Abstract
The RAS protooncogene plays a central role in regulation of cell proliferation, and point mutations leading to oncogenic activation of Ras occur in a large number of human cancers. Silencing of tumor suppressor genes by DNA methyltransferase 1 (Dnmt1) is essential for oncogenic cellular transformation by Ras, and Dnmt1 is over-expressed in numerous human cancers. Here we provide new evidence that the pleiotropic Regulator of G protein Signaling (RGS) family member RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated degradation of Dmnt1 and promoting apoptosis. Employing mouse embryonic fibroblasts (MEFs) from wild type (WT) and RGS6−/− mice, we found that oncogenic Ras induced up-regulation of RGS6, which in turn blocked Ras-induced cellular transformation. RGS6 functions to suppress cellular transformation in response to oncogenic Ras by down regulating Dnmt1 protein expression leading to inhibition of Dnmt1-mediated anti-apoptotic activity. Further experiments showed that RGS6 functions as a scaffolding protein for both Dnmt1 and Tip60 and is required for Tip60-mediated acetylation of Dnmt1 and subsequent Dnmt1 ubiquitylation and degradation. The RGS domain of RGS6, known only for its GAP activity toward Gα subunits, was sufficient to mediate Tip60 association with RGS6. This work demonstrates a novel signaling action for RGS6 in negative regulation of oncogene-induced transformation and provides new insights into our understanding of the mechanisms underlying Ras-induced oncogenic transformation and regulation of Dnmt1 expression. Importantly, these findings identify RGS6 as an essential cellular defender against oncogenic stress and a potential therapeutic target for developing new cancer treatments.
Keywords: RGS protein, Ras, cellular transformation, Dnmt1, Tip60, RGS domain, apoptosis
Introduction
RGS6 is in the RGS protein family, the members of which function as GTPase activating proteins (GAPs) for Gα subunits to negatively regulate heterotrimeric G protein signaling(1-3). Although aberrant G protein activation has been linked to the initiation and progression of various cancers(4), the role of RGS proteins in oncogenesis remains unexplored. A link between RGS6 and cancer was first provided when a RGS6 single nucleotide polymorphism (SNP) leading to increased RGS6 translation was found to be associated with a significant reduction in bladder cancer risk in humans, especially smokers(5). Based upon this finding, we undertook studies to examine the role of RGS6 in carcinogenesis as a potential novel tumor suppressor. We found that RGS6 dramatically inhibited growth and induced apoptosis in breast cancer cells, and that RGS6 down regulation correlated with increasing breast tumor grade in human patient samples(6). We further discovered that RGS6 is required for the ability of the chemotherapeutic agent doxorubicin to activate the ATM-p53-apoptosis pathway in MEFs and cancer cells(7). Importantly, these actions of RGS6 were independent of its ability to interact with or inactivate G proteins, identifying a novel signaling activity for a member of the RGS protein family.
Here we show that RGS6 is induced by oncogenic Ras and blocks Ras-induced cellular transformation by a novel mechanism involving Dnmt1. This work was inspired by our previous finding that RGS6 forms complexes with Dnmt1 indirectly by binding to DMAP1 (Dnmt1-associated protein)(8). However, the physiological significance of the RGS6-Dnmt1 association remained unknown. Canonically, Dnmt1 functions to maintain genomic DNA-methylation patterns in proliferating cells(9). It also methylates CpG islands in promoter regions, an important mechanism for silencing gene expression(10). Increasing evidence suggests that Dnmt1-dependent, DNA methylation-mediated silencing of tumor suppressor genes is essential for tumor development and progression, as well as cellular transformation induced by oncogenes, such as Ras(11-17). Although increased Dnmt1 expression has been observed in a variety of cancers and occurs in tumors harboring Ras mutations(18-24), the mechanism underlying over-expression of Dnmt1 in cancers remains unknown. Here, we identify an essential role for RGS6 in modulating Dnmt1 protein levels by scaffolding Dnmt1 and Tip60 and promoting Tip60-dependent Dnmt1 acetylation, leading to Dnmt1 ubiquitylation and degradation. This study provides new insights into our understanding of the mechanism underlying Ras-induced transformation and identifies novel signaling actions for an RGS protein family member.
Results and Discussion
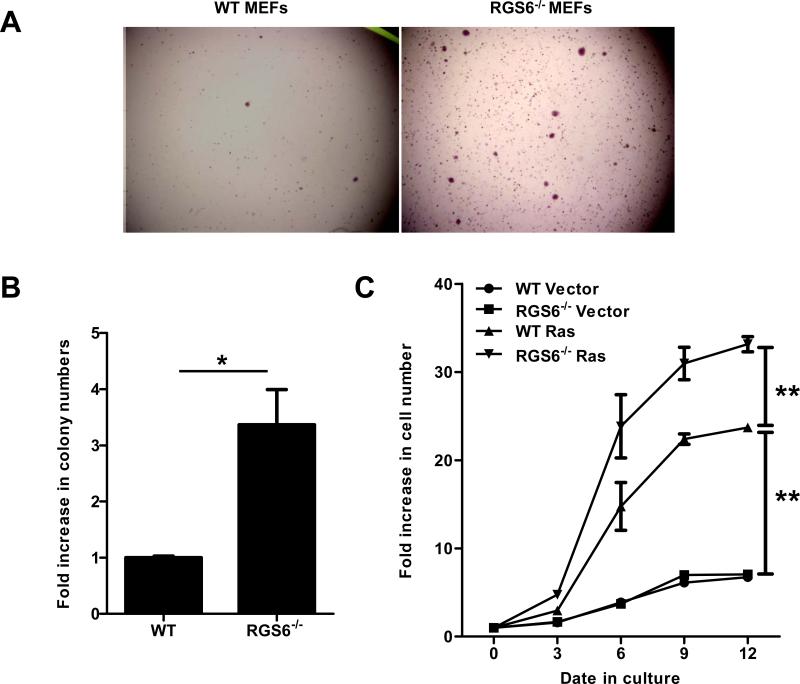
To determine the role of RGS6 in Ras-induced oncogenic transformation, we compared Ras-induced colony formation in soft agar (anchorage-independent growth) by WT and RGS6−/− MEFs. To prevent p53-dependent, irreversible cellular senescence(25) and ensure significant Ras transformation efficiency, MEFs were co-infected with two viruses expressing oncogenic HRas(G12V) and dominant negative p53(R175H). H-Ras(G12V)-induced colony formation was greatly enhanced in RGS6−/− vs. WT MEFs (Figs. 1A,1B,S1A), demonstrating that RGS6 inhibits Ras-induced oncogenic transformation. In addition, Ras-induced cell proliferation was significantly enhanced in RGS6−/− MEFs (Fig. 1C). Subsequent studies showed that HRas(G12V) induced a robust up-regulation of RGS6 protein levels in MEFs (Fig. 2A). Thus, RGS6 is induced by Ras and functions as a key negative regulator of Ras-induced cellular transformation and proliferation. These findings provide the first evidence linking a RGS protein family member to oncogene-induced transformation. Recently we reported that RGS6 induction by doxorubicin is required for activation of the ATM-p53-apoptosis pathway(7) and that RGS6 promotes apoptosis by p53-independent mechanisms in cancer cells(6). The ability of RGS6 to inhibit Ras-induced transformation was p53-independent as it occurred in cells expressing dominant negative p53. Together our results suggest that RGS6, like p53, is induced both by genotoxic and oncogenic stimuli and, likely via its pro-apoptotic actions, represents a critical cellular defender against oncogene-induced cellular transformation and subsequent tumorigenesis.
Fig. 1.
RGS6 blocks Ras-induced oncogenic cellular transformation. H-Ras(G12V)-induced colonies formed in soft agar by WT or RGS6−/− MEFs were quantified. Representative images are shown in (A) and numbers of colonies per well from three independent experiments are quantified and shown in (B). *, p<0.005 (student's t-test). All statistical analyses were performed with GraphPad Prism Software. Numbers of colonies formed by WT MEFs are set as 1. Growth rates of WT or RGS6−/− MEFs expressing either vector or H-Ras(G12V) were measured and shown in (C). Two-way ANOVA revealed a significant effect of genotype (F3,40 = 618.37; p<0.0001) and time (F4,40 = 561.27; p<0.0001). **, p<0.0001 (bonferroni multiple comparisons). Control viruses and viruses expressing H-Ras(G12V) or p53(R175H) were generated by transfection of 293T cells with empty pLWZ-Hygro or pBabe-Puro vector, or pLWZ-p53(R175H) or pBabe-H-Ras(G12V), together with packaging plasmids pAdv and Ψ2 via a standard Ca2+ phosphate transfection protocol with viruses harvested 24 h post-transfection. WT or RGS6−/− MEFs were incubated with virus-containing medium for 48 hrs and subjected to selection with 1.5 μg/μl Puromycin and 75 μg/μl Hygromycin for 72 hrs. After selection, MEFs were subjected to a soft agar assay according to a standard protocol, or were plated in triplicate in 6-well culture plates (1x104 cells/well) and cell numbers determined at 3, 6, 9 and 12 days after plating. Data are presented as mean ± S.E.M.
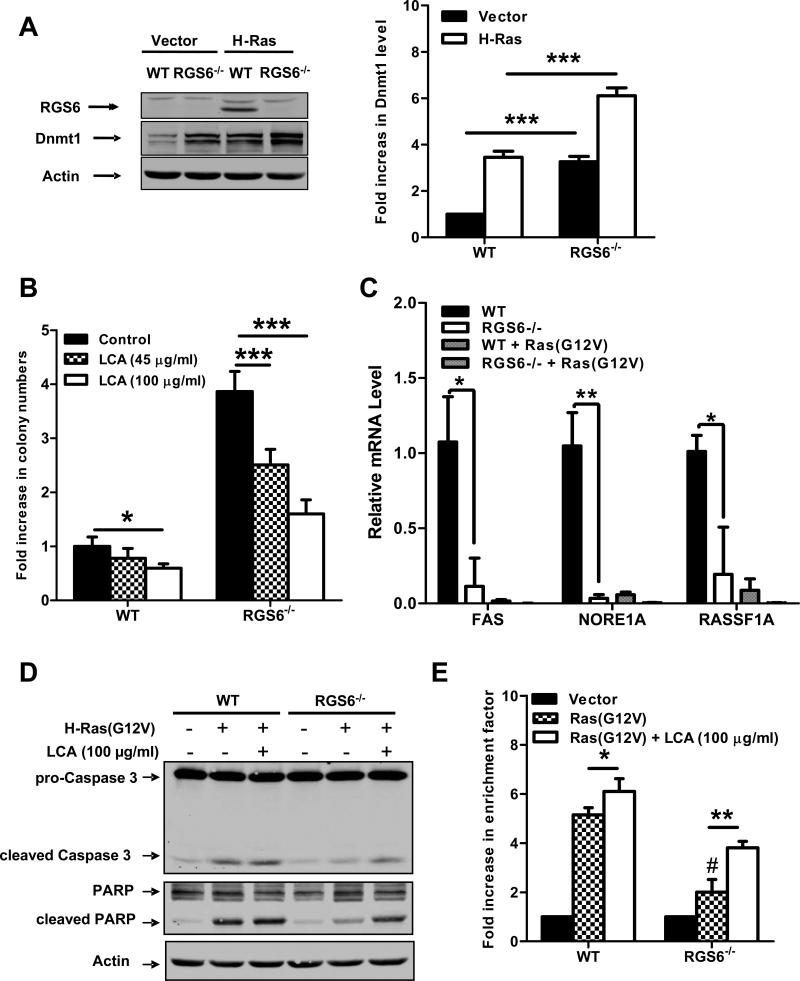
Fig. 2.
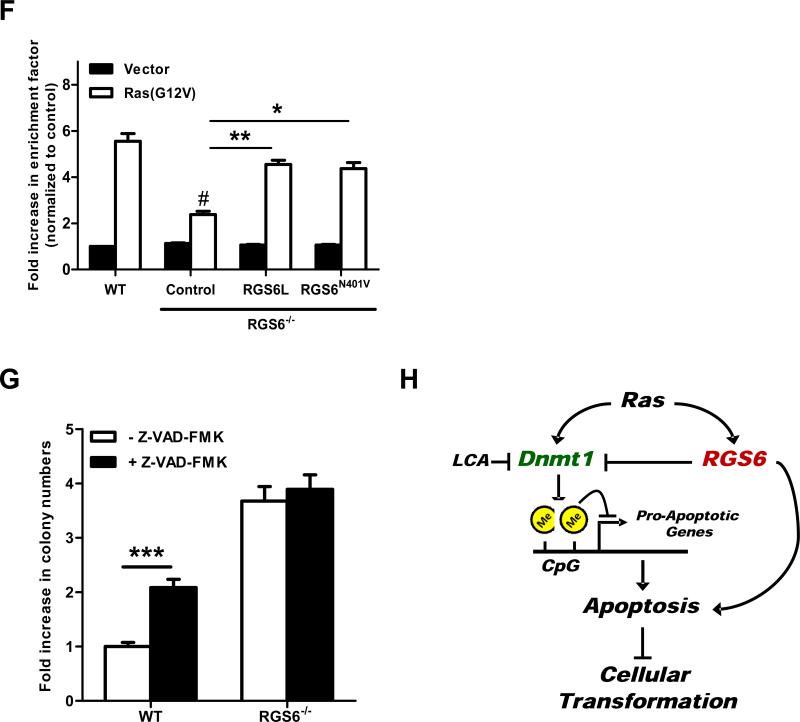
RGS6 is induced by Ras and blocks Ras-induced transformation by down-regulating Dnmt1 and suppressing Dnmt1-mediated apoptosis. (A) Representative immunoblots (left panel) of RGS6 and Dnmt1 and summary data (right panel) of Dnmt1 levels in MEFs expressing control vectors or H-Ras(G12V) /p53(R175H). Two-way ANOVA revealed a significant effect of genotype (F1,8 = 354.05; p<0.0001) and H-Ras infection (F3,40 = 304.21; p<0.0001) on Dnmt1 expression. H-Ras expressing MEFs were prepared as described in Fig 1. MEFs were grown in normal culture medium for 2 weeks after antibiotics selection. Cell lysates were then prepared and subjected to immunoblotting as previously described(7). Antibodies used were: RGS6 (prepared in the lab(7), 1:5000); Dnmt1 (Santa Cruz, 1:1000); Actin (Sigma, 1:2000). (B) LCA blocks enhancement of Ras-induced colony formation by RGS6−/− MEFs. There was a significant effect of genotype (F1,40 = 521.02; p<0.0001) and drug treatment (F2,40 = 89.92; p<0.0001). Ras-induced colony formation in soft agar by WT or RGS6−/− MEFs was performed in the presence and absence of the indicated concentrations of LCA as described in Fig. 1. (C) Loss of RGS6 promotes silencing of pro-apoptotic Dnmt1 target genes FAS, NORE1A, and RASSF1A. For each gene there was a significant effect (p<0.05) of genotype and transfection condition by two-way ANOVA. H-Ras(G12V) was transiently transfected into MEFs using Lipofectamine 2000 as described(7). mRNA from control and Ras(G12V) transfected MEFs was prepared using a Qiagen RNeasy Mini kit and first stand cDNA synthesis was performed with SuperScript III (Invitrogen, CA). Real time PCR was carried out using iQ™ SYBR® Green Supermix (Bio-Rad, CA) according to the manufacturer's protocol. PCR primers used are outlined in Table S1 and 18s rRNA was used as an internal control to normalize RNA levels. Ras-induced caspase 3 and PARP cleavage (D) and apoptosis (E) is impaired in RGS6−/− MEFs and partly restored by treatment with LCA. For apoptosis assay two-way ANOVA revealed a significant effect of genotype (F1,12 = 127.24; p<0.0001) and drug treatment (F2,12 = 207.68; p<0.0001). Apoptosis is expressed as a fold increase in enrichment factor (cytoplasmic nucleosomes, i.e. cytoplasmic histone-associated DNA fragments) measured with the Roche Cell Death Detection Kit. Apoptosis was analyzed 72 hours after H-Ras(G12V) transfection. (F) RGS6L and RGS6LN401V rescue restores the impaired Ras(G12V)-induced apoptotic response (methods described in legend of Fig. 2E) in RGS6−/− MEFs. Two-way ANOVA revealed a significant effect of RGS6 transfection condition (F3,16 = 23.13; p<0.0001). Cells were cotransfected with and without H-Ras and indicated RGS6 construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. (G) Inhibition of apoptosis enhances HRas(G12V)/ p53(R175H)-induced colony formation in WT but not RGS6−/− MEFs (n = 9). Two-way ANOVA revealed a significant effect of genotype (F1,32 = 1047.69; p<0.0001) and drug treatment (F1,32 = 87.83; p<0.0001). Soft agar colony assays in WT and RGS6−/− MEFs were performed in the presence and absence of pan-caspase inhibitor Z-VAD-FMK (40 μM, Sigma) as described in Fig. 1. (G) Schematic diagram illustrating the role of RGS6 in Ras-induced oncogenic transformation. Oncogenic Ras induces RGS6 which directly induces apoptosis and suppresses Dnmt1 protein expression, thereby impairing Dnmt1 silencing of apoptotic genes. *, p<0.05; **, p<0.01; ***, p<0.001; #, p<0.001 vs. WT + H-Ras (bonferroni multiple comparisons). Unless otherwise indicated all experiments were performed in triplicate. Data are presented as mean ± S.E.M.
Silencing of tumor suppressor genes by Dnmt1 is essential for oncogenic transformation by Ras. The possibility that RGS6 negatively regulates Ras-induced transformation by inhibiting Dnmt1 was suggested by our previous finding that RGS6 complexes with Dnmt1 indirectly via DMAP1(8). While interaction with RGS6 was found to inhibit the transcriptional repressor activity of DMAP1(8), we did not investigate the effect of RGS6 on Dnmt1 activity or stability. Given these observations, we hypothesized that RGS6 might suppress cellular transformation by inhibiting Dnmt1. We first examined Dnmt1 protein levels in vector and H-Ras(G12V)-expressing WT and RGS6−/− MEFs. Consistent with previous reports(26, 27), H-Ras(G12V) induced up-regulation of Dnmt1 protein levels in both WT and RGS6−/− MEFs (Fig. 2A). Remarkably, loss of RGS6 led to up-regulation of Dnmt1 like that induced by Ras in WT MEFs (Fig. 2A,S1B), and Ras-induced up-regulation of Dnmt1 was further enhanced in RGS6−/− MEFs. This effect of RGS6 on Dnmt1 expression appears to occur primarily by post-transcriptional mechanisms as loss of RGS6 had little or no effect on Dnmt1 mRNA levels (Fig. S2). These findings provide new evidence that RGS6 is a powerful negative regulator of basal and Ras-induced Dnmt1 protein levels. This experiment prompted the question of whether the robust up-regulation of Dnmt1 in RGS6−/− MEFs is causally linked to the enhanced Ras-induced transformation in these cells.
To address this question, we investigated the effect of Dnmt1 inhibition on Ras-induced oncogenic transformation and apoptosis in WT and RGS6−/− MEFs. Dnmt1 is a multidomain enzyme, which is highly activated by deletion of the replication foci targeting sequence (28). The activated form of Dnmt1 was subjected to a high throughput screen using a fluorigenic assay, which identified laccaic acid A (LCA), an anthraquinone natural product, as a direct DNA-competitive inhibitor of Dnmt1(29). We reasoned that if loss of RGS6 promotes oncogenic transformation due to Dnmt1 up-regulation, LCA should impair this response. Figure 2B shows that LCA exerted a significant dose-dependent inhibition of Ras-induced colony formation in RGS6−/− MEFs while slightly reducing colony formation in WT MEFs at the highest dose used. Similar results were obtained with the non-selective Dnmt1 inhibitor 5-Aza-2’-deoxycytidine (5-Aza-dC)(30-32), though 5-Aza-dC did not affect colony formation in WT MEFs at doses sufficient to significantly impair transformation of RGS6−/− cells (Fig. S3A). These results suggest that RGS6 prevents Ras-induced oncogenic transformation through its ability to inhibit Dnmt1.
Dnmt1 silences genes encoding pro-apoptotic proteins that normally function to protect against oncogene-induced transformation. RASSF1A and NORE1A are pro-apoptotic Ras effectors that mediate oncogenic Ras-induced apoptosis(33). Products of other genes targeted by Dnmt1-mediated gene silencing, such as Fas(34) and Par4(27), are also pro-apoptotic. Dnmt1 inhibitors(35, 36) or Dnmt1 knockdown(37) induces apoptosis in cancer cells. Therefore, up-regulation of Dnmt1, as observed in RGS6−/− cells, would likely impair Ras-induced apoptosis and promote oncogenic transformation. Consistent with the increase in Dnmt1 protein, expression of pro-apoptotic Dnmt1 target genes FAS, NORE1A, and RASSF1A was significantly lower in RGS6−/− MEFs (Fig. 2C). Ras(G12V) transfection suppressed expression of these genes to levels observed in RGS6−/− MEFs under basal conditions (Fig. 2C) directly correlating with the observed trend in Dnmt1 protein levels (Fig. 2A). We then compared Ras-induced apoptosis in WT and RGS6−/− MEFs in the presence and absence of LCA and 5-AzadC. H-Ras(G12V) promoted apoptosis in WT MEFs as shown by cleavage of caspase 3 and PARP (Fig. 2D,S3B) and increases in cytoplasmic histone-associated DNA fragments (Fig. 2E,S3C). Importantly, we observed a significant impairment in Ras-induced apoptosis in RGS6−/− MEFs that was partially restored by treatment with LCA (Fig. 2D, 2E) or 5-Aza-dC (Fig. S3B, S3C). The related but inactive anthraquinone UI1055 did not affect Ras-induced apoptosis in cells of either genotype (Fig. S4). Also, re-expression of RGS6L or the GAP-deficient RGS6(N401V) mutant restored the Ras-induced apoptotic response in RGS6−/−MEFs (Fig. 2F). Thus, RGS6 prevents Ras-induced apoptosis by Dnmt1-dependent and G protein-independent mechanisms.
The inability of LCA to completely restore Ras-induced apoptosis in RGS6−/− MEFs could be due to incomplete inhibition of Dnmt1 or reflect the contribution of direct apoptotic actions of RGS6. Indeed, we have shown that Ras induces RGS6 (Fig 2A), and our previous studies demonstrated that RGS6 possesses potent pro-apoptotic activity(6). Given that RGS6 is required for Ras-induced apoptosis, we determined the importance of apoptosis in suppression of Ras-induced transformation by RGS6. Figure 2G shows that Z-FAD-FMK, a pan caspase inhibitor, greatly increased H-Ras(G12V)-induced colony formation in soft agar in WT but not RGS6−/− MEFs. These findings indicate that apoptosis prevents Ras-induced colony formation and that this response is absent in RGS6−/− MEFs. Together, our results support a model in which RGS6 up-regulation by Ras blocks Ras-induced transformation via suppression of Dnmt1 protein expression and inhibition of Dnmt1-mediated anti-apoptotic activity as well as through the possible direct, pro-apoptotic actions of RGS6 (Fig. 2H).
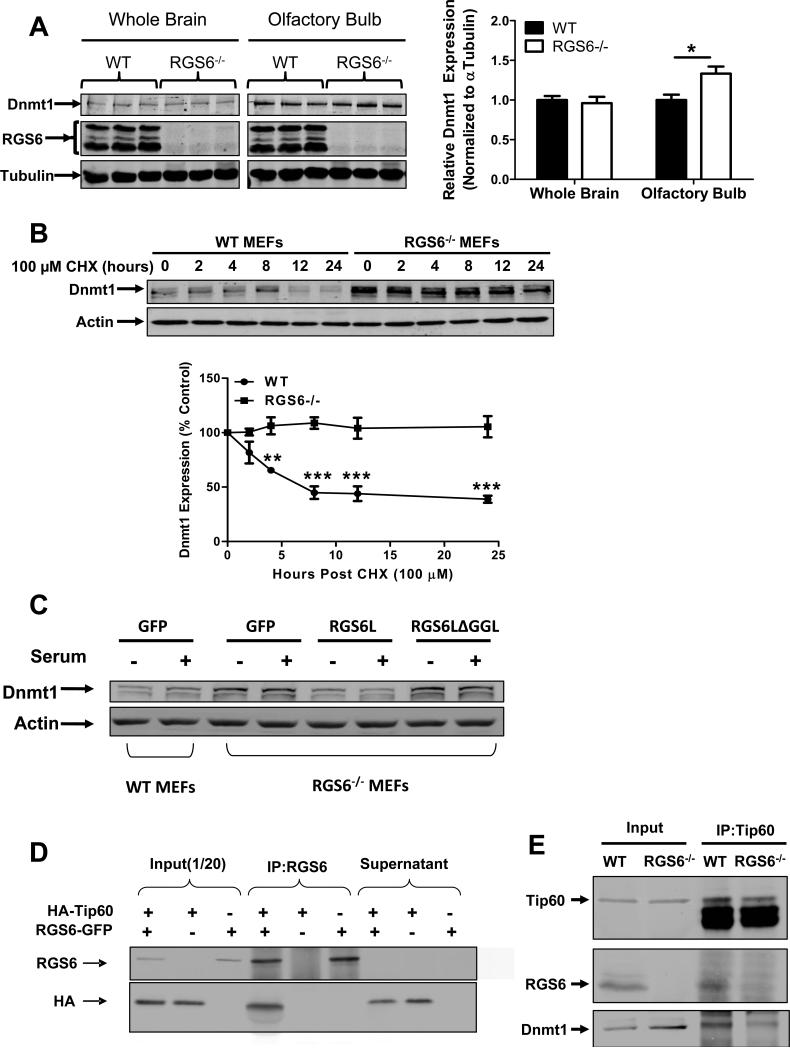
Dnmt1 expression levels were unaltered in RGS6−/− whole brain tissue lysates. However, a small but significant increase in Dnmt1 protein levels was detected in specific brain regions including olfactory bulb (Fig. 3A). The olfactory bulb is one site of adult neurogenesis(38) implying RGS6-mediated suppression of Dnmt1 expression is particularly important in replicating cell types. In other studies, we have shown that carcinogen-induced Dnmt1 up-regulation is potentiated in RGS6−/− mice(39). Together, these findings indicate that RGS6 also controls Dnmt1 protein expression in vivo.
Fig. 3.
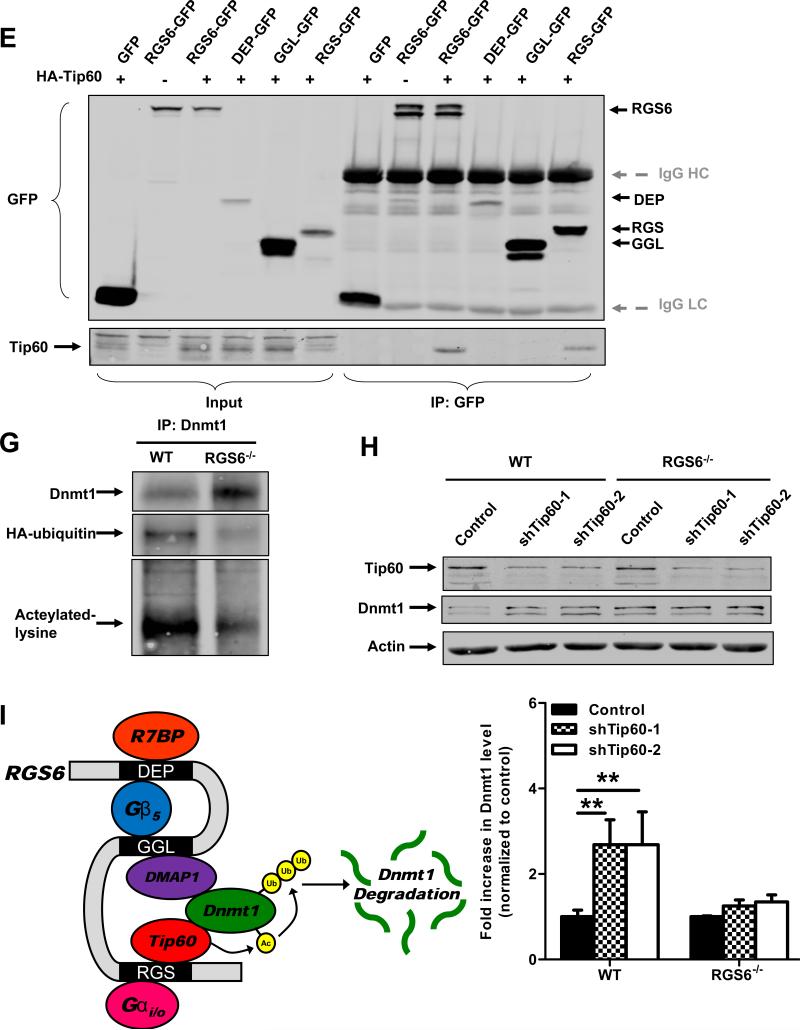
RGS6 promotes degradation of Dnmt1 by scaffolding Dnmt1 and Tip60. (A) Dnmt1 protein levels are increased in RGS6−/− olfactory bulb but not whole brain tissue lysates (n = 6). 40 μg of protein were subjected to SDS-PAGE and immunoblotting. Representative immunoblots (left panel) and quantification (right panel) are shown. *, p<0.05 (student's t-test). (B) Dnmt1 is stabilized in RGS6−/− MEFs. WT and RGS6−/− MEFs were treated with CHX (100μM) for indicated time, lysed, and probed for Dnmt1 and Actin (loading control). Dnmt1 levels from triplicate experiments were quantified and expressed as % control levels. Two-way ANOVA revealed a significant effect of genotype (F1,24 = 130.55; p<0.0001) and time(F5,24 = 5.99; p=0.001). (C) Transfection of full length RGS6L but not RGS6LΔGGL (Fig. S5) suppressed Dnmt1 protein expression in RGS6−/− MEFs. (D) Co-immunoprecipitation of RGS6 and Tip60 in COS-7 cells co-transfected with RGS6-GFP and HA-Tip60. Cell lysates were subjected to immunoprecipitation with anti-RGS6 and probed for RGS6 and HA. (E) Co-immunoprecipitation of endogenous RGS6 and Tip60 in vivo. Whole brain lysates were subjected to immunoprecipitation with anti-Tip60 and probed for RGS6, Tip60 and Dnmt1. (F) The RGS domain of RGS6 was sufficient to mediate interaction between RGS6 and Tip60. GFP-RGS6 proteins were efficiently immunoprecipitated by anti-GFP from transfected COS-7 cell lysates despite some differences in their expression and precipitates were probed for GFP and Tip60. A schematic of truncation mutants used is depicted in Fig. S5. IgG heavy chains (HC) and light chains (LC) are labeled. (G) Loss of RGS6 impairs acetylation and ubiquitylation of Dnmt1. WT and RGS6−/− MEFs were transfected with pcDNA3-His-ubiquitin via Lipofectamine 2000 according to the manufacturer's protocol. Cell lysates were prepared and subjected to immunoprecipitation with anti-Dnmt1. Immunoprecipitates were probed for Dnmt1, His and acetylated-lysine. (H) Tip60 knockdown with shRNAs increases the Dnmt1 protein level in WT MEFs but not in RGS6−/− MEFs. 293T cells were co-transfected with envelope plasmid pVSV-G, packaging plasmid PAX2, and empty viral vector or viral DNA containing shTip60-1 or shTip60-2 (Open Biosystems) by electroporation as described(44). Twenty four hours after transfection, virus-containing culture medium was collected. WT and RGS6−/− MEFs were incubated with virus-containing medium for 48 hours. Representative immunoblot (upper panel) and summary of multiple experiments illustrating effects of Tip60 knockdown on Dnmt1 protein levels in WT and RGS6−/− MEFs (lower panel) are shown. Two-way ANOVA revealed a significant effect of genotype (F1,12 = 23.05; p=0.0004) and Tip60 knockdown (F2,12 = 11.84; p=0.0014). (F) Hypothetical model depicting RGS6 scaffolding of Tip60 and Dnmt1 to facilitate Tip60 acetylation of Dnmt1 and subsequent Dnmt1 polyubiquitylation and degradation. Tissue lysates were prepared and quantified as described previously(45). Immunoblotting and transfections were performed as previously described(7). Antibodies used were: RGS6 (prepared in the lab(7), 1:5000); HA (Santa Cruz, 1:1000); Tip60 (Santa Cruz N17, 1:1000); Dnmt1 (Santa Cruz, 1:1000); GFP (Santa Cruz, 1:2000); His (Santa Cruz, 1:1000); Acetylated-lysine (Cell signaling, 1:5000); Actin (Sigma, 1:2000); Tubulin (Millipore, MA; 1:5000). **, p<0.01; ***, p<0.001 (bonferroni multiple comparisons). Unless otherwise indicated all experiments were performed in triplicate and representative results are shown. Data are presented as mean ± S.E.M.
We then undertook experiments to elucidate the mechanisms underlying regulation of Dnmt1 protein levels by RGS6. Our findings suggest that RGS6 negatively regulates basal Dnmt1 protein levels by post-transcriptional mechanisms (Figs. 2A, S2). Indeed analysis of Dnmt1 protein half-life in cyclohexamide (CHX) treated MEFs revealed a remarkable stabilization of Dnmt1 in RGS6−/− cells (Fig. 3B). In fact, no detectable loss of Dnmt1 expression was seen even after inhibition of protein synthesis for 24 hours in the absence of RGS6 (Fig. 3B). Dnmt1 stability is regulated by acetylation-dependent ubiquitylation and deubiquitylation primarily controlled by Tip60, an acetyltransferase, which, through direct acetylation, drives ubiquitylation and degradation of Dnmt1(37). We hypothesized that RGS6 might recruit Dnmt1 and Tip60 to mediate acetylation-driven degradation of Dnmt1 based upon two intriguing findings. First, we found previously that RGS6 complexes with Dnmt1 through association with DMAP1, an interaction mediated by the GGL (Gγ subunit like) domain of RGS6(8). This association between RGS6 and DMAP1 is necessary for control of Dnmt1 stability because re-expression of full length RGS6, but not a mutant lacking the GGL domain in RGS6−/− MEFs restored Dnmt1 expression to WT levels (Fig. 3C). Second, Axin, an atypical member of the RGS protein family, functions as a scaffold for Tip60 and p53 to facilitate p53 acetylation(40). Indeed, HA-Tip60 can be co-immunoprecipitated with RGS6-GFP demonstrating that Tip60 and RGS6 form a stable complex when co-expressed in cells (Fig. 3D). To determine whether this association occurs in vivo, we performed co-immunoprecipitation studies in whole mouse brain lysates where both proteins are natively expressed. Lysates from WT and RGS6−/− mice were subjected to immunoprecipitation with anti-Tip60 and probed for RGS6, Tip60 and Dnmt1. Consistent with our findings in transfected cells, endogenous RGS6 and Tip60 form a complex (Fig. 3E). Importantly, the amount of Dnmt1 that co-immunoprecipitated with Tip60 was greatly reduced in RGS6−/− lysates. This latter finding indicates that RGS6 is an important scaffolding partner for Dnmt1 and Tip60.
We next investigated which of RGS6's functional domains mediates its association with Tip60. In addition to the RGS domain, conferring its ability to bind and enhance the GTPase activity of Gα subunits, RGS6 possesses DEP (Dishevelled, Egl-10, Pleckstrin homology) and GGL domains necessary for protein-protein interactions including GGL domain-dependent binding to Gβ5 and DMAP1(8, 41, 42). To identify the domains of RGS6 necessary for Tip60 association, we compared the ability of GFP-tagged RGS6 and each of its functional domains to coimmunoprecipitate HA-Tip60 in transfected cells. Both full length RGS6 and its RGS domain alone, but neither its N-terminal DEP-containing nor GGL domains, co-immunoprecipitated Tip60 (Fig. 3F). These results show that the RGS domain of RGS6 is sufficient to mediate Tip60-RGS6 association. Thus, Tip60 binds directly or indirectly to the RGS domain of RGS6 located adjacent to the GGL domain where Dnmt1 is bound via DMAP1.
The crystal structure of the closely related RGS9:Gβ5 complex revealed that the GGL domain is sandwiched between the RGS domain and Gβ5(43). Moreover, our finding that DMAP1 binding to the GGL domain of RGS6 is not competitive with Gβ5(8) suggests that the DMAP1 binding surface is between the GGL and RGS domain interface. Thus, Tip60 association with the RGS domain of RGS6 would be on the same surface where we believe Dnmt1 is bound to RGS6, a speculation supported by our finding that RGS6 is required for Tip60-mediated Dnmt1 degradation (Fig. 3H). Interestingly, the RGS domain of Axin was not sufficient for Tip60 binding(40). We suspect that RGS6 and Axin bind Tip60 differently, in part because Axin possesses an RGS “like” domain that differs structurally from the RGS domain of canonical RGS proteins including RGS6. Our findings reveal an entirely novel function of the RGS domain, whose only previously known function was to bind and accelerate the GTPase activity of Gα subunits. These results show that the RGS domain of RGS6 functions to scaffold Tip60 near its GGL domain-bound Dnmt1 substrate and raise the intriguing possibility that RGS6 promotes Tip60 mediated Dnmt1 acetylation leading to ubiquitylation and degradation of Dnmt1.
To test the above possibility, we examined the role of RGS6 in Dnmt1 acetylation and ubiquitylation and Tip60-dependent Dnmt1 degradation. First, we compared Dnmt1 acetylation and ubiquitylation in immunoprecipitates from WT and RGS6−/− MEFs transfected with Hisubiquitin. Acetylation and ubiquitylation of Dnmt1 were greatly reduced in RGS6−/− vs WT MEFs, consistent with the much greater level of Dnmt1 in RGS6−/− immunoprecipitates (Fig. 3G). We next tested the role of RGS6 in Tip60-mediated degradation of Dnmt1 by Tip60 knockdown studies in WT and RGS6−/− MEFs. Figure 3H shows that Tip60 knockdown with two distinct Tip60 shRNAs greatly up-regulated the low Dnmt1 levels in WT MEFs. In contrast, knockdown of Tip60 had no effect on Dnmt1 expression in RGS6−/− MEFs. These observations confirm that RGS6 has an essential role in Tip60-mediated degradation of Dnmt1. Moreover, they indicate that Tip60 mediates the suppression of Dnmt1 protein levels by RGS6. These novel findings support a model in which RGS6 promotes association between Dnmt1 and Tip60 by acting as a scaffold protein, thereby mediating Tip60 acetylation of Dnmt1 leading to Dnmt1 ubiquitylation and degradation (Fig. 3I).
In summary, we provide evidence for a new and crucial role for RGS6 in suppression of oncogenic transformation. RGS6 blocks Ras-induced transformation in part by strongly down regulating Dnmt1, a protein that mediates silencing of pro-apoptotic and tumor suppressor genes. Our finding that RGS6 functions as a scaffold for Tip60 and Dnmt1 to mediate Tip60 acetylation and subsequent degradation of Dnmt1 provides new understanding of the mechanisms responsible for regulating Dnmt1 expression and activity. That Tip60 associates with RGS6 via its RGS domain provides the first evidence for a novel function of the RGS domain beyond its role in G protein regulation. Our findings suggest that loss of RGS6, as we observed during breast cancer progression(6), may be a mechanism underlying up-regulation of Dnmt1 and increases in DNA methylation during carcinogenesis. Indeed, we recently demonstrated a large increase in carcinogen-induced Dnmt1 expression in RGS6−/− mouse mammary glands compared to WT tissue(39). Our demonstration of an essential role for RGS6 in blocking oncogenic transformation by promoting Dnmt1 degradation identify RGS6 as a potential therapeutic target for the treatment of human cancers.
Supplementary Material
Acknowledgments
We thank Dr. John Koland for his careful reading of and useful suggestions for this manuscript, and Sara Reed for identifying effective Tip60 shRNAs for our assays.
Financial Support: This project was supported by National Cancer Institute grants and contracts CA161882 (RAF), HHSN261200433000C (CB), CA090367 (DEQ), and American Cancer Society (PF-11-141-01 (RLF)).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272(7):3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 2.Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J Biol Chem. 1998;273(3):1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 3.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 4.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 5.Berman DM, Wang Y, Liu Z, Dong Q, Burke LA, Liotta LA, et al. A functional polymorphism in RGS6 modulates the risk of bladder cancer. Cancer Res. 2004;64(18):6820–6826. doi: 10.1158/0008-5472.CAN-04-1916. [DOI] [PubMed] [Google Scholar]
- 6.Maity B, Yang J, Huang J, Askeland RW, Bera S, Fisher RA. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J Biol Chem. 2011;286(2):1409–1419. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Yang J, Maity B, Mayuzumi D, Fisher RA. Regulator of G protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Cancer Res. 2011;71(20):6310–6319. doi: 10.1158/0008-5472.CAN-10-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Fisher RA. RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J Biol Chem. 2004;279(14):14120–14128. doi: 10.1074/jbc.M309547200. [DOI] [PubMed] [Google Scholar]
- 9.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 10.Leonhardt H, Cardoso MC. DNA methylation, nuclear structure, gene expression and cancer. J Cell Biochem. Suppl. 2000;35:78–83. doi: 10.1002/1097-4644(2000)79:35+<78::aid-jcb1129>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 11.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33(3):163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 12.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim Biophys Acta. 2004;1704(2):87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66(17):8342–8346. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- 14.Ordway JM, Williams K, Curran T. Transcription repression in oncogenic transformation: common targets of epigenetic repression in cells transformed by Fos, Ras or Dnmt1. Oncogene. 2004;23(21):3737–3748. doi: 10.1038/sj.onc.1207483. [DOI] [PubMed] [Google Scholar]
- 15.Patra SK. Ras regulation of DNA-methylation and cancer. Exp Cell Res. 2008;314(6):1193–1201. doi: 10.1016/j.yexcr.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Patra SK, Szyf M. DNA methylation-mediated nucleosome dynamics and oncogenic Ras signaling: insights from FAS, FAS ligand and RASSF1A. Febs J. 2008;275(21):5217–5235. doi: 10.1111/j.1742-4658.2008.06658.x. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry WS, Nelkin BD, Celano P, Yen RW, Falco JP, Hamilton SR, et al. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci USA. 1991;88(8):3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He S, Wang F, Yang L, Guo C, Wan R, Ke A, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS One. 2011;6(11):e27684. doi: 10.1371/journal.pone.0027684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Nakagawa T, Kanai Y, Saito Y, Kitamura T, Kakizoe T, Hirohashi S. Increased DNA methyltransferase 1 protein expression in human transitional cell carcinoma of the bladder. J Urol. 2003;170(6.1):2463–2466. doi: 10.1097/01.ju.0000095919.50869.c9. [DOI] [PubMed] [Google Scholar]
- 21.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105(4):527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 22.Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164(2):689–699. doi: 10.1016/S0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96(7):403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu YM, Huang Q, Lin J, Hu Y, Chen J, Lai MD. Expression of human DNA methyltransferase 1 in colorectal cancer tissues and their corresponding distant normal tissues. Int J Colorectal Dis. 2007;22(6):661–666. doi: 10.1007/s00384-006-0224-4. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 26.Pakneshan P, Szyf M, Rabbani SA. Methylation and inhibition of expression of uPA by the RAS oncogene: divergence of growth control and invasion in breast cancer cells. Carcinogenesis. 2005;26(3):557–564. doi: 10.1093/carcin/bgi009. [DOI] [PubMed] [Google Scholar]
- 27.Pruitt K, Ulku AS, Frantz K, Rojas RJ, Muniz-Medina VM, Rangnekar VM, et al. Ras-mediated loss of the pro-apoptotic response protein Par-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J Biol Chem. 2005;280(24):23363–23370. doi: 10.1074/jbc.M503083200. [DOI] [PubMed] [Google Scholar]
- 28.Syeda F, Fagan RL, Wean M, Avvakumov GV, Walker JR, Xue S, et al. The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J Biol Chem. 2011;286(17):15344–15351. doi: 10.1074/jbc.M110.209882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fagan RL, Cryderman DE, Wallrath L, Brenner C. Laccaic Acid A is a direct, DNA-competitive inhibitor of DNA methyltransferase 1. J Biol Chem. doi: 10.1074/jbc.M113.480517. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81(22):6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31(34):3827–3844. doi: 10.1038/onc.2011.552. [DOI] [PubMed] [Google Scholar]
- 32.Maslov AY, Lee M, Gundry M, Gravina S, Strogonova N, Tazearslan C, et al. 5-Aza-2′-deoxycytidine-induced genome rearrangements are mediated by DNMT1. Oncogene. 2012;31(50):5172–5179. doi: 10.1038/onc.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22(56):8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 34.Santourlidis S, Warskulat U, Florl AR, Maas S, Pulte T, Fischer J, et al. Hypermethylation of the tumor necrosis factor receptor superfamily 6 (APT1, Fas, CD95/Apo-1) gene promoter at rel/nuclear factor kappaB sites in prostatic carcinoma. Mol Carcinog. 2001;32(1):36–43. doi: 10.1002/mc.1062. [DOI] [PubMed] [Google Scholar]
- 35.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24(23):3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 36.Chen M, Shabashvili D, Nawab A, Yang SX, Dyer LM, Brown KD, et al. DNA methyltransferase inhibitor, zebularine, delays tumor growth and induces apoptosis in a genetically engineered mouse model of breast cancer. Mol Cancer Ther. 2012;11(2):370–382. doi: 10.1158/1535-7163.MCT-11-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M, Gao J, Du YQ, Gao DJ, Zhang YQ, Li ZS, et al. Reduction of pancreatic cancer cell viability and induction of apoptosis mediated by siRNA targeting DNMT1 through suppression of total DNA methyltransferase activity. Mol Med Report. 2010;3(4):699–704. doi: 10.3892/mmr_00000320. [DOI] [PubMed] [Google Scholar]
- 38.Seaberg RM, van der Kooy D. Adult rodent neurogenic regions: the ventricular subependyma contains neural stem cells, but the dentate gyrus contains restricted progenitors. J Neurosci. 2002;22(5):1784–1793. doi: 10.1523/JNEUROSCI.22-05-01784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maity B, Stewart A, O'Malley Y, Askeland RW, Sugg SL, Fisher RA. Regulator of G Protein Signaling 6 (RGS6) is a novel suppressor of breast tumor initiation and progression. Carcinogenesis. doi: 10.1093/carcin/bgt128. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Lin S, Wang X, Lian G, Lu Z, Guo H, et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol. 2009;11(9):1128–1134. doi: 10.1038/ncb1927. [DOI] [PubMed] [Google Scholar]
- 41.Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280(7):5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 42.Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. Identification of the Gbeta5-RGS7 protein complex in the retina. Biochem Biophys Res Commun. 1998;249(3):898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- 43.Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat Struct Mol Biol. 2008;15(2):155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, Chatterjee TK, Fisher RA. RGS6 interacts with SCG10 and promotes neuronal differentiation. Role of the G gamma subunit-like (GGL) domain of RGS6. J Biol Chem. 2002;277(40):37832–37839. doi: 10.1074/jbc.M205908200. [DOI] [PubMed] [Google Scholar]
- 45.Maity B, Stewart A, Yang J, Loo L, Sheff D, Shepherd AJ, et al. Regulator of G protein signaling 6 (RGS6) protein ensures coordination of motor movement by modulating GABAB receptor signaling. J Biol Chem. 2012;287(7):4972–4981. doi: 10.1074/jbc.M111.297218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.