Abstract
The discovery of disease-specific biomarkers in oral fluids has revealed a new dimension in molecular diagnostics. Recent studies have reported the mechanistic involvement of tumor cells derived mediators, such as exosomes, in the development of saliva-based mRNA biomarkers. To further our understanding of the origins of disease-induced salivary biomarkers, we here evaluated the hypothesis that tumor-shed secretory lipidic vesicles called exosome-like microvesicles (ELMs) that serve as protective carriers of tissue-specific information, mRNAs, and proteins, throughout the vasculature and bodily fluids. RNA content was analyzed in cell free-saliva and ELM-enriched fractions of saliva. Our data confirmed that the majority of extracellular RNAs (exRNAs) in saliva were encapsulated within ELMs. Nude mice implanted with human lung cancer H460 cells expressing hCD63-GFP were used to follow the circulation of tumor cell specific protein and mRNA in the form of ELMs in vivo. We were able to identify human GAPDH mRNA in ELMs of blood and saliva of tumor bearing mice using nested RT-qPCR. ELMs positive for hCD63-GFP were detected in the saliva and blood of tumor bearing mice as well as using electric field-induced release and measurement (EFIRM). Altogether, our results demonstrate that ELMs carry tumor cell–specific mRNA and protein from blood to saliva in a xenografted mouse model of human lung cancer. These results therefore strengthen the link between distal tumor progression and the biomarker discovery of saliva through the ELMs.
Introduction
Human saliva is a clear, slightly acidic (pH 6.0–7.0) biofluid known to contain a diverse range of proteins [1], over 3000 unique mRNAs [1], [2], [3], and 700 bacterial species [4]. In recent years, a number of investigations evaluating the constituency of oral fluid have discovered that it may actually reflect an individual's physiological condition. In fact, multiple groups have identified saliva-based proteomic, transcriptomic, and microbiological markers for Sjögren's syndrome, inflammatory bowel disease, and even cancers [5], [6], [7], [8], [9], [10], [11], [12], [13]. Although promising, the novelty of using saliva as an effective evaluator of local and systemic health has not found widespread acceptance. Significant skepticism remains regarding how these unique molecular indicators are developed in saliva.
Exosomes are small, lipid-bound, spherical structures measuring approximately 30 to 100 nm in diameter [14], [15]. Randomly formed from the invagination of intracellular vesicles, exosomes often contain biologically active host cell lipids, proteins, miRNAs, mRNAs, ncRNAs, and other cellular constituents [14], [16], [17], [18], [19], [20], [21]. Microvesicles containing similar host cell biomolecules and heterogeneous in size may also be formed by the budding-off the cellular membrane [22], [23]. Majority of vesicles isolated from body fluids are referred as exosomes based on their exosomal protein markers [24], [25]. However, the microvesicles do be co-isolated using current available purification method [24], [25]. We, therefore, collectively refer these vesicles as exosome-like microvesicles (ELMs) here. ELMs are known to shed continuously from multiple cell types including: hematopoietic, intestinal epithelial, Schwann, fat, neuronal, fibroblasts, and several tumor cell lines [26]. Many types of cancer cells release ELMs and tumor-derived ELMs carry a wide range of nucleic acids, including miRNA, mRNA, ncRNA and DNA [21]. ELMs containing these nucleic acids have been shown to reflect the genetic status of tumor, and be able to travel to distant site and transfer their cargo to the recipient cells and to induce phenotypic changes [27], [28], [29]. Previous investigations have revealed that tumors are often the primary source of circulating membrane vesicles and increased amount of tumor derived protein, RNA and DNA were found in the blood of cancer patients [30], [31], [32], [33].
In addition to their presence in blood, ELMs are also present in urine, saliva, breast milk, malignant and pleural effusions, synovial fluid, epididymal fluid, and amniotic fluid [21], [26]. Therefore, tissue-specific exosomes with their constituent tissue-specific biomarkers can serve as a biomarker source for the diagnosis, prognosis, and monitoring of disease [34], [35], [36].
Recent evidence has emerged describing a role for ELMs in the processes that govern the induction of discriminatory salivary biomarkers [17], [37], [38]. However, the mechanisms underpinning the etiology and biogenesis of saliva-based biomarkers have not been clearly explained. Understanding how these markers come to exist in oral fluids will both shed light on the body's capacity for extracellular communication and help credential salivary biomarkers as an acceptable mode for personalized medical assessment.
In this study, we demonstrated that exRNA in cell-free saliva were largely encapsulated and protected from degradation by ELMs. A novel human lung cancer mouse xenograft model in which implanted human lung cancer H460 cells consistently express exosomal marker hCD63-GFP was developed to study whether ELMs contribute to the emergency of discriminatory salivary biomarkers during systemic disease progression. We were able to identify hCD63-GFP positive ELMs and human GAPDH mRNA in hCD63 positive ELMs in the blood and saliva of tumor bearing mice. Our evidence indicates that ELMs play a critical role in this process by acting to protect and transport tumor-specific molecular information throughout the vasculature, as well as in bodily fluids. This study represents a substantial discovery, describing the induction of discriminatory saliva-based biomarkers, Understanding these phenomena may facilitate the development of novel strategies for diagnostics, monitoring, and therapeutics.
Materials and Methods
Ethics Statement
Human saliva samples were obtained from healthy volunteers under the institutional review board protocol (IRB#10-000431) approved by the University of California Los Angeles IRB. Written informed consent forms were obtained from all participants.
All mouse procedures were approved by the UCLA Animal Research Committee in compliance with the Association for Assessment and Accreditation of Laboratory Care (AAA-LAC) International.
Saliva Collection and Salivary ELMs Isolation
Unstimulated saliva samples were obtained and processed according to previously established protocols [2]. ELMs were isolated using Exoquick precipitation solution (System Biosciences, Inc.). Three hundred microliters of cell-free saliva was thoroughly mixed with an equal volume Exoquick solution, incubated at 4°C overnight, and centrifuged at 1500×g for 15 min at 4°C. The pellets were then resuspended in water [39] and used for RNA isolation and immunoblotting via electron microscopy (EM).
Isolation of RNA from Saliva and ELMs
Cell-free saliva, salivary ELMs, and ELM-depleted saliva supernatant were treated with RNase cocktail (final concentration 100 U/ml) with or without 1% Triton X-100 (Tx) at room temperature for 20 min. RNA was extracted from these processed samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The isolated RNA was quantified using the RiboGreen RNA quantification Kit (Invitrogen) and analyzed by reverse transcription PCR (RT-PCR) followed by quantitative PCR (qPCR).
Cell Culture
The human non-small cell lung cancer cell line H460 and mouse Lewis lung cancer cell line LLC1 were obtained from ATCC. Cells were grown in DMEM medium with glutamax-I supplemented with 10% (v/v) FBS and Pen/Strep in an atmosphere of 95% air and 5% CO2 at 37°C. H460 cells were transfected with the plasmid encoding hCD63-GFP, pCT-CD63-GFP (SBI, USA). Stable H460-hCD63-GFP cells were selected and maintained in puromycin-containing medium at 1 µg/ml. Dimethyl amiloride (DMA) (Sigma, MO) was used to inhibit ELM secretion in H460-hCD63-GFP cells [40]. Cells were treated with 1 µmol or 10 µmol DMA for 48 h, and the amount of ELMs secreted into medium in the presence of DMA was determined and compared to untreated controls using an acetylcholinesterase activity assay. The trypan blue exclusion assay was used to determine the cytotoxity of DMA using a Beckman Coulter Vi-Cell (Beckman Coulter Inc., CA).
Western blotting
H460 hCD63-GFP and H460 cells were grown in serum-free DMEM supplemented with 1% non-essential amino acids for 48 h. ELMs were collected from the conditioned media and lysed along with both H460 hCD63-GFP and H460 cells. The H460 hCD63-GFP and H460 cell and ELM lysates (5 µg protein) were denatured at 95°C for 10 min and loaded onto 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a nitrocellulose membrane and probed with rabbit anti-GFP HRP-conjugated antibody (Invitrogen). ELMs were isolated from human saliva using exoquick precipitation (System Biosciences, Inc.). Exoquick precipitated pellets were lysed and then separated by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and probed with rabbit anti-CD63 (Santa Cruz Biotechnology, Santa Cruz, CA) followed by an incubation with a secondary antibody (Invitrogen).
Isolation and Quantification of ELMs from Condition Medium
ELMs were prepared using differential ultracentrifugation methods, as described previously [41]. Briefly, cell-free conditioned media was centrifuged at 10,000×g for 30 min at 4°C, then at 100,000×g for 2 h at 4°C. The ELM pellet was washed and resuspended in PBS. The Amplex Red acetylcholinesterase assay kit was used to measure the total amount of ELMs according to the manufacturer's protocol (Invitrogen, USA).
Characterization of ELMs Using Electron Microscopy
Isolated ELMs were loaded onto carbon-coated grids; fixed in 2% paraformaldehyde; washed in PBS; and immunolabeled with anti-hCD63 (BD Biosciences, US) or anti-CopGFP primary antibodies (Evrogen, Moscow, Russia) and goat anti-mouse or rabbit IgG coupled to 10-nm or 15-nm gold particles (Sigma, US). The grids were post-fixed in 2.5% glutaraldehyde, washed, and contrasted with 2% uranyl acetate, as previously described [42]. The ELMs were then examined with a JEOL 100GX transmission electron microscope (JEOL USA, Inc. Peabody, MA) (Electron Microscopy Service Center, Brain Research Institute, UCLA).
Mouse Lung Cancer Xenograft Model
Male athymic BALB/c nude mice were obtained from Charles River (MA, USA) and weighed 20–22 g at the start of the experiments. The mice were housed in sterilized filter-topped cages and maintained in sterile conditions. A total of 1×106 H460 hCD63-GFP cells, or 100 µl saline were injected into the left chest cavity of nude mice. Briefly, mice were anesthetized using 1–3% isoflurane in oxygen from a precision vaporizer. Mice were placed in the right lateral decubitus position. One hundred microliters of cells or saline were injected slowly into the left intercostal space at the dorsal mid-axillary line just below the inferior border of the scapular using a 30-gauge needle attached to a 1-ml syringe. The needle was advanced approximately 5 mm through the chest wall into the pleural space. After injection, the needle was retracted and mice were turned to the left lateral decubitus position for recovery. Some tumor-bearing mice were treated with daily intrapleural injection of 1 µmol/kg DMA (Sigma) or PBS control for 1 week before sacrifice [43].
Collection of Mouse Saliva, Blood, and Tumor Tissue
Twenty days after tumor implantation, saliva was collected and mice were sacrificed. Mild anesthesia was induced by intramuscular (IM) injection of 1 µl/kg body weight of a solution containing 60 mg/ml ketamine (Phoenix Scientific, St. Joseph, MO) and 8 mg/ml xylazine (Phoenix Scientific, CA). To stimulate saliva secretion, mice were subcutaneously injected between the ears with pilocarpine (0.05 mg pilocarpine/100 g body weight). Collection was completed in 20 min using a micropipette, and samples were immediately placed in pre-chilled 1.5-ml microcentrifuge tubes. Samples were centrifuged at 3500 rpm for 30 min at 4°C and supernatant was collected and stored at −80°C until analysis. Blood was collected in BD vacutainer tubes containing clot activator (BD Biosciences, CA) and centrifuged at 1000×g for 10 min after mice were sacrificed. Salivary glands and tumor tissues were removed from mice, snap-frozen in liquid nitrogen, and stored at −80°C.
Detection of hCD63/GFP-Positive ELMs Using EFIRM Technology
The human hCD63/GFP-positive ELMs were quantified using immuno-adsorption and an electrochemical sensor detection technology, called electric field-induced release and measurement (EFIRM) [44], [45]. Briefly, saliva, serum or conditioned media were centrifuged at 1000 rpm at 4°C for 10 min. The cell-free supernatant was further centrifuged at 3500 rpm at 4°C for 30 min, to eliminate cellular debris. Five microliters of streptavidin-coated magnetic beads (Invitrogen, USA) were mixed with 1 µl of biotinylated anti-human CD63 (hCD63) antibody (Ancell, USA) on a HulaMixer Sample Mixer (Invitrogen) for 30 min at room temperature. Then, 1 ml of conditioned medium, or 10 µl of saliva or serum diluted in 990 µl of casein-PBS (Invitrogen, USA), was incubated with the anti-hCD63-conjugated magnetic beads for 2 h at room temperature to form ELMs–magnetic bead complexes. Beads were then washed twice with casein-PBS and incubated with rabbit anti-GFP HRP-conjugated polyclonal antibody for 1 h at room temperature. After incubation, the beads were washed twice with casein-PBS and accumulated onto an electrochemical sensor by applying a magnetic field [44], [45].
Confocal Microscopy
The tumor sections were examined using a Leica TCS-SP2 confocal microscope with Plan APO 63×1.4 NA oil objective lens and LCS confocal software.
RT-qPCR Assay
RT-PCR followed by separate qPCR (hereafter termed RT-qPCR) was performed to detect salivary mRNAs. Multiplex RT-PCR preamplification of three mRNAs was performed using a SuperScript III platinum qRT-PCR System (Invitrogen) with a pool of outer primer sets, and conducted using a GeneAmp PCR-System 9700 (Applied Biosystems) with a fixed thermal-cycling program. SYBR Green qPCR was performed to quantitatively detect the expression levels of salivary mRNAs. The qPCR sample was prepared by combining 2× qPCR Mastermix (Applied Biological Materials), inner primers (900 nmol/l; Table S1 in Supporting information S1), and 2 µl cDNA template. The total volume of each reaction was 10 µl, adjusted with nuclease-free water. The qPCR associated with melting-curve analysis was conducted using an AB-7500HT System (Applied Biosystems) with a fixed thermal-cycling program. Each gene was tested in triplicate for all samples, including the negative control, in which the cDNA template was the product of the RT-PCR preamplification negative control.
Human GAPDH Nested RT-PCR Detection
hCD63-positive ELMs from mouse saliva or blood were adsorbed onto anti-human anti-CD63 antibody-coated magnetic beads, as previously described [44], [45]. Beads were washed twice with PBS and resuspended in 10 µl TE buffer (Ambion, USA). Human GAPDH RNA was reverse transcribed using 4 µl of ELM-bead complex and the SuperScript III One-Step RT-PCR System (Invitrogen, USA). Nested PCR reactions for human GAPDH were performed using the following primer sets:
5′-TCAAGTGGGGCGATGCTGGC-3′ and 5′-TGGGGGCATCAGCAGAGGGG-3′, 5′-GCTGGCGCTGAGTACGTCGT-3′ and 5′-CCTGCAAATGAGCCCCAGCC-3′. PCR clean-up was performed using ExoSAP-IT after the initial RT-PCR reaction (Affymetrix, CA, USA). Native -PAGE (15%) was used to reveal the 72-bp human GAPDH PCR product.
Statistical Analysis
All statistical analyses were conducted using the IBM SPSS Statistics version 21; mean values, and standard deviations were calculated using descriptive statistics. One-way analysis of variance, two-sample t tests were used for testing the difference between expression values. P values <0.05 were considered significant. Post-hoc comparisons were conducted by using Tukey's analysis. Data are expressed as mean ± SD.
Results
Salivary ExRNAs are Predominantly Contained Within ELMs
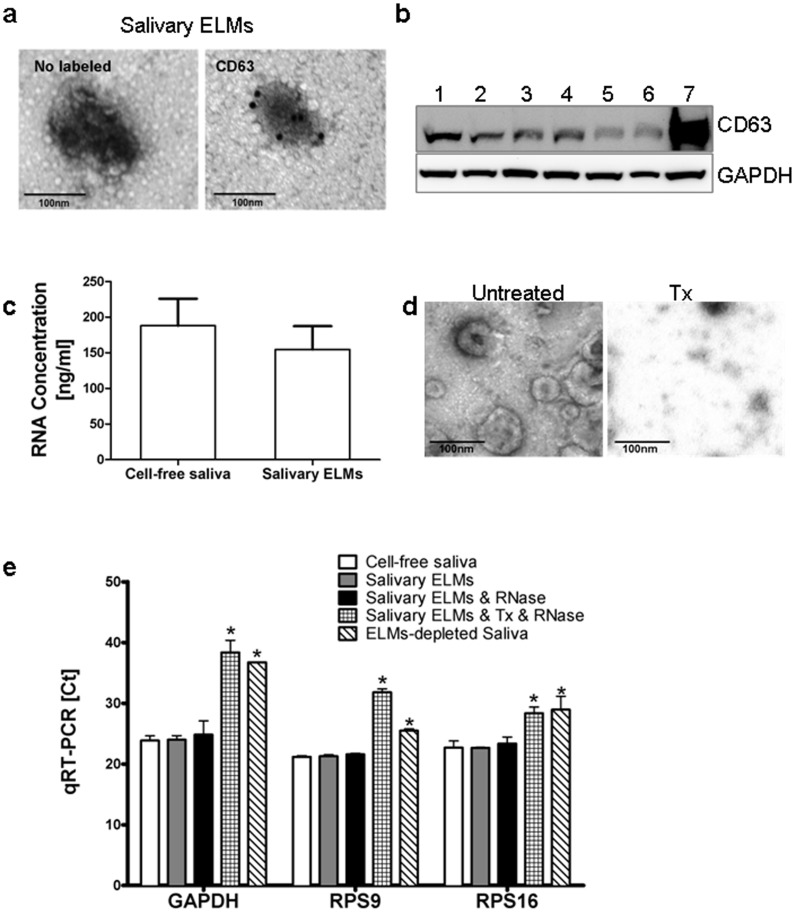
Previous studies have demonstrated that cell-free saliva supernatant contains more than 3000 different mRNA species [1], [2], [3], and the exRNAs remain stable unless they are exposed to detergents [46]. Lipidic vesicles, exosomes, protected the breakdown of exRNAs in saliva [42]. To evaluate whether all exRNAs in saliva are subject to this type of vesicular protection, we compared exRNAs in cell-free saliva and salivary ELMs. We first isolated salivary ELMs from the cell-free saliva of seven healthy subjects, which were characterized by the expression of exosomal surface marker CD63 (Fig. 1a, b). exRNAs in salivary ELMs and cell-free salivary were quantified and found to range from from 74 to 300 ng/ml and 87 to 350 ng/ml, respectively, with no statistically significant difference (Fig. 1c). Consistent with previous observation [42], the exRNAs in salivary ELMs is protected from RNase degradation. Quantitative PCR analysis of extracted RNAs revealed no notable differences in C t values of the reference genes GAPDH, ß-actin, or RPS9 between cell-free saliva, salivary ELMs, and salivary ELMs with RNase treatment (Fig. 1e). 1% Triton X-100 (Tx) solution compromised structural integrity of salivary ELMs (Fig. 1d). ELMs with Triton X-100 plus RNase treatment or ELMs-depleted saliva exhibited substantially higher C t values of reference genes GAPDH, ß-actin, or RPS9 (Fig. 1e) These results indicate that exRNAs in cell-free saliva are largely encased within salivary ELMs.
Figure 1. Salivary RNAs reside within salivary ELMs.
(a) Representative electron microscopy images of salivary ELMs, and anti-CD63 immunogold-labeled ELMs in human saliva. (b) Western blot analysis of salivary ELMs using antibodies against human CD63 and GAPDH. Lane 1 to 7 are protein extracts of the salivary ELM pellets obtained from seven donors using Exoquick precipitation. (c) Ribogreen RNA quantitation from seven donors. Error bars represent means ± SD. (d) Representiave electron microscopy images of salivary ELMs and salivary ELMs treated with Tx. (e) RT-qPCR results of three reference genes from the following samples: cell-free saliva; salivary ELMs; salivary ELMs treated with RNase; salivary ELMs treated with Tx and RNase and ELM-depleted saliva. Data are expressed as means ± SD. *P<0.05, statistically significant difference from the saliva group.
Next, we sought to compare the transcriptome content of salivary ELMs vs. cell-free saliva using microarray expression analysis (Affymetrix HU133 plus 2.0). Approximately 2938 and 2040 mRNA transcripts were detected in cell-free saliva and isolated salivary ELMs respectively. Categorizing each group ontologically revealed similar mRNA profiles (Fig. S1a in Supporting information S1). Eight mRNAs were abundant across each data set (Fig. S1b in Supporting information S1). These overlapping molecular characteristics suggest that the transcriptomic constituency of saliva may exclusively reside within ELMs, a structure whose integrity is critical for the stability of salivary RNA.
Establishment of an ELM-Labeling Xenograft Lung Cancer Mouse Model
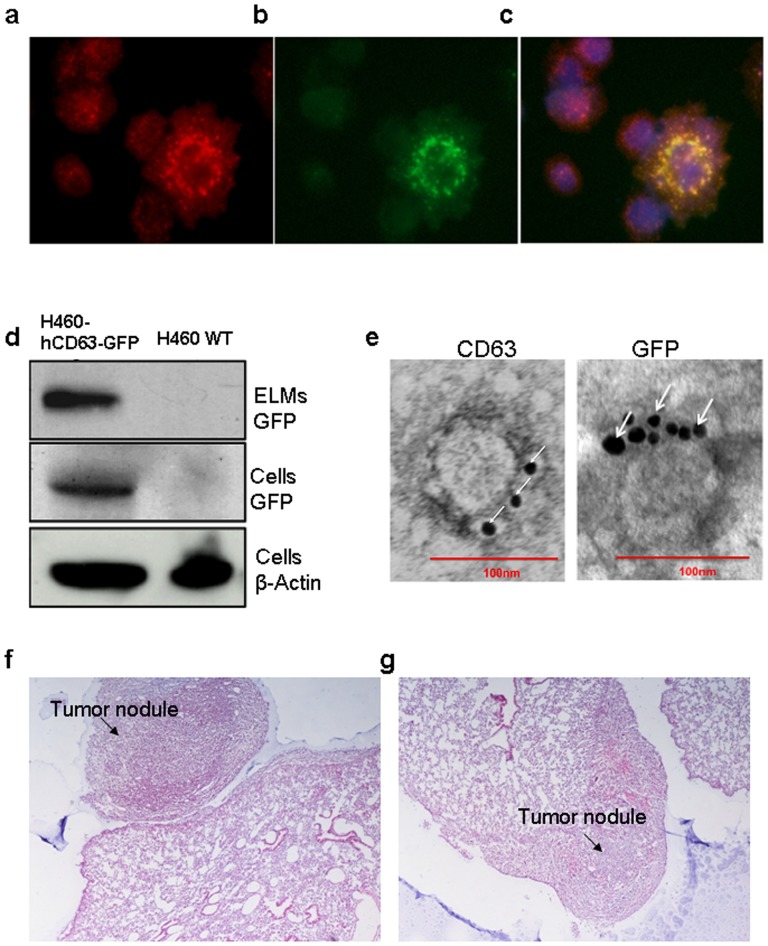
To help determine whether ELMs function as a conduit for long-range transportation of tissue-specific biomolecules, we labeled and tracked tumor-derived ELMs in vivo. We implanted nude mice with H460 human non-small cell lung cancer (hNSCLC) cells that stably expressed a well-known exosomal surface protein, hCD63 [20] fused to GFP (Fig. 2a–d). Prior to implantation, GFP-labeled H460-derived ELMs were confirmed by Western blotting and immuno-electron microscopy (Fig. 2d, e). The presence of tumors 20 days after implantation were confirmed by H&E section showing tumor nodule attached to the pleural surface and tumor nodule in the lung parenchyma (Fig. 2f, g). Together, these results confirm the establishment of human lung cancer xenograft mouse model with trackable tumor cell derived ELMs.
Figure 2. Establishment of a human lung cancer xenograft mouse model.
To generate our xenograft lung cancer mouse model, H460 human lung cancer cells that stably express hCD63-GFP were orthotopically injected into male athymic BALB/c nude mice. CD63 immunolabeled (a, red) and GFP-postive (b, green) H460 CD63-GFP cells are shown. (c) Merge image of CD63, GFP, and DAPI staining. (d) Anti-CD63 and anti-GFP Western blot of ELMs and cell lysates from H460 CD63-GFP and H460 cells. (e) Electron microscopy images of anti-CD63-labeled and anti-GFP-labeled ELMs isolated from the conditioned medium of H460 CD63-GFP cells. Scale bar = 100 nm. (f, g) Nude mice were intrapleurally injected with 1×106 H460 hCD63-GFP cells. H&E staining of pulmonary tumor tissue shows (f) tumor nodule attached to the pleural surface and (g) tumor nodule in the lung parenchyma.
Tumor-Specific mRNA in Blood and Salivary ELMs from Tumor-Bearing Mice
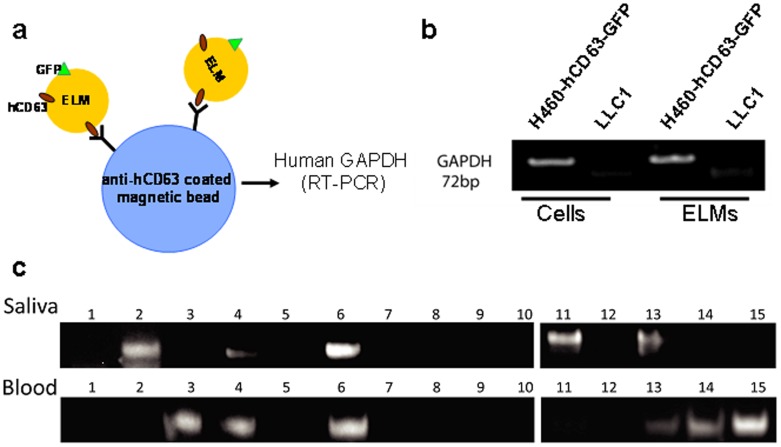
With our in vivo model in place, we chose to explore its ability to determine the capacity of ELMs to shuttle tumor-specific mRNAs throughout the body. To effectively employ this model, we found it imperative to identify ELM-derived transcriptomic indicators of our implanted tumor cells. Given that GAPDH mRNA is abundant in ELMs [47], we elected to pursue it as a potential marker of ELMs exRNA transport. Using anti-hCD63 magnetic beads, we captured hCD63-positive ELMs from both the blood and the saliva of mice implanted with human H460-hCD63-GFP cells (Fig. 3a). To assess the specificity of our primers, we compared human GAPDH RT-PCR results on RNA extracted from H460-hCD63-GFP cells, murine Lewis lung carcinoma LL2/LLC1 cells, and their respective ELMs. Nested RT-PCR revealed that only H460 hCD63-GFP cells and their ELM derivatives were positive for human GAPDH mRNA (Fig. 3b). These results suggest that human GAPDH mRNA is an acceptable marker to detect H460 hCD63-GFP tumor cell-specific mRNA carried by hCD63-positive ELMs.
Figure 3. Detection of human GAPDH mRNA in tumor-bearing mice.
(a) Illustration of anti-hCD63-coated magnetic beads used to isolate hCD63-positive ELMs from saliva, and blood. (b) This native PAGE analysis depicts the 72-bp nested RT-PCR products for human GAPDH from H460-hCD63-GFP cells, LLC1 cells, and (c) the blood and saliva of 15 H460-hCD63-GFP tumor-bearing or control mice.
Next, we evaluated the efficacy of human GAPDH mRNA as an acceptable indicator of ELM-mediated exRNA transportation in our mouse model. Previous works report that mice that have been intrapleurally implanted with 1×106 H460 cells subsequently developed lung tumor nodules within 20 days post-injection [48]. Accordingly, human GAPDH was detected in the blood and salivary ELMs of experimental mice within 20 days of implantation (Fig. 3c and g). 40% and 33% of all tumor-bearing mice carried the human GAPDH mRNA in their blood and salivary ELMs respectively (Fig. 3c; Table 1). These findings suggest that ELMs present in blood and saliva hold tissue-specific, viable, transcriptomic information, and support the hypothesis that ELMs might serve as potential targets of biomarker discovery and development.
Table 1. Detection of human GAPDH mRNA in saliva and blood from mice bearing hCD63-GFP tumors.
| Mouse ID | Blood | Saliva |
| 1 | ND | ND |
| 2 | ND | D |
| 3 | D | ND |
| 4 | D | D |
| 5 | ND | ND |
| 6 | D | D |
| 7 | ND | ND |
| 8 | ND | ND |
| 9 | ND | ND |
| 10 | ND | ND |
| 11 | ND | D |
| 12 | ND | ND |
| 13 | D | D |
| 14 | D | ND |
| 15 | D | ND |
Detection of Tumor-Specific Pprotein in the Blood and Salivary ELMs of Tumor-Bearing Mice
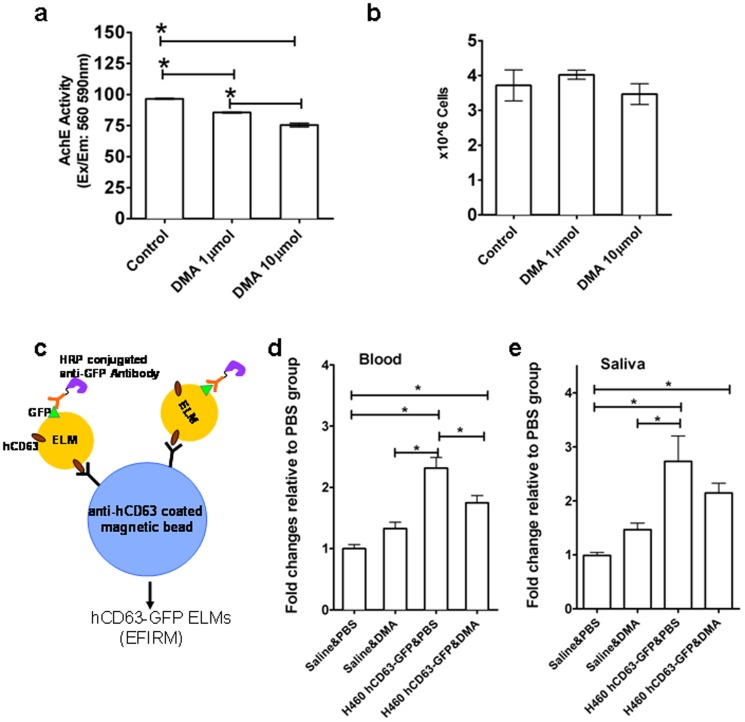
After demonstrating that ELMs contain tissue-specific exRNA molecules, we further probed these microvesicles to determine the likelihood that they contain additional molecular constituents. We focused on the proteomic content of hCD63-GFP-positive ELMs from saliva and blood. In doing so, we employed EFIRM technology (Fig. 4c) [45] to evaluate the blood and salivary hCD63-GFP-positive ELMs from H460 hCD63-GFP tumor-bearing mice exposed to either the ELMs secretion inhibitor dimethyl amiloride (DMA) [43] or PBS. Compared with PBS, DMA-treated H460 hCD63-GFP cells exhibited a non-cytotoxic, dose-dependent decrease in the ELM concentration of conditioned media (Fig. 4a, b). These results were echoed in vivo when, after 7 days of DMA or PBS treatment, the concentration of blood hCD63-GFP-positive ELMs was significantly decreased in DMA-treated mice compared with control animals (Fig. 4d). Salivary hCD63-GFP-positive ELMs was lower in DMA treated mice but not significant (Fig. 4e).
Figure 4. hCD63-GFP-positive ELMs in saliva and blood from tumor-bearing mice.
(a) Acetylcholinesterase quantification of ELMs from the conditioned media of H460-hCD63-GFP cells treated with 1 µmol or 10 µmol DMA for 48 h. (b) Trypan blue exclusion assay of H460-hCD63-GFP cells after 48 h DMA treatment. (c) Illustration of the measurement of hCD63-positive ELMs in saliva and blood using EFIRM technology, allowing the detection of hCD63-GFP positive ELMs by electrochemical sensors. Samples of (d) blood and (e) saliva from control or tumor-bearing mice treated with PBS or DMA for 1 week and assayed for hCD63-GFP-positive ELMs using EFIRM technology. N = 9–11 per group; *P<0.05, statistically significant differences.
Collectively, these outcomes suggest a role for ELMs as conduits of tissue-specific molecular information. Furthermore, this finding highlights the concerted utility of EFIRM as a diagnostic tool and saliva as a diagnostic medium.
Discussion
Discovering saliva-based biomarkers of oral and systemic disease [1], [2] has suggested the possibility of a paradigm shift in the field of molecular diagnostics. Identifying and validating disease-specific molecules in oral fluids could be of great interest to scientists and clinicians alike, and may facilitate the development of early disease diagnostic procedures and screening programs worldwide. Despite the inherent significance here, the mechanism by which markers of distal pathologies come to exist in salivary secretions currently eludes us. Here we show that small secretory lipidic vesicles called ELMs may have a substantial role in contributing to this phenomenon. We demonstrate that ELMs, secreted from distal tissues, encase and protect cell-specific RNAs and proteins within extracellular environments, including blood and saliva. Overall, our results suggest that ELMs play a significant part in transmitting biochemical information from remote cell populations to the oral cavity.
In agreement with our evidence, a recent publication showed that seven validated pancreatic cancer-specific mRNA markers and GAPDH have been detected in mouse salivary and blood-derived exosomes in a pancreatic cancer mouse model, and some of these mRNA markers were abolished when tumor cell exosome biogenesis was blocked [38]. Another interesting study found that mice bearing melanoma tumors have overlapping transcriptomic signatures in two tissues: the tumor itself, and the salivary gland [49]. Yet, it was concluded that salivary transcriptomic regulation was achieved through tumor cell-released mediators, such as growth factors and other inducers [49].
However, whether the induction of salivary molecular signatures were disease specific biomolecules directly shuttled into saliva through ELMs, or the result of a synergistic interplay between salivary glands and tumor-derived mediators, including growth factor and ELMs, remain largely unknown. Our mouse model was injected with human H460-hCD63-GFP lung cancer cells, which subsequently produced species-specific CD63 proteins and GAPDH mRNA, the very markers we detected and reported in blood and saliva. Accordingly, identification of these unique markers in distant tissues and fluids most likely happens after their excretion from xenograft hosts. Moreover, the appearance of saliva-based protein marker in our model showed a small decrease upon inhibition of ELM secretion, detracting from theories suggesting other means of regulation (Fig. 4e).
We however could not completely exclude the possibility that some of them were from metastatic tumor cells. To the best of our knowledge, no evidence of oral metastasis was seen in this widely used NCI-H460 human tumor xenograft model by previous publication [31], [48]. In addition, we did not observe lung metastasis at oral cavity by gross examination in this study. Therefore, we are inclined to conclude that H460-hCD63-GFP tumor-derived ELMs played a formative role in the generation of salivary biomarkers in our model.
Our present results in combination with previous reports demonstrate that ELMs thwart the enzymatic degradation of RNAs by providing a protective milieu [42], [50], [51], [52]. However, previous publication noted an ancillary mechanism of extracellular RNA transport by insinuating that circulating RNAs are associated with and sheltered from destruction by large protein complexes called ribonucleoproteins [53]. We have previously shown that approximately 30% of most common salivary exRNAs contain AU-rich elements (ARE) in their 3′ untranslated regions, which complex with ARE binding protein and protect salivary exRNAs from degradation [54]. In this study, 69% exRNAs identified from saliva of healthy individuals were present in salivary ELMs (Fig.S1a in Supporting information S1). We therefore speculate that most salivary exRNAs reside within ELMs. However, we cannot rule out the possibility of ribonucleoprotein contamination in our ELM samples. Our method of isolation employed the commercially available Exoquick kit to precipitate ELMs from bodily fluids [39], a technology that is relatively new and has not been thoroughly characterized [39]. Therefore, we cannot conclude that our ELM preparations were ribonucleoprotein-free, which may have affected our results. To overcome this issue, future experiments should employ ultracentrifugation-based ELM isolation protocols. Although this procedure requires larger volumes of fluid [42], it is the gold standard for purifying microvesicles and should quell any contamination concerns.
As mentioned previously, a number of tumor cells commonly shed ELMs into the peripheral circulation [55], [56]. It has also been demonstrated that ELMs exist within salivary secretions and carry transcriptomic information [42]. Although the origin of salivary ELMs remains unknown, here we provide evidence for their etiology by illustrating that peripherally secreted ELMs traverse the vasculature and land on oral cavity. We reveal this phenomenon by verifying that existence of seven salivary mRNA markers [57] in the malignant pleural effusion derived ELMs under same clinical setting (Table S2 in Supporting information S1). Four of these markers (BRAF [58], EGFR [59], LZTS1 [60], and FGF19 [61], [62]) have been directly correlated with lung cancer development and progression. These findings implicate ELMs as carriers of tissue-specific biomolecules throughout the vasculature and within bodily fluids.
To effectively investigate our hypothesis, we found it most convincing to pursue an animal model whose ELM constituents would be undeniably discriminatory. Using xenograft mice for this purpose is an ideal way to establish the fundamentals of our proposed mechanism, as implanted H460A cells should release species-specific ELMs that contain human molecular information. Identifying these markers in murine blood and saliva provides sufficient credence for the concept of ELM-based biomarker discovery using saliva.
Despite the strength of our design, a number of issues must be considered when determining the collective significance of the experimental outcomes. First, although we revealed species-specific ELM protein and transcript in the blood and saliva of our xenograft mouse model, we cannot justifiably conclude that they are identical. In other words, the structure and molecular constituency of ELMs respectively derived from blood or saliva may be fundamentally unique. Blood-based ELMs and their contents may be altered prior to their subsequent introduction into the oral cavity. Although our observations are in agreement with previous reports [47], we concede that the data presented here does not eliminate or definitively describe the interplay of ELMs within salivary gland tissues. Comprehensive proteomic and transcriptomic characterizations of hCD63-GFP–positive ELMs found in bodily fluids are needed to shed light on these issues. Future studies using patient-derived lung cancer xenograft mouse model containing a variety of genetic aberrations, and comprehensively proteomic and transcriptomic characterizations of blood and saliva ELMs could further our understanding of the role of ELMs in the circulation of tumor-specific bimolecules as well as the biogenesis of salivary biomarkers.
Second, detecting ELMs in oral fluids presented a substantial obstacle. Current assays, including Western blotting and acetylcholinesterase assay, proved unreliable owing to the small volumes of blood and saliva samples that are obtainable from murine subjects. Our solution, EFIRM, involves two antibodies that bind to non-overlapping hCD63-GFP fusion protein [45]. Because ELMs are commonly characterized by their CD63 membrane-bound proteins, there is an inherent concern of non-specific binding by murine ELMs to anti-human CD63 magnetic beads. To address this issue, we tested the specificity of EFIRM and found that mouse salivary ELMs produced a signal similar to background [45]. This outcome substantiates the specificity of this technique, and more importantly supports our aforementioned EFIRM data indicating the existence of human-specific molecules in the saliva and blood of our xenograft mouse model.
When the data is considered in its totality, one interesting value presents itself as somewhat of an aberration. Two tumor-bearing mice had human GAPDH mRNA in their saliva, but not their blood. While we speculate that distant pathologies secrete ELMs into the vasculature, these data insinuate that might not be the case. One possible explanation for this inconsistency may be the location of the tumorigenic cells. As it turns out, sputum (a respiratory expectorate used in lung cancer biomarker analysis [63], [64]) might be a means of bypassing the circulatory system. In this scenario, cancer cells or microvesicles could travel to the oral cavity via the pharynx, leaving the blood absent of disease-specific markers. This eventuality may be enhanced by the anatomical position of the neoplasm within the lungs. Hence, a lack of blood-derived lung cancer markers would not disqualify our model; on the contrary, it would strengthen the suggested efficacy of oral fluids as a diagnostic medium. In any case, additional investigations will be required to delineate the communicative interaction between the oral cavity and distant diseases.
Although our report does not definitively explain the etiology of salivary biomarkers, here we describe the detection of tumor cell-specific molecules in ELMs derived from saliva. Overall, our data suggest that ELMs released from distant tissues are involved in this process by serving as protective conduits of biochemical information. Considering this information, we put forth the idea that saliva is an effective source of discriminatory biomarkers and suggest that salivary ELMs are instrumental in their presentation.
Supporting Information
Supporting information, figure and tables.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data can be found at the GEO database. Accession “GSE50700” is now open to public. The link is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50700.
Funding Statement
This study was supported by a Ruth L. Kirschstein National Research Service Award (grant T32-DE07296) and the National Institute of Dental and Craniofacial Research/National Institutes of Health (grants UO1-DE017790 and RO1-DE15970). The project described was also supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health (grant UL1TR000124). It was also supported by the California Tobacco-Related Diseases Research Program (TRDRP) 20PT-0032, 21RT-0112 and also DoD grant LS110207. Support was also received from a Faculty Seed Grant from the Univerity of California, Los Angeles School of Dentistry (grant 441901-69749-FWEI-FY11DR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hu S, Wang J, Meijer J, Ieong S, Xie Y, et al. (2007) Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum 56: 3588–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Y, St John MA, Zhou X, Kim Y, Sinha U, et al. (2004) Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 10: 8442–8450. [DOI] [PubMed] [Google Scholar]
- 3. Lee YH, Wong DT (2009) Saliva: an emerging biofluid for early detection of diseases. Am J Dent 22: 241–248. [PMC free article] [PubMed] [Google Scholar]
- 4. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, et al. (2010) The Human Oral Microbiome. Journal of Bacteriology 192: 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, et al. (2010) Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol 48: 2965–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, et al. (2010) Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138: 947–e941 – [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang L, Xiao H, Karlan S, Zhou H, Gross J, et al. (2010) Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One 5: e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Streckfus C, Bigler L, Dellinger T, Dai X, Kingman A, et al. (2000) The Presence of Soluble c-erbB-2 in Saliva and Serum among Women with Breast Carcinoma: A Preliminary Study. Clinical Cancer Research 6: 2363–2370. [PubMed] [Google Scholar]
- 9. Brinkmann O, Wong DT (2011) Salivary transcriptome biomarkers in oral squamous cell cancer detection. Advances in clinical chemistry 55: 21–34. [DOI] [PubMed] [Google Scholar]
- 10. Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, et al. (2012) Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol 227: 658–667. [DOI] [PubMed] [Google Scholar]
- 11. Farrell JJ ZL, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61: 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu S, Vissink A, Arellano M, Roozendaal C, Zhou H, et al. (2011) Identification of autoantibody biomarkers for primary Sjögren's syndrome using protein microarrays. PROTEOMICS 11: 1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee Y-H, Kim J, Zhou H, Kim B, Wong D (2012) Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. Journal of Molecular Medicine 90: 427–434. [DOI] [PubMed] [Google Scholar]
- 14. Thery C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9: 581–593. [DOI] [PubMed] [Google Scholar]
- 15. Yang C, Robbins PD (2011) The Roles of Tumor-Derived Exosomes in Cancer Pathogenesis. Clinical and Developmental Immunology 2011: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dimov I, Jankovic Velickovic L, Stefanovic V (2009) Urinary Exosomes. TheScientificWorldJOURNAL 9: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lotvall J, Valadi H (2007) Cell to Cell Signalling via Exosomes Through esRNA. Cell Adhesion & Migration 1: 156–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Putz U, Howitt J, Doan A, Goh C-P, Low L-H, et al. (2012) The Tumor Suppressor PTEN Is Exported in Exosomes and Has Phosphatase Activity in Recipient Cells. Sci Signal 5 ra70-. [DOI] [PubMed] [Google Scholar]
- 19. Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2: 569–579. [DOI] [PubMed] [Google Scholar]
- 20. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- 21. Vlassov AV, Magdaleno S, Setterquist R, Conrad R (2012) Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta (BBA) - General Subjects 1820: 940–948. [DOI] [PubMed] [Google Scholar]
- 22. Cocucci E, Racchetti G, Meldolesi J (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol 19: 43–51. [DOI] [PubMed] [Google Scholar]
- 23. Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, et al. (2005) Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci 118: 2849–2858. [DOI] [PubMed] [Google Scholar]
- 24. Webber J, Clayton A (2013) How pure are your vesicles? J Extracell Vesicles 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Record M, Subra C, Silvente-Poirot S, Poirot M (2011) Exosomes as intercellular signalosomes and pharmacological effectors. Biochemical Pharmacology 81: 1171–1182. [DOI] [PubMed] [Google Scholar]
- 27. Hess C, Sadallah S, Hefti A, Landmann R, Schifferli JA (1999) Ectosomes released by human neutrophils are specialized functional units. J Immunol 163: 4564–4573. [PubMed] [Google Scholar]
- 28. MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, et al. (2001) Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity 15: 825–835. [DOI] [PubMed] [Google Scholar]
- 29. Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, et al. (2001) Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med 7: 297–303. [DOI] [PubMed] [Google Scholar]
- 30. Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 10: 42–46. [DOI] [PubMed] [Google Scholar]
- 31. Taylor DD, Lyons KS, Gercel-Taylor C (2002) Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecol Oncol 84: 443–448. [DOI] [PubMed] [Google Scholar]
- 32. Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, et al. (2009) High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One 4: e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, et al. (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9: e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bland J (2008) The Importance of Functional Biomarkers in the Management of Chronic Illness. Altern Ther Health Med 14: 18–21. [PubMed] [Google Scholar]
- 35. Bossuyt PM (2011) THe thin line between hope and hype in biomarker research. JAMA: The Journal of the American Medical Association 305: 2229–2230. [DOI] [PubMed] [Google Scholar]
- 36. Frangogiannis NG (2012) Biomarkers: hopes and challenges in the path from discovery to clinical practice. Translational Research 159: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lasser C, Seyed Alikhani V, Ekstrom K, Eldh M, Torregrosa Paredes P, et al. (2011) Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. Journal of Translational Medicine 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, et al.. (2013) Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. [DOI] [PMC free article] [PubMed]
- 39. Taylor DD, Zacharias W, Gercel-Taylor C (2011) Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 728: 235–246. [DOI] [PubMed] [Google Scholar]
- 40. Sreekumar PG, Kannan R, Kitamura M, Spee C, Barron E, et al. (2010) alphaB crystallin is apically secreted within exosomes by polarized human retinal pigment epithelium and provides neuroprotection to adjacent cells. PLoS One 5: e12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, et al. (1996) B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, et al. (2010) Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One 5: e8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, et al. (2010) Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest 120: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei F, Patel P, Liao W, Chaudhry K, Zhang L, et al. (2009) Electrochemical sensor for multiplex biomarkers detection. Clin Cancer Res 15: 4446–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei F, Yang J, Wong DT (2013) Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron 44C: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee YH, Zhou H, Reiss JK, Yan X, Zhang L, et al. (2011) Direct saliva transcriptome analysis. Clin Chem 57: 1295–1302. [DOI] [PubMed] [Google Scholar]
- 47. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kraus-Berthier L, Jan M, Guilbaud N, Naze M, Pierre A, et al. (2000) Histology and sensitivity to anticancer drugs of two human non-small cell lung carcinomas implanted in the pleural cavity of nude mice. Clin Cancer Res 6: 297–304. [PubMed] [Google Scholar]
- 49. Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, et al. (2009) Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One 4: e5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, et al. (2010) Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 4: 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P (2011) Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park NJ, Li Y, Yu T, Brinkman BM, Wong DT (2006) Characterization of RNA in saliva. Clin Chem 52: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, et al. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Palanisamy V, Park NJ, Wang J, Wong DT (2008) AUF1 and HuR proteins stabilize interleukin-8 mRNA in human saliva. J Dent Res 87: 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. De la Pena H, Madrigal JA, Rusakiewicz S, Bencsik M, Cave GWV, et al. (2009) Artificial exosomes as tools for basic and clinical immunology. Journal of Immunological Methods 344: 121–132. [DOI] [PubMed] [Google Scholar]
- 56. Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, et al. (2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360: 295–305. [DOI] [PubMed] [Google Scholar]
- 57. Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, et al. (2012) Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 69: 3341–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pao W, Girard N (2011) New driver mutations in non-small-cell lung cancer. Lancet Oncol 12: 175–180. [DOI] [PubMed] [Google Scholar]
- 59. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 60. Nonaka D, Fabbri A, Roz L, Mariani L, Vecchione A, et al. (2005) Reduced FEZ1/LZTS1 expression and outcome prediction in lung cancer. Cancer Res 65: 1207–1212. [DOI] [PubMed] [Google Scholar]
- 61. Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, et al. (2008) Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene 27: 85–97. [DOI] [PubMed] [Google Scholar]
- 62. Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, et al. (2011) Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 19: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Petty TL (2000) The early identification of lung carcinoma by sputum cytology. Cancer 89: 2461–2464. [DOI] [PubMed] [Google Scholar]
- 64. Gray RD, Duncan A, Noble D, Imrie M, O'Reilly DS, et al. (2010) Sputum trace metals are biomarkers of inflammatory and suppurative lung disease. Chest 137: 635–641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information, figure and tables.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data can be found at the GEO database. Accession “GSE50700” is now open to public. The link is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50700.