Abstract
Background
The short stature homeodomain transcription factors SHOX and SHOX2 play key roles in limb formation. To gain more insight into genes regulated by Shox2 during limb development, we analyzed expression profiles of WT and Shox2−/− mouse embryonic limbs and identified the T-Box transcription factor Tbx4 as a potential downstream target. Tbx4 is known to exert essential functions in skeletal and muscular hindlimb development. In humans, haploinsufficiency of TBX4 causes small patella syndrome, a skeletal dysplasia characterized by anomalies of the knee, pelvis, and foot.
Results
Here, we demonstrate an inhibitory regulatory effect of Shox2 on Tbx4 specifically in the forelimbs. We also show that Tbx4 activates Shox2 expression in fore- and hindlimbs, suggesting Shox2 as a feedback modulator of Tbx4. Using EMSA studies, we find that Tbx4/TBX4 is able to bind to distinct T-box binding sites within the mouse and human Shox2/SHOX2 promoter.
Conclusions
Our data identifies Tbx4 as a novel transcriptional activator of Shox2 during murine fore- and hindlimb development. Tbx4 is also regulated by Shox2 specifically in the forelimb bud possibly via a feedback mechanism. These data extend our understanding of the role and regulation of Tbx4 and Shox2 in limb development and limb associated diseases.
Introduction
Vertebrate limb buds initially consist of proliferating mesenchymal cells enveloped by ectoderm that emerge from the lateral plate mesoderm (LPM). As outgrowth continues, progenitor cells from the LPM differentiate into bones, tendons, and some of the vasculature whereas muscles are formed by migrating precursors derived from adjacent somites (Pearse et al., 2007). Skeletal elements are formed by mesenchymal condensations, which differentiate into chondrocytes and are later replaced by bone (Kronenberg, 2003). The correct patterning of the developing limbs requires a coordinated network of signaling molecules interlinked by feedback loops and their targets. The proximodistal outgrowth and patterning is controlled by the apical ectodermal ridge (AER) via a positive FGF feedback loop resulting in the formation of stylopod, zeugopod, and autopod (Mariani and Martin, 2003; Zeller et al., 2009; Duboc and Logan, 2011).
Transcriptional regulatory genes orchestrate the expression of numerous target genes important for limb patterning (Mariani and Martin, 2003). Various defects in limb formation arise from mutations in these genes and genes of the Hox and T-box transcription factor families provide prominent examples. Simultaneous mutations of Hoxa11/Hoxd11 in mice, e.g., lead to a severe shortening of the ulna and radius and Hoxa10/Hoxc10/Hoxd10-deficient mice develop shortened femurs (Davis et al., 1995; Wellik and Capecchi, 2003; Boulet and Capecchi, 2004). Mutations in T-box transcription factor genes lead to various human syndromes associated with limb malformations, including small patella syndrome (TBX4) and Holt-Oram syndrome (TBX5) (Basson et al., 1997; Li et al., 1997; Bongers et al., 2004).
During limb development, Tbx4 and its paralog Tbx5 show almost exclusive expression patterns with Tbx4 mainly expressed in the hindlimb and Tbx5 expression restricted to the forelimb (Chapman et al., 1996; Gibson-Brown et al., 1996; Naiche et al., 2011). The expression in their respective limb fields suggests that Tbx4 and Tbx5 might have a role in determining limb-type identity (Duboc and Logan, 2011). Tbx5 and Tbx4 also have well-described roles in initiation and initial outgrowth of the fore- and hindlimb buds. Due to an insufficient establishment of the Fgf signaling loop between the LPM and the overlying ectoderm, mice deficient for Tbx4 or Tbx5 do not form limb buds properly. Fgf10 was shown to be a target of both transcription factors, but in contrast to Tbx5, Tbx4 is not required exclusively for Fgf10 expression (Ng et al., 2002; Agarwal et al., 2003; Naiche and Papaioannou, 2003; Rallis et al., 2003).
The short stature homeobox-containing gene SHOX and its paralog SHOX2 encode two members of paired related homeodomain transcription factors with crucial functions during embryonic development. SHOX was identified as a gene controlling human growth as mutations and deletions lead to the short stature and skeletal deformities associated with Leri-Weill dyschondrosteosis (LWD) and Langer mesomelic dysplasia (LMD) (Belin et al., 1998; Shears et al., 1998; Schiller et al., 2000; Zinn et al., 2002; Benito-Sanz et al., 2005). Moreover, SHOX defects have been identified in the non-syndromic isolated forms of short stature with a prevalence of 5–17% in geographically different populations (Chen et al., 2009; Rosilio et al., 2012). A characteristic clinical feature of LWD and LMD patients is a mesomelic shortening of the zeugopod elements (the forearms and lower legs) as well as a typical malformation of the forearms, termed Madelung deformity. The role of SHOX in the etiology of short stature suggests crucial functions in proximodistal limb formation and bone development.
A paralog of the SHOX gene, SHOX2, has an identical homeo-domain (60 amino acids) and shows an overall similarity on the amino acid level of 65% (Blaschke et al., 1998; Semina et al., 1998). Although SHOX2 has not been linked to any human phenotype so far, analysis of Shox2-deficient mouse models revealed that Shox2 also plays a key role in limb development, where it controls neural, muscular, and skeletal processes. Both conditional and conventional knockout of Shox2 lead to a dramatic shortening of the stylopod elements of the limbs due to delayed chondrocyte maturation and differentiation (Cobb et al., 2006; Yu et al., 2007). In addition, loss of Shox2 function causes altered muscular development and innervation defects in the proximal forelimbs (Vickerman et al., 2011). Different genes in limb development including Runx2/3, Ihh, and Bmp4 have been shown to be regulated by Shox2 (Cobb et al., 2006; Yu et al., 2007; Vickerman et al., 2011), while Hoxa11 and Hoxd11 act upstream of Shox2 to regulate chondrocyte differentiation (Gross et al., 2012).
To further elucidate Shox2-dependent signaling pathways during limb development, we searched for new Shox2 target genes using microarray expression profiling. Tbx4 was found to be dynamically regulated by Shox2 in the forelimb, but not in the hindlimb, which raised our interest. In addition, Tbx4 was identified as a novel transcriptional activator of Shox2 in both fore-and hindlimbs, strongly suggesting that Shox2 acts as a feedback modulator of Tbx4 during limb development.
Results
Tbx4 Expression Is Increased in the Developing Forelimbs of Shox2-Deficient Mice
To identify Shox2-regulated genes during limb development, we used our previously generated Shox2 knockout mouse model (Blaschke et al., 2007), depicting a severe shortening of mutant (Shox2−/−) fore- and hindlimbs (Fig. 1) consistent with other Shox2 knockout mouse models (Cobb et al., 2006; Yu et al., 2007). To uncover novel Shox2 transcriptional target genes in limbs, gene expression was compared in wildtype (WT) and Shox2 mutants, first using pooled fore- and hindlimb tissue at stage E12.5 and subsequently forelimb tissue at stage E11.5 by microarray analysis. Among differentially regulated putative candidate genes (see Suppl. Table S1), the well-known transcription factor Tbx4 was upregulated in the limbs of Shox2−/− embryos at both developmental stages, E11.5 and E12.5. As Tbx4 has essential functions in skeletal and muscular development of the hindlimbs, similar to Shox2, we chose it for further analysis.
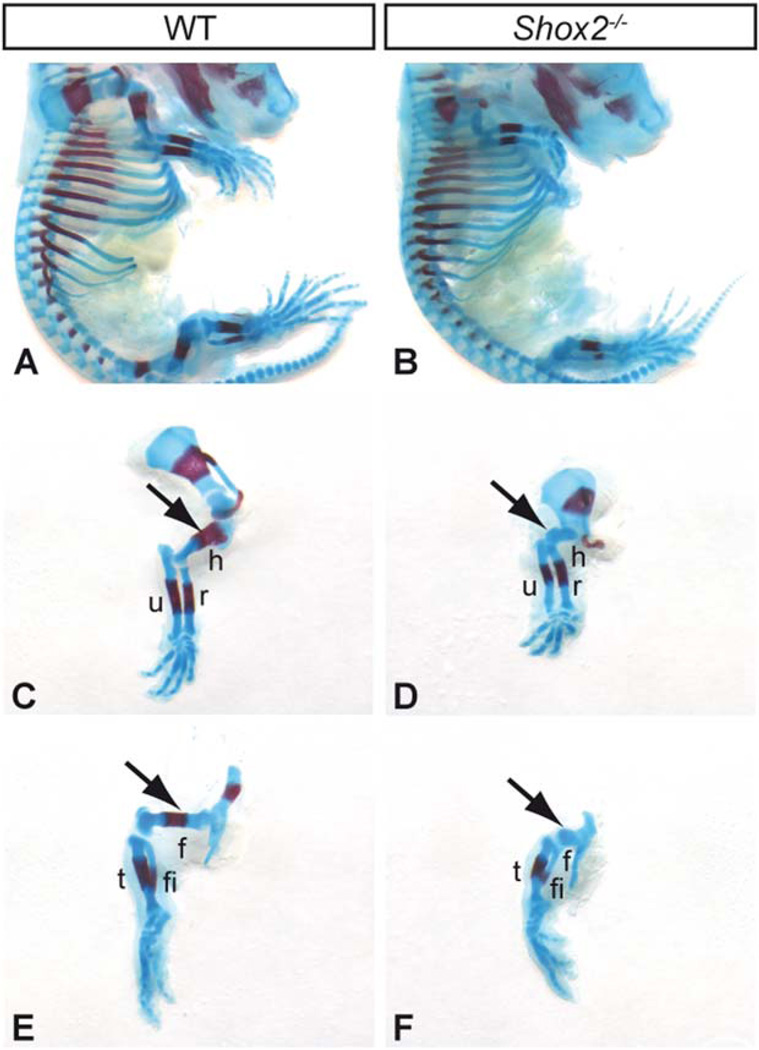
Fig. 1.
Shox2 deficiency results in shortened limbs. Alizarin red (bone, red) - Alcian blue (cartilage, blue) staining on E16.5 WT (A) and Shox2 mutant (B) embryos revealed a severe shortening of Shox2 mutant fore-(D) and hindlimbs (F) compared to the wildtype (C, E). The proximal skeletal elements humerus and femur are particularly affected and show no ossification (arrows). The more distal parts of the Shox2−/− limbs (radius, ulna, tibia, and fibula) are only slightly shortened compared to the wildtype. h, humerus; r, radius; f, femur; fi, fibula; t, tibia; u, ulna.
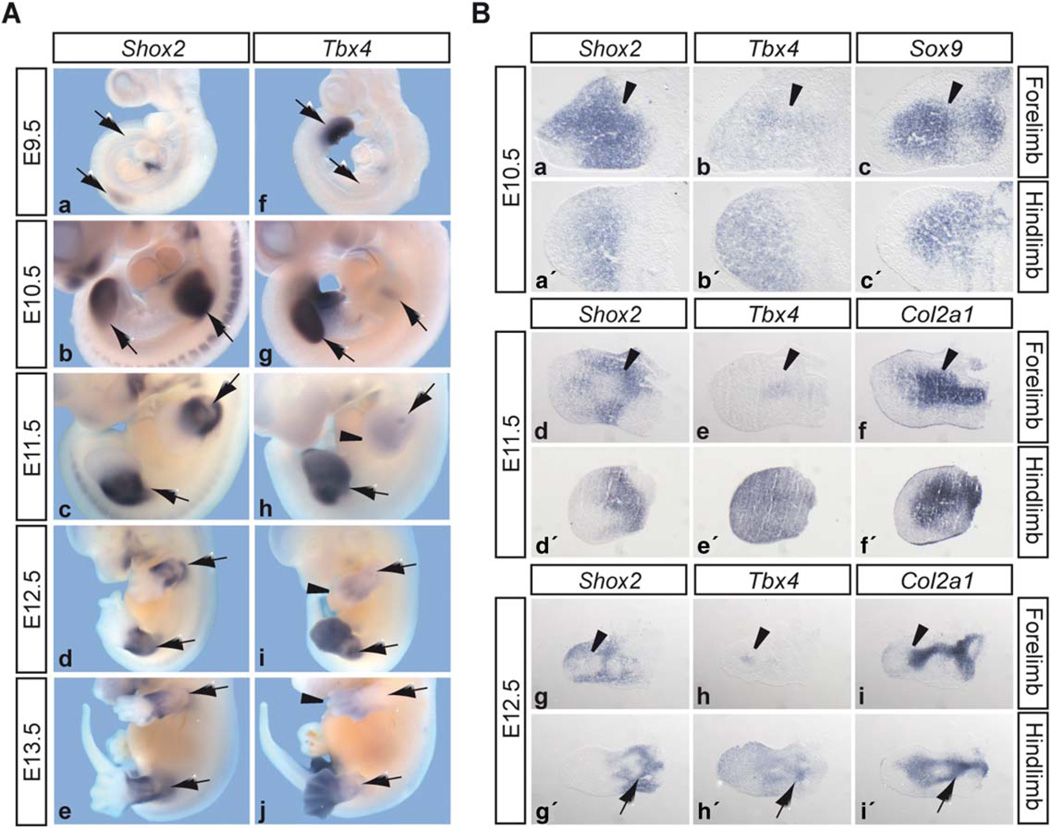
As a first step in validating Tbx4 as a putative target gene of Shox2, we compared the expression of both genes in mouse embryos of different developmental stages by in situ hybridization (Fig. 2). Shox2 expression starts at the onset of limb outgrowth at E9.5 in the forelimb (Fig. 2A a). During later stages, Shox2 is broadly expressed in fore- and hindlimbs, at E10.5 throughout the whole limb buds and from E11.5 onwards restricted to the proximal part of the limbs (Fig. 2A b–e). As shown previously (Naiche et al., 2011), Tbx4 is only transiently expressed in the forelimb, starting at E10.5 in a discrete localized region of the proximal limb bud and decreasing at E11.5 (Fig. 2A g, h). Then, beginning at E11.5, Tbx4 is expressed more diffusely in the distal part of the forelimb and proceeds distally between E12.5 and E13.5 (Fig. 2A h–j). In the hindlimb field, strong Tbx4 expression can be observed before hindlimb outgrowth, at E9.5, in the lateral plate mesoderm (LPM) (Fig. 2A f). Expression continues from E10.5 until E12.5 in the entire hindlimb bud, becoming more distally restricted at E13.5 (Fig. 2A g–j). To analyze Shox2 and Tbx4 expression in more detail, we performed in situ hybridization on adjacent limb sections from different developmental stages (Fig. 2B). The distinct proximal Tbx4 expression domain of the E10.5 forelimb is consistent with that of Sox9, a marker for condensing mesenchyme, and overlaps with the broader Shox2 expression domain (Fig. 2B a–c, arrowheads). From E11.5 onwards, the specific Tbx4-expressing region corresponds to a Col2a1-positive cartilaginous element of the dorsal forelimb (Fig. 2B d–i, arrowheads) and the expression patterns of Shox2 and Tbx4 are mutually exclusive. In the E10.5 and E11.5 hindlimb, Shox2 and Tbx4 expression broadly overlaps throughout the proximal limb mesenchyme (Fig. 2B a′–f′) and at E12.5 both are expressed in areas surrounding the proximal cartilaginous elements (Fig. 2B g′–i′, arrows).
Fig. 2.
Shox2 and Tbx4 expression during murine limb development. A: Whole mount in situ hybridization on mouse embryos of different developmental stages (limbs or limb developing regions are indicated by arrows). Shox2 expression starts at E9.5 in the forelimb buds (a), is expressed throughout fore- and hindlimb buds at E10.5 (b), and from E11.5–E13.5 restricted to the proximal part of the limbs (c–e). Tbx4 hindlimb expression starts at E9.5 in the lateral plate mesoderm (f), continues throughout E10.5–E12.5 (g–i), and restricts distally at E13.5 (j). In the forelimbs, distinct Tbx4 staining is visible at E10.5-E12.5 (g–i). In addition, a diffuse Tbx4 expression can be seen at E11.5 (h, arrowhead), which again restricts distally from E12.5 onward (i, j, arrowheads). B: In situ hybridization on 12 µm adjacent limb sections of different developmental stages. Dorsal sections of the forelimb (a–i) and medial sections of the hindlimbs (a′-i′) are presented. The Tbx4 expression domain of the forelimb and the corresponding Shox2 expression are indicated at E10.5 (a, b), E11.5 (d, e), and E12.5 (g, h) by arrowheads. Sox9 and Col2a1 were used as markers for condensing mesenchyme, and cartilaginous skeletal elements, respectively. Hindlimb expression of Shox2 (g′) and Tbx4 (h′) surrounding the cartilaginous elements at E12.5 are indicated by arrows.
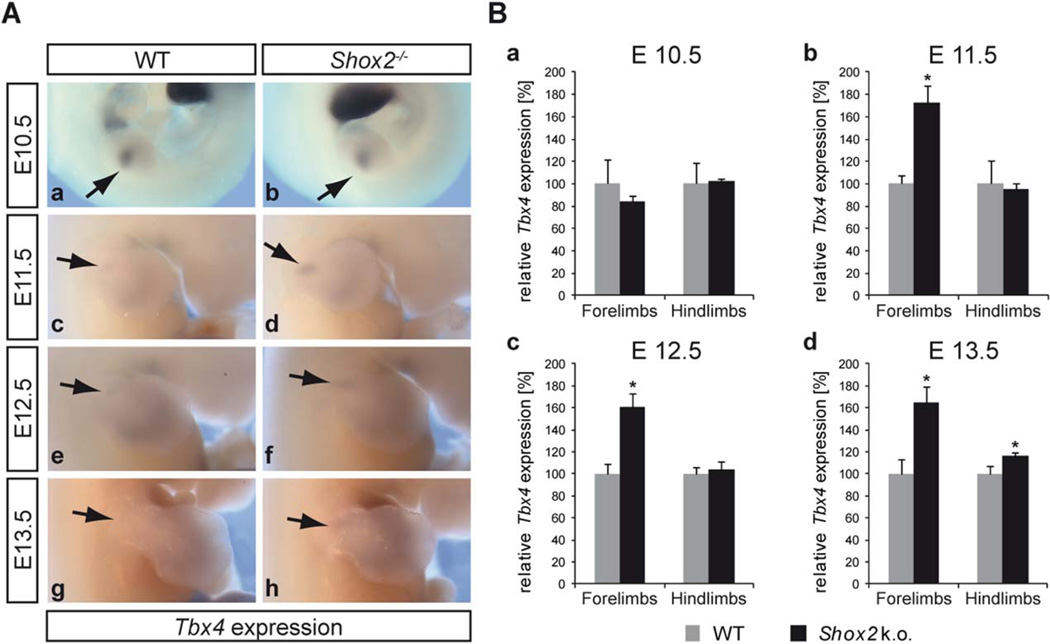
To test whether Tbx4 expression depends on Shox2 in the developing limbs, we carried out whole mount in situ hybridization (WISH) on WT and Shox2−/− embryos at 4 different developmental stages (Fig. 3A). These studies revealed that at E11.5, E12.5, and E13.5, Tbx4 is upregulated in the specific dorsal Tbx4-expressing region of the mutant forelimb (Fig. 3A d, f, h). In all stages, hindlimb expression seems unaffected (data not shown). To quantify these findings, qRT-PCR was carried out using reverse transcribed RNA from separately dissected E10.5–E13.5 fore- and hindlimb tissue (Fig. 3B). As Tbx4 is expressed specifically in the proximal region of the forelimb, only this part of the limb was dissected and used for the experiments. In accordance with the WISH results, no increase in Tbx4 expression was detected at E10.5, but a significant increase in Tbx4 expression in mutant forelimbs could be detected at E11.5, E12.5, and E13.5 (Fig. 3B b–d). Hindlimb expression was again largely unaffected at E10.5 to E12.5 and only slightly increased at E13.5 in Shox2−/− hindlimbs (Fig. 3B a–d). Thus, our analyses show that Shox2 has an inhibitory effect on Tbx4 expression in the forelimbs during various developmental stages.
Fig. 3.
Tbx4 is upregulated in Shox2−/− forelimbs. A: Whole mount in situ hybridisation on WT and Shox2−/− embryos using a Tbx4 RNA probe shows no altered expression of Tbx4 in E10.5 Shox2−/− forelimbs (b), but an upregulation of Tbx4 in E11.5 (d), E12.5 (f), and E13.5 (h) forelimbs compared to the WT (a, c, e, g), indicated by arrows; n = 3 independent stainings for every stage, each performed with 2–3 littermates per genotype. B: Quantification by qRT-PCR using WT and Shox2−/− limb tissue. Shox2 deficiency results in a significant increase of Tbx4 mRNA (∼60– 70%) in E11.5 (b), E12.5 (c), and E13.5 (d) forelimbs. Hindlimb expression is largely unaffected at E10.5 to E12.5 (a–c) and only slightly elevated at E13.5 (d). Significance is indicated by asterisks: *P ≤ 0.05; n = 3 independent experiments for every stage, each performed with 2–7 littermates per genotype; grey bars indicate relative Tbx4 mRNA levels in WT, black bars relative Tbx4 mRNA levels in Shox2−/−.
Tbx4 Regulates Shox2 Expression in Developing Fore-and Hindlimbs
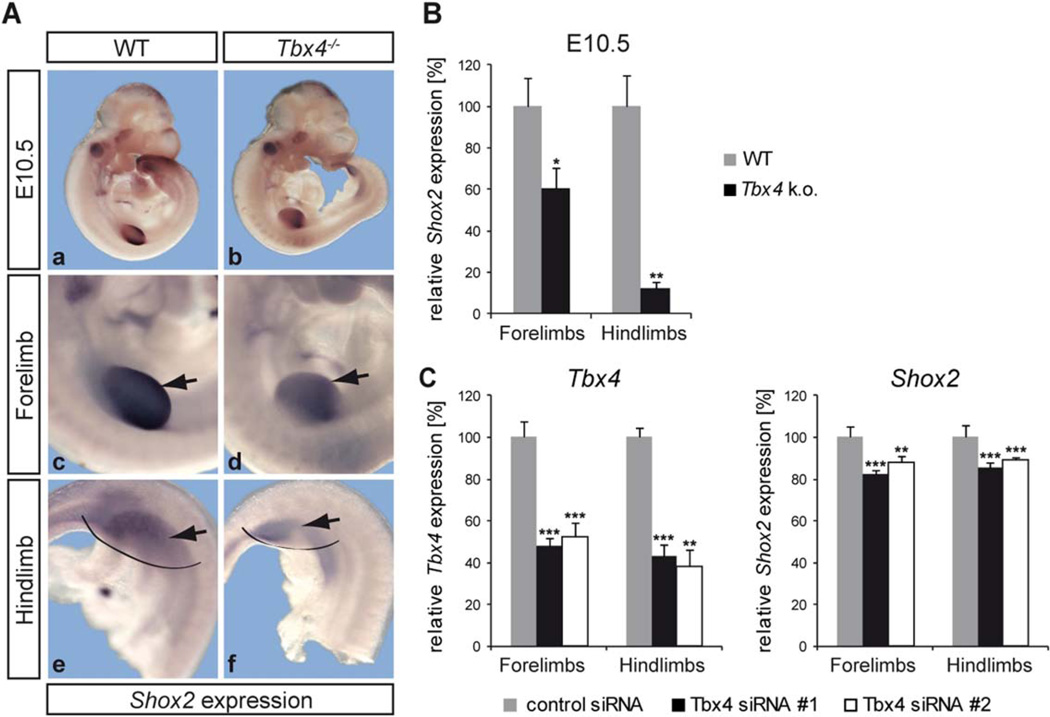
Considering that Tbx4 is expressed earlier than Shox2 in the limbs and that the regulation of Tbx4 by Shox2 primarily affects the forelimb, we asked whether Tbx4 may also act as an upstream regulator of Shox2, which in turn signals back via a feedback mechanism. To investigate this possibility, we compared Shox2 expression at E10.5 (when Tbx4 expression starts in the forelimb) in WT and Tbx4−/− embryos. We can demonstrate that Shox2 expression is reduced in both fore- and hindlimbs of Tbx4−/− embryos and that there is a stronger effect in the hindlimbs (Fig. 4A d, f). Quantification by qRT-PCR using separately dissected fore-and hindlimbs of E10.5 Tbx4-deficient mice revealed a significant reduction of Shox2 expression in the fore- and hindlimb (Fig. 4B).
Fig. 4.
Shox2 is downregulated in Tbx4−/− fore- and hindlimbs. A: Whole mount in situ hybridization on E10.5 WT and Tbx4−/− embryos using a Shox2 RNA probe shows a decreased Shox2 expression in both fore- (d) and hindlimbs (f) of Tbx4−/− embryos (indicated by arrows). N = 7; limbs in a, b are magnified in c–f. B: Results were confirmed by qRT-PCR using WT and Tbx4−/− limb tissue. Tbx4 knockout leads to a ∼40% decrease of Shox2 mRNA in forelimbs and a ∼85% decrease in hindlimbs; *P <0.05, **P <0.01; n = 3 independent experiments performed with 2–3 pooled embryos of 2 litters; grey bars indicate relative Shox2 mRNA levels in WT, black bars relative Shox2 levels in Tbx4−/−C:Tbx4 knockdown in primary fore- and hindlimb cells. Transfection with 2 different Tbx4 siRNAs (#1, #2), in parallel with a control siRNA, results in a ∼50–60% reduction of Tbx4 mRNA levels after 24 hr (left), leading to a ∼10–20% downregulation of Shox2 (right) in fore- and hindlimb cells; **P ≤ 0.01, ***P ≤ 0.001; n = 10 for siRNA #1, n = 7 for siRNA #2; grey bars show relative Tbx4/Shox2 mRNA levels after control siRNA transfection; black and white bars relative Tbx4/Shox2 mRNA levels after Tbx4 siRNA transfection.
Tbx4 null mice initiate but do not continue hindlimb outgrowth and die at E10.5 due to chorioallantoic fusion defects (Naiche and Papaioannou, 2003). To address whether reduced Shox2 expression may be caused by apoptosis, we carried out TUNEL analysis on WT and Tbx4−/− embryos at stage E10.5 and revealed that the apoptotic effect is minor (3.2% increase of apoptotic cells in the mutant forelimb and 9% in the mutant hindlimb; data not shown). In addition, siRNA-mediated knockdown experiments in primary embryonic fore- and hindlimb cells isolated from WT embryos showed that Shox2 is significantly down-regulated in fore- and hindlimb cells independent of apoptosis or aberrant limb formation (Fig. 4C). A nonspecific effect of the control siRNA on Tbx4 or Shox2 expression could be excluded (data not shown). Together, these data strongly support that Tbx4 acts as a transcriptional activator of Shox2.
Tbx4 Binds the Shox2 Promoter
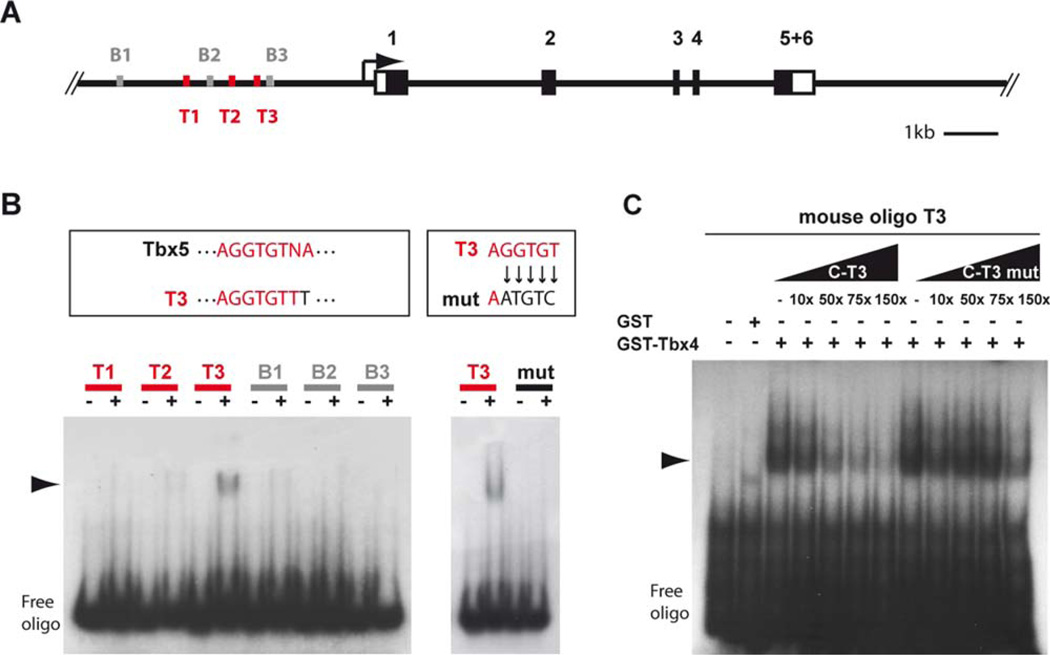
To investigate the transcriptional regulation of Shox2 by Tbx4 on the promoter level, electrophoretic mobility shift assays (EMSA) were carried out. A ∼4 kb genomic upstream region of the murine Shox2 transcriptional start site was examined for T-box binding sites by the MatInspector software tool (Genomatix Software GmbH) (Quandt et al., 1995; Cartharius et al., 2005). Specific Tbx4 binding sites are not known so far, but 3 binding sites for Brachyury, another T-box transcription factor, could be detected within a region of ∼3 kb. In addition, we identified 3 different sequences similar to the known TBX5-binding site (A/G)GGTGT(C/T/G)(A/G) (Ghosh et al., 2001) (Fig. 5A). As Tbx4 and Tbx5 are the most closely related of all known T-box proteins with an overall amino acid sequence homology of 52% and almost identical T-domains with 95% identity, we speculated that both proteins may share similar binding specificities, comparable to findings where Tbx4 was shown to be able to bind to the TBX5 binding site in the human ANF promoter (Arora et al., 2012). To determine if Tbx4 is able to interact directly with T-box binding sites in the murine Shox2 promoter, EMSAs were performed using 6 different oligonucleotides containing the 3 Tbx5-like (T1–T3) and 3 Brachyury (B1–B3) binding sites (Fig. 5B). We show a specific binding of purified GST-tagged Tbx4 protein to the Tbx5-like oligo T3 but not to T1 or T2. There is also no binding to the Brachyury oligos B1–B3. To define the precise binding site, several nucleotides of the Tbx5 core binding sequence were mutated, which abolishes Tbx4 binding (Fig. 5B). In addition, competition assays show that excess of unlabelled Tbx5-like 3 (C-T3) decreases binding capability of GST-Tbx4 to labelled oligo Tbx5-like 3 (T3), whereas excess of mutated Tbx5-like 3 (C-T3 mut) has no effect (Fig. 5C).
Fig. 5.
2Tbx4 binds the murine Shox2 promoter. A: Schematic illustration of the murine Shox2 gene (black boxes represent coding exons, white boxes UTRs) harbouring 3 binding sites similar to Tbx5 sites (T1–T3, red) and 3 Brachyury binding sites (B1–B3, grey) upstream of the transcriptional start site. B: EMSA shows specific binding of GST-Tbx4 fusion protein to an oligonucleotide containing T3 but not to T1, T2, and B1–B3 containing oligos. T3 differs from the known Tbx5 site in 1 nucleotide. Mutation of 5 nucleotides within the Tbx5 related core sequence of T3 inhibits binding of GST-Tbx4. Shift is marked by an arrowhead. C: Competition EMSA shows that excess (10-, 50-, 75-, and 150-fold molar) of unlabelled T3 (C-T3) decreases binding of GST-Tbx4 to labelled T3, whereas excess of mutated T3 (C-T3 mut) has no effect. Shift is marked by an arrowhead.
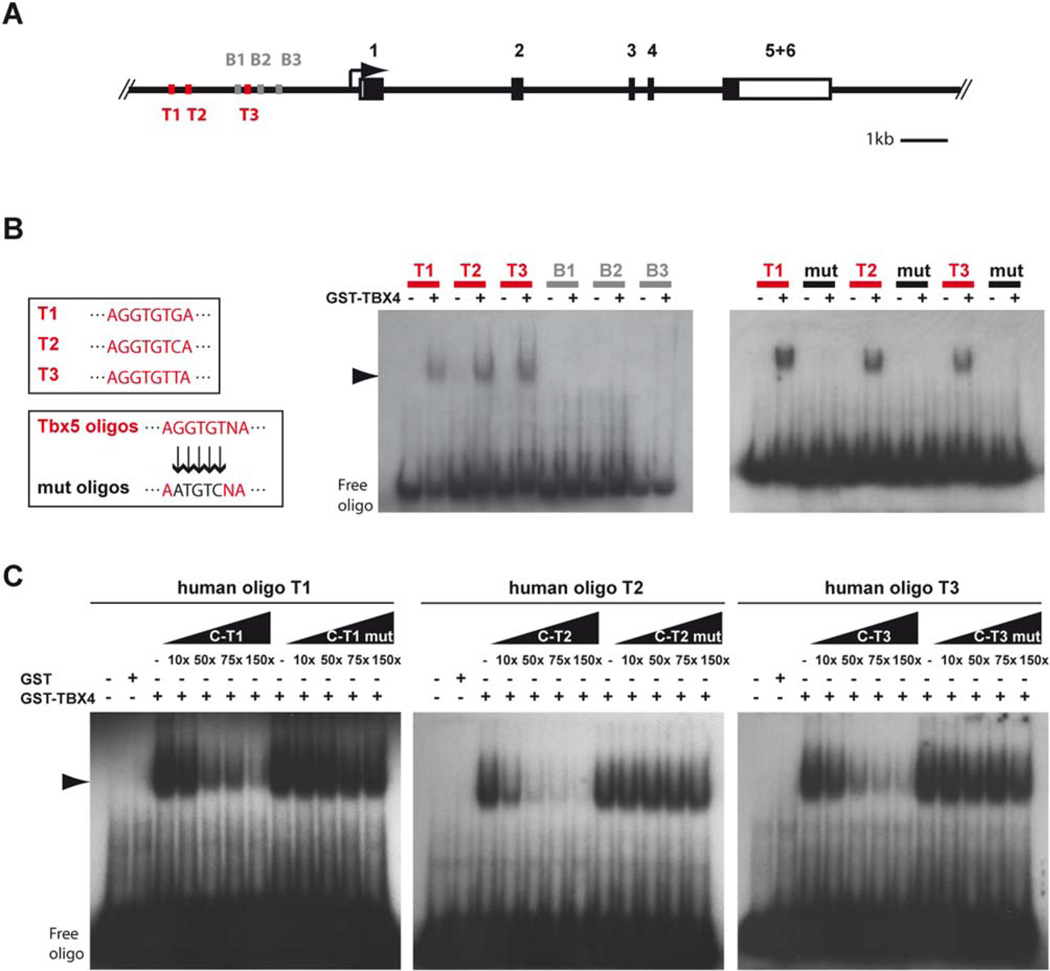
Mutations in the TBX4 and TBX5 gene cause developmental syndromes associated with limb deformities in humans, known as small patella (TBX4) and Holt-Oram (TBX5) syndrome (Basson et al., 1997; Li et al., 1997; Bongers et al., 2004; McDermott et al., 2004). We investigated whether binding capacities of Tbx4 on the Shox2 promoter also apply to the human system. In addition to 3 BRACHYURY binding sites (B1–B3), the human SHOX2 promoter contains 3 sites matching known TBX5 binding elements (T1–T3, Fig. 6A). Human GST-tagged TBX4 protein interacts with all the 3 TBX5 binding sites (Fig. 6B) and binding is reduced upon addition of increasing amounts of unlabelled TBX5 binding site containing oligos (C-T1–3, Fig 6C). Consistent with the mouse data, there is no binding to the 3 BRACHYURY binding sites (Fig. 6B). Thus, TBX4/Tbx4 is able to bind the SHOX2/Shox2 promoter in both human and mouse.
Fig. 6.
TBX4 binds the human SH0×2 promoter. A: Schematic illustration of the human SHOX2 gene (black boxes represent coding exons, white boxes UTRs) harbouring 3 TBX5 (T1–T3, red) and 3 BRACHYURY (B1-B3, grey) binding sites upstream of the transcriptional start site. B: EMSA shows specific binding of GST-TBX4 fusion protein to oligonucleotides containing T1–T3 but not to B1–B3 binding sites. Mutation of 5 nucleotides within the TBX5 core sequence of T1, T2, and T3 inhibits the binding of GST-TBX4. Shift is marked by an arrowhead. C: Competition EMSA shows that excess (10-, 50-, 75-, and 150-fold molar) of unlabelled T1 (C-T1), T2 (C-T2), and T3 (C-T3) decreases binding of GST-TBX4 to labelled T1, T2, and T3, whereas excess of mutated T1 (C-T1 mut), T2 (C-T2 mut), and T3 (C-T3 mut) has no effect. Shift is marked by an arrowhead.
Discussion
To extend our knowledge of Shox2 functions during limb development, we searched for novel genes regulated by Shox2. We identified Tbx4 as a particularly attractive candidate that has essential functions in skeletal and muscular development of the hindlimbs, similar to Shox2. Making use of two different knockout mouse models, we validated a regulatory link of Tbx4 and Shox2 in developing limbs. Shox2 has a negative regulatory effect on Tbx4 specifically in proximal forelimbs at developmental stages E11.5 to E13.5, but not in hindlimbs. This regulation can be matched to a distinct Tbx4 expression domain within the cartilage of the dorsal forelimb where Shox2 expression is normally excluded. Our data also revealed that Tbx4 activates Shox2 in both fore- and hindlimbs, implicating Shox2 as a possible feedback modulator of Tbx4 in the forelimb. The loss of the distinct proximal Tbx4 expression in the forelimb results in a general downregulation of Shox2 within the entire forelimb bud. To explain this observation, one could speculate that localized Tbx4 expression in the WT forelimb induces upregulation of Shox2 in nearby cells via a diffusible signaling molecule. In the absence of Tbx4, these cells fail to upregulate Shox2 and stay in a more primitive state, which appears as a downregulation of Shox2. Considering equal expression of Shox2 in fore- and hindlimbs, the observed forelimb restricted regulation of Tbx4 by Shox2 was surprising. Interestingly, in a different study, SHOX, the highly related paralog of SHOX2, has been shown to completely rescue the Shox2−/− phenotype only in the forelimb, while the hindlimb defect persisted (Liu et al., 2011). A forelimb-specific corepressor of Shox2 acting in a temporal manner could explain this inhibitory effect on Tbx4 expression from E11.5 onwards in the forelimb but not in the hindlimb. Together, these data strongly suggest that the genetic environment of limb-type-specific cofactors is important to mediate differential Shox2 effects in fore-and hindlimbs.
One interesting aspect is the biological significance of the Shox2 regulatory effect on Tbx4 in the defined proximal forelimb domain. Lineage tracing experiments revealed that Tbx4- expressing cells of this specific domain contribute mostly to the tendons around the elbow and to the periphery of the bone [Naiche et al., 2011). Tbx4-deficient mice had no apparent defects in gross forelimb morphology, yet the development of Tbx4 mutant forelimb tendons was never examined in detail (Naiche and Papaioannou, 2007). The distinct expression of Tbx4 in the forelimbs of mouse embryos was not found in Zebrafish pectoral fin buds or Xenopus forelimbs and only very low levels were seen during a single stage (stage 29) in the chicken wing (Gibson-Brown et al., 1998; Logan et al., 1998; Ruvinsky et al., 2000; Takabatake et al., 2000). These species-dependent expression differences could be the consequence of an enhancer element, which was shown previously to drive Tbx4 forelimb expression in the mouse and is known to be poorly conserved in the other species (Menke et al., 2008). The regulation of Tbx4 by Shox2 may therefore contribute to the development of distinct tendons and bone elements particularly in the developing mammalian forelimb.
We have provided evidence that Tbx4 regulates Shox2. Tbx4 is already strongly expressed in the lateral plate mesoderm at E9.5 prior to hindlimb bud outgrowth, when Shox2 expression begins. We show that Tbx4 is an activator of Shox2 in both limb types, which is in contrast to the forelimb-specific inhibition of Tbx4 by Shox2. In addition to its early function in limb outgrowth, Tbx4 is also crucial for the development of hindlimb skeletal elements as well as muscle and tendons during a second later phase of limb formation (Naiche and Papaioannou, 2003, 2007; Hasson et al., 2010). The deletion of Tbx4 in mouse shortly after hindlimb initiation leads to abnormal pelvises and severely hypoplastic femurs. This phenotype resembles the drastically shortened humerus and femur as well as the mildly abnormal pelvic girdle caused by Shox2 deficiency (Cobb et al., 2006; Yu et al., 2007) (Fig. 1). In addition to the skeletal malformations, the conditional loss of Tbx4 results in disturbed limb muscle patterning, size, and orientation (Hasson et al., 2010). Interestingly, altered muscle patterning with reorientated and abnormal muscle bundles was also reported in Shox2-deficient mice (Vickerman et al., 2011), suggesting that Tbx4 and Shox2 share critical functions during limb development. Compared to the Tbx4 mutant phenotype, however, in the Shox2 mutant fewer muscles are affected and defects are restricted to the proximal part of the limbs. Thus, the loss of either gene, Tbx4 or Shox2, leads to a phenotype in skeletal and muscular elements.
The gene pair Tbx4 and Tbx5 originated from a common ancestral gene by tandem duplication and both genes have almost identical T-box domains (Agulnik et al., 1996). Overlapping binding and regulative properties are therefore conceivable. We have shown that Tbx4 is able to bind to a sequence motif very similar to known Tbx5 binding motifs within the murine Shox2 promoter. A specific interaction of TBX4 with three TBX5 binding sites in the human promoter region has also been demonstrated, suggesting that the regulation of SHOX2 by TBX4 also plays a role in human limb development and limb-associated diseases. Tbx4 is capable of compensating for a loss of Tbx5 function in the forelimbs (Minguillon et al., 2005) and both proteins transactivate the same reporter containing T-box binding elements via a shared activator domain (Ouimette et al., 2010). Considering this and the fact that Tbx5 acts as an upstream regulator of Shox2 during heart development (Puskaric et al., 2010), one can hypothesize that Tbx5 is the Tbx4 corresponding regulator of Shox2 in the forelimb controlling muscle and skeleton development. Simultaneously, Tbx4 activates Shox2, probably in a feedback loop, contributing to forelimb tendon and bone formation.
During limb development, only a small number of genes are differentially expressed in either fore- or hindlimbs including specific members of the T-box family (Gibson-Brown et al., 1996). The vast majority of genes playing a role in limb formation including Shox2 are equally expressed in both limb structures (Blaschke et al., 1998; Semina et al., 1998; Zeller et al., 2009). Our study demonstrates that genes expressed at similar abundance in both limb types (e.g. Shox2) can be activated by genes distinctly expressed in fore- and hindlimbs (e.g. Tbx4). In turn, a gene expressed in both fore- and hindlimb can have a regulatory effect on a target gene specifically expressed in only one limb type.
Experimental Procedures
Mice and Tissue Collection
Mice used were Shox2−/− (Blaschke et al., 2007) and Tbx4−/− (Naiche and Papaioannou, 2003). For maximum litter size, we crossed the Shox2−/− mice into the CD-1 outbred strain. Breeding and genotyping was performed as previously described (Naiche and Papaioannou, 2003; Blaschke et al., 2007) and detection of the mating plug was considered 0.5 days post conception (E0.5). For E12.5 microarray analysis, isolated fore- and hindlimbs were combined; for E11.5 microarray analysis and quantitative RT-PCR (qRT-PCR), they were analyzed separately. Limb buds from littermates of the same genotype were pooled and total RNA was purified using TRIzol® (Invitrogen, Carlsbad, CA) extraction. For in situ hybridization, isolated embryos or limbs were fixed in 4% paraformaldehyde at 4° C overnight, dehydrated in methanol and stored at −20° C (whole mount in situ hybridization), or incubated in 30% sucrose at 4° C overnight and embedded in tissue freezing medium (Jung) (section in situ hybridization).
Histological Stainings
Alcian Blue/Alizarin Red stainings of E16.5 skeletal preparations were performed according to standard protocols (Nagy et al., 2003). An Alcian Blue solution containing 150 mg Alcian Blue 8 GX (Sigma, St. Louis, MO)/l in 95% Ethanol, 20% acetic acid, and an Alizarin Red solution containing 50 mg Alizarin Red (Sigma)/l and 10 g KOH/l was used.
Microarray Hybridization and Analysis
Gene expression profiling was carried out using oligonucleotide arrays of the MoGene 2.0 ST (E11.5)- and Mouse Genome 430 2.0 (E12.5)-type from Affymetrix (Santa Clara, CA) according to the manufacturer’s protocol. For E11.5, forelimb RNA from 3–4 embryos of 2 different litters was used for hybridization to 2 arrays per genotype (WT and Shox2−/−). For E12.5, combined fore- and hindlimb RNA from 3–4 embryos of 2 different litters was pooled and used for hybridization to 1 array per genotype. RNA quality was confirmed by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and 200 ng was used to generate biotinylated ssDNA followed by hybridization to arrays. Statistical comparisons of WT and Shox2−/− chip data were performed using the software package JMP Genomics, version 4.0 from SAS (SAS Institute, Cary, NC). Values of perfect-matches were log transformed, quantile normalised, and fitted with log-linear mixed models, with probe ID and genotype considered to be constant. A custom CDF version 11 (E12.5) and version 17 [E11.5) with UniGene-based gene definitions was used to annotate the arrays. The microarray data were deposited in the NCBI GEO database with accession number GSE51523 (for E11.5 arrays) and GSE41945 (for E12.5 arrays).
DNA Constructs
To generate RNA antisense probes for in situ hybridization, 607 bp (Tbx4, accession number NM_011536.2), 654 bp (Col2a1, accession number NM_031163.3), and 637 bp (Sox9, accession number NM_011448.4) of the mRNA sequence were amplified by PCR using the primers Tbx4 ISH for2/rev2, Col2a1 ISH for/rev, Sox9 ISH for/rev (Table 1) and mouse E12.5 hindlimb cDNA. The PCR products were then subcloned into the pSTBlue1 vector (Novagen, Madison, WI). The plasmid pCR-S-Og12 for generation of the Shox2 mRNA antisense probe was described previously (Blaschke et al., 1998). GST-Tbx4/GST-TBX4 expression vectors for EMSA were generated by amplifying the mouse/human Tbx4/TBX4 coding sequence (accession number NM_018488.2/NM_011536.2) using cDNA from E12.5 primary mouse hindlimb cells and normal human dermal fibroblasts (NHDF), respectively. Subsequently, Tbx4/TBX4 was subcloned via BamHI/HindIII (mouse) and HindIII/XhoI (human) into pET-41a(+) vector (Novagen). All primer sequences used for cloning are listed in Table 1.
TABLE 1.
Oligonucleotides
| Name | Sequence (5’-3’) | Application | |
|---|---|---|---|
| mHprt1 qRT for | TCCTCCTCAGACCGCTTTT | qRT-PCR | |
| mHprt1 qRT rev | CCTGGTTCATCATCGCTAATC | ||
| mSdha qRT for | CATGCCAGGGAAGATTACAAA | ||
| mSdha qRT rev | GTTCCCCAAACGGCTTCT | ||
| mShox2 qRT for | ACCAATTTTACCCTGGAACAAC | ||
| mShox2 qRT rev | TCGATTTTGAAACCAAACCTG | ||
| mTbx4 qRT for | GCATGAGAAGGAGCTGTGG | ||
| mTbx4 qRT rev | TTACCTTGTAGCTGGGGAACA | ||
| mTbx4 ISH for2 | CCGACGAGAACAATGCTTTT | ISH probe cloning | |
| mTbx4 ISH rev2 | GCCCCGAACTCGAGTACATA | ||
| mCol2a1 ISH for | GCCAAGACCTGAAACTCTGC | ||
| mCol2a1 ISH rev | TTCGCAATGGATTGTGTTGT | ||
| mSox9 ISH for | CTGAAGGGCTACGACTGGAC | ||
| mSox9 ISH rev | CATTGACGTCGAAGGTCTCA | ||
| mTbx4 pET41a+ BamHI for | CGCGGGATCCATGCTGCAGGATAAGGGCCTGTC | pET41a+ - mTbx4 cloning | |
| mTbx4 pET41a+ HindIII rev | CGCGAAGCTTGTCCATCGGTCCAGTTCTCCA | ||
| hTBX4 pET41a+ HindIII for | TGGAGAACTGGACTGACGGACAGTCGACCGCG | pET41a+ - hTBX4 cloning | |
| hTBX4 pET41a+ Xhol rev | TGGAGAACTGGACTGACGGACTCGAGCGCG | ||
| mTbx51ike_EMSA_for1 | T1 | GGGAAATTGGTTAGTTCCTGTTGAGCGAGGTGGAAATTCGCCCAACAATAATAAATCATGGGA | |
| mTbx51ike_EMSA_rev1 | GGGTCCCATGATTTATTATTGTTGGGCGAATTTCCACCTCGCTCAACAGGAACTAACCAATTT | ||
| mTbx51ike_EMSA_for2 | T2 | GGGGGGATATAAGAGGCTAACAGAAGAGGTGTATGTTTCCATATGTGAAAAGTTCATAAAGAA | |
| mTbx51ike_EMSA_rev2 | GGGTTCTTTATGAACTTTTCACATATGGAAACATACACCTCTTCTGTTAGCCTCTTATATCCC | ||
| mTbx51ike_EMSA_for3 | T3 | GGGACCAGGCTTCTAGCAGGGGCTATTTGGGAGGTGTTTACGAATATGTATTTCGTCATAGAG | |
| mTbx51ike_EMSA_rev3 | GGGCTCTATGACGAAATACATATTCGTAAACACCTCCCAAATAGCCCCTGCTAGAAGCCTGGT | ||
| Mut_mTbx51ike_EMSA_for3 | T3-mut | GGGACCAGGCTTCTAGCAGGGGCTATTTGGGAATGTCTTTACGAATATGTATTTCGTCATAGAG | |
| Mut_mTbx51ike_EMSA_rev3 | GGGCTCTATGACGAAATACATATTCGTAAAGACATTCCCAAATAGCCCCTGCTAGAAGCCTGGT | ||
| mBrachy_EMSA_for1 | B1 | GGGTACGTAGCTACTTCACTGGGAGCTTTTACATCTAAGTTCAAAGATGACAACTCGAATACC | |
| mBrachy_EMSA_rev1 | GGGGGTATTCGAGTTGTCATCTTTGAACTTAGATGTAAAAGCTCCCAGTGAAGTAGCTACGTA | ||
| mBrachy_EMSA_for2 | B2 | GGGCTAGATACTAAGGTTCAGATTTCCACAGAGATTTGAGATGGAACTCACTTCCAGTATAGG | |
| mBrachy_EMSA_rev2 | GGGCCTATACTGGAAGTGAGTTCCATCTCAAATCTCTGTGGAAATCTGAACCTTAGTATCTAG | ||
| mBrachy_EMSA_for3 | B3 | GGGAAAGCAAGAGTTCATGTACTTCAATAATACCTAAATGTTTAATGCAATCGCTGTTAACCA | |
| mBrachy_EMSA_rev3 | GGGTGGTTAACAGCGATTGCATTAAACATTTAGGTATTATTGAAGTACATGAACTCTTGCTTT | ||
| TBX5_EMSA_for1 | T1 | GGGGCATCACCTGGGTCCAGACCTTTGGGCTCACACCTCCCGTTGACGTGCAAGCGCCGCAGTCC | |
| TBX5_EMSA_rev1 | GGGGGACTGCGGCGCTTGCACGTCAACGGGAGGTGTGAGCCCAAAGGTCTGGACCCAGGTGATGC | ||
| TBX5_EMSA_for2 | T2 | GGGAATCTGCATCGTAGGAAGTTTTGTTTATGACACCTACCCGAAACAAAATTGGTTATCTCACG | |
| TBX5_EMSA_rev2 | GGGCGTGAGATAACCAATTTTGTTTCGGGTAGGTGTCATAAACAAAACTTCCTACGATGCAGATT | ||
| TBX5_EMSA_for3 | T3 | GGGTTTTTGAGTTATTTGTGAATATGTATTTAACACCTCAGTTTGAGAGCACCTTGAGCACCGTA | |
| TBX5_EMSA_rev3 | GGGTACGGTGCTCAAGGTGCTCTCAAACTGAGGTGTTAAATACATATTCACAAATAACTCAAAAA | ||
| Mut_TBX5_EMSA_for1 | T1-mut | GGGGCATCACCTGGGTCCAGACCTTTGGGCTCGACATTCCCGTTGACGTGCAAGCGCCGCAGTCC | |
| Mut_TBX5_EMSA_rev1 | GGGGGACTGCGGCGCTTGCACGTCAACGGGAATGTCGAGCCCAAAGGTCTGGACCCAGGTGATGC | ||
| Mut_TBX5_EMSA_for2 | T2-mut | GGGAATCTGCATCGTAGGAAGTTTTGTTTATGGACATTACCCGAAACAAAATTGGTTATCTCACG | |
| Mut_TBX5_EMSA_rev2 | GGGCGTGAGATAACCAATTTTGTTTCGGGTAATGTCCATAAACAAAACTTCCTACGATGCAGATT | ||
| Mut_TBX5_EMSA_for3 | T3-mut | GGGTTTTTGAGTTATTTGTGAATATGTATTTAGACATTCAGTTTGAGAGCACCTTGAGCACCGTA | |
| Mut_TBX5_EMSA_rev3 | GGGTACGGTGCTCAAGGTGCTCTCAAACTGAATGTCTAAATACATATTCACAAATAACTCAAAAA | ||
| BRACHY_EMSA_for1 | B1 | GGGAAGCTCAGTAGATGAAATATGACTTCAAACCTAAAAGTCATGTACTTAAAAAGGCAAACAA | |
| BRACHY_EMSA_rev1 | GGGTTGTTTGCCTTTTTAAGTACATGACTTTTAGGTTTGAAGTCATATTTCATCTACTGAGCTT | ||
| BRACHY_EMSA_for2 | B2 | GGGAAAGCAAGTACTGATAACAATGTATTCATACCTAAAAGCTATAGCTGCGGAAAAATTATAGA | |
| BRACHY_EMSA_rev2 | GGGTCTATAATTTTTCCGCAGCTATAGCTTTTAGGTATGAATACATTGTTATCAGTACTTGCTTT | ||
| BRACHY_EMSA_for3 | B3 | GGGGCCACACTTAGAGGTATATTTGTTTACACACTTAAATATAAC ATCATATGTAAATATTCTTT | |
| BRACHY_EMSA_rev3 | GGGAAAGAATATTTACATATGATGTTATATTTAAGTGTGTAAACAAATATACCTCTAAGTGTGGC | ||
In Situ Hybridization
Riboprobe generation and whole mount in situ hybridization on mouse embryos were performed as reported (Harland, 1991). Digoxigenin-labeled antisense (as) and sense (s) RNA was synthesized from the plasmids pCR-S-Og12 (Shox2, linearized by SacI(as)/XhoI (s) and transcribed using T7 (as)/T3 (s) polymerase), pSTBlue1-Tbx4 (linearized by MluI (as)/HindIII (s) and transcribed using SP6 (as)/T7 (s) polymerase), pSTBlue1-Col2a1 (linearized by KpnI (as)/HindIII (s) and transcribed using SP6 (as)/T7 (s) polymerase) and pSTBlue1-Sox9 (linearized by HindIII (as)/KpnI (s) and transcribed using T7 (as)/SP6 (s) polymerase). Section in situ hybridization on adjacent 12 µm cryosections was performed as described previously (Decker et al., 2011).
cDNA Synthesis and Quantitative RT-PCR
1 µg of total RNA was transcribed into cDNA by SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen). qRT-PCR was performed using the Applied Biosystems 7500 Real-Time PCR System and SYBR Green ROX dye (Thermo Scientific, Waltham, MA). Each of the samples was analyzed in duplicate and relative mRNA levels were assessed according to the delta-delta Ct method (Pfaffl, 2001) by normalization to succinate dehydrogenase complex subunit A (Sdha) and hypoxanthine phosphoribosyltransferase 1 (Hprt1). All qRT-PCR primer sequences are listed in Table 1.
Apoptosis Detection
Apoptotic cells in the limb region of E10.5 WT and Tbx4−/− embryos were visualized by TUNEL staining using an in situ cell death detection kit (TMR red, Roche, Indianapolis, IN) according to the manufacturer’s instructions. TUNEL staining was performed on transverse 10 µm cryosections of E10.5 embryos and sections were counterstained using Hoechst (Invitrogen); 3–8 different fore- and hindlimb sections per embryo from in total 4 embryos per genotype (WT and Tbx4−/−, n=4) were used. Apoptotic cells and Hoechst-stained cells were counted using the analyze particles tool from ImageJ and the number of apoptotic cells was normalized to the whole cell number.
Limb Cell Culture and Transfection
Limb buds (fore- and hindlimbs separately) were dissected at E12.5, dissociated mechanically (using forceps) and subsequently enzymatically (using 0.5% Trypsin-EDTA from Gibco, Gaithersburg, MD), and plated on cell culture dishes coated with 0.1% gelatine in PBS. Cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium; Gibco) containing 10% FBS (fetal bovine serum gold; PAA), 1% penicillin/streptomycin (Gibco), 1% L-glutamine (Gibco), and 1% NEAA (non-essential amino acids; Gibco) at 37°C, 5% CO2 and 95% humidity. For knockdown experiments, cells were seeded in 6-well plates and transfected with 40 pmol/well of Tbx4 silencer select siRNA (Invitrogen) #1 (ID S18204), siRNA #2 (ID 74781), or control siRNA by using RNAiMax (Invitrogen). Cells were harvested 24 hr after transfection for qRT-PCR.
Electrophoretic Mobility Shift Assay (EMSA)
To identify T-box binding sites within the Shox2/SHOX2 promotor, a ∼4 kb genomic upstream region of the transcriptional start site was examined using the MatInspector software tool (Genomatix Software GmbH) (Quandt et al., 1995; Cartharius et al., 2005). The murine Shox2 promoter region contained 3 Brachyury binding sites with the core sequence T(G/C)ACACCT/AGGTGTGAAATT (Kispert and Hermann, 1993; Ghosh et al., 2001) (Brachyury 1 [−4,856 bp/−4,841 bp], Brachyury 2 [−3,158 bp/−3,143 bp], Brachyury 3 [−2,089 bp/−2,074 bp]) and additionally we identified 3 different sequences similar to known Tbx5 binding sites with the core sequence (A/G)GGTGT(C/T/G)(A/G) (Ghosh et al., 2001) (Tbx5-like 1 [−3,623 bp/−3,616 bp], Tbx5-like 2 [−2,707 bp/−2,700 bp] and Tbx5-like 3 [−2,341 bp/−2,348 bp]). The human SHOX2 promoter region contained 3 BRACHYURY (BRACHYURY 1 [−2,598 bp/−2,583 bp], BRACHYURY 2 [−2,227 bp/−2,211 bp], BRACHYURY 3 [−1,745 bp, −1,730 bp]) and 3 TBX5 (TBX5 1 [−4,054 bp/−4,049 bp], TBX5 2 [−3,690bp/−3,685 bp] and TBX5 3 [−2,482 bp/−2,477 bp]) binding sites.
EMSA was performed as reported previously (Schneider et al. 2005). For the binding reaction, 32P-labelled, 60-bp double stranded mouse Shox2 oligonucleotides (Oligo T1 [−3,647 bp/−3,576 bp]; Oligo T2 [−2,730 bp/−2,671 bp]; T3 [−2,376 bp/−2,317 bp]; B1 [−4,879 bp/−4,820 bp]; B2 [−3,180 bp/−3,121 bp; B3 [−2,111 bp/2,052 bp]; Table 1) and human SHOX2 oligonucleotides (Oligo T1 [−4,084 bp/−4,023 bp]; Oligo T2 [−3,719 bp/−3,658 bp]; Oligo T3 [−2,511 bp/−2,450 bp]; Oligo B1 [−2,620 bp/−2,560 bp]; Oligo B2 [−2,249 bp/−2,188 bp]; Oligo B3 [−2,511 bp/−2,450 bp]; Table 1), were used together with purified, bacterially expressed recombinant GST-Tbx4/GST-TBX4 protein.
Statistical Analysis
Results of qRT-PCR are presented as mean ± SEM from at least 3 independent experiments. Experimental group comparisons were performed using the Student’s t-test and differences were considered significant if P < 0.05.
Supplementary Material
Acknowledgments
We thank Maria Muciek and Carsten Sticht for help with the microarray analysis and Roland Knopf for animal care.
Grant sponsor: Deutsche Forschungsgemeinschaft; Grant number: RA 380/12-1.
Footnotes
Additional Supporting Information may be found in the online version of this article.
The authors declare there was no conflict of interest.
References
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Agulnik SI, Garvey N, Hancock S, Ruvinsky I, Chapman DL, Agulnik I, Bollag R, Papaioannou V, Silver LM. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–254. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Del Alcazar CM, Morrisey EE, Naiche LA, Papaioannou VE. Candidate gene approach identifies multiple genes and signaling pathways downstream of tbx4 in the developing allantois. PLoS One. 2012;7:e43581. doi: 10.1371/journal.pone.0043581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Belin V, Cusin V, Viot G, Girlich D, Toutain A, Moncla A, Vekemans M, Le Merrer M, Munnich A, Cormier-Daire V. SHOX mutations in dyschondrosteosis (Leri-Weill syndrome) Nat Genet. 1998;19:67–69. doi: 10.1038/ng0198-67. [DOI] [PubMed] [Google Scholar]
- Benito-Sanz S, Thomas NS, Huber C, Gorbenko del Blanco D, Aza-Carmona M, Crolla JA, Maloney V, Rappold G, Argente J, Campos-Barros A, Cormier-Daire V, Heath KE. A novel class of Pseudoautosomal region 1 deletions downstream of SHOX is associated with Leri-Weill dyschondrosteosis. Am J Hum Genet. 2005;77:533–544. doi: 10.1086/449313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Monaghan AP, Schiller S, Schechinger B, Rao E, Padilla-Nash H, Ried T, Rappold GA. SHOT, a SHOX-related homeobox gene, is implicated in craniofacial, brain, heart, and limb development. Proc Natl Acad Sci USA. 1998;95:2406–2411. doi: 10.1073/pnas.95.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Scholer H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Bongers EM, Duijf PH, van Beersum SE, Schoots J, Van Kampen A, Burckhardt A, Hamel BC, Losan F, Hoefsloot LH, Yntema HG, Knoers NV, van Bokhoven H. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development. 2004;131:299–309. doi: 10.1242/dev.00936. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Freeh K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. Matlnspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chen J, Wildhardt G, Zhong Z, Roth R, Weiss B, Steinberger D, Decker J, Blum WF, Rappold G. Enhancer deletions of the SHOX gene as a frequent cause of short stature: the essential role of a 250 kb downstream regulatory domain. J Med Genet. 2009;46:834–839. doi: 10.1136/jmg.2009.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J, Dierich A, Huss-Garcia Y, Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc Natl Acad Sci USA. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Decker E, Durand C, Bender S, Rodelsperger C, Glaser A, Hecht J, Schneider KU, Rappold G. FGFR3 is a target of the homeobox transcription factor SHOX in limb development. Hum Mol Genet. 2011;20:1524–1535. doi: 10.1093/hmg/ddr030. [DOI] [PubMed] [Google Scholar]
- Duboc V, Logan MP. Regulation of limb bud initiation and limb-type morphology. Dev Dyn. 2011;240:1017–1027. doi: 10.1002/dvdy.22582. [DOI] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10:1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, Papaioannou VE. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev. 1996;56:93–101. doi: 10.1016/0925-4773(96)00514-x. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–2509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- Gross S, Krause Y, Wuelling M, Vortkamp A. Hoxa11 and hoxd11 regulate chondrocyte differentiation upstream of runx2 and shox2 in mice. PLoS One. 2012;7:e43553. doi: 10.1371/journal.pone.0043553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, Mohun TJ, Logan MP. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev Cell. 2010;18:148–156. doi: 10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Hermann BG. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993;12:4898–4899. doi: 10.1002/j.1460-2075.1993.tb06179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson Dl, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen CH, Espinoza-Lewis RA, Jiao Z, Sheu I, Hu X, Lin M, Zhang Y, Chen Y. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J Biol Chem. 2011;286:17029–17038. doi: 10.1074/jbc.M111.234252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Simon HG, Tabin C. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development. 1998;125:2825–2835. doi: 10.1242/dev.125.15.2825. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Martin GR. Deciphering skeletal patterning: clues from the limb. Nature. 2003;423:319–325. doi: 10.1038/nature01655. [DOI] [PubMed] [Google Scholar]
- McDermott DA, Fong JC, Basson CT. Holt-Oram Syndrome. Seattle: Gene Reviews; 2004. [Google Scholar]
- Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- Minguillon C, Del Buono J, Logan MP. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell. 2005;8:75–84. doi: 10.1016/j.devcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo. Cold Spring Harbor, NY: Cold Spring Habor Laboratory Press; 2003. [Google Scholar]
- Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allan-tois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Arora R, Kania A, Lewandoski M, Papaioannou VE. Identity and fate of Tbx4-expressing cells reveal developmental cell fate decisions in the allantois, limb, and external genitalia. Dev Dyn. 2011;240:2290–2300. doi: 10.1002/dvdy.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JK, Kawakami Y, Buscher D, Raya A, Itoh T, Koth CM, Rodriguez Esteban C, Rodriguez-Leon J, Garrity DM, Fishman MC, Izpisua Belmonte JC. The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development. 2002;129:5161–5170. doi: 10.1242/dev.129.22.5161. [DOI] [PubMed] [Google Scholar]
- Ouimette JF, John ML, L’Honore A, Gifuni A, Drouin J. Divergent transcriptional activities determine limb identity. Nat Commun. 2010;1:35. doi: 10.1038/ncomms1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse RV, 2nd, Scherz PJ, Campbell JK, Tabin CJ. A cellular lineage analysis of the chick limb bud. Dev Biol. 2007;310:388–400. doi: 10.1016/j.ydbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, Rappold G. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum Mol Genet. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. Tbx5 is required for fore-limb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Rosilio M, Huber-Lequesne C, Sapin H, Carel JC, Blum WF, Cormier-Daire V. Genotypes and phenotypes of children with SHOX deficiency in France. J Clin Endocrinol Metab. 2012;97:E1257–E1265. doi: 10.1210/jc.2011-3460. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Oates AC, Silver LM, Ho RK. The evolution of paired appendages in vertebrates: T-box genes in the zebrafish. Dev Genes Evol. 2000;210:82–91. doi: 10.1007/s004270050014. [DOI] [PubMed] [Google Scholar]
- Schiller S, Spranger S, Schechinger B, Fukami M, Merker S, Drop SL, Troger J, Knoblauch H, Kunze J, Seidel J, Rappold GA. Phenotypic variation and genetic heterogeneity in Leri-Weill syndrome. Eur J Hum Genet. 2000;8:54–62. doi: 10.1038/sj.ejhg.5200402. [DOI] [PubMed] [Google Scholar]
- Schneider KU, Marchini A, Sabherwal N, Roth R, Niesler B, Marttila T, Blaschke RJ, Lawson M, Dumic M, Rappold G. Alteration of DNA binding, dimerization, and nuclear translocation of SHOX homeodomain mutations identified in idiopathic short stature and Leri-Weill dyschondrosteosis. Hum Mutat. 2005;26:44–52. doi: 10.1002/humu.20187. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter RS, Murray JC. A new human homeobox gene OGI2X is a member of the most conserved homeobox gene family and is expressed during heart development in mouse. Hum Mol Genet. 1998;7:415–422. doi: 10.1093/hmg/7.3.415. [DOI] [PubMed] [Google Scholar]
- Shears DJ, Vassal HJ, Goodman FR, Palmer RW, Reardon W, Superti-Furga A, Scambler PJ, Winter RM. Mutation and deletion of the pseudoautosomal gene SHOX cause Leri-Weill dyschondrosteosis. Nat Genet. 1998;19:70–73. doi: 10.1038/ng0198-70. [DOI] [PubMed] [Google Scholar]
- Takabatake Y, Takabatake T, Takeshima K. Conserved and divergent expression of T-box genes Tbx2-Tbx5 in Xenopus. Mech Dev. 2000;91:433–437. doi: 10.1016/s0925-4773(99)00329-9. [DOI] [PubMed] [Google Scholar]
- Vickerman L, Neufeld S, Cobb J. Shox2 function couples neural, muscular and skeletal development in the proximal fore-limb. Dev Biol. 2011;350:323–336. doi: 10.1016/j.ydbio.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu H, Yan M, Yang J, Long F, Muneoka K, Chen Y. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev Biol. 2007;306:549–559. doi: 10.1016/j.ydbio.2007.03.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Wei F, Zhang L, Elder FF, Scott CI, Jr, Marttila P, Ross JL. Complete SHOX deficiency causes Langer mesomelic dysplasia. Am J Med Genet. 2002;110:158–163. doi: 10.1002/ajmg.10422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.