Abstract
Growth of the maize (Zea mays) endosperm is tightly regulated by maternal zygotic and sporophytic genes, some of which are subject to a parent-of-origin effect. We report here a novel gene, maternally expressed gene1 (meg1), which shows a maternal parent-of-origin expression pattern during early stages of endosperm development but biallelic expression at later stages. Interestingly, a stable reporter fusion containing the meg1 promoter exhibits a similar pattern of expression. meg1 is exclusively expressed in the basal transfer region of the endosperm. Further, we show that the putatively processed MEG1 protein is glycosylated and subsequently localized to the labyrinthine ingrowths of the transfer cell walls. Hence, the discovery of a parent-of-origin gene expressed solely in the basal transfer region opens the door to epigenetic mechanisms operating in the endosperm to regulate certain aspects of nutrient trafficking from the maternal tissue into the developing seed.
INTRODUCTION
Double fertilization in flowering plants results in the formation of two very different structures—the embryo and endosperm (Nawaschin, 1898; Guignard, 1899; Kiesselbach, 1949). The embryo is diploid, whereas the endosperm is a triploid structure in most angiosperms, containing two maternal genomes and one paternal genome, a combination that is held to be essential for correct seed development (Lin, 1984; Scott et al., 1998).
In maize (Zea mays), as in most other species, the endosperm initially develops as a coenocyte and subsequently follows a program of cellularization requiring close coordination between nuclear division and cell wall formation (Olsen, 2001; Sørensen et al., 2002; Dickinson, 2003). After cellularization, the maize endosperm differentiates a range of tissues that assume specialized roles within the developing seed. The basal endosperm transfer layer (BETL) is responsible for the uptake of nutrients from the maternal tissue; the embryo surrounding region acts as a nutritive and protective layer investing the embryo; and the starchy endosperm accumulates storage proteins and carbohydrates, whereas the aleurone is involved in the breakdown and mobilization of storage products upon germination (Lopes and Larkins, 1993; Becraft, 2001; Olsen, 2001). Apart from acting to accumulate and process nutrients for eventual transfer to the developing embryo and seedling, the endosperm is held to assume both a regulatory function during embryogenesis (Lopes and Larkins, 1993) and play a pivotal role in the control of seed size (Lin, 1982; Birchler and Hart, 1987; Kermicle and Alleman, 1990; Scott et al., 1998). The endosperm may also be involved in speciation, for it has been proposed to prevent wide hybridization by acting as a postzygotic barrier to seed development (Cooper and Brink, 1942; Gutiérrez-Marcos et al., 2003).
Both embryo and endosperm develop within the ovule integuments, and although little is known of the interactions that take place between these tissues, it is likely that a coordinated interplay between the sporophytic and gametophytic tissues is a crucial component of seed formation (Lopes and Larkins, 1993). The extent to which maternal tissue is essential for this process is unclear because somatic embryogenesis and endosperm development can occur in vitro in the absence of maternal tissue (Zimmerman et al., 1989; Kranz et al., 1998). There is, however, clear genetic evidence that female sporophytic and gametophytic genes govern early endosperm development (Chaudhury and Berger, 2001; Evans and Kermicle, 2001; Grini et al., 2002; Garcia et al., 2003); for instance, a number of female-gametophytic mutations in Arabidopsis thaliana have been shown to severely affect development of the seed, particularly the endosperm (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kohler et al., 2003). The subsequent molecular characterization of these mutations revealed the existence of a set of proteins (MEDEA [Grossniklaus et al., 1998; Luo et al., 1999], FIS2 [Luo et al., 1999], and FIE [Ohad et al., 1999]) that are closely related to the Drosophila melanogaster Polycomb-group (PcG) proteins. In plants, as in flies, these PcG proteins aggregate into complexes (Kohler et al., 2003) that are required for the establishment of the anterior–posterior axis in the endosperm (Sørensen et al., 2001) and repression of precocious embryo and endosperm development until fertilization (Luo et al., 2000; Spillane et al., 2000). Interestingly, the expression of these genes in the endosperm is restricted to the maternal alleles by a mechanism conventionally termed genomic imprinting (Kinoshita et al., 1999; Luo et al., 2000; Grossniklaus et al., 2001). Although genomic imprinting is well characterized in mammals (Reik and Walter, 2001), it remains poorly understood in plants, with only few imprinted loci reported to date (reviewed in Alleman and Doctor, 2000; Walbot and Evans, 2003).
In maize, the majority of imprinted loci are subject to a parent-of-origin pattern of expression at a particular allele, otherwise known as allele-specific imprinting; examples include α-tubulin3 and α-tubulin4 (Lund et al., 1995a, 1995b), the R locus controlling aleurone pigmentation (Kermicle, 1970), and dzr1 that regulates accumulation of 10-kD zeins (Chaudhuri and Messing, 1994). By contrast, only a few maize loci, namely fie1, fie2 (Danilevskaya et al., 2003; Gutiérrez-Marcos et al., 2003), and nrp1 (Guo et al., 2003), display gene-specific imprinting, as has been reported in Arabidopsis for MEDEA, FIE, FIS2 (reviewed in Baroux et al., 2002), and FWA (Kinoshita et al., 2004). Whereas allele-specific imprinting occurs in later stages of maize endosperm development (Alleman and Doctor, 2000), gene-specific imprinting usually only occurs during early stages of development (for examples, see Kinoshita et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000; Danilevskaya et al., 2003; Gutiérrez-Marcos et al., 2003). The only exceptions so far identified are fie1 (Danilevskaya et al., 2003; Gutiérrez-Marcos et al., 2003) in maize and FIS2 (Luo et al., 2000) in Arabidopsis, which remain imprinted throughout endosperm development.
To explore the nature and extent of this parent-of-origin gene expression in the maize endosperm, we performed a molecular screen to identify genes that exhibit either a maternal or paternal pattern of expression (Gutiérrez-Marcos et al., 2003). We report here the characterization of a novel gene belonging to a family of sequences predominantly expressed in the endosperm. maternally expressed gene1 (meg1) is preferentially expressed through the maternal allele during early endosperm development but at later stages is expressed from both parental alleles. Interestingly, meg1 encodes a small, glycosylated, Cys-rich polypeptide exclusively localized within the labyrinthine walls of the BETL. The discovery of meg1 reveals the existence of a previously unknown class of BETL-specific proteins, while adding significantly to a rapidly increasing group of sequences exhibiting either constitutive or transient parent-of-origin transcriptional regulation during early endosperm development.
RESULTS
Identification and Cloning of the meg Gene Family
A genomic screen based on allelic message display (AMD) was designed to identify endosperm transcripts showing parent-of-origin patterns of expression (Gutiérrez-Marcos et al., 2003). For AMD, we used RNA isolated from endosperms that were dissected from four reciprocally crossed parental inbred lines. This analysis resulted in the identification of several fragments exhibiting monoallelic maternal expression, one of which was termed meg1 (Figure 1). The full-length meg1 cDNA and three other similar but nonidentical cDNAs, consequently termed meg2, meg3, and meg4, were identified using the gel-purified DNA fragment to screen a 7 d after pollination (DAP) endosperm cDNA library. After searching the maize genome database, an additional two ESTs showing partial similarity to the C terminus of the translated MEG1 protein sequence were identified and named meg5 and meg6. We further identified by database search several ESTs from wheat (Triticum aestivum) and barley (Hordeum vulgare) with open reading frames displaying strong similarity to the meg gene family.
Figure 1.
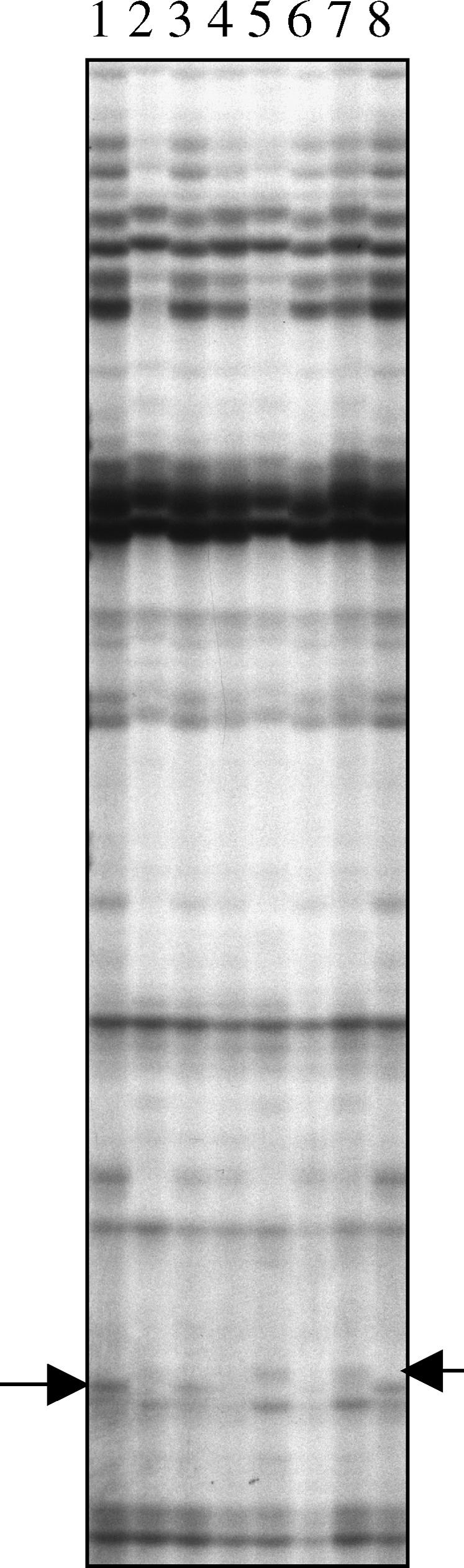
Autoradiograph of an AMD Gel.
Arrows highlight maternal allelic expression of meg1. F2 selfed (lane 1); Mo17 selfed (lane 2); F2 × Mo17 (lane 3); Mo17 × F2 (lane 4); A69Y selfed (lane 5); F2 selfed (lane 6); A69Y × F2 (lane 7); F2 × A69Y (lane 8).
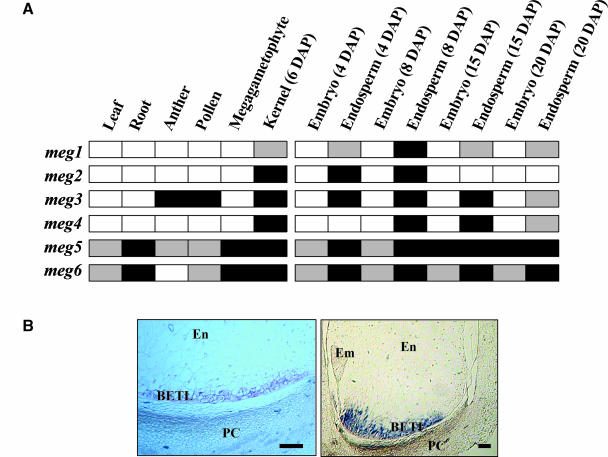
The expression patterns of meg1 and the other meg cDNAs were investigated by RT-PCR using gene-specific oligonucleotide pairs (Figure 2A). meg1 transcript was only present in endosperm samples from 4 to 20 DAP, indicating that its expression is endosperm specific. meg2 and meg4 also showed a similar pattern of expression. By contrast, meg3 was expressed in endosperm as well as in anther and pollen samples, whereas meg5 and meg6 were found to be expressed in most tissues tested (Figure 2A).
Figure 2.
Expression Analysis of meg1 and Related Sequences.
(A) RT-PCR of meg1 and five related cDNAs using RNA samples isolated from a range of tissues and amplified with gene-specific primers (see supplemental data online). Dark and light coloration indicate high and intermediate signal, respectively, and open boxes indicate no expression.
(B) mRNA localization of meg1 in developing seeds. Left, 4 DAP; right, 12 DAP. Em, embryo; En, endosperm; PC, pedicel. Scale bars = 100 μm.
In situ hybridization of meg1 was performed on kernel sections at various stages of development with a gene-specific probe. No signal was detected with the sense probe (data not shown), but the antisense probe generated a strong signal found only in the transfer cells from 4 DAP and showing maximum expression at 10 to 12 DAP (Figure 2B). Expression declined thereafter and was absent in >25 DAP kernel sections (data not shown).
DNA gel blot analysis showed that ∼4 to 5 copies of meg1 are present in most maize inbred lines, teosintes, and other grasses (data not shown). Subsequent restriction fragment length polymorphism analysis using a population of immortalized F2 maize lines enabled us to map the meg1 gene cluster to the short arm of chromosome 7, between markers csu13 and bngl1200.
meg1 Has a Maternal Parent-of-Origin Pattern of Expression
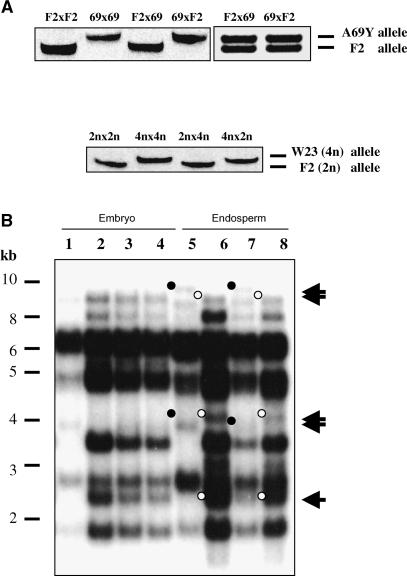
The maternal parent-of-origin expression pattern of meg1 was further investigated by allele-specific RT-PCR using oligonucleotides designed to detect polymorphisms in the 3′ untranslated region of the gene between inbred lines F2 and A69Y, and W23 and other standard inbred lines. The results confirmed exclusive expression of the meg1 maternal allele at early stages of endosperm development (i.e., from 4 DAP; Figure 3A). Surprisingly, we found that at later stages (12 DAP) meg1 expression became biallelic (Figure 3A). To study the effects of altering the maternal to paternal genomic ratio in the endosperm on meg1 expression, reciprocal crosses between diploid and tetraploid inbred lines were performed. Allele-specific RT-PCR showed that the meg1 parent-of-origin expression pattern remained unaltered (Figure 3A).
Figure 3.
Parent-of-Origin Expression and Methylation Analysis of meg1.
(A) Allele-specific RT-PCR analysis of meg1 in the endosperm. Top left, meg1 sequence polymorphism detected in endosperms from selfed and reciprocally crossed F2 and A69Y (69) lines. Note that endosperms resulting from reciprocal crosses show expression of the maternal meg1 allele at 4 DAP. Top right, meg1 expression is biparental in 12 DAP endosperms resulting from F2 and A69Y reciprocal crosses. Bottom, meg1 sequence polymorphism in F2 diploid (2n) and W23 tetraploid (4n) endosperms and showing monoallelic maternal expression in 4 DAP endosperms resulting from reciprocal interploidy crosses.
(B) Methylation analysis of meg1 in 6 DAP embryos and endosperms. Embryo samples (lanes 1 to 4): W22 selfed (lane 1); A69Y selfed (lane2); W22 × A69Y (lane 3); A69Y × W22 (lane 4). Endosperm samples (lanes 5 to 8): W22 selfed (lane 5); A69Y selfed (lane 6); A69Y × W22 (lane 7); W22 × A69Y (lane 8). Closed circles, W22-specific fragment. Open circles, A69Y-specific fragment.
Maternal meg1 Alleles Are Hypomethylated in the Endosperm
Because meg1 is subject to a parent-of-origin pattern of expression in the endosperm, we hypothesized that the male and female alleles could be differentially methylated. We tested our hypothesis by examining the methylation status of meg1 parental alleles. Genomic DNA obtained from embryo and endosperm samples harvested at 6 DAP was digested with a methylation-insensitive restriction enzyme (HindIII) and a methylation-sensitive enzyme (AvaII) in separate reactions. These samples were subsequently analyzed by DNA gel blot hybridization using a gene-specific probe. No parental profiles were observed in the endosperm when samples were digested with HindIII (data not shown), whereas differences were observed between parental meg1 polymorphic alleles when these samples were digested with AvaII (Figure 3B). Two W22-specific fragments (∼4.0 and 8.5 kb) and two A69Y-specific fragments (∼2.7 and 4.5 kb) were present in endosperm samples only when W22 and A69Y lines were used as a pollen source, respectively, but not when used as females, indicating that meg1 maternal alleles are hypomethylated. These data point to a correlation between methylation status of a given meg1 allele and its expression in the endosperm.
MEG Proteins Contain Conserved Cys-Rich Motifs
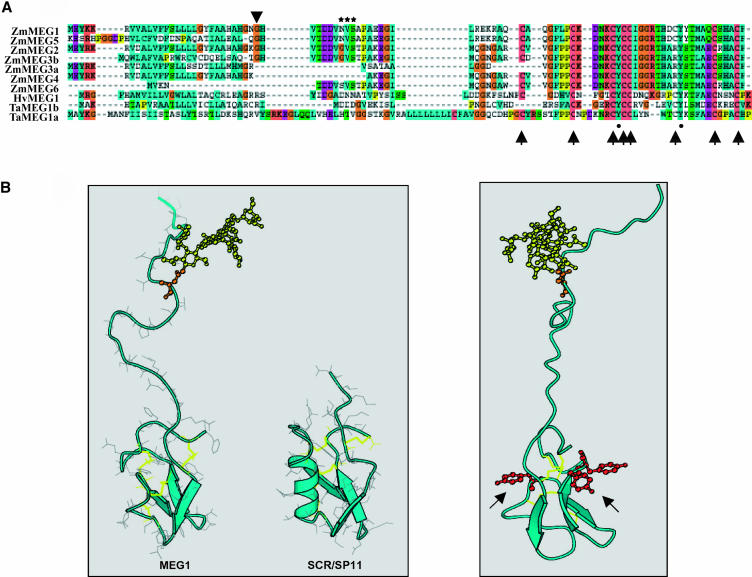
The full-length meg1 cDNA contained an open reading frame of 88 amino acids (Figure 4A), encoding a predicted 9794-D protein with a pI of 7.5. The MEG1 polypeptide contains a hydrophobic N-terminal region with characteristics of a 27–amino acid signal peptide (von Heijne, 1986). Detailed analysis identified a putative cleavage site between His-26 and Glu-27, thus producing a polypeptide containing 61 amino acids, with a predicted molecular mass of 6730 D and pI of 6.2. A protein alignment of MEG1 and the other MEG proteins (i.e., MEG2 to MEG6) revealed the presence of a highly conserved Cys-rich domain in the C-terminal portion (Figure 4A). Further, a conserved amino acid motif comprising eight Cys and two Tyr residues were present in most MEG proteins as well as in the related proteins from T. aestivum and H. vulgare (Figure 4A). A further interesting feature common to most maize MEG proteins is a conserved group of amino acids with high homology to a glycosylation sequon (Mellquist et al., 1998; Wormald and Dwek, 1999), located in the predicted cleaved polypeptide (Figure 4A).
Figure 4.
MEG1 Protein Analysis.
(A) Amino acid conservation of predicted MEG polypeptides in maize (Zm), barley (Hv), and wheat (Ta). Arrowhead indicates putative cleavage site of transit peptides. Stars denote the putative glycosylation sequon. Arrows mark the positions of conserved Cys, whereas closed circles highlight the conserved Tyr residues. Note that MEG3 has two predicted polypeptides (MEG3a and MEG3b).
(B) Homology-based model of MEG1 structure. Left, comparison of the predicted model of MEG1 and the crystal structure of SCR/SP11. Disulfide bonds are shown in yellow; single N-glycan is shown as a yellow ball-and-stick model. β-Strands are represented by broad arrows and the α helix by a helical ribbon. Right, spatial distribution of conserved Tyr residues on MEG1 predicted structure, shown as red balls and sticks (arrows).
To better understand the function of the conserved Cys and Tyr residues in the native MEG1 protein, we generated a sequence homology–based model. In developing this model, the pattern of disulfide bonds in MEG1 was assumed to be similar to that described for other proteins containing eight conserved Cys residues, such as SP11 and plant defensins (Thomma et al., 2002; Mishima et al., 2003). The molecular architecture of this group of proteins consists of a small α/β structure with extensive loop regions, held together by four disulfide bonds (Thomma et al., 2002). When the sequence of MEG1 was aligned with that of SP11, deletions were revealed in the α-helical region between Cys 2 and 3, in addition to an almost complete deletion of the region containing the hypervariable loop between Cys 3 and 4 and a significant insertion between Cys 6 and 7 (Figure 4B). It is interesting to note that despite these disparities, the extended loop between Cys 6 and 7 in MEG1 occupies a similar structural domain to that of the loop between Cys 3 and 4 in SP11 and plant defensins (Figure 4B). Importantly, in our predicted structure for MEG1, the conserved Tyr residues are located in accessible positions at the surface of the protein (Figure 4B).
The MEG1 Protein Is Localized to the Wall Ingrowths of the Basal Endosperm Transfer Cells
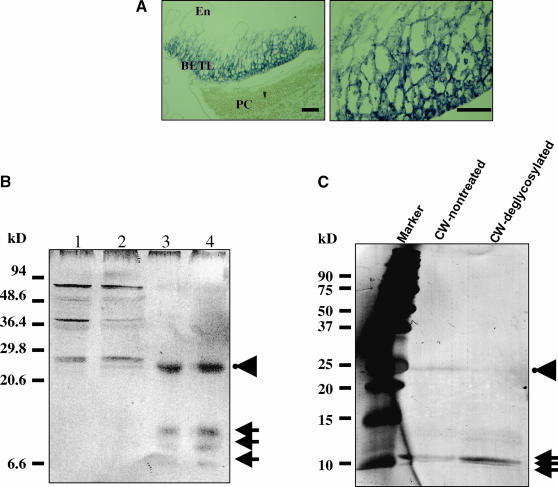
To determine the localization of MEG1 protein in maize endosperm, we raised a polyclonal antiserum using a synthetic peptide for the N terminus of the putatively processed MEG1 polypeptide. Immunolocalization was performed using the purified antiserum, which detected MEG1 protein adjacent to the cell wall ingrowths of basal endosperm transfer cells (Figure 5A). To obtain biochemical evidence of protein localization to the cell wall, protein extracts from 10 DAP endosperms were fractionated as described by Serna et al. (2001) and analyzed by immunoblotting (Figure 5B). We found that a number of proteins recognized by the antibody with predicted molecular masses ranging from ∼20 to 50 kD were present in the cytoplasmic fractions. In cell wall preparations, three proteins of ∼4 to 10 kD gave the greatest signal intensity, suggesting that the putative cleaved MEG1 polypeptide is located in the cell wall (Figure 5B). Unexpectedly, an ∼25-kD protein was also detected in the same preparation (Figure 5B). To test whether this protein corresponded to a glycosylated form of MEG1, we treated isolated proteins obtained from cell wall fractions of 10 DAP endosperms with exoglycosidases and subsequently detected MEG1 proteins by immunoblotting. After exoglycosidase treatment of the cell wall protein fraction, we found a clear reduction in the amount of this 25-kD protein, accompanied by an increase in the amount of 4- to 10-kD protein detected by the antibody (Figure 5B), thus demonstrating that MEG1 proteins are present in a glycosylated form in the transfer cell walls.
Figure 5.
MEG1 Protein Localization.
(A) Immunolocalization of MEG1 proteins. Left, 12 DAP basal endosperm; right, magnification of basal transfer cells showing cell wall localization of MEG1. En, endosperm; PC, pedicel. Scale bars = 100 μm.
(B) Protein gel blot of proteins isolated from 10 DAP endosperms after subcellular fractionation in a 20% SDS-PAGE. Cytoplasmic fraction I (lane 1); cytoplasmic fraction II (lane 2); cell wall fraction I (lane 3); cell wall fraction II (lane 4). Arrows, proteins detected with anti-MEG1 antibody. Arrowhead denotes the presence of an ∼25-kD protein(s).
(C) Protein gel blot of MEG1 proteins before and after treatment with exoglycosidases. Proteins were separated on a 12% SDS-PAGE. Arrowhead, putative glycosylated form of MEG1 detected in a partially purified cell wall fraction. Arrows, nonglycosylated MEG1 protein(s).
The meg1 Promoter Is Transcriptionally Activated by ZmMRP1 in Vitro
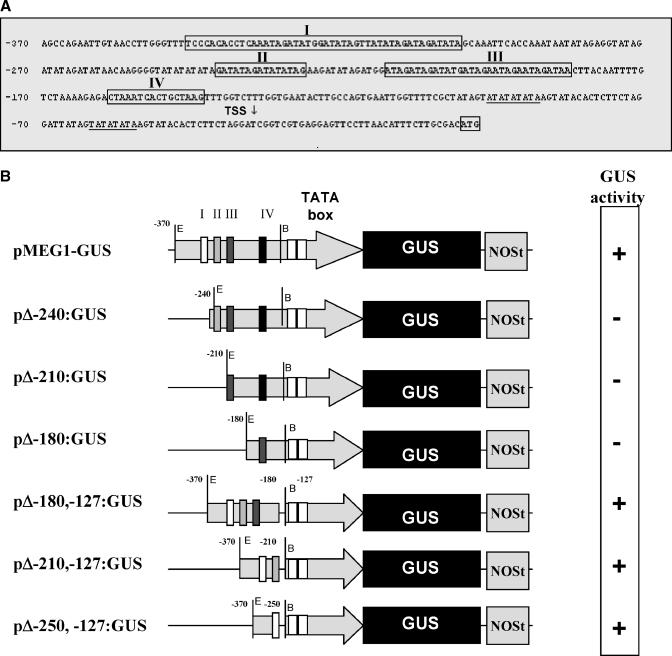
The meg1 promoter sequence was obtained from a maize genomic BAC library (O'Sullivan et al., 2001). Interestingly, through a sequence comparison analysis, we noticed that the full-length promoter region of the meg1 (Figure 6A) shared conserved regions among promoters belonging to known genes that are expressed in maize basal endosperm transfer cells (Yang et al., 1999; Sevilla-Lecoq et al., 2003). It has recently been shown that an endosperm transfer cell–specific transcription factor, ZmMRP1, is able to transactivate expression of several of these BETL-specific promoters (Gomez et al., 2002). Therefore, to investigate whether ZmMRP1 was also capable of transactivating the meg1 promoter, we transformed tobacco (Nicotiana tabacum) protoplasts with a transcriptional fusion construct containing the full-length meg1 promoter fused to the uidA reporter gene and a nopaline synthase (NOS) terminator (denominated pMEG1-GUS), together with a 35S-driven ZmMRP1 transcriptional fusion construct (termed pMON-MRP1). Preliminary data revealed that pMEG1-GUS was strongly transactivated by pMON-MRP1 (data not shown). To define the promoter sequence recognized by ZmMRP1, we generated a deletion series of the meg1 promoter. These fragments were individually fused to uidA, and each construct was used to cobombard etiolated maize leaves with pMON-MRP1. Transactivation was confirmed by the presence of β-glucuronidase (GUS) staining. We found that by removing 120 bp of the distal-most portion of the promoter (−370 to −250 region), transactivation by pMON-MRP1 was disabled (Figure 6B). To determine whether the presence of this 120-bp minimal promoter region permitted transactivation by pMON-MRP1, either alone or in combination with other regions of the meg1 promoter, deletions were performed in the reverse orientation. From this analysis, the 120-bp minimal promoter region (−370 to −250) in combination with the putative TATA box region (−127 to 1) emerged as sufficient to confer transactivation of the meg1 promoter by pMON-MRP1 (Figure 6B). It therefore follows that ZmMRP1 may activate meg1 expression by direct interaction with the −370 to −250 domain of the promoter, a sequence that is conserved among promoters of other genes expressed in the BETL.
Figure 6.
Analysis of the meg1 Promoter.
(A) Promoter sequence of meg1 showing four conserved regions also present in the promoters of BETL1, BETL4, and AE1. Putative TATA motifs are underlined. Transcription start site (TSS) is labeled with an arrow, and the first codon (ATG) is boxed.
(B) Deletion analysis of the meg1 promoter. Left, schematic representations of each promoter-deletion construct used for cobombardment with pMON-MRP1. Right, results obtained from the cobombardment assay; plus sign indicates the presence of GUS staining (transactivation); minus sign denotes absence of GUS staining (lack of transactivation).
Expression of pMEG1-GUS in BETL Cells Is Dependent on Parental Inheritance
To determine whether the full-length promoter region of meg1 fused to the uidA reporter could also drive transgene expression in the endosperm transfer cells, we stably transformed maize plants with the pMEG1-GUS transcriptional fusion construct. Five independent transgenic lines (0757-1C, 0757-2E, 0757-2F, 0757-2D, and 0757-2L) were selected on the basis of high levels of transgene expression, as determined by histochemical staining of GUS. GUS staining was only ever observed in BETL endosperm tissue and absent in all other plant tissues tested (data not shown). To confirm the stable pattern of transgene expression in the endosperm, these lines were backcrossed as females with pollen from A188 standard inbreds for four consecutive generations.
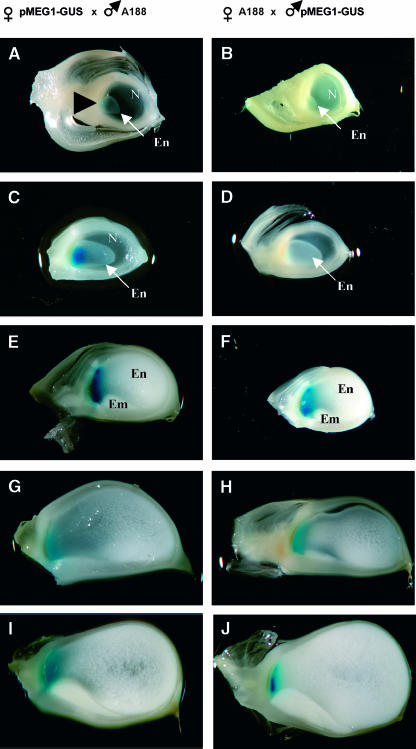
Two of these independent transgenic lines (0757-2E and 0757-2D) were subsequently selected at random and reciprocally outcrossed (i.e., used as females and males) with wild-type A188 plants to investigate any possible changes in transgene expression associated with parental origin. In both instances, 200 to 280 kernels were isolated at different developmental time points and histochemically stained for GUS. Strikingly, the timing at which GUS staining became apparent differed significantly depending on whether the transgene was transmitted maternally or paternally; when plants carrying the pMEG1-GUS transgene were either self-pollinated or outcrossed as females, GUS staining was present in transfer cells from 4 DAP (i.e., at a time when the endosperm becomes fully cellular), whereas when plants were outcrossed as males (i.e., with the transgene transmitted paternally through pollen), GUS accumulation was evident only after 10 DAP (Figure 7). The apparent delay in expression of the paternally transmitted transgene was confirmed at the transcriptional level by RNA gel blot analysis (data not shown). In all cases a maximum level of GUS staining was attained at 10 to 12 DAP (Figure 7), thereafter declining until 25 DAP, when it was no longer detectable.
Figure 7.
Pattern of Maternal and Paternal pMEG1-GUS Expression during Endosperm Development.
Saggital section of kernels resulting from reciprocal crosses between A188 and transgenic plants carrying a transgene with the promoter sequence of meg1 fused to uidA and analyzed by GUS staining. Arrowhead marks region of GUS staining. Em, embryo; En, endosperm; N, nucellus.
(A), (C), (E), (G), and (I) Seeds from plants carrying the pMEG1-GUS transgene after crossing with A188 pollen.
(B), (D), (F), (H), and (J) Seeds from A188 plants crossed with pollen from transgenic plants bearing the pMEG1-GUS transgene.
(A) and (B) 4 DAP.
(C) and (D) 6 DAP.
(E) and (F) 10 DAP.
(G) and (H) 15 DAP.
(I) and (J) 20 DAP.
Taken together, these data strongly suggest that the full-length promoter of meg1 is not only able to confer transgene expression in the BETL but that it is also able to regulate expression of the transgene differentially, depending upon whether it is maternally or paternally transmitted.
DISCUSSION
meg1 Shows a Transient Parent-of-Origin Pattern of Expression in the Endosperm
Gene expression in the triploid endosperm is unusual in that, for some sequences, inherited alleles are expressed according to their parental origin (reviewed in Baroux et al., 2002; Walbot and Evans, 2003). Dosage is certainly responsible for much of this differential expression (Birchler, 1993), and it has recently been shown that allelic dosage accounts for the expression pattern of the majority of endosperm genes in maize (Guo et al., 2003). Evidence is, however, accumulating that a variety of other mechanisms can regulate allele-specific gene activity in the endosperm. These range from temporal asymmetry in expression of parental alleles (Springer et al., 2000; Vielle-Calzada et al., 2000) to a complete silencing of the paternal allele (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kinoshita et al., 1999). To determine the extent to which postfertilization gene expression in the maize endosperm is regulated by these mechanisms, a screen was developed to identify endosperm genes expressed from either maternal or paternal alleles (Gutiérrez-Marcos et al., 2003). We report here that meg1 is expressed maternally only during early endosperm development but is expressed from both parental alleles at later stages.
The mechanism for which the maternal alleles of meg1 are transcribed while the paternal allele is transiently silenced is not fully understood. It is possible that a general suppression of the paternal genome, which has been reported for Arabidopsis by Vielle-Calzada et al. (2000), may be responsible. However, evidence is now accumulating that some maize, and indeed some Arabidopsis genes, are transcribed shortly after transmission through the pollen (Weijers et al., 2001; Scholten et al., 2002).
More feasible, perhaps, is the preferential expression of the maternal meg1 allele simply resulting from a dosage effect reflecting the 2:1 maternal to paternal allelic balance in the endosperm. However, in this case meg1 monoallelic expression would be expected to be constitutively and not transiently regulated, as reported for other dosage-dependent sequences (Guo et al., 2003). Deviations in timing and level of dosage-dependent allelic expression do, however, occur frequently in maize endosperm among different inbred lines and have been attributed to heterochronic allelic variation (Guo et al., 2003). However, for the parental inbred lines tested, we did not find evidence of variation in meg1 parent-of-origin allelic expression (Figure 3A).
Alternatively, the paternally inherited allele may be silenced by an epigenetic mechanism similar to that already reported for both Arabidopsis and maize (reviewed in Alleman and Doctor, 2000; Baroux et al., 2002). The monoallelic maternal expression of meg1 is gene specific, and, thus, similar to that reported for several developmentally important genes, including MEDEA, FIS2, FIE, and FWA in Arabidopsis (Ohad et al., 1996; Kinoshita et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000; Kinoshita et al., 2004) and fie2 and nrp1 in maize (Danilevskaya et al., 2003; Guo et al., 2003; Gutiérrez-Marcos et al., 2003).
The molecular basis of this epigenetic control of gene expression in the endosperm has yet to be fully established. In Arabidopsis, maternal allelic expression of MEDEA and FWA is induced by DEMETER (Choi et al., 2002; Kinoshita et al., 2004), but little is known of the mechanism(s) responsible for parent-of-origin expression of genes in maize and, indeed, other sequences in Arabidopsis. In mammals, most genes showing this parent-of-origin imprinting exhibit a parental asymmetry in methylation (Reik and Walter, 2001). Similar studies in maize revealed that some imprinted loci displayed a strong correlation between hypomethylation of maternally inherited alleles and maternal allelic expression in the endosperm (Lund et al., 1995a, 1995b; Alleman and Doctor, 2000). Our data also suggest that meg1 maternal alleles may be hypomethylated in the endosperm during early development. A strong correlation thus seems to exist between methylation status and allelic gene expression; whether this methylation asymmetry involves only a few genes or is the result of a more global, genome-wide mechanism operating in the maize endosperm remains to be determined. Interestingly, Kinoshita et al. (2004) and Xiao et al. (2003) have demonstrated that the expression of imprinted FWA and MEDEA from maternal alleles in the Arabidopsis endosperm is dependent on the maintenance of a CpG DNA methyltransferase activity, which maintains a silenced, methylated state of the paternal allele.
Transient parent-of-origin expression such as we describe here for meg1 is reminiscent of that reported for some imprinted genes in Arabidopsis (MEDEA and FIE) and maize (fie2). Importantly, the full-length promoter of meg1 is capable of driving transgene expression in an identical manner to that of the endogenous meg1 gene, both temporally and according to its parental origin. Similarly in Arabidopsis, it has been reported that the expression of transgenic reporters fused to the promoters of imprinted MEDEA and FIE genes display a parent-of-origin expression pattern during early endosperm development, followed by a reactivation of the paternally inherited transgene before cellularization of the endosperm (Luo et al., 2000; Yadegari et al., 2000). Why some genes should commence biallelic transcription at midstage in endosperm development is not yet known. In maize, however, it may not be a coincidence that the point at which both parental alleles of meg1 (and fie2) are expressed coincides with the onset of endoreduplication (Schweizer et al., 1995; Larkins et al., 2001; Dilkes et al., 2002). Endoreduplication has been associated with a decrease in histone 1 and posttranscriptional changes in high mobility group I/Y proteins (Zhao and Grafi, 2000), events that may lead to alterations in chromatin structure and/or changes in DNA methylation and perhaps to transcriptional activation of previously silenced paternal sequences.
MEG1 Is Specific of the Basal Endosperm Transfer Region
Several endosperm transfer cell–specific genes have been identified in maize, many of which belong to large gene families such as the BETLs and BAPs (Hueros et al., 1995; Serna et al., 2001). Whereas mRNA in situ and promoter-GUS fusion expression data revealed that meg1 is specifically located in the basal transfer region of the endosperm from 4 to 20 DAP, sequence analysis at the nucleotide and amino acid level revealed no homology to other known proteins, thus identifying MEG1 as a novel transfer cell–specific protein. MEG1 is a small polypeptide that is either localized in or adjacent to the massive, labyrinthine cell wall ingrowths of the transfer cells. The majority of transfer cell–specific proteins reported to date are small and either secreted into the pedicel region or localized to the BETL cell walls (Hueros et al., 1995; Serna et al., 2001). An unusual feature of MEG1 proteins is the presence of a conserved Cys-rich motif in the C-terminal region and a putative N-terminal glycosylation signal (Mellquist et al., 1998; Wormald and Dwek, 1999). This Cys-rich motif, which consists of eight Cys residues and two Tyr residues, is conserved in all the MEG proteins and bears strong resemblance to Cys-rich domains identified in a number of other plant proteins (Domingo et al., 1999; Schopfer et al., 1999). Cys-rich domains are believed to generate protein conformations that can expose side chain residues (Berg and Shi, 1996), such as the Tyr residues found in MEG1. Although yet to be confirmed for MEG1, the exposure of Tyr residues in synthetic proteins has been shown to facilitate binding to cell wall components (reviewed in Cassab, 1998).
Unlike other maize transfer cell–specific proteins, such as the BETL family, MEG1-like proteins are present also in other grasses, suggesting that aspects of MEG1 function are conserved among the cereals. The molecular configuration and location of MEG1 protein in maize suggests that it either plays a structural role in the basal endosperm transfer region, given its tight association with the cell wall ingrowths, or a defensive function against pathogens, which has been hypothesized for the BETL1 and BETL3 proteins (Hueros et al., 1995, 1999a). Alternatively, because a number of Cys-rich proteins such as the pollen determinant of sporophytic incompatibility in Brassica, SCR/SP11 (Schopfer et al., 1999), function as signaling molecules, MEG1 may play a role in signaling, perhaps operating as part of a cross talk between the endosperm and the maternal tissue. The posttranslational modification of MEG1 by N-linked glycosylation may further support this inference because in plants, protein glycosylation is required for a variety of biological functions, including cell–cell communication and signaling (Cheung et al., 1996; Wilson, 2002).
Although there is clear evidence that endosperm development is under strong maternal genetic control (Felker et al., 1985; Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kohler et al., 2003), the interactions between the developing seed and the surrounding maternal tissues remain enigmatic (reviewed in Lopes and Larkins, 1993). The BETL region is in direct contact with the maternal pedicel tissue and is established early in endosperm development (Costa et al., 2003). Further, the transfer cells that comprise this tissue are sites of nutrient translocation (reviewed in Becraft, 2001; Olsen, 2001; Thompson et al., 2001), and it is reasonable to expect that evolutionary pressure has resulted in significant maternal control over their activity. In this context, it is interesting that only the maternal meg1 allele is transcribed during BETL differentiation (Becker et al., 1999). Recently, another gene has been identified, which is both expressed in the basal region of the maize endosperm and exhibits a similar parent-of-origin pattern of expression (Magnard et al., 2003). This discovery of mechanisms regulating maternal expression of transfer cell–specific genes increases the likelihood that resource allocation to the endosperm is maternally modulated, which has implications for seed size and, ultimately, yield. This evidence that the female parent can regulate nutrient supply to the seed strengthens the view that paternally and maternally derived genes compete over resources in the progeny (Haig and Westoby, 1989), an hypothesis that has also been supported by experiments involving intraspecific interploidy crosses (Lin, 1984; Haig and Westoby, 1991; Scott et al., 1998). After such crosses in maize, both gene expression in the BETL and its cellular structure are greatly disrupted (Charlton et al., 1995; Gutiérrez-Marcos et al., 2003).
meg1 represents a novel gene that is specifically expressed in the endosperm transfer cell region and is subject to a transient parent-of-origin effect. Although the mechanism responsible for this pattern of allelic expression is not yet well understood, our findings contribute to an increasing body of data that point to the existence of a group of transfer cell–specific genes whose expression is under maternal control. This level of control would, of course, provide the maternal parent with the opportunity to distribute resources effectively, taking into account overall nutrient levels within the plant and other local and environmental factors.
METHODS
Plant Material and Growth Conditions
Maize (Zea mays) diploid inbred lines W22, F2, B73, and Mo17 and tetraploid inbred W23 (Maize Genetics Cooperation Stock Center, Urbana, IL) were glasshouse grown at Oxford University and in Jealott's Hill (Syngenta, Berkshire, UK) between 1996 and 2002, under the following regime: 16-h daylength (supplemented with metal halide lamps at 250 mmols, when required) at 22°C to 28°C during the day, and at 16°C to 20°C at night. Humidity levels were set at ∼40% to 50% daytime and 60% to 70% at night. Kernels were harvested at 4 to 25 DAP. Embryos and endosperms were isolated and pooled, then frozen in liquid nitrogen and stored at −80°C.
Allelic Message Display
Total endosperm RNA was extracted from plants that were either selfed or reciprocally crossed among W22, F2, B73, and Mo17 inbred lines and used for AMD-PCR according to a modified protocol of Hagiwara et al. (1997). Briefly, reverse-transcribed RNA was used as source material for PCR (HIEROGLYPH kit; Genomix-Beckman, Fullerton, CA), with labeled [α-33P]dATP. A combination of 24 random oligonucleotides and 10 degenerated poly(T) primers (240 combinations) was used to carry out PCR reactions on endosperm samples, and products were analyzed in a semiautomatic Genomix LR DNA sequencing system. After exposure to film (Biomax MR; Kodak, London, UK), candidate bands were excised from the gel, PCR amplified, and subcloned into a suitable plasmid vector.
Identification of meg1 cDNA and Related Sequences
Full-length cDNAs were obtained after screening a 7 DAP maize endosperm cDNA library (Hueros et al., 1999b). Among the 500,000 plaques screened, five cDNA clones were identified and sequenced at the biochemistry sequencing unit (Oxford University). After screening an F2 BAC library (O'Sullivan et al., 2001), six genomic fragments that hybridized with a meg1 probe were subcloned into pBluescript II KS+ (Stratagene, La Jolla, CA) and sequenced.
Gene Expression Analysis
Tissue-specific expression of meg1 and other meg sequences was assessed by RT-PCR (see supplemental data online for primer details). For mRNA localization, in situ hybridization was performed on developing kernels according to a published method (Jackson, 1991), with minor modifications (Costa et al., 2003).
For allele-specific RT-PCR, we sequenced meg1 alleles from several inbred lines (diploid A188, B73, Mo17, F2, W22, and tetraploid W23), which exhibited 99.5% identity (data not shown). To differentiate between meg1 alleles, an amplified and cleaved polymorphic sequences technique was used (Neff et al., 1998). RT-PCR analysis was performed with primers (MEG1 [5′-TGCTGCTCATGCGCATGGGGCTG-3′] and MEG1HpaI [5′-TTGTATATAAAAACAGTGATGTTAA-3′]), and PCR products were subsequently digested with HpaI to generate the following fragments: 177 bp in F2 and 198 bp in A69Y or W23 standard inbred lines. The glutathione synthase1 gene was amplified as a control (see supplemental data online).
Immunolocalization
Polyclonal antiserum was raised in rabbit against a synthetic peptide (N-APAEEGILREKRAQC-C) and affinity purified with an immobilized peptide using a Sulpholink coupling gel system (Pierce, Rockford, IL). Maize kernels were fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.2, for 12 to 24 h depending on the tissue volume. Samples were dehydrated in an ethanol series and wax embedded. Sections were deparaffinized and blocked in 1% BSA in PBS (10 mM sodium phosphate and 150mM NaCl, pH 7.4) for 30 min at room temperature and incubated overnight with anti-MEG1 antiserum or preimmune serum (both diluted 1:500). The immunoreactions were detected using an alkaline phosphatase-coupled secondary antibody (Sigma, St. Louis, MO; diluted 1:1000) and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as substrate.
Subcellular Fractionation of Proteins
Fractionation of subcellular components of endosperms (obtained from a standard F2 inbred) was performed as described by Serna et al. (2001). Proteins were separated by 20% SDS-PAGE according to Laemmli (1970) and electroblotted onto a polyvinylidine difluoride membrane. Proteins were immunodetected using an enhanced chemiluminescence method (Amersham, Buckinghamshire, UK).
Glycosylation Analysis
A partially purified 10 DAP endosperm cell wall fraction was incubated in the presence of 1 unit of β-N-acetylhexosaminidase (New England Biolabs, Hitchin, UK) and 4 units of α-mannosidase (New England Biolabs) in 40 μL of 100 mM sodium acetate, pH 5.0, for 48 h at 37°C. Proteins were fractionated by 12% SDS-PAGE and immunodetected as above.
Protein Modeling
Molecular modeling was performed on a Silicon Graphics Fuel workstation using the programs InsightII and Discover (Accelrys, San Diego, CA). The C-terminal region of MEG1 (residues 47 to 88) was modeled based on the crystal structure of SP11 (Mishima et al., 2003). The sequence similarity between this region of MEG1 and SP11 is not high, except for the conserved presence of eight Cys residues. The N-terminal region of MEG1 (residues 27 to 46) was modeled as a random chain in the absence of any structural data or sequence similarity with proteins of known structure. In the absence of any sequence data for the glycan, a typical plant glycan, Xylβ1-2(Manα1-3)(Manα1-6)Manβ1-4GlcNacβ1-4(Fucα1-3)GlcNAc (Wilson, 2002), was added to Asn-36. The N-linked glycan structure was built using average crystallographic torsion angles for the glycosidic linkages (Wormald et al., 2002; Petrescu et al., 2004), and the conformation of the Asn-GlcNAc linkage was based on average crystallographic values (Petrescu et al., 2004).
Generation and Analysis of Transgenic Lines
The promoter region of meg1 was isolated by PCR using two specific oligonucleotides (pMEG1-GUS primers; see supplemental data online) and subcloned into pGEM-T easy vector (Promega, Madison, WI). After digestion with EcoRI and ClaI, the 371-bp fragment was fused to the uidA sequence and the NOS terminator of the pSLJ4K1 vector (Jones et al., 1992) to generate the PROZmMEG1-1-uidA-NOS transcriptional fusion, denoted pMEG1-GUS. Embryogenic type II calli were transformed with the construct and regenerated as described previously (Bonello et al., 2000). Plants were genotyped for the presence of the transgene via PCR using specific oligonucleotides (GUS.FOR and GUS.REV; see supplemental data online), backcrossed to wild-type A188 plants for four successive generations, and then bulked by selfing. Histochemical analysis of transgenic lines was performed as described previously (Costa et al., 2003). Briefly, stained kernels were hand-sectioned, fixed then dehydrated to 70% ethanol, and digitally imaged.
Promoter Deletion Analysis
The serial deletion analysis was performed by PCR amplification of different regions of the meg1 promoter, using multiple oligonucleotide pairs (see supplemental data online), then subcloned into pGEM-T easy vector (Promega). Constructs were digested with EcoRI and BstXI, and fragments were subcloned into the pMEG1-GUS construct predigested with EcoRI and BstXI. By following this approach, each deletion was subcloned immediately upstream of the putative TATA boxes identified in the meg1 promoter.
The ZmMRP-1 coding region was isolated by PCR using oligonucleotides MRP.FOR (5′-GGATCCATGAATCCCAACTTCAACAGTG-3′) and MRP.REV (5′-GAATTCTTATCGGTTATATATCTGGCTCTCC-3′). PCR fragments were subcloned in pGEM-T easy (Promega), digested with BamHI/EcoRI, and the 327-bp fragment was subcloned into pMON30049 (Pang et al., 1996) to generate the construct pMON-MRP1.
Plasmid DNA was isolated by the QIAprep midi kit (Qiagen, Hilden, Germany) and coated onto tungsten (M10) particles according to Klein et al. (1992). For transient transformation, Hi-II maize seeds were surface-sterilized and germinated in the dark. Etiolated leaves (2 cm wide) were sectioned into 1 to 2 cm–long pieces and cobombarded with each meg1 promoter deletion construct and pMON-MRP1 using a Bio-Rad Biolistic PDS-1000/He device (Hertfordshire, UK). Gold particles (0.6 nm; Bio-Rad) were coated with the DNA plasmid mixture, including 2.5 g of pMON-MRP1 derived plasmid and 2.5 g of each meg1 promoter deletion construct. Tissues were positioned 6 cm from the microcarrier stopping screen, itself located 5 cm below the 6.2 MPa rupture disc. After bombardment, samples were incubated in the dark on MS solid media containing 100 mg/L of myo-inositol, 2 g/L of Gln, 30 g/L of sucrose, and MS vitamins (Sigma, Poole, UK) for 24 h at 26°C. A minimum of three independent experiments were conducted for each promoter deletion made. Transcriptional activation was confirmed by GUS staining the leaf discs according to Jefferson (1989), with modifications. Leaf tissue was stained in a solution containing 0.5 mg/mL of X-glucuronide (Clontech, Palo Alto, CA), 0.5 mM phosphate buffer, pH 7.0, 0.1% Triton X-100, and 20% (v/v) methanol.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY536120 (meg1 genomic locus), AY536121 (meg1 cDNA), AY536122 (meg2 cDNA), AY536123 (meg3 cDNA), AY536124 (meg4 cDNA), AY536125 (meg5 cDNA), and AY536126 (meg6 cDNA).
Supplementary Material
Acknowledgments
We thank Qing Zhang for help with sequencing, Suzanne O'Shea and David Fitter for technical assistance, and Christine Surman for greenhouse assistance. We thank John Baker for help with imaging, Andy Greenland and all at Syngenta-JHIRS for advice and use of plant growth facilities, and Keith Edwards and Donal O'Sullivan for use of their F2 BAC library. We thank Tom Brutnell for stimulating discussions and the Maize Genetics Cooperation Stock Center (Urbana, IL) for providing the inbred lines. We also thank M. Shirakawa for providing the pdb file for SP11, before deposition in the Protein Data Bank. Research was funded by Oxford University, EU Framework IV and V (MAZE) initiatives.
Online version contains Web-only data.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jose Gutiérrez-Marcos (jose.gutierrez@plants.ox.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019778.
References
- Alleman, M., and Doctor, J. (2000). Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol. Biol. 43, 147–161. [DOI] [PubMed] [Google Scholar]
- Baroux, C., Spillane, C., and Grossniklaus, U. (2002). Genomic imprinting during seed development. Adv. Genet. 46, 165–214. [DOI] [PubMed] [Google Scholar]
- Becker, H., Hueros, G., Maitz, M., Varotto, S., Serna, A., and Thompson, R.D. (1999). Domains of gene expression in developing endosperm. In Fertilization in Higher Plants, M. Cresti, G. Cai, and A. Moscatelli, eds (Heidelberg, Germany: Springer-Verlag), pp. 361–375.
- Becraft, P.W. (2001). Cell fate specification in the cereal endosperm. Semin. Cell Dev. Biol. 12, 387–394. [DOI] [PubMed] [Google Scholar]
- Berg, J.B., and Shi, Y. (1996). The galvanization of biology: A growing appreciation for the roles of zinc. Science 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27, 181–204. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A., and Hart, J.R. (1987). Interaction of endosperm size factors in maize. Genetics 117, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello, J.F., Opsahl-Ferstad, H.G., Perez, P., Dumas, C., and Rogowsky, P.M. (2000). Esr genes show different levels of expression in the same region of maize endosperm. Gene 246, 219–227. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 281–309. [DOI] [PubMed] [Google Scholar]
- Charlton, W.L., Keen, C.L., Merriman, C., Lynch, P., Greenland, A.J., and Dickinson, H.G. (1995). Endosperm development in Zea mays: Implications of gametic imprinting and paternal excess in regulation of transfer layer development. Development 121, 3089–3097. [Google Scholar]
- Chaudhuri, S., and Messing, J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. USA 91, 4867–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., and Berger, F. (2001). Maternal control of seed development. Semin. Cell Dev. Biol. 12, 381–386. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., Zhan, X.Y., Wang, H., and Wu, H.M. (1996). Organ-specific and Agamous-regulated expression and glycosylation of a pollen tube growth-promoting protein. Proc. Natl. Acad. Sci. USA 93, 3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. [DOI] [PubMed] [Google Scholar]
- Cooper, D.C., and Brink, R.A. (1942). The endosperm as a barrier to interespecific hybridization in flowering plants. Science 95, 75–76. [DOI] [PubMed] [Google Scholar]
- Costa, L.M., Gutiérrez-Marcos, J.F., Greenland, A.J., Brutnell, T.P., and Dickinson, H.G. (2003). The globby1 (glo1–1) mutation disrupts nuclear and cell division in the developing maize seed causing aberrations in endosperm cell fate and tissue differentiation. Development 130, 5009–5017. [DOI] [PubMed] [Google Scholar]
- Danilevskaya, O.N., Hermon, P., Hantke, S., Muszynski, M.G., Kollipara, K., and Ananiev, E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, H.G. (2003). Plant cell cycle: Cellularisation of the endosperm needs spätzle. Curr. Biol. 13, R146–R148. [DOI] [PubMed] [Google Scholar]
- Dilkes, B.P., Dante, R.A., Coelho, C., and Larkins, B.A. (2002). Genetic analyses of endoreduplication in Zea mays endosperm: Evidence of sporophytic and zygotic maternal control. Genetics 160, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo, C., Sauri, A., Mansilla, E., Conejero, V., and Vera, P. (1999). Identification of a novel peptide motif that mediates cross-linking of proteins to cell walls. Plant J. 20, 563–570. [DOI] [PubMed] [Google Scholar]
- Evans, M.M., and Kermicle, J.L. (2001). Interaction between maternal effect and zygotic effect mutations during maize seed development. Genetics 159, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felker, F.C., Peterson, D.M., and Nelson, O.E. (1985). Anatomy of immature grains of eight maternal effect shrunken endosperm barley mutants. Am. J. Bot. 72, 248–256. [Google Scholar]
- Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jurgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, E., Royo, J., Guo, Y., Thompson, R., and Hueros, G. (2002). Establishment of cereal endosperm expression domains: Identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 14, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini, P.E., Jurgens, G., and Hulskamp, M. (2002). Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162, 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Spillane, C., Page, D.R., and Kohler, C. (2001). Genomic imprinting and seed development: endosperm formation with and without sex. Curr. Opin. Plant Biol. 4, 21–27. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA a Polycomb group gene in Arabidopsis. Nature 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Guignard, L. (1899). Sur les antherozoides et la double copulation sexualle chez les vegetaux angiospermes. CR Acad. Sci. Paris 128, 864–871. [DOI] [PubMed] [Google Scholar]
- Guo, M., Rupe, M.A., Danilevskaya, O.N., Yang, X., and Hu, Z. (2003). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36, 30–44. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos, J.F., Pennington, P.D., Costa, L.M., and Dickinson, H.G. (2003). Imprinting in the endosperm: A possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara, Y., Hirai, M., Nishiyama, K., Kanazawa, I., Ueda, T., Sakaki, Y., and Ito, T. (1997). Screening for imprinted genes by allelic message display: Identification of a paternally expressed gene impact on mouse chromosome 18. Proc. Natl. Acad. Sci. USA 94, 9249–9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D., and Westoby, M. (1989). Parent specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155. [Google Scholar]
- Haig, D., and Westoby, M. (1991). Genomic imprinting in the endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333, 1–13. [Google Scholar]
- Hueros, G., Gomez, E., Cheikh, N., Edwards, J., Weldon, M., Salamini, F., and Thompson, R.D. (1999. b). Identification of a promoter sequence from the BETL1 gene cluster able to confer transfer-cell-specific expression in transgenic maize. Plant Physiol. 121, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueros, G., Royo, J., Maitz, M., Salamini, F., and Thompson, R.D. (1999. a). Evidence for factors regulating transfer cell-specific expression in maize endosperm. Plant Mol. Biol. 41, 403–414. [DOI] [PubMed] [Google Scholar]
- Hueros, G., Varotto, S., Salamini, F., and Thompson, R.D. (1995). Molecular characterization of BET1, a gene expressed in the endosperm transfer cells of maize. Plant Cell 7, 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. (1991). In situ hybridisation in plants. In Molecular Plant Biology: A Practical Approach, M. McPherson, ed (Oxford, UK: Oxford University Press), pp. 163–174.
- Jefferson, R.A. (1989). The GUS reporter gene system. Nature 342, 837–838. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kermicle, J.L. (1970). Dependence of the R-mottled aleurone phenotype in maize on the mode of sexual transmission. Genetics 66, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L., and Alleman, M. (1990). Gametic imprinting in maize in relation to the angiosperm life cycle. Dev. Suppl., 9–14. [PubMed]
- Kiesselbach, T.A. (1949). The Structure and Reproduction of Corn. (Lincoln, NE: University of Nebraska Press).
- Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S., Fischer, R.L., and Kakutani, T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303, 521–523. [DOI] [PubMed]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11, 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, T.M., Arentzen, R., Lewis, P.A., and Fitzpatrick-McElligott, S. (1992). Transformation of microbes, plants and animals by particle bombardment. Biotechnology (NY) 10, 286–291. [DOI] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, E., von Wiegen, P., Quader, H., and Lorz, H. (1998). Endosperm development after fusion of isolated, single maize sperm and central cells in vitro. Plant Cell 10, 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52, 183–192. [PubMed] [Google Scholar]
- Lin, B.-Y. (1982). Association of endosperm reduction with parental imprinting in maize. Genetics 100, 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.-Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, G., Ciceri, P., and Viotti, A. (1995. a). Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 8, 571–581. [DOI] [PubMed] [Google Scholar]
- Lund, G., Messing, J., and Viotti, A. (1995. b). Endosperm-specific demethylation and activation of specific alleles of alpha-tubulin genes of Zea mays L. Mol. Gen. Genet. 246, 716–722. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard, J.-L., Lehouque, G., Massonneau, A., Frangne, N., Heckel, T., Gutiérrez-Marcos, J.F., Perez, P., Dumas, C., and Rogowsky, P.M. (2003). ZmEBE genes show a novel, continuous expression pattern in the central cell before fertilization and in specific domains of the resulting endosperm after fertilization. Plant Mol. Biol. 53, 821–836. [DOI] [PubMed] [Google Scholar]
- Mellquist, J.L., Kasturi, L., Spitalnik, S.L., and Shakin-Eshleman, S.H. (1998). The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37, 6833–6837. [DOI] [PubMed] [Google Scholar]
- Mishima, M., Takayama, S., Sasaki, K., Jee, J.G., Kojima, C., Isogai, A., and Shirakawa, M. (2003). Structure of the male determinant factor for Brassica self-incompatibility. J. Biol. Chem. 278, 36389–36395. [DOI] [PubMed] [Google Scholar]
- Nawaschin, S.G. (1898). Resultate einer revision der befruchtungsvorgaenge bei Lilium martagon und Fritillaria tenella. Bull. Acad. Imp. Sci. St. Petersburg 9, 377–382. [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.A. (2001). Endosperm Development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267. [DOI] [PubMed] [Google Scholar]
- O'Sullivan, D.M., Ripoll, P.J., Rodgers, M., and Edwards, K.J. (2001). A maize bacterial artificial chromosome (BAC) library from the European flint inbred line F2. Theor. Appl. Genet. 103, 425–432. [Google Scholar]
- Pang, S.Z., DeBoer, D.L., Wan, Y., Ye, G., Layton, J.G., Neher, M.K., Armstrong, C.L., Fry, J.E., Hinchee, M.A., and Fromm, M.E. (1996). An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol. 112, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrescu, A.J., Milac, A.L., Petrescu, S.M., Dwek, R.A., and Wormald, M.R. (2004). Statistical analysis of the protein environment of N-glycosylation sites: Implications for occupancy, structure, and folding. Glycobiology 14, 103–114. [DOI] [PubMed]
- Reik, W., and Walter, J. (2001). Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Scholten, S., Lorz, H., and Kranz, E. (2002). Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J. 32, 221–231. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Schweizer, L., Yerk-Davis, G.L., Phillips, R.L., Srienc, F., and Jones, R.J. (1995). Dynamics of maize endosperm development and DNA endoreduplication. Proc. Natl. Acad. Sci. USA 92, 7070–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341. [DOI] [PubMed] [Google Scholar]
- Serna, A., Maitz, M., O'Connell, T., Santandrea, G., Thevissen, K., Tienens, K., Hueros, G., Faleri, C., Cai, G., Lottspeich, F., and Thompson, R.D. (2001). Maize endosperm secretes a novel antifungal protein into adjacent maternal tissue. Plant J. 25, 687–698. [DOI] [PubMed] [Google Scholar]
- Sevilla-Lecoq, S., Deguerry, F., Matthysrochon, E., Perez, P., Dumas, C., and Rogowsky, P.M. (2003). Analysis of ZmAE3 upstream sequences in maize endosperm and androgenic embryos. Sex. Plant Reprod. 16, 1–8. [Google Scholar]
- Sørensen, M.B., Chaudhury, A.M., Robert, H., Bancharel, E., and Berger, F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11, 277–281. [DOI] [PubMed] [Google Scholar]
- Sørensen, M.B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jurgens, G., Somerville, C., Lepiniec, L., and Berger, F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129, 5567–5576. [DOI] [PubMed] [Google Scholar]
- Spillane, C., MacDougall, C., Stock, C., Koehler, C., Vielle-Calzada, J.P., Nunes, S.M., Grossniklaus, U., and Goodrich, J. (2000). Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10, 1535–1538. [DOI] [PubMed] [Google Scholar]
- Springer, P.S., Holding, D.R., Groover, A., Yordan, C., and Martienssen, R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127, 1815–1822. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Cammue, B.P., and Thevissen, K. (2002). Plant defensins. Planta 216, 193–202. [DOI] [PubMed] [Google Scholar]
- Thompson, R.D., Hueros, G., Becker, H., and Maitz, M. (2001). Development and functions of seed transfer cells. Plant Sci. 160, 775–783. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1986). A new method for predicting signal sequence cleavage site. Nucleic Acids Res. 14, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V., and Evans, M.M. (2003). Unique features of the plant life cycle and their consequences. Nat. Rev. Genet. 4, 369–379. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Geldner, N., Offringa, R., and Jurgens, G. (2001). Seed development: Early paternal gene activity in Arabidopsis. Nature 414, 709–710. [DOI] [PubMed] [Google Scholar]
- Wilson, I.B. (2002). Glycosylation of proteins in plants and invertebrates. Curr. Opin. Struct. Biol. 12, 569–577. [DOI] [PubMed] [Google Scholar]
- Wormald, M.R., and Dwek, R.A. (1999). Glycoproteins: Glycan presentation and protein-fold stability. Struct. Fold. Des. 7, R155–R160. [DOI] [PubMed] [Google Scholar]
- Wormald, M.R., Petrescu, A.J., Pao, Y.L., Glithero, A., Elliott, T., and Dwek, R.A. (2002). Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 102, 371–386. [DOI] [PubMed] [Google Scholar]
- Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J.J., Goldberg, R.B., Pennell, R.I., and Fischer, R.L. (2003). Imprinting of the MEA polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5, 891–901. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G., Salamini, F., Thompson, R., and Hueros, G. (1999). Novel basal endosperm transfer cell layer (betl) specific genes. In Max Planck Gesellschaft, Patent Cooperation Treaty WO9950427.
- Zhao, J., and Grafi, G. (2000). The high mobility group I/Y protein is hypophosphorylated in endoreduplicating maize endosperm cells and is involved in alleviating histone H1-mediated transcriptional repression. J. Biol. Chem. 275, 27494–27499. [DOI] [PubMed] [Google Scholar]
- Zimmerman, J.L., Apuya, N., Darwish, K., and O'Carroll, C. (1989). Novel regulation of heat shock genes during carrot somatic embryo development. Plant Cell 1, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.