL15 have been shown to affect expression of AtGA2ox6 (Figures 1D and 1F; Wang et al., 2002). The increased accumulation of AtGA2ox6 mRNA in response to AGL15 was not an indirect effect resulting from feed-forward control (reviewed in Hedden and Phillips, 2000; Olszewski et al., 2002) because endogenous biologically active GA concentrations were lower in 35S:AGL15 plants than in the wild type (Figure 4G). Additionally, many aspects of ectopic expression of 35S:AGL15 can be explained by increased expression of AtGA2ox6, and, in some cases, a knockdown mutation of AtGA2ox6 partially alleviated the phenotype conferred by the 35S:AGL15 transgene. Finally, as shown in Wang et al. (2002) and Figure 1D, regulatory sequences of AtGA2ox6 conferred regulation in response to AGL15, and a specific cis-acting element responsible for this regulation was identified. Thus, the evidence is very strong that AGL15 directly regulates expression of a gene encoding a GA 2-oxidase. GAs have profound effects on plant growth and development and AtGA2ox6's expression pattern, and the phenotypes of altered expression indicate an important role in embryogenesis, seed development, and germination.
METHODS
Plant Material
Arabidopsis thaliana ecotype Wassilewskija seeds were surface-sterilized and sown on MS germination medium (Invitrogen, Carlsbad, CA) with 50 μg/mL of kanamycin for transgenic seed. After chilling at 4°C for 2 d, seeds were transferred to 22°C under a 23-h-light/1-h-dark cycle. Seedlings were transplanted 7 to 10 d later to ProMix BX (Premier Brands, Rivière-du-Loup, Canada). Plants were grown under a 16-h-light (∼110 μmol m−2 s−1)/8-h-dark regime.
Enrichment Test for in Vivo Binding by AGL15
Enrichment tests for in vivo binding of AGL15 to a DNA fragment containing the CArG motif from the AtGA2ox6 regulatory region were performed as described in Wang et al. (2002). Primers specific for AtGA2ox6 (5′-TCCATACGGTTGACAAATG-3′ and 5′-CCTAGTTTGAAAATGACTAGACTG-3′), as well as primers that amplify a DNA fragment from the coding region of UBQ6 (5′-GGTGCTAAGAAGAGGAAGAAT-3′ and 5′-CTCCTTCTTTCTGGTAAACGT-3′), were used in multiplex PCR on total (input) DNA and on DNA fragments selected by ChIP using AGL15-specific antiserum. ChIP experiments were performed as in Wang et al. (2002) and were followed by a second immunoprecipitation of AGL15-DNA fragments as described in Weinmann et al. (2001). Briefly, protein A-Sepharose beads (Sigma, St. Louis, MO) were washed after the first immunoprecipitation as described in Wang et al. (2002), and elution was performed in 50 mM NaHCO3 and 1% SDS by agitating on a Vortex-Genie 2, vortex setting 3 (Fisher Scientific, Pittsburgh, PA), for 30 min at room temperature. The beads were removed by centrifugation and 5 volumes of immunoprecipitation buffer (Wang et al., 2002) added to the eluant along with another aliquot of anti-AGL15 serum. After incubation overnight at 4°C with gentle mixing on a rotating wheel, the sample was centrifuged in a microcentrifuge at top speed for 2 min at 4°C, and the supernatant moved to a new tube with protein A-Sepharose. The remainder of the second immunoprecipitation was identical to the description in Wang et al. (2002). Quantitation was performed using ImageQuant TL (Amersham Biosciences, Piscataway, NJ).
ChIP was also performed using Brassica napus cv Tower seeds containing heart to early bent-cotyledon stage embryos. Oligonucleotide primers were designed to the 5′ region of a putative Brassica ortholog of AtGA2ox6 (5′-GTGGCAATAATTTGGTTTTTCAA-3′ and 5′-TGTTGAAGAAGGTAAACCCATGT-3′) and to a Brassica β-tubulin (5′-CGAGAGGATCACAGCAATACAG-3′ and 5′-GGATCCATTCCACAAAGTAGGA-3′) for use in enrichment tests.
Electrophoretic Mobility Shift Assay
A DNA fragment corresponding to the AtGA2ox6 regulatory region (from −389 to −343) and containing a putative but noncanonical CArG motif (−372 to −361) was generated using two partially complementary oligonucleotides and extension by PCR. The two oligonucleotides were as follows: oligonucleotide 1, 5′-CTTGTACAACTTAAGATCCAATTTAATGGAGATA-3′, and oligonucleotide 2, 5′-TGACATGCATGTCTATCTCCATTAAATTGGATCT-3′. A mutated version of the DNA fragment was created using oligonucleotide 3 (same as oligonucleotide 1 except that the CC was changed to TT) and oligonucleotide 4 (same as oligonucleotide 2 except that the GG was changed to AA). The wild-type and mutant DNA fragments were labeled with 32P-dATP and EMSA performed as described previously (Wang et al., 2002).
Generation of GUS Reporter Lines and Activity Assays
AtGA2ox6:GUS transgenic plants, in which the reporter gene GUS was under the control ∼2.3 kb of the 5′ regulatory region of AtGA2ox6, were as described in Wang et al. (2002). To generate the mAtGA2ox6:GUS reporter plant lines, all approaches were the same except that the CC was changed to TT within a CArG motif located at −372 to −361 using oligonucleotides 3 and 4 described above and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Transgenic lines containing a single insert were identified and were crossed to AGL15 constitutively expressing plants that contained a single hemizygous copy of 35S:AGL15. The F1 siblings with or without the 35S:AGL15 transgene can be easily separated visually at the seedling stage, using petiole and hypocotyl length, and cotyledon epinasty (Fernandez et al., 2000). GUS activities of individual seedlings in subsequent generations were measured by MUG assays using a Hoefer DynaQuant 200 fluorometer (Hoefer Scientific Instruments, San Francisco, CA) as described in Gallagher (1992). Histochemical staining for GUS activity was as in Fernandez et al. (2000), followed in some cases by clearing of the seeds in Hoyer's solution as described in Schwartz et al. (1994).
AtGA2ox6 Gain-of-Function and Loss-of-Function Plants
Confirmation of the coding region of AtGA2ox6 was described previously (Wang et al., 2002). Two different constructs were generated to drive constitutive expression of AtGA2ox6. One construct corresponded to the cDNA, whereas the other included all of the introns (i.e., genomic version). The cDNA was obtained by reverse transcription from developing siliques, followed by PCR amplification as described previously (Wang et al., 2002) and insertion into the SmaI site of pBluescript II KS+ (pBSII KS+) (Stratagene) to produce pBSII KS-AtGA2ox6. The insert was excised using XbaI (5′ end of the gene) and SalI and ligated to pBIMC that was digested with XbaI (35S promoter side) and XhoI. pBIMC is a derivative of pBI121 in which the GUS gene is replaced by a multiple cloning site (provided by Deane Falcone, University of Kentucky). The gene with introns was obtained by PCR amplification from Arabidopsis genomic DNA and ligation into pBIMC as above. All constructs were confirmed by sequencing. The plasmids were introduced into Agrobacterium tumefaciens strain GV3101, and Arabidopsis plants were transformed as described previously (Wang et al., 2002).
To obtain loss-of-function plants, both the Alpha and Basta T-DNA insertion populations (Arabidopsis Knockout Facility, University of Wisconsin–Madison) were screened for insertion alleles of AtGA2ox6 according to the instructions on the facility's Web site (http://www.biotech.wisc.edu/Arabidopsis). A primer corresponding to the T-DNA left border JL-202 (Krysan et al., 1999) and two gene-specific primers located at the 5′ and 3′ regions of the AtGA2ox6 gene were used to identify potential mutant alleles. The AtGA2ox6 specific primers to screen the T-DNA population were 5′-GCTTTATTCATCTTGGCAATAATTTTGGT-3′ and 5′-AACAATCCAACAATCATATTTTCTTTTTCAA-3′. The primers used for RT-PCR are identical to those used to obtain the cDNA (Wang et al., 2002).
The transgene for complementation tests consisted of ∼2.3 kb 5′ of the start codon, the entire coding region including introns, and ∼0.8 kb 3′ of the stop codon. The fragment was generated by PCR amplification from wild-type Arabidopsis Wassilewskija genomic DNA, cloned into pCAMBIA1300, and introduced into A. tumefaciens GV3101 and ga2ox6-1 homozygous plants. Transformants were selected on media containing both kanamycin and hygromycin.
In Vitro GA 2-Oxidase Activity Assay
The coding region of AtGA2ox6 was excised from the pBSII KS-AtGA2ox6 construct described above using XhoI and HindIII, and inserted into the pMal-c2x expression vector (New England Biolabs, Beverly, MA). Escherichia coli strain BL21-RIL (Stratagene) was transformed with the pMal-c2x-AtGA2ox6 fusion construct and expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside to a culture with OD600 of 0.5. After 3 h at room temperature, the cells were harvested and processed using CelLytic B-II bacterial cell lysis/extraction reagent (Sigma) following the manufacturer's instructions. The presence of the maltose binding protein-AtGA2ox6 fusion protein in the soluble portion of the cell lysate was confirmed by SDS-PAGE.
The GA 2-oxidase activity assay was performed by incubating 80 μL of soluble fraction of the E. coli cell lysate with 200 ng of [17,17-2H2]GA1, GA4, or GA20 (purchased from Lew N. Mander, Australian National University, Canberra, Australia) in a final reaction volume of 200 μL containing 50 mM Tris-HCl, pH 7.5, 0.5 mM FeSO4, 5 mM 2-ketoglutarate, and 5 mM ascorbate. The pH of the reaction was reconfirmed after the GA was added. After incubating at 30°C for 2 h, the reaction was stopped by addition of 1.8 mL of 0.1 N HCl to lower the pH to between 2 and 3. The products were extracted twice with 2 mL of ethyl acetate (saturated with 1% acetic acid), dried under vacuum, resuspended in 100 μL of methanol, and derivatized to methylester-trimethylsilyl ether (Me-TMSi). Derivatization was performed first with 10 to 20 μL of methanol and 50 to 100 μL of fresh CH2N2 and incubated at room temperature for 30 min. After solvents were eliminated under N2, the sample was freeze-dried, followed by treatment with 50 μL of a mixture of dry pyridine and N,O-bistrimethylsilyl trifluoroacetamide (1:2) plus 1% trimethylchlorosilane (Pierce Chemical, Rockford, IL) and heated for 30 min at 80°C. One microliter was injected in splitless mode onto a DB-5ms column (30 m, 0.25 mm ID × 0.25 μm film thickness; Agilent/J&W Scientific, Folsom, CA), fitted in a gas chromatograph (Trace GC; ThermoFinnigan, San Jose, CA) with a capillary direct interface to a mass spectrometer PolarisQ ion trap system (ThermoFinnigan). The GC temperature program was 50°C (1-min hold) to 150°C at 20°C min−1, then to 280°C at 6°C min−1, using helium as a carrier gas at 1 mL min−1. For product identification, full scan spectra analysis of [17,17-2H2]GAs were monitored at the retention times of authentic [17,17-2H2]GA standards.
Quantification of Endogenous GA Concentrations
Rosette leaves were harvested from plants 5 weeks after the completion of germination, flash frozen in liquid N2, and freeze-dried. Approximately 500 mg dry weight was homogenized in liquid N2, extracted with 80% methanol and 1% acetic acid at 4°C overnight, and 10 ng of each of the following GAs, [17, 17-2H2]GA1, GA4, GA8, GA9, GA19, GA20, and GA34 (L. Mander, Australian National University, Adelaide, Australia), were added as internal standards and allowed to equilibrate for 30 min. GA extraction and purification was as described in Volmaro et al. (1998), except the second HPLC using a Nucleosil (N[CH3]2) column was omitted, and no bioassay was performed to locate GA presence. Instead, fractions from 10 to 30 min were collected from the C18 reverse phase column (μBondapak, 300 × 3.9 mm; Waters Associates, Milford, MA) at a flow rate of 2 mL min−1; from 0 to 10 min with 10% methanol in 1% acetic acid, from 10 to 40 min with 10% to 73% methanol in 1% acetic acid, from 40 to 50 min with 73% methanol in 1% acetic acid, and from 50 to 60 min with 100% methanol (conditions based on experience of R. Bottini with the same column). After solvent evaporation under low pressure, samples were converted to the Me-TMSi derivatives, and 1 μL was injected in splitless mode in a GC-MS as described above. In selected ion monitoring mode, the parent ion and at least four other characteristic ions (Hedden, 1987; Talon et al., 1990) for both the deutero-isotope and the endogenous compound were monitored. The identity of endogenous GAs was established by coincidence of the relative intensities of the five characteristic ions and the retention time for the endogenous compound with those of the deutero-GAs used as internal standards. By comparison of the peak area for the parent ion of the endogenous GA versus the parent ion of the corresponding deutero GA standard, the amount of free acid GA was calculated from triplicate samples using Xcalibur software (ThermoFinnigan).
RNA Gel Blot Analysis
RNAs used for expression pattern analysis were extracted from various organs of 5-week-old plants as described previously (Wang et al., 2002). To stage siliques, flowers were tagged on the day that they opened and siliques collected at the appropriate time afterward. 35S:AtGA2ox6 plants were assessed for the amount of expression of the transgene by extracting total RNA from 12-d-old seedlings grown on germination media. Total RNA was subjected to electrophoresis in a 1% agarose gel in the presence of 0.5 M formaldehyde and then transferred to Zeta-Probe GT blotting membrane (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. For slot blots, 1 μg of mRNA (obtained using the PolyATract mRNA isolation system; Promega, Madison, WI) was applied to Zeta-Probe GT blotting membrane using a vacuum manifold (Bio-Rad). Filters were probed with 32P-labeled DNA probe specific for AtGA2ox6 or for β-2 Tubulin (Snustad et al., 1992) at 68°C in 0.5 M NaHPO4, pH 7.2, and 7% SDS for 16 h, followed with two 30-min washes at 68°C in 40 mM KHPO4, pH 7.2, and 5% SDS and one 30-min wash in 40 mM NaHPO4, pH 7.2, and 1% SDS at 68°C. Washed filters were exposed to a storage phosphor screen (Molecular Dynamics, Sunnyvale, CA) and signal detected on a PhosphorImager (Model 445SI; Molecular Dynamics).
RT-PCR Analysis
RNA was isolated from imbibed seeds or establishing seedlings using the hot borate method (Wilkins and Smart, 1996) or TRIzol reagent (Life Technologies, Rockville, MD) respectively. To test for feed-forward regulation, 3-week-old plants were sprayed with 100 μM GA3 or GA4, and rosette leaves collected 16 h afterward for total RNA isolation. Semiquantitative RT-PCR was performed to evaluate the AtGA2ox6 expression amounts as described previously (Wang et al., 2002). Quantitation was performed using ImageQuant TL.
Germination Assays
The seeds used for germination assays were harvested from mature plants of different genotypes grown in the same tray under the conditions described above. To ensure the plants matured at the same time, 35S:AtGA2ox6 and 35S:AGL15 seeds were germinated 1 to 2 weeks earlier depending on the particular line. All the germination experiments were performed in 4-cm Petri dishes on two layers of germination paper (Stults Scientific Eng., Springfield, IL) soaked with 4 mL of water, different concentrations of GA4+7, or paclobutrazol. After 6 d of incubation under continuous light at 25°C, the percentage of seeds completing germination was assessed. In some experiments, seeds were chilled at 4°C for 3 d after imbibition and before incubation at 25°C. Each line was tested in triplicate, with ∼100 seeds per plate.
Seedling Liquid Culture System
The liquid culture system was essentially as described by Mordhorst et al. (1998) with further details provided by Harding et al. (2003), except that GA3 or paclobutrazol was added to the media in some experiments.
Statistical Analysis
All comparisons of the effect of the mutations, transgenes, or treatments were performed using analysis of variance (the analysis of variance procedure of SAS, 1999) when the number of replications was equal. If replication number was not equal, the general linear model was used (SAS, 1999). In either instance, if the test indicated that there were significant differences among means, Tukey's mean separation test (SAS, 1999) was used to distinguish among them.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At1g02400, B7033271, BH741917, B7081538, and AF258790.
Acknowledgments
We thank Weining Tang, Lynnette Dirk, and Randy Dinkins for critical reading of the manuscript, as well as members of the University of Kentucky Seed Biology Group for helpful suggestions. We also acknowledge two anonymous reviewers for critical review of and suggestions on the manuscript. We thank Donna Fernandez and Melissa Lehti-Shiu (University of Wisconsin–Madison) for providing seeds for the constitutively expressing AGL15 plants and for the agl15 plants, Deane Falcone for the pBIMC vector, Qilong Xu and Cong Zhu (University of Kentucky, Lexington) for materials for RT-PCR experiments, and Abbott Biochemicals (Chicago, IL) for the kind gift of the GA4+7. In addition, we thank Jack Goodman at the University of Kentucky Mass Spectrometry Facility for obtaining mass spectrometry data, Neil Fannin for assistance with the HPLC, and Lowell Bush and Harold Burton (University of Kentucky) for use of lab facilities for a portion of this work. We also acknowledge Ruben Bottini (University of Cuyo, Argentina) for training in quantitation of GAs. This research was supported by the National Science Foundation (Grant IBN-9984274 to S.E.P.) and by the University of Kentucky. This article (03-06-077) is published with the approval of the Director of the Kentucky Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sharyn Perry (sperr2@uky.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021261.
References
- Andrew, D.J., and Scott, M.P. (1992). Downstream of the homeotic genes. New Biol. 4, 5–15. [PubMed] [Google Scholar]
- Arnone, M.I., and Davidson, E.H. (1997). The hardwiring of development: Organization and function of genomic regulatory systems. Development 124, 1851–1864. [DOI] [PubMed] [Google Scholar]
- Burgeff, C., Liljegren, S.J., Tapia-Lopez, R., Yanofsky, M.F., and Alvarez-Buylla, E.R. (2002). MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214, 365–372. [DOI] [PubMed] [Google Scholar]
- Callis, J., Raasch, J.A., and Vierstra, R.D. (1990). Ubiquitin extension proteins of Arabidopsis thaliana: Structure, localization, and expression of their promoters in transgenic tobacco. J. Biol. Chem. 265, 12486–12493. [PubMed] [Google Scholar]
- Carey, M., and Smale, S.T. (2000). Confirming the functional importance of a protein-DNA interaction. In Transcriptional Regulation in Eukaryotes: Concepts, Strategies and Techniques. (New York, NY: Cold Spring Harbor Laboratory Press), pp. 291–317. [DOI] [PubMed]
- Chen, J.T., and Chang, W.C. (2003). Effects of GA3, ancymidol, cycolcel and paclobutrazol on direct somatic embryogenesis of Oncidium in vitro. Plant Cell Tissue Organ Cult 72, 105–108. [Google Scholar]
- Fernandez, D.E., Heck, G.R., Perry, S.E., Patterson, S.E., Bleecker, A.B., and Fang, S.-C. (2000). The embryo MADS domain factor AGL15 acts postembryonically: Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett, J.W., and Wasserman, W.W. (2000). Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol. 11, 19–24. [DOI] [PubMed] [Google Scholar]
- Flanagan, C.A., and Ma, H. (1994). Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 26, 581–595. [DOI] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992). Chapter 3: Quantitation of GUS activity by fluorometry. In GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, S.R. Gallagher, ed (San Diego, CA: Academic Press), pp. 47–59.
- Graba, Y., Aragnol, D., and Pradel, J. (1997). Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays 19, 379–388. [DOI] [PubMed] [Google Scholar]
- Harding, E.W., Tang, W., Nichols, K.W., Fernandez, D.E., and Perry, S.E. (2003). Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hays, D.B., Yeung, E.C., and Pharis, R.P. (2002). The role of gibberellins in embryo axis development. J. Exp. Bot. 53, 1747–1751. [DOI] [PubMed] [Google Scholar]
- Heck, G.R., Perry, S.E., Nichols, K.W., and Fernandez, D.E. (1995). AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P. (1987). Gibberellins. In Principles and Practice of Plant Hormone Analysis, L. Rivier and A. Crozier, eds (London, UK: Academic Press), pp. 9–110.
- Hedden, P. (2002). Gibberellin metabolism and its regulation. J. Plant Growth Regul. 20, 317–318. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Hedden, P., Phillips, A.L., Rojas, M.C., Carrera, E., and Tudzynski, B. (2002). Gibberellin biosynthesis in plants and fungi: A case of convergent evolution? J. Plant Growth Regul. 20, 319–331. [DOI] [PubMed] [Google Scholar]
- Hutchinson, M.J., KrishnaRaj, S., and Saxena, P.K. (1997). Inhibitory effect of GA3 on the development of thidiazuron-induced somatic embryogenesis in geranium (Pelargonium x hortorum Bailey) hypocotyl cultures. Plant Cell Rep. 16, 435–438. [DOI] [PubMed] [Google Scholar]
- Krysan, P., Young, J., and Sussman, M. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst, A.P., Hartog, M.V., Tamer, M.K.E., Laux, T., and de Vries, S.C. (2002). Somatic embryogenesis from Arabidopsis shoot apical meristem mutants. Planta 214, 829–836. [DOI] [PubMed] [Google Scholar]
- Mordhorst, A.P., Voerman, K.J., Hartog, M.V., Meijer, E.A., van Went, J., Koornneef, M., and de Vries, S.C. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K.-i., Allis, C.D., and Grewal, S.I.S. (2001). Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.-C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrej, V., Navratilova, B., and Lebeda, A. (2002). Influence of GA3 on the zygotic embryogenesis of Cucumis species in vitro. Biologia (Bratisl.) 57, 523–525. [Google Scholar]
- Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Orlando, V., Strutt, H., and Paro, R. (1997). Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Parkin, I., Sharpe, A., Keith, D., and Lydiate, D. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38, 1122–1131. [DOI] [PubMed] [Google Scholar]
- Perry, S.E., Lehti, M.D., and Fernandez, D.E. (1999). The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 120, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., Nichols, K.W., and Fernandez, D.E. (1996). The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell 8, 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Rider, S.D., Henderson, J.T., Jerome, R.E., Edenberg, H.J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., Iwahori, S., and Matsuoka, M. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125, 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS. (1999). Statistical Analysis Systems, Version 8. (Cary, NC: SAS Institute).
- Schomburg, F.M., Bizzell, C.M., Lee, D.J., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, B.W., Yeung, E.C., and Meinke, D.W. (1994). Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120, 3235–3245. [DOI] [PubMed] [Google Scholar]
- Scott, D.B., Jin, W., Ledford, H.K., Jung, H.-S., and Honma, M.A. (1999). EAF1 regulates vegetative-phase change and flowering time in Arabidopsis. Plant Physiol. 120, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, M.F., and Rao, S. (2002). Of chips and ChIPs. Science 296, 666–669. [DOI] [PubMed] [Google Scholar]
- Snustad, D.P., Haas, N.A., Kopczak, S.D., and Silflow, C.D. (1992). The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. Plant Cell 4, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Reid, J.B., and Kamiya, Y. (1997). Gibberellins are required for embryo growth and seed development in pea. Plant J. 12, 1329–1338. [Google Scholar]
- Talon, M., Koornneef, M., and Zeevaart, J. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87, 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., and Perry, S.E. (2003). Binding site selection for the plant MADS-domain protein AGL15: An in vitro and in vivo study. J. Biol. Chem. 278, 28154–28159. [DOI] [PubMed] [Google Scholar]
- Theissen, G. (2001). Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96, 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuji, Y., and Kuriyama, K. (2003). Involvement of gibberellin and cytokinin in the formation of embryogenic cell clumps in carrot (Daucus carota). J. Plant Physiol. 160, 133–141. [DOI] [PubMed] [Google Scholar]
- Volmaro, C., Pontin, M., Luna, V., Baraldi, R., and Bottini, R. (1998). Blue light control of hypocotyl elongation in etiolated seedlings of Lactuca sativa (L.) cv. Grand Rapids related to exogenous growth regulators and endogenous IAA, GA3, and abscisic acid. Plant Growth Regul. 26, 165–173. [Google Scholar]
- Wang, H., Tang, W., Zhu, C., and Perry, S.E. (2002). A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS-domain protein that preferentially accumulates in embryos. Plant J. 32, 831–843. [DOI] [PubMed] [Google Scholar]
- Weinmann, A.S., Bartley, S.M., Zhang, T., Zhang, M.Q., and Farnham, P.J. (2001). Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21, 6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, T.A., and Smart, L.B. (1996). Isolation of RNA from plant tissue. In A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis, P.A. Krieg, ed (New York: Wiley-Liss), pp. 21–41.
- Wyrick, J.J., and Young, R.A. (2002). Deciphering gene expression regulatory networks. Curr. Opin. Genet. Dev. 12, 130–136. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2002). Gibberellins and light-stimulated seed germination. J. Plant Growth Regul. 20, 369–376. [DOI] [PubMed] [Google Scholar]