Abstract
Vein patterns in leaves and cotyledons form in a spatially regulated manner through the progressive recruitment of ground cells into vascular cell fate. To gain insight into venation patterning mechanisms, we have characterized the cotyledon vascular pattern2 (cvp2) mutants, which exhibit an increase in free vein endings and a resulting open vein network. We cloned CVP2 by a map-based cloning strategy and found that it encodes an inositol polyphosphate 5′ phosphatase (5PTase). 5PTases regulate inositol (1,4,5) triphosphate (IP3) signal transduction by hydrolyzing IP3 and thus terminate IP3 signaling. CVP2 gene expression is initially broad and then gradually restricted to incipient vascular cells in several developing organs. Consistent with the inferred enzymatic activity of CVP2, IP3 levels are elevated in cvp2 mutants. In addition, cvp2 mutants exhibit hypersensitivity to the plant hormone abscisic acid. We propose that elevated IP3 levels in cvp2 mutants reduce ground cell recruitment into vascular cell fate, resulting in premature vein termination and, thus, in an open reticulum.
INTRODUCTION
Modern dicots have an interconnected vascular network that transports essential nutrients in a source-to-sink manner. This differs from the primitive vein pattern in which conducting tissue formed simple dendritic (tree-like) formations (Roth-Nebelsick et al., 2001). The closed reticulum pattern that characterizes the venation of dicot leaves is thought to have been derived from a primitive open conduit through positive selection because closed conduits have more efficient transport capacity and provide improved structure and stability to the leaf (Roth-Nebelsick et al., 2001).
During leaf development, veins form within a field of subepidermal ground cells in the form of procambium in an event that is developmentally coordinated with lamina formation (Dengler, 2001). Vein orders are formed progressively and join in a hierarchical manner (Telfer and Poethig, 1994; Nelson and Dengler, 1997; Kinsman and Pyke, 1998; Candela et al., 1999). The prominent midvein forms first, in parallel with the emerging leaf primordium and in continuity with vasculature of the central axis. Subsequently, secondary or lateral veins diverge from the midvein to extend toward leaf margins as the lamina expands laterally. As expansion continues, the minor veins, composed of tertiary, quaternary, and intramarginal veins, intercalate between and join the lower vein orders. Quaternary veins generally appear as freely terminating veinlets, defined as strands that are joined at only one end, presumably because ground cells have lost competence or signals for vascular cell fate. Before the differentiation of veins, their incipient paths can be recognized first as a path of ground cells with responsiveness of auxin-sensitive reporters (Mattsson et al., 2003) and then anatomically as files of elongated procambial cells that are visually distinct from ground cells. Several markers can identify procambial files, including Athb8, Athb20, and VH1 (Baima et al., 1995; Clay and Nelson, 2002; Kang and Dengler, 2002; Mattsson et al., 2003). The evident temporal and spatial coordination of procambial and mesophyll cell differentiation implicates continuous cell-to-cell communication along the path of vein recruitment and differentiation.
The directional transport of auxin appears to play a role in establishing the longitudinal polarity and strand continuity associated with vein formation (Sachs, 1991). The effects of exogenously applied auxin transport inhibitors and the pattern of activation of DR5 auxin-responsive reporter genes suggest that auxin acts as a positional cue for vein forming events (Mattsson et al., 1999, 2003; Sieburth, 1999). In addition, biochemical support for the polar flow of auxin is provided by the identification of the putative auxin efflux carriers, the asymmetrically localized PIN proteins (Galweiler et al., 1998). Venation pattern defects, such as loss of vascular cell polarity and of uniform cell files, are observed in mutants with defects in several auxin related genes. These mutants include those that were identified on the basis of defective embryo patterning, such as monopteros, bodenlos, and gnom/emb30 (Berleth and Jurgens, 1993; Shevell et al., 1994; Busch et al., 1996; Przemeck et al., 1996; Hardtke and Berleth, 1998; Hamann et al., 1999, 2002), or of an alteration in auxin response such as auxin resistant6 (Hobbie et al., 2000). Although the gene products of the vascular patterning mutants scarface (Deyholos et al., 2000) and lopped1/tornado1 (Carland and McHale, 1996; Cnops et al., 2000) have not been identified yet, physiological analysis implicates aberrant auxin response or transport.
Other mutant studies imply roles for cytokinin (Mahonen et al., 2000; Inoue et al., 2001), small peptides (Casson et al., 2002), brassinosteroids (Szekeres et al., 1996; Choe et al., 1999), and sterols (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Carland et al., 2002; Souter et al., 2002) in vein patterns. There are phenotypic abnormalities shared between sterol and auxin mutants that include polarity defects in vascular cells and/or embryogenesis. It has been proposed that sterol defects influence the membrane localization of auxin carriers (Carland et al., 2002; Grebe et al., 2003), although the relationship between sterols and auxin-facilitated polarizing events has not been firmly established. Recently, sterol methyltransferase1orc mutants were shown to have mislocalized PIN protein, providing a link between sterols and the auxin polarity maintaining machinery (Willemsen et al., 2003). Furthermore, hydra mutants exhibit enhanced auxin response (Souter et al., 2002).
The cotyledon vascular pattern2 (cvp2) and forked (fkd1) mutants appear to end vascular strand propagation prematurely, resulting in an unclosed reticulum with open secondary and higher order veins (Carland et al., 1999; Steynen and Schultz, 2003). Both cvp2 and fkd1 mutants were isolated in genetic screens for vascular patterning mutants without obvious defects in plant growth or morphology. Neither mutant exhibits defects in vascular cell morphology or polarity, as do many of the auxin and sterol mutants described above. Although both mutants show a loss of vein anastomoses with many vein orders, they do show distinct differences. Consistent with the wild-type appearance of cvp2, no perturbations in auxin content, response, or transport were detected, and cvp2 has an increase in cotyledon lateral veins. By contrast, fkd1 exhibits reduced auxin response, an unaltered number of cotyledon lateral veins, and elongated first rosette leaves. Leaf morphological alterations are subtle in fkd1 and absent in cvp2, supporting the hypothesis that the major veins influence leaf shape more than the minor veins do (Dengler and Kang, 2001). The characterization of cvp2 mutants led us to postulate a role for CVP2 in the perception or transduction of a signal critical for the propagation of procambial strand formation.
Here, we report that the vascular patterning gene CVP2 encodes an inositol polyphosphate 5′ phosphatase (5PTase). 5PTases have been shown in animal systems (Majerus et al., 1999) and in plants (Berdy et al., 2001; Sanchez and Chua, 2001; Perera et al., 2002) to regulate inositol signal propagation by their ability to cleave a 5′ phosphate from various inositol stereoisomers. In this manner, they are negative regulators of inositol (1,4,5) triphosphate (IP3) signal transduction and of other second messengers, such as Ca2+. In agreement with the proposed role of 5PTases, we show that there are increased levels of IP3 in cvp2 mutants and that mutants are hypersensitive to abscisic acid (ABA). CVP2 is expressed in developing vascular cells in several plant organs. CVP2 is the only member of the 15 5PTases in Arabidopsis thaliana that has been identified by mutation. Its phenotype suggests a role for IP3 signaling in an early stage of vascular patterning. We propose that the open vascular network in cvp2 mutants is attributable to unregulated CVP2-mediated IP3 production that elicits upregulation of otherwise tightly controlled response genes, presumably by the inappropriate release of Ca2+. Misregulation of these genes prevents the specification of ground cells into vascular cell fate during vascular strand propagation.
RESULTS
cvp2 Mutants Have an Open Vein Network
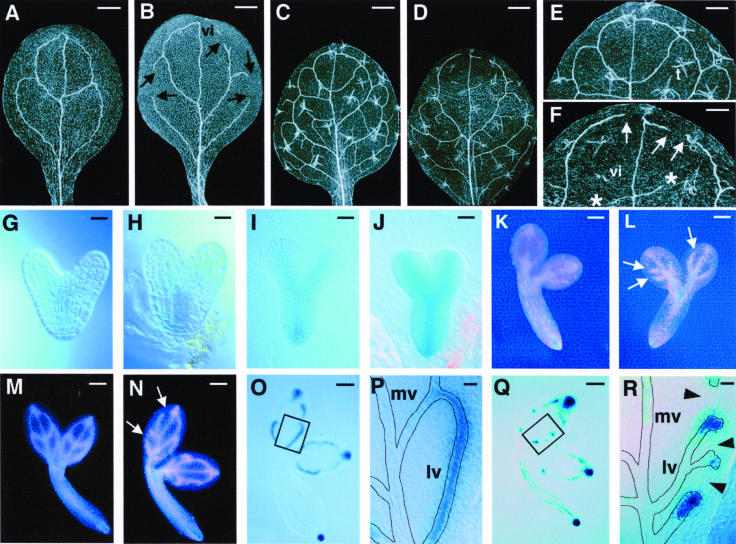
A detailed description of the cvp2 mutant phenotype was previously presented (Carland et al., 1999). Wild-type cotyledons have a closed reticulum generally consisting of three to four closed loops (Figure 1A). By contrast, cvp2 mutant cotyledons exhibit an open reticulum, characterized by an increase in free vein endings (Figure 1B). Short stretches of veins, known as vascular islands because they appear to form in isolation from other veins, appear frequently in cvp2 cotyledons. Unlike wild-type rosette leaves in which secondary veins join apical veins, in cvp2 leaves, secondary veins usually do not meet previously formed veins distally (Figures 1C to 1F). This is particularly apparent with apical secondary veins. Wild-type tertiary veins bridge secondary veins to form a connected conduit. The corresponding tertiary veins in cvp2 mutant leaves do not span secondary veins and appear to terminate prematurely. In contrast with wild-type quaternary veins, which form freely ending veinlets, the corresponding veins in cvp2 mutant leaves appear as vascular islands and fail to form vein junctions. A comparison of wild-type and cvp2 lateral vein development revealed that similar to wild-type veins, cvp2 mutant veins show progressive decrease in girth but terminate prematurely (Carland et al., 1999), suggesting that CVP2 promotes vascular cell proliferation and prevents premature vein termination.
Figure 1.
CVP2 Acts Early in Procambial Patterning during Embryogenesis.
(A) to (F) Cleared specimens viewed with dark-field optics. Wild-type cotyledon (A), cvp2 cotyledon (B), wild-type leaf (C), and cvp2 leaf (D) are shown. Higher magnification of (C) and (D) are shown in (E) and (F), respectively. With dark-field optics, trichomes (t) appear translucent with three spikes.
(G) to (N) Athb8:GUS embryo expression. Wild-type heart (G), cvp2 heart (H), wild-type torpedo (I), cvp2 torpedo (J), wild-type walking stick (K), cvp2 walking stick (L), wild-type bent cotyledon (M), and cvp2 bent cotyledon (N) are shown. Dark-field images are more sensitive than Nomarski optical images and show weak GUS expression as pink and strong GUS expression as blue.
(O) to (R) DR5:GUS expression of bent cotyledon stage embryo. Wild-type embryo (O), wild-type proximal lateral vein at a higher magnification of the boxed region in (O) to show uniform DR5:GUS expression (P), cvp2 embryo (Q), and cvp2 proximal lateral vein at a higher magnification of the boxed region in (Q) to show short stretches of DR5:GUS expression in cvp2 mutants (R). Arrowheads designate absence of DR5:GUS. In wild-type embryos, 369 out of 394 lateral veins showed continuous DR5:GUS expression. In cvp2 embryos, 33 out of 330 lateral veins showed continuous DR5:GUS expression. Roots of embryos in (O) and (Q) were distorted while separating cotyledons. Vein patterns are outlined in (P) and (R).
Arrows indicate free vein endings of secondary veins; asterisks indicate free vein endings of tertiary veins. Dark-field optics were used for (A) to (F) and (K) to (N); Nomarski optics were used for (G) to (J) and (O) to (R). vi, vascular island; mv, midvein; lv, lateral vein. Bars = 500 μM in (A) to (D), 250 μM in (E) and (F), 10 μM in (G), (H), (P), and (R), 25 μM in (I) and (J), 50 μM in (K) and (L), and 80 μM in (M), (N), (O), and (Q).
Procambial Cell-Specific Expression of Athb8 Is Dependent on CVP2 Function
To determine the stage at which CVP2 acts in vein formation, we used the procambial cell-specific marker Athb8 (Baima et al., 1995; Kang and Dengler, 2002) as a reporter construct to monitor procambial cell patterning during embryogenesis because of the simplicity of the cotyledon venation pattern. A line with a single copy Athb8:β-glucuronidase (GUS) transgene was crossed to cvp2 to generate a homozygous cvp2/Athb8:GUS line. Athb8:GUS staining was centrally localized, corresponding to future sites of vascular development, in both wild-type and cvp2 heart and torpedo embryos, and was consistent with the unaffected midvein and root vasculature observed in cvp2 seedlings (Figures 1G to 1J). In both wild-type and cvp2 walking-stick stage embryos, Athb8:GUS was visible in the apical loop (Figures 1K and 1L). However, in the wild type, this loop was closed, and in cvp2, the open vein network was just beginning to appear. At the bent cotyledon stage of wild-type embryos, staining was restricted to procambial cells forming the four continuous loops observed in mature embryos and cotyledons (Figure 1M), whereas in cvp2 embryos, staining revealed an open network, anticipating the vascular patterning defects observed in cvp2 mutant cotyledons (Figure 1N). This confirmed our earlier observations of procambial cells in unstained embryos that CVP2 was acting early in procambial strand formation during embryogenesis.
Procambial Cell Patterning in cvp2 Mutant Embryos Is Coincident with DR5:GUS Expression
We used the DR5:GUS synthetic reporter construct (Ulmasov et al., 1997) to map auxin-responsive cells in cvp2 mutant embryos. This auxin-responsive reporter has been used to infer local auxin levels (Sabatini et al., 1999) and can serve as an early indicator of preprocambial cell patterning during vein formation in developing leaves (Mattsson et al., 2003). DR5:GUS appeared in a progressively formed pattern that foreshadowed all vein orders as they appear in a developing leaf. To determine if the establishment of preprocambial formation was affected in cvp2 mutant embryos, we generated a cvp2/DR5:GUS line and examined DR5:GUS histochemical staining in embryos. The reporter gene was expressed in incipient vascular cells, before detectable vascular cell morphological changes, and was not expressed in fully elongated procambial cells, consistent with previous reports (Mattsson et al., 2003). As shown in Figures 1O and 1P, DR5:GUS marked secondary veins as a continuous file of cells in wild-type bent cotyledon stage embryos but was no longer expressed in the elongated procambial cells of the midvein. Distal secondary veins showed diminishing DR5:GUS expression as these cells elongated. Similar to the wild type, there was either no or very weak GUS histochemical staining in the elongated procambium of cvp2 midvein and secondary veins (Figures 1Q and 1R). By contrast, cvp2/DR5:GUS mutant embryos at the same stage exhibited a discontinuous pattern characterized by short stretches of DR5:GUS-expressing cells that appeared to be vein termini. There was no evidence of DR5:GUS cells between these preprocambial foci, suggesting that CVP2 acts before the acquisition of preprocambial identity (Figure 1R, arrowheads).
CVP2 Encodes a 5PTase
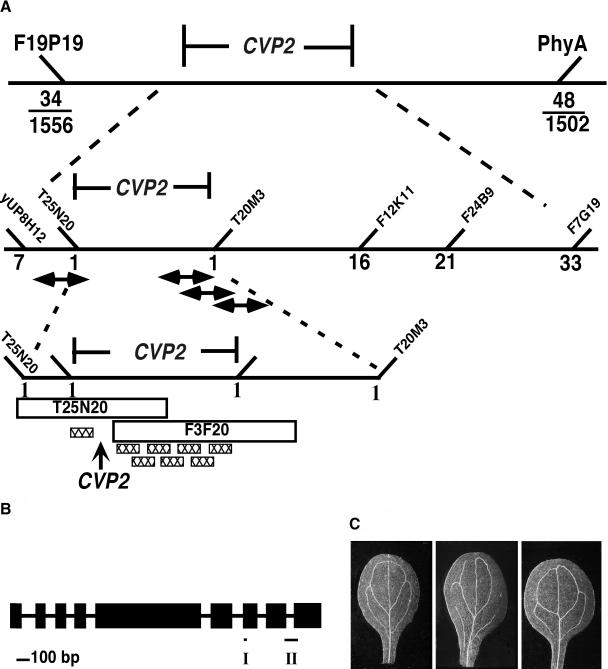
CVP2 was mapped to the top of chromosome one between the polymorphic markers F19P19 and PhyA (Figure 2A). Breakpoint analysis with flanking polymorphic markers, which we identified from known markers on the restriction fragment length polymorphism map or from the limited genomic sequence that was available at the time, localized CVP2 to ∼270 kb. Using probes derived from candidate BAC clones, we identified transformation-competent artificial clones (TAC) (Liu et al., 1999) that were within the CVP2 region but did not span the CVP2 region in its entirety. Positive TAC clones were introduced into the cvp2 mutant, and because transgenic plants harboring full-length TAC clones failed to complement cvp2, we were able to further refine the map position of CVP2 to overlapping BACs T25N20 and F3F20. Additional potential CVP2-containing clones derived from a BAC F3F20 library and a binary cosmid library (Schulz et al., 1994) that were assayed for cvp2 complementation failed to rescue cvp2. There were four annotated genes in the CVP2 region that were not represented in those libraries, including T25N20.12, which was mutant in cvp2-1. Sequencing of the cvp2-1 allele of gene T25N20.12 revealed a single nucleotide substitution in the gene encoding a previously uncharacterized 5PTase (At1g05470; Figure 2B). Phenotypic analysis of six cvp2 alleles (cvp2-1 through cvp2-6) showed that all mutant alleles had identical phenotypes, suggesting that they represented null alleles. Sequence analysis of four alleles (cvp2-1, cvp2-2, cvp2-3, and cvp2-4) revealed single nucleotide changes consistent with the production of a nonfunctional protein (Figure 2B).
Figure 2.
Map-Based Cloning of CVP2.
(A) Scheme showing the map-based cloning of CVP2. CVP2 was localized to the top of chromosome one, represented by a solid horizontal line, using the flanking markers PhyA and F19P19. For PhyA and F19P19, the fractions indicate the number of recombinants over the total number of meiotic events. The numbers of recombinants for additional markers are shown below the diagonal line. The name of the BAC clone (45° angle) is given for a specific polymorphic marker that was derived from the BAC. Double-headed arrows represent TACs that were identified in the region by hybridization studies. Hatched boxes represent some of the clones that were found to reside in the vicinity of CVP2 and assayed for cvp2 complementation. Because none of these clones rescued the cvp2 mutant phenotype, CVP2 was localized to one of four genes on BAC T25N20 that were not represented in the libraries.
(B) CVP2 gene structure. Solid vertical bars represent exons. Exon/intron junctions were determined by comparing a CVP2 cDNA isolated from seedling tissue with the genomic sequence in the database. Mutations identified in four cvp2 alleles are given. Specifically, cvp2-1 has a G-to-A nucleotide substitution that substitutes a Gly residue for Asp at amino acid 567; cvp2-2 contains a C-to-T nucleotide substitution that changes an Ala to a Val in the PAWCDRIL (amino acid 535) site of domain II; cvp2-3 contains a G-to-A nucleotide substitution that changes a Trp to a stop codon at amino acid 144; cvp2-4 (diepoxybutane mutant) has an A-to-T nucleotide substitution at the fourth exon/intron junction (amino acid 136), which is predicted to cause a splicing error resulting in a misread message for 45 residues before termination. Short horizontal bars represent 5PTase signature motifs, domains I and II.
(C) Complementation of cvp2 with 5PTase. A genomic clone of At1g05470 containing the coding region and 1.5 kb upstream of the translational start codon was introduced into cvp2-1 mutants. Cleared specimens were viewed with dark-field optics. Left, the wild type; center, cvp2-1; right, cvp2-1/At1g05470.
There were discrepancies in the exon/intron annotation of CVP2, which is based on software predictions because the CVP2 cDNA had not been identified. The observation that a CVP2 cDNA is not represented in the collection of 140,000 ESTs suggested that CVP2 is a rare message. In agreement with this low level of expression, we were unable to detect a CVP2 transcript by RNA gel blot hybridization using 10 μg of RNA from various organs and from hormonal-induced (cytokinin, ethylene, brassinosteroid, auxin, and gibberellic acid) seedlings (data not shown). Furthermore, attempts to amplify full-length CVP2 cDNA by RT-PCR using RNA templates from various wild-type tissues failed. To identify the CVP2 cDNA, we conducted RT-PCR using transgenic seedlings overexpressing the CVP2 genomic clone as template RNA. Nested PCR resulted in an amplified product of 1.83 kb. Sequencing of this cDNA, which encodes a 609–amino acid protein, revealed the exon/intron structure displayed by the National Center for Biotechnology Information (NCBI) (Figure 2B). CVP2 identity was verified upon full complementation of the cvp2 vascular patterning defect with a genomic clone encompassing the coding region and flanking 1.5 kb upstream region (Figure 2C), although overexpression of CVP2 cDNA or of a genomic clone with the 35S promoter of Cauliflower mosaic virus did not rescue cvp2 vein patterns, indicating sensitivity to 5PTase levels. Overexpression of CVP2 cDNA or of a genomic clone in the wild type did not yield a vascular patterning defect.
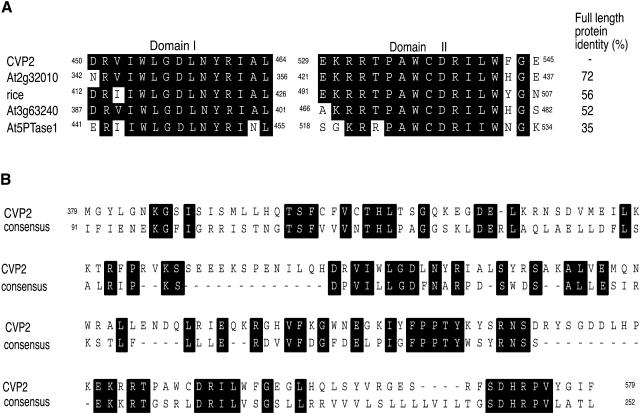
5PTases cleave the phosphate in the 5′ ring position of the water-soluble I(1,4,5)-P3 and I(1,3,4,5)-P4 and/or the lipids PI(4,5)-P2 and PI(3,4,5)-P3 (Majerus et al., 1999). In mammalian systems, 5PTases are classified into four groups (Types I, II, III, and IV) based on sequence homology and substrate specificity that is usually dependent upon the 5PTase subcellular localization (Matzaris et al., 1998; Majerus et al., 1999). For example, the cytosolic/peripheral membrane phosphoinositide phosphatases, the synaptojanins, hydrolyze both soluble and insoluble substrates and are classified as Type II 5PTases. Computer analysis suggested that only Type I and Type II 5PTases are represented in Arabidopsis, maize (Zea mays), rice (Oryza sativa), tomato (Lycopersicon esculentum), and Medicago truncatula (Berdy et al., 2001). Phylogenetic analysis positioned CVP2 and eight other At5PTases as Type I (Berdy et al., 2001), which in mammalian systems have been shown to be the most active 5PTases (Majerus et al., 1999). Mammalian Type I 5PTases hydrolyze IP3 and IP4, and because IP3 allows the release of internally stored Ca2+, they are hypothesized to terminate Ca2+ signaling (Connolly et al., 1985). Both At5PTase1 and At5PTase2 have been shown to use IP3 and IP4 as substrates (Berdy et al., 2001; Sanchez and Chua, 2001). At5PTase1 is unable to hydrolyze the lipid-linked inositol phospholipids, thus confirming its identity as Type I 5PTase (Berdy et al., 2001). Like all 5PTases, CVP2 contains the conserved domains I and II (Connolly et al., 1985). CVP2 is closely related to two other Arabidopsis 5PTases, At2g32010 and At3g63240, sharing 72 and 52% identities, respectively (Figure 3A). Homology of CVP2 to other Type I At5PTases drops off dramatically to 30 to 36% identities. In this regard, it is significant that a monocot rice 5PTase is more closely related to CVP2 than CVP2 is to other At5PTases.
Figure 3.
CVP2 Is a Member of the Endonuclease/Exonuclease/Phosphatase Family.
(A) Alignment of CVP2 with other plant 5PTases. The accession number for the rice 5PTase is AA037964.
(B) CVP2 is aligned to a consensus sequence for endonuclease/exonuclease/phosphatase family members according to the NCBI conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd). The C-terminal part of the CVP2 protein is shown. Members with accession numbers for which the consensus sequence was derived include the following: S. pombe synaptojanin, 1I9Y_A; leptospira hemolytic protein, AAB68647; S. pombe RNA nuclease, CAB42372; C. elegans reverse transcriptase, CAB07490; catabolite repressor protein, ACC44428; Homo sapiens Type I 5PTase, Q14642; bovine DNase I, 1DNK_A; Escherichia coli Exonuclease III, 1AKO; and human endonuclease I 1DEB_B. Alignments were conducted using MacVector 7.0 (Oxford Molecular, Madison, WI). Identical residues are shaded black.
The x-ray crystal structure of Type II 5PTase synaptojanin from Schizosaccharomyces pombe revealed an endonuclease fold originally identified in DNase I (Tsujishita et al., 2001). Protein alignment software showed that Types I and II 5PTases, including CVP2, src homology2 (SH2)-containing inositol phosphates, and other proteins involved in diverse signaling pathways, have a structural similarity to Mg2+-dependent endonucleases (Figure 3B) through the conservation of catalytic residues and a four-layered α/β sandwich motif that is a structural domain with two predominantly antiparallel sheets positioned between two α-helical layers (Dlakic, 2000). Other members of the endonuclease/exonuclease/phosphatase family include major human apurinic/apyrimidinic endonuclease, yeast carbon catabolite repressor protein, which is involved in transcriptional activation and cell cycle regulation, and cytolethal distending toxin B, which causes cell cycle arrest (Dlakic, 2000). The presence of the nuclease catalytic core suggests that this diverse class of proteins has similar cleavage mechanisms but distinct substrate specificity.
CVP2 Is Expressed in Developing Vascular Cells of Many Different Organs
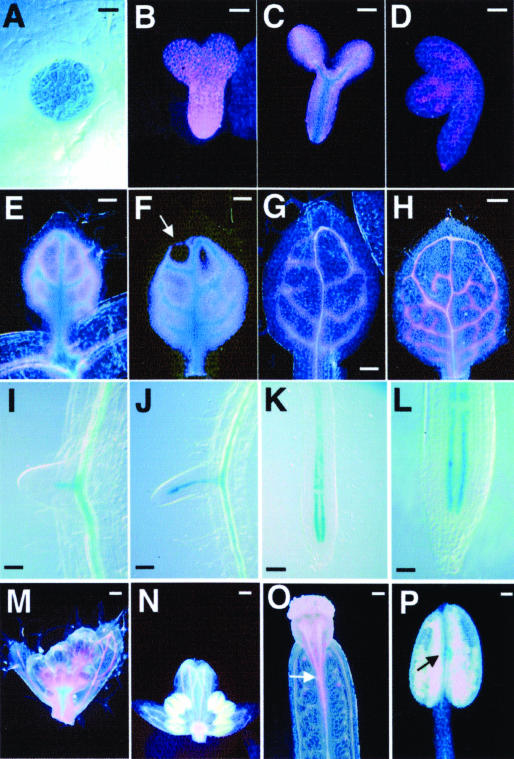
cvp2 mutations affect the vascular pattern of all foliar organs (Carland et al., 1999), suggesting that CVP2 is required in numerous organs. Because CVP2 was expressed at low levels, we examined its expression pattern throughout development using GUS reporter gene expression (see Methods). During early stages of embryogenesis, CVP2:GUS was broadly expressed throughout globular and early torpedo stages (Figures 4A and 4B). At the late torpedo stage, CVP2:GUS was strongly expressed in developing vascular cells and weakly expressed in surrounding cells (Figure 4C). By the walking-stick stage, CVP2:GUS was restricted to incipient vascular cells that delineated the apical loop and midvein of the cotyledon and root vasculature (Figure 4D). A similar pattern was observed during leaf development (Figures 4E to 4H). CVP2:GUS was strongly expressed in procambium and weakly expressed in areole cells in emerging leaves (Figures 4E and 4F). As leaves continued to expand, CVP2:GUS was limited to developing vascular cells (Figures 4G and 4H). Procambium-specific CVP2:GUS histochemical staining extended in parallel with the elongating root (Figures 4I and 4J) until root maturity, when CVP2:GUS was localized to two immature vascular cell files flanking the central core in the root differentiation zone (Figures 4K and 4L). During inflorescence development, CVP2:GUS expression was broad in young floral buds and gradually became restricted to the procambium of cauline leaves, sepals, petals, gynoecia, and anthers (Figures 4M to 4P). In summary, diminishing CVP2:GUS expression occurs in conjunction with vascular cell fate determination in developing organs.
Figure 4.
CVP2 Expression by GUS Histochemical Staining.
(A) to (D) Embryos at globular (A), torpedo (B), late torpedo (C), and walking-stick stages (D).
(E) to (H) Sequential series of developing first rosette leaf.
(E) Young leaf to show strong CVP2 expression in procambial cells and weaker expression in areoles.
(F) Slightly more mature leaf to show the decrease in weak areole CVP2 expression at leaf apex (arrow) reflecting the basipetal maturation of the leaf.
(G) More advanced leaf that demonstrates CVP2 expression in all developing veins.
(H) CVP2 expression is restricted to developing veins in fully developed leaf. Note the absence of GUS staining in the apical loop in which vascular cells have already differentiated as tracheary elements and appear as white cells.
(I) to (L) Root expression.
(I) and (J) CVP2 is expressed in the procambial core of the emerging lateral root.
(K) and (L) CVP2 expression is limited to root tip, specifically to two procambial cell files.
(M) to (P) Floral expression of CVP2. CVP2 is broadly expressed in inflorescence (M) and then restricted to developing floral organ veins, such as those in the sepal (N), gynoecium (O), and anther (P).
Dark-field optics were used for (B) to (H) and (M) to (P); Nomarski optics were used for (A) and (I) to (L). Bars = 10 μM in (A), 25 μM in (B), (I), and (L), 40 μM in (C), 50 μM in (D), (E), (F), (J), (K), and (P), 60 μM in (G), 100 μM in (H), (M), and (O), and 200 μM in (N).
CVP2 Has Elevated IP3 Levels and Increased Sensitivity to ABA
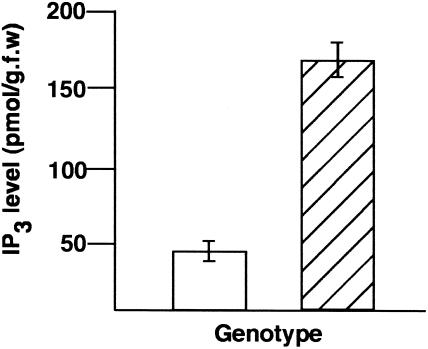
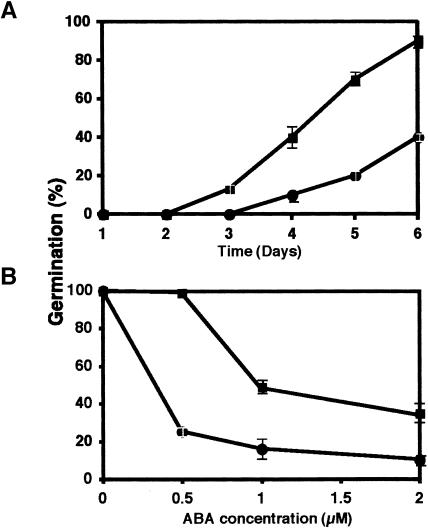
Transgenic manipulation of 5PTase levels alters IP3 levels (Berdy et al., 2001; Sanchez and Chua, 2001; Perera et al., 2002). If CVP2 is regulating IP3 levels, then one would anticipate higher IP3 levels in cvp2 mutants because IP3 would not be hydrolyzed to the inactive form inositol (1,4) diphosphate (IP2). Measurement of IP3 levels showed greater than a threefold increase in cvp2-1 seedlings compared with the wild type (Figure 5). Modulation of IP3 levels has been shown to affect ABA signaling. Transgenic plants overexpressing At5PTase I and II both showed ABA insensitivity (Sanchez and Chua, 2001; Burnette et al., 2003). Similarly, loss-of-function mutations in the FIERY1 (FRY1) gene, an inositol polyphosphate 1-phosphatase that shows low level IP3 catabolism, exhibits ABA hypersensitivity that is associated with elevated IP3 levels (Xiong et al., 2001). To address ABA response in cvp2, dose–response experiments were conducted, measuring seed germination as a function of ABA level (Figures 6A and 6B). On media with no hormone, wild-type and cvp2 germination rates were indistinguishable, suggesting that cvp2 is not sensitive to endogenous ABA levels. On ABA-containing media, cvp2 showed lower levels of seed germination compared with the wild type, indicating hypersensitivity to exogenous ABA. cvp2 root growth also showed increased sensitivity to ABA. After 4 d of growth on 10 μM ABA, wild-type root growth was 41 ± 2.4% inhibited. However, cvp2 root growth was inhibited by 56 ± 4.3%. In contrast with fry1, which exhibited elevated responses to additional stresses, such as high osmotic levels, cvp2 seedlings appeared to demonstrate wild-type responses to NaCl and mannitol (data not shown), suggesting that aberrant cvp2 IP3 signaling specifically affects ABA response rather than a general stress response.
Figure 5.
IP3 Levels Are Elevated in cvp2 Mutants.
Experiments were done in triplicate and error bars indicate standard error. Open, the wild type; hatched, cvp2. g.f.w., grams of fresh weight.
Figure 6.
cvp2 Response to ABA.
Seed germination was measured on increasing doses of ABA for 6 d. Wild-type and cvp2 seed germination on 2 μM ABA over a 6-d period (A) and on day 4 with increasing amounts of ABA (B). Squares, the wild type; circles, cvp2. Values represent triplicate experiments, and error bars show standard error.
DISCUSSION
Dicots have a vascular pattern composed of reiterating vein branching. The mechanisms that guide this developmental process are not well understood. We previously reported on the cvp2 mutant, which is characterized by a reduction in cell recruitment into vascular tissue but otherwise normal vascular cell morphology. Consistent with its wild-type appearance, cvp2 exhibits no defects in auxin biosynthesis, response, or transport. Based on our characterization, we postulated an early role for CVP2 in the perception or transduction of a signal that regulates vascular strand propagation. Mutations in the 5PTase encoded by CVP2 result in higher IP3 levels and revert the normally closed reticulum of Arabidopsis to a more primitive open reticulum. CVP2 represents the only member of this gene family identified by mutation. This finding provides the first evidence of a role for the highly conserved signal transduction pathway, IP3 signaling, in vascular patterning.
IP3-Facilitated Release of Calcium Transients and Development
In mammalian systems, IP3-mediated Ca2+ influx, controlled by 5PTases, is involved in diverse cellular processes, such as regulation of gene transcription, secretion, ovulation, cytoskeletal rearrangements, and cell cycle (Berridge, 1993, 1995). As has been shown in animal and plant systems, cells respond to oscillations in Ca2+ that are differentially perceived and spatially restricted. Type I 5PTases, which use IP3 and IP4 as substrates, are primary regulators of IP3-facilitated Ca2+release (Speed et al., 1996). For example, the underexpression of a Type I 5PTase by antisense methods in rat kidney cells failed to terminate IP3 signaling events and resulted in the spontaneous release of Ca2+ without external stimulation. A 2.6-fold increase in IP3 was associated with a 4.1-fold increase in basal intracellular Ca2+ and resulted in transformed cells characterized by more rapid growth and the ability to form colonies on soft agar and tumors in nude mice (Speed et al., 1999). On the organismal level, a distinct number of calcium oscillations regulates the egg-to-embryo transition in mice (Ducibella et al., 2002). Initiation of transition requires the fewest Ca2+ transients, and progression requires prolonged exposure to Ca2+ transients. In plants, there are established Ca2+ parameter-regulated developmental processes exemplified by stomatal closure. A specific Ca2+ signature with a defined frequency of Ca2+ oscillations controls the amount of stomatal closure, such that an increase or decrease in the frequency of Ca2+ oscillations prevents stomatal closure (Allen et al., 2001). In this manner, a single signaling molecule can drive multiple events by the specific Ca2+ requirement of the process (Berridge et al., 2003).
CVP2-Mediated IP3 Signal Transduction Regulates Vein Patterns
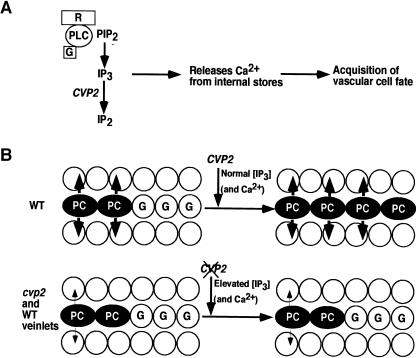
Based on the role of animal 5PTases to act as second messengers and regulate incremental Ca2+ release in response to a primary stimulus (Berridge, 1993), we propose a revised model specific for the involvement of CVP2 in IP3-regulated vascular patterning (Figure 7A). A membrane-bound receptor perceives a primary stimulus and triggers a G protein–linked phospholipase C to cleave the phospholipid, phosphotidylinositol (4,5) bisphosphate (PIP2), into diacylglycerol and IP3. IP3 releases Ca2+ from internal stores in a tightly controlled manner to elicit the initialization of ground cells into vascular cells. CVP2 acts as a negative regulator of IP3 signaling by hydrolyzing IP3 into the inactive form IP2. During the extension of wild-type procambial strands, CVP2 regulates IP3 levels and the release of Ca2+, instructing ground cells to acquire vascular cell identity (Figure 7B). In cvp2 mutants, the absence of functional CVP2 causes excessive IP3 signaling, leading to persistent Ca2+ release, possibly without oscillations, and to an aberrant response, thereby reducing the number of ground-to-procambial cell transitions. This scenario is analagous to events during stomatal closure in which Ca2+ is required for stomatal closure; however, persistent Ca2+ exposure disrupts stomatal movement. This defect is restricted to vascular cell proliferation (vein girth) and continuity (vein length) and thus does not affect vein initiation in most vein orders, suggesting that CVP2 is not the limiting factor in vein development. We propose that wild-type veinlet formation, whose vein endings normally terminate in the areole, show a similar cvp2 Ca2+ response profile. The use of Ca2+ imaging tools, such as chameleon indicators, will enable us to visualize elevated Ca2+ levels at the cellular level.
Figure 7.
Model for the Role of CVP2.
(A) Hydrolyzing activity of CVP2. This model is based on animal systems in which the role of 5PTases is better understood. In response to as yet unidentified external stimulus, a membrane receptor activates phospholipase C (PLC)/G protein complexes (G) to breakdown PIP2 into IP3 (and diacylgylcerol). Acting as a second messenger, IP3 amplifies the primary signal by releasing Ca2+ from internal stores. The downstream biological response for the appropriate Ca2+ parameters is acquisition of vascular cell fate. CVP2 serves to hydrolyze IP3 into the inactive form, inositol (1,4) diphosphate (IP2), and thus terminates IP3 signal transduction.
(B) Progessing procambial strand (PC) encounters ground cells (G). In the wild type, CVP2 would act to prevent sustained Ca2+ oscillations by hydrolyzing IP3 into IP2. In this manner, the correct Ca2+ signature is perceived by downstream Ca2+-dependent target genes, resulting in the recruitment of ground cells into vascular cell fate. In cvp2 foliar organs and in freely terminating wild-type veinlets, because of the absence of CVP2 hydrolytic activity, IP3 levels are elevated, resulting in sustained Ca2+ release. The inappropriate Ca2+ levels reduce ground cell specification into procambial cells, presumably by misregulated Ca2+ response genes, and the veins prematurely terminate. The decrease in size and number of arrows from the procambial cells indicates that the radial proliferation of cvp2 veins is also affected.
How might misregulated cvp2 response genes result in an open reticulum? One possibility is that cvp2 Ca2+ response prevents the stimulation of Ca2+-dependent effectors to act as cell-to-cell communicators for ground-to-procambial cell transition. An alternative model is based on the role of members of the endonuclease/exonuclease/phosphatase family in other systems as cell cycle regulators. In cvp2 developing veins, ground cells lose vascular cell competence before completion of all vein orders formed during leaf development. In cvp2 mutants, excessive Ca2+ release may cause aberrant cell cycling, resulting in either accelerated lamina differentiation or delayed procambial strand formation, in either case affecting the competence of cells to respond to inductive signals. The use of well-defined procambial and cell cycle markers will assist in distinguishing these possibilities. These models are based on the novel role of IP3 signaling in vascular patterning and are in agreement with CVP2 acting independently from other vascular patterning genes.
There are two models proposed for venation patterns. One model, the diffusion-reaction prepattern hypothesis, proposes that an initially homogenous field can be patterned by the interplay between a long-range diffusible inhibitor and an autocatalytic molecule (Meinhardt, 1996). The second model, known as the canalization of signal flow hypothesis, proposes that the patterning of aligned, polar vascular cells is a consequence of the directional flow of auxin (Sachs, 1991). The finding that the open reticulum formed in cvp2 is because of impaired IP3 signal transduction neither supports nor refutes either model. Basing the phenotype on the diffusion-reaction model, elevated IP3 levels and its subsequent effectors in cvp2 may act antagonistically with the reinforcing mechanism or synergistically with the inhibitor. With regards to the canalization model, auxin may not be able to be transported from one cvp2 procambial cell to an adjacent cell because of defective transport capacity as a result of elevated IP3 signaling. However, auxin itself has been reported to rapidly induce IP3 signaling (Ettlinger and Lehle, 1988). Furthermore, unlike many other venation patterning mutants, cvp2 mutants exhibit no auxin-associated defects, and based on double mutant combinations, appear to act independently from mutants with auxin-related vascular patterning defects, such as cvp1 (Carland et al., 1999) and monopteros (F. Carland and T. Nelson, unpublished data). Elucidating the cvp2 open vein network awaits the identification of additional components in CVP2-mediated IP3 signal transduction.
The venation patterns of all foliar organs are affected in cvp2 mutants, corresponding to the CVP2 expression pattern. However, CVP2 is also expressed in the root, which is unperturbed in cvp2 mutants. This exception may be attributable to redundancy with the two closely related At5PTases that also show root expression (F. Carland and T. Nelson, unpublished data). Analysis of CVP2 promoter-driven At5PTases and double mutant analysis should reveal overlapping functions among the At5PTases. Alternatively, because CVP2 disruption appears to only affect vein continuity in lateral vein orders, it is possible that the root vasculature would not show a phenotype as a result of different regulation than that observed in foliar organs because root vasculature is maintained by fixed cell files and not reticulated. This may be analogous to the midvein of foliar organs, which is also unaffected in cvp2 mutants. In light of this result, it is interesting that the monocot rice, which has parallel vein patterns, has a 5PTase closely related to CVP2. In fact, CVP2 shares more identities to this rice 5PTase than to most of the other Arabidopsis 5PTases. It is tempting to speculate a role for this 5PTase in the extension of transverse veins in monocot leaves. Similar to cvp2, the rice mutant radicleless1 exhibits premature termination of commissural veins, but in contrast with cvp2 mutants, it is also affected in auxin response (Scarpella et al., 2003).
cvp2 represents the only reported mutation in the At5PTases. Only null mutants were identified, suggesting that weaker alleles do not have a detectable phenotype. The only other mutation reported in a Type I 5PTase in an intact organism is in Caenorhabditis elegans (Bui and Sternberg, 2002). The targeted deletion of a Type I 5PTase in C. elegans resulted in a developmental abnormality causing aberrent ovulation, suggesting that this 5PTase is a negative regulator of IP3 signaling required during ovulation. However, the misexpression of C. elegans Type I 5PTase did not cause a phenotype. Similarly, the gain-of-function version of CVP2 did not affect vascular patterning in the wild type or in cvp2 mutants. Misexpression of CVP2 and CVP2-like genes by inducible promoters or cell-specific promoters may yield additional information on the phenotype of reduced IP3 levels predicted by the role of CVP2 as a negative regulator of IP3 signaling.
IP3 Signaling and ABA Response
Several genes have been implicated in ABA response, including those involved in small ubiquitin-like modifier conjugation, farnesylation, signaling, and stress response (Cutler et al., 1996; Pei et al., 1998; Xiong et al., 2001; Jung et al., 2002; Kwak et al., 2002; Lois et al., 2003). In many but certainly not all cases, gain-of-function versions of ABA response genes, either through mutation or through overexpression, exhibited insensitivity to ABA, and conversely, recessive alleles demonstrated increased sensitivity to ABA. For example, dominant ABA insensitve1 (abi1) mutants showed resistance to ABA, whereas loss-of-function abi1 alleles are hypersensitive to ABA (Gosti et al., 1999). IP3 regulators have also been shown to exhibit similar profiles in ABA response. FRY1/SAL1 encodes a bifunctional protein with inositol polyphosphate 1-phosphatase and 3′(2′),5′ bisphosphate nucleotidase activities (Murguia et al., 1995; Quintero et al., 1996; Xiong et al., 2001). Further testing of substrate specificity also suggested a role for FRY1 in hydrolyzing IP3, and indeed, fry1 plants had elevated IP3 levels (Xiong et al., 2001). fry1 mutants were also hypersensitive to ABA and showed elevated activation of several ABA- and stress-responsive genes. Experiments directed at transgenic manipulation of 5PTases in plants included dexamethasone-induced overexpression of At5PTase II and overexpression of human Type I 5PTase and At5PTase I (Berdy et al., 2001; Sanchez and Chua, 2001; Perera et al., 2002). In the cases tested, elevated levels of 5PTase resulted in decreased IP3 levels and in ABA insensitivity. In the case of human Type I 5PTase overexpression, PIP2 levels were also reduced. Misexpression of the ABA-induced At5PTase I caused a delay in additional ABA-induced genes (Burnette et al., 2003). These results suggested that At5PTase I acts to terminate ABA signaling. There were no reports of vein pattern defects in the fry1 or At5PTase overexpressors, and at this time, the causal relationship between CVP2-regulated ABA signaling and venation patterns is unclear, but the altered ABA response in cvp2 mutants is consistent with other members of IP3 regulators.
Angiosperms have a closed reticulum that is thought to have arisen from the primitive open vein network for redundancy in transport in case of plant injury and for more efficient transport (Roth-Nebelsick et al., 2001). Freely ending veinlets may be the remnants of this open reticulum that are now confined to higher vein orders. We speculate that IP3 signaling played a role in evolution of open to closed vascular networks, although it may not be the primary determinant because of the second messenger role. The principal regulator of a closed reticulum may be the stimulus that is perceived. Using CVP2 as a reference point, we can proceed to identify downstream targets through microarray technology and in reverse to elucidate the upstream components. For example, CVP2 itself is likely to be tightly regulated based on the rareness of message and on the highly temporally and spatially restricted nature of its expression pattern. Furthermore, CVP2 acts early in defining those cells that will participate in procambial strand formation. Identification of upstream regulators and downstream targets will aid in elucidating the role of CVP2-mediated IP3 signaling in venation patterns.
METHODS
Plant Materials and Growth Conditions
cvp2-1, cvp2-2, and cvp2-3 were identified from an ethyl methanesulfonate–mutagenized population (Carland et al., 1999) of the Columbia ecotype (Col). cvp2-4, cvp2-5, and cvp2-6 were identified in a diepoxybutane-mutagenized population of Col (N. Clay and T. Nelson, unpublished data). The cvp2-1 allele was used in all studies unless otherwise noted. DR5:GUS and Athb8:GUS are single-copy transgenes in Col. Only plants homozygous for the reporter gene construct and cvp2-1 were used in this study. Seed was surface sterilized and plated on MS media (Sigma, St. Louis, MO)/0.75% agar supplemented with 1% sucrose and B5 vitamins, pH 5.7. After a 2 to 3 d of cold treatment in the dark, the plates were transferred to growth chambers with continuous light at 21°C. Seven-day-old light-grown seedlings were transplanted to 2:1 Metro Mix 200:vermiculite and grown with similar light and temperature conditions. To test cvp2 response to ABA, seed was plated on media containing 0, 0.5, 1.0, and 2.0 μM ABA, cold treated (4°C) for 3 d, and transferred to light. Seed germination was scored at 24-h intervals for 6 d. Seed was considered germinated when the radicle had fully emerged from the seed coat. cvp2 response assays to NaCl and mannitol were conducted as reported (Xiong et al., 2001). For root inhibition studies, wild-type and cvp2 seeds were grown on MS media for 4 d and transferred to media with and without 10 μM ABA. Root growth in the presence of ABA was measured after 4 d of vertical growth and expressed as a percentage of root growth in the absence of hormone. Experiments were done in triplicate.
Histology and GUS Staining
Foliar tissue was cleared and prepared for photography as previously described (Carland et al., 1999). To visualize GUS staining, we used a staining solution composed of 100 mM phosphate buffer, pH 7.0, 1 mM EDTA, pH 8.0, 2 mM β-mercaptoethanol, 100 μg/mL of chloramphenicol, 0.01% Triton X-100, 2 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide, and 5 mM potassium ferricyanide and ferrocyanide. Embryos were dissected from ovules before staining to ensure an even distribution of substrate. After vacuum infiltration of GUS stain, samples were placed at 37°C for 4 to 6 h. GUS stain was replaced with fixative (3:1 ethanol:acetic acid) and placed at 4°C overnight. Specimens were dehydrated with 70 and 100% ethanol at 4°C, cleared in 10% NaOH at 37°C for 3 to 4 h, and mounted in 50% glycerol. Specimens were viewed shortly after mounting and photographed with a Zeiss Axiophot microscope (Jena, Germany) using dark-field and Nomarski optics. Following this method, there was minimal GUS diffusion.
Positional Cloning of CVP2
cvp2-1 (Col) was crossed to Landsberg erecta to generate a large mapping population (n = 804 plants). Plants were scored at the seedling stage by removal of a cotyledon that had been cleared in fixative. Seedlings were then transplanted to soil and allowed to self-fertilize, yielding F3 seed. F3 seed was scored by clearing ∼30 seedlings from each F2 plant. DNA was isolated as described by Carland et al. (2002), and PCR was performed with standard conditions. Novel polymorphic markers were identified by three methods. (1) A gene on the restriction fragment length polymorphism map in the vicinity of CVP2 was used as a marker on genomic DNA gel blots or converted to a PCR-based polymorphic marker. (2) As BAC DNA sequence became available on the Arabidopsis Information Resource Web site (http://www.arabidopsis.org/), small segments of DNA were amplified from Col and Landsberg erecta ecotypes and sequenced. Sequences were analyzed for base pair changes that could be converted to a polymorphism by cleaved-amplified polymorphic sequence or derived cleaved-amplified polymorphic sequence methods. (3) BAC DNA sequences were scanned by eye for regions of low complexity (i.e., AT-rich regions) that were tested for use as small nucleotide polymorphisms. Three libraries were screened for cvp2 complementing clones. Screening of the TAC library was performed as previously described (Carland et al., 2002). A binary cosmid chapter library (Schulz et al., 1994) was screened with amplified PCR products from the CVP2 spanning region. Secondary and tertiary screens were conducted on positive chapters until a single clone had been identified. Restriction enzyme and sequence analysis confirmed the map position of the cosmid. Single clones were electroporated into Agrobacterium tumefaciens, and positive clones were chosen as small tetracycline (2 μg/mL) resistant colonies (larger colonies were identified as spontaneous mutants). A third library was prepared from Sau3A1 partially digested BAC F3F20 DNA on which CVP2 was thought to reside. Size-selected (8 to 12 kb) DNA was isolated by low melt agarose followed by β-agarase digestion according to the manufacturer's instructions (New England Biolabs, Beverly, MA) and ligated to BamH1-digested pCAMBIA 2300 (CAMBIA, Canberra, Australia). Map positions of clones were determined by fingerprinting methods and confirmed by DNA sequence of clone ends. After electroporation of clones into A. tumefaciens, they were introduced into Arabidopsis thaliana genotypes using the floral dip method (Clough and Bent, 1998).
Plasmid Constructions
A CVP2 genomic clone was isolated from BAC T25N20 DNA by PCR using the primers T12.9 (5′-GGTTTTGGCAATTTGTATCCC-3′) and T12.1 (5′-GTCCTAATCTGTCGGTTTGGTG-3′) and cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) to generate the construct p1791. p1791 was digested with EcoRI and ligated to pCAMBIA2300 to construct p1851. Fifty-seven independent transformants of p1851 in cvp2 were assessed for complementation of the vascular patterning defect. Approximately 15% did not show complementation, presumably because of position effects, and were not analyzed further. CVP2:GUS construct was designed by amplifying Col DNA with T12.9 and T12.10 (5′-GCTTTTAAATTCCATGAAGATGGGC-3′) and cloning the amplified product into pCR2.1. This construct (p1751) was digested with BamH1 and XbaI and ligated to pBI101 (Clontech, Palo Alto, CA) that had been digested with XbaI and BamHI. Multiple independent lines were analyzed for CVP2:GUS expression and showed similar expression profiles. A line with a single transgene was selected for detailed analysis. Overexpression of a genomic clone of CVP2 that spanned the coding region was constructed by amplifying this region with the primers T12-21 (5′-GAAGATCTTCGCCCATCTTCATGGAATTTAAAAGC-3′) and T12-22 (5′-GAACTAGTCCGCGAATTTGTGTGTTTCTAG-3′) that were engineered with BglII and SpeI sites, respectively. After digestion, the DNA was ligated to XbaI- and BamHI-digested pZP35 (Hadjukiewicz et al., 1994) DNA to generate the construct p1972. We experienced difficulty in amplifying CVP2 cDNA from several different tissue sources using RT-PCR. To isolate CVP2 cDNA, RNA was prepared from Col/p1972 seedlings for use as template RNA, with random hexamers as downstream primers. A first round of RT-PCR using several primer pairs did not yield a product; therefore, a second round of PCR was performed using the nested primers T12-24 (5′- GAATTTCTGAAATGAAGCTGGC-3′) and T12-13 (5′-CTAGAAGAAGCTGAGCTCGG-3′). This PCR yielded a single product of ∼1.8 kb whose identity was confirmed with sequence analysis. CVP2 cDNA was subcloned into a plant transformation vector as described above.
All PCR-based clones were sequenced for verification.
IP3 Assays
IP3 assays were conducted using the radioreceptor method (Amersham TRK1000; Piscataway, NJ) according to the manufacturer's instructions with several modifications.
Briefly, 2 g of 8-d-old media-grown seedlings were ground in liquid nitrogen. Then, 0.4 mL of 20% perchloric acid was added for extraction of IP3 from the cells. Samples were placed on ice for 20 min and sonicated for 15 s with a duty cycle of 50 at a medium setting. After centrifugation, the supernatant was titrated to pH 7.5. Samples were centrifuged to remove precipitate, and extracts were frozen until use. Counts were assayed using a scintillation counter programmed to measure H3. Following the manufacturer's suggestions, all controls were performed, and a dilution series was conducted to ensure no inhibitors were present. Recovery yields were also measured.
Sequence data from this article have been deposited with the GenBank/EMBL data libraries under accession numbers AA037964, AAB68647, CAB42372, CAB07490, ACC44428, and Q14642.
Acknowledgments
We would like to thank Hsu-Liang Hsieh and Genki Suzuki (formerly of Yale University) for advice in library construction, Nicole Kho for providing cvp2-4, cvp2-5, and cvp2-6, members of the laboratory of Timothy Nelson for valuable discussions, and Neil McHale (Connecticut Agricultural Experiment Station) for critical review of the manuscript. We are grateful to Jane Murfett and Tom Guilfoyle (University of Missouri, Columbia) and Nancy Dengler (University of Toronto) for providing DR5:GUS and Athb8:GUS transgenic lines, respectively. We acknowledge the ABRC for providing TAC and BAC clones. This research was supported by National Science Foundation Grant IBN-0110730 to T.N.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Timothy Nelson (timothy.nelson@yale.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021030.
References
- Allen, G.J., Chu, S.P., Harrington, C.L., Schumacher, K., Hoffmann, T., Tang, Y.Y., Grill, E., and Schroeder, J.I. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Berdy, S.E., Kudla, J., Gruissem, W., and Gillaspy, G.E. (2001). Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol. 126, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth, T., and Jurgens, G. (1993). The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Berridge, M.J. (1993). Inositol trisphosphate and calcium signalling. Nature 361, 315–325. [DOI] [PubMed] [Google Scholar]
- Berridge, M.J. (1995). Calcium signalling and cell proliferation. Bioessays 17, 491–500. [DOI] [PubMed] [Google Scholar]
- Berridge, M.J., Bootman, M.D., and Roderick, H.L. (2003). Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. [DOI] [PubMed] [Google Scholar]
- Bui, Y.K., and Sternberg, P.W. (2002). Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol. Biol. Cell 13, 1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette, R.N., Gunesekera, B.M., and Gillaspy, G.E. (2003). An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol. 132, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, M., Mayer, U., and Jurgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation of gene GNOM: Gene structure and intragenic complementation. Mol. Gen. Genet. 250, 681–691. [DOI] [PubMed] [Google Scholar]
- Candela, H., Martinez-Laborda, A., and Micol, J.L. (1999). Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev. Biol. 205, 205–216. [DOI] [PubMed] [Google Scholar]
- Carland, F.M., Berg, B.L., FitzGerald, J.N., Jinamornphongs, S., Nelson, T., and Keith, B. (1999). Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 11, 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14, 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., and McHale, N.A. (1996). LOP1: A gene involved in auxin transport and vascular patterning in Arabidopsis. Development 122, 1811–1819. [DOI] [PubMed] [Google Scholar]
- Casson, S.A., Chilley, P.M., Topping, J.F., Evans, I.M., Souter, M.A., and Lindsey, K. (2002). The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14, 1705–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C.P., Gregory, B.D., Ross, A.S., Tanaka, A., Yoshida, S., Tax, F.E., and Feldmann, K.A. (1999). The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221. [PMC free article] [PubMed] [Google Scholar]
- Clay, N.K., and Nelson, T. (2002). VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 14, 2707–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cnops, G., Wang, X., Linstead, P., Van Montagu, M., Van Lijsebettens, M., and Dolan, L. (2000). Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127, 3385–3394. [DOI] [PubMed] [Google Scholar]
- Connolly, T.M., Bross, T.E., and Majerus, P.W. (1985). Isolation of a phosphomonoesterase from human platelets that specifically hydrolyzes the 5-phosphate of inositol 1,4,5-trisphosphate. J. Biol. Chem. 260, 7868–7874. [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Dengler, N. (2001). Regulation of vascular development. J. Plant Growth Regul. 20, 1–13. [Google Scholar]
- Dengler, N., and Kang, J. (2001). Vascular patterning and leaf shape. Curr. Opin. Plant Biol. 4, 50–56. [DOI] [PubMed] [Google Scholar]
- Deyholos, M.K., Cordner, G., Beebe, D., and Sieburth, L.E. (2000). The SCARFACE gene is required for cotyledon and leaf vein patterning. Development 127, 3205–3213. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., Li, H., Zhou, W., Whoriskey, W.J., Nes, W.D., and Fink, G.R. (2000). STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic, M. (2000). Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 25, 272–273. [DOI] [PubMed] [Google Scholar]
- Ducibella, T., Huneau, D., Angelichio, E., Xu, Z., Schultz, R.M., Kopf, G.S., Fissore, R., Madoux, S., and Ozil, J.P. (2002). Egg-to-embryo transition is driven by differential responses to Ca2+ oscillation number. Dev. Biol. 250, 280–291. [PubMed] [Google Scholar]
- Ettlinger, C., and Lehle, L. (1988). Auxin induces rapid changes in phosphatidylinositol metabolites. Nature 331, 176–178. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H.J., Rook, M.B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387. [DOI] [PubMed] [Google Scholar]
- Hadjukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Baurle, I., Kientz, M., and Jurgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jurgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., McGovern, M., Hurwitz, L.R., Pierro, A., Liu, N.Y., Bandyopadhyay, A., and Estelle, M. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127, 23–32. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jang, J.C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Jung, J.Y., Kim, Y.W., Kwak, J.M., Hwang, J.U., Young, J., Schroeder, J.I., Hwang, I., and Lee, Y. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14, 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., and Dengler, N. (2002). Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216, 212–219. [DOI] [PubMed] [Google Scholar]
- Kinsman, E.A., and Pyke, K.A. (1998). Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Moon, J.H., Murata, Y., Kuchitsu, K., Leonhardt, N., DeLong, A., and Schroeder, J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14, 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois, L.M., Lima, C.D., and Chua, N.H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15, 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen, A.P., Bonke, M., Kauppinen, L., Riikonen, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus, P.W., Kisseleva, M.V., and Norris, F.A. (1999). The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 274, 10669–10672. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- Matzaris, M., O'Malley, C.J., Badger, A., Speed, C.J., Bird, P.I., and Mitchell, C.A. (1998). Distinct membrane and cytosolic forms of inositol polyphosphate 5-phosphatase II. Efficient membrane localization requires two discrete domains. J. Biol. Chem. 273, 8256–8267. [DOI] [PubMed] [Google Scholar]
- Meinhardt, H. (1996). Models of biological pattern formation: Common mechanism in plant and animal development. Int. J. Dev. Biol. 40, 123–134. [PubMed] [Google Scholar]
- Murguia, J.R., Belles, J.M., and Serrano, R. (1995). A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267, 232–234. [DOI] [PubMed] [Google Scholar]
- Nelson, T., and Dengler, N. (1997). Leaf vascular pattern formation. Plant Cell 9, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Perera, I.Y., Love, J., Heilmann, I., Thompson, W.F., and Boss, W.F. (2002). Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant Physiol. 129, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck, G.K., Mattsson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J., Garciadeblas, B., and Rodriguez-Navarro, A. (1996). The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2'),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Nebelsick, A., Uhl, D., Mosbrugger, V., and Kerp, H. (2001). Evolution and function of leaf venation architecture: A review. Ann. Bot. 87, 553–566. [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sachs, T. (1991). Cell polarity and tissue patterning in plants. Dev. Suppl. 1, 83–93. [Google Scholar]
- Sanchez, J.P., and Chua, N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Rueb, S., and Meijer, A.H. (2003). The RADICLELESS1 gene is required for vascular pattern formation in rice. Development 130, 645–658. [DOI] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jurgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., and Jurgens, G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31, 61–73. [DOI] [PubMed] [Google Scholar]
- Schulz, B., Bennett, M., Dilkes, B., and Feldmann, K. (1994). T-DNA tagging in Arabidopsis: Cloning by gene disruption. In Plant Molecular Biology Manual K3. (Belgium: Kluwer Academic Publishers), pp. 1–14.
- Shevell, D.E., Leu, W.M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E. (1999). Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 121, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). hydra Mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed, C.J., Little, P.J., Hayman, J.A., and Mitchell, C.A. (1996). Underexpression of the 43 kDa inositol polyphosphate 5-phosphatase is associated with cellular transformation. EMBO J. 15, 4852–4861. [PMC free article] [PubMed] [Google Scholar]
- Speed, C.J., Neylon, C.B., Little, P.J., and Mitchell, C.A. (1999). Underexpression of the 43 kDa inositol polyphosphate 5-phosphatase is associated with spontaneous calcium oscillations and enhanced calcium responses following endothelin-1 stimulation. J. Cell Sci. 112, 669–679. [DOI] [PubMed] [Google Scholar]
- Steynen, Q.J., and Schultz, E.A. (2003). The FORKED genes are essential for distal vein meeting in Arabidopsis. Development 130, 4695–4708. [DOI] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1994). Leaf development in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 379–401.
- Tsujishita, Y., Guo, S., Stolz, L.E., York, J.D., and Hurley, J.H. (2001). Specificity determinants in phosphoinositide dephosphorylation: Crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105, 379–389. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, B., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]