Abstract
SUMO, a small ubiquitin-related modifier, is known to covalently attach to a number of nuclear regulatory proteins such as p53, IκB, promyelocytic leukemia protein and c-Jun. The sumoylation reaction is catalyzed by the SUMO protease, which exposes the C-terminal active glycine residue of the nascent SUMO, the heterodimeric SUMO activating enzyme, the SUMO conjugating enzyme, Ubc9, and SUMO protein ligases, in a manner similar to ubiquitinylation. Identification of SUMO-regulated proteins is hampered by the fact that many sumoylated proteins are present at a level below normal detection limit. This limitation was overcome by either in vivo overexpression of Myc-SUMO or in vitro sumoylation with excess biotin–SUMO and Ubc9. Sumoylated proteins so obtained were affinity purified or isolated by immunoprecipitation. The isolated sumoylated proteins were identified by sequence analysis using mass spectrometric methods. Results reveal that several heterogeneous nuclear ribonucleoproteins (hnRNPs), zinc finger proteins, and nuclear pore complex proteins were sumoylated. The sumoylation of hnRNP A1, hnRNP F, and hnRNP K were confirmed in vivo by coimmunoprecipitation. In view of the facts that hnRNPs have been implicated in RNA splicing, transport, stability, and translation, our findings suggest that sumoylation could play an important role in regulating mRNA metabolism.
Reversible covalent modification of proteins is a widely used regulatory mechanism for transmitting biological signals and for regulating the activity, biosynthesis, and degradation of major enzymes. This is due, in part, to its enormous capacity for integrating biological information and for signal amplification (1, 2). In addition to modification with low molecular weight modifiers, such as phosphorylation, nucleotidylation, and acetylation, ubiquitin is a well documented posttranslational protein modifier. Ubiquitinylation involves the covalent attachment of the C terminus of ubiquitin to the ε-amino moiety of a specific lysyl residue in modified proteins. In general, polyubiquitinylation tends to associate with the 26 S proteasome-mediated protein degradation, whereas monoubiquitinylation has been reported to be involved in receptor endocytosis, protein sorting, subnuclear trafficking, meiosis, and chromatin remodeling (3–5). To date, there are >10 known ubiquitin-like proteins that have been shown to ligate to other target protein molecules. Among them, small ubiquitin-related modifier (SUMO) is the most studied modifier. It possesses only an 18% identity in sequence homology to ubiquitin; nevertheless, its 3D structure is very similar to that of ubiquitin. SUMO has been shown to ligate to numerous proteins and modulate their translocation, activity, or stability (6–9). In view of the fact that, in mammalian cells, there are two more SUMO homologs, SUMO2 and SUMO3, additional functions of protein sumoylation have yet to be identified.
Similar to ubiquitinylation, covalent attachment of SUMO to its target proteins requires three or four enzymes, namely: (i) SUMO protease that cleaves the C terminus of the nascent SUMO to expose the C-terminal glycine residue; (ii) the heterodimeric SUMO-activating enzyme that separates the adenylation and thio-esterification functions into the SAE1 (Aos1 in yeast) and the SAE2 (Uba2 in yeast) subunits, respectively; (iii) Ubc9, a SUMO-conjugating enzyme that ligates directly to its protein substrate, such as RanGAP1; and (iv) an E3-like SUMO ligase (10, 11). In contrast to the large number of ubiquitin ligases that have been found, three SUMO E3s have been identified so far. Mammalian protein inhibitors of activated STAT (PIAS) act as SUMO E3s (12) for LEF1, p53, and c-Jun. RanBP2 is a nuclear pore complex-associated protein that has SUMO E3 activity toward Sp100 and histone deacetylase (13, 14). Pc2, a polycomb group protein, is a SUMO E3 that acts on C-terminal binding protein (15). All three E3s have RING-like domains and act as nonenzymatic adapter proteins that recruit the substrate proteins, SUMO, and Ubc9. Several different SUMO hydrolases have been characterized that specifically cleave the isopeptide bonds between SUMO and its protein substrates. These hydrolases likely participate in determining in vivo steady state levels of sumoylated proteins (16–18).
Most of SUMO substrate proteins found so far are important regulatory proteins, such as RanGAP1, PCNA, IκB, p53, c-jun, topoisomerases, promyelocytic leukemia protein, mitogen activated protein kinase kinase 1 (MEK1), and transcription factor Sp3. Many SUMO substrates are transcription factors or cofactors (8). The transcriptional synergy control sites map to the sumoylation consensus signature motifs, ψKX(E/D), ψ stands for a large hydrophobic amino acid residue (19). Sumoylation clearly plays a regulatory role in transcription.
Eukaryotic RNA transcripts must contain information that specifies their synthesis, splicing, nuclear export, subcellular localization, stability, silencing, and translation. An important theme has emerged in the past few years that much of the information is provided by the family of proteins collectively referred to as heterogeneous nuclear ribonucleoproteins (hnRNPs). Most hnRNPs shuttle between cytoplasm and nucleus and participate in posttranscriptional RNA processing steps in the RNA metabolism from transcription to translation (20). hnRNPs associate with the transcripts, pre-mRNA, and mRNA, and exhibit structural and functional diversity and dynamics (20, 21). The best studied hnRNPs are from human and are designated from A1 (34 kDa) to U (120 kDa). Sumoylation of hnRNPs could provide a versatile regulatory mechanism for the regulation of RNA transcript processing. Whether sumoylation of hnRNPs occurs has not been reported. We used proteomic approaches to identify SUMO substrates by mass spectrometry, and found several hnRNPs, zinc finger proteins, and nuclear pore proteins as SUMO substrate candidates in vivo and in vitro. Sumoylation of these proteins opens the likelihood of the regulation of RNA metabolism at the transcriptional as well as posttranscriptional levels by sumoylation.
Materials and Methods
Materials. Human embryonic kidney 293 Tet-On cell line, pTRE2hyg2-Myc, pTRE2hyg2-Myc-luciferase (Luc), hygromycin, and doxycycline (Dox) were purchased from BD Biosciences/Clontech. Antibiotic G418, goat anti-mouse IgG FITC conjugate, protein A agarose, anti-β-actin antibody, protease inhibitor cocktails, and 4′-6-diamidino-2-phenylindole hydrochloride (DAPI) were purchased from Sigma. Anti-Myc, anti-SUMO, anti-hnRNP A1, anti-RanGAP1, anti-RanBP2 antibodies, and agarose conjugated with anti-Myc antibody were purchased from Santa Cruz Biotechnology. Anti-hnRNP K antibody was a gift from K. Bomsztyk (University of Washington Medical Center, Seattle). Anti-hnRNP F was a gift from D. Black (University of California, Los Angeles). Sequencing-grade trypsin was purchased from Promega.
Plasmid Construction. Escherichia coli expression plasmids for biotin–SUMO and His-Ubc9 were described as in ref. 22. The cDNA encoding the processed SUMO(1–97) with GlyGly at the C terminus was amplified by PCR and inserted into pTRE2hyg2-Myc as an NheI/ClaI fragment to construct a pTRE2hyg2-Myc-SUMO plasmid.
Cell Culture, Transfections, and Immunofluorescence. HEK 293 Tet-On cells were cultured in DMEM supplemented with 10% Tet-system approved FBS (BD Biosciences/Clontech) and 100 μg/ml of antibiotic G418. pTRE2hyg2-Myc-SUMO and pTRE2hyg2-Myc-Luc were transfected into HEK 293 Tet-On cells by using FuGENE 6 (Roche Molecular Biochemicals). Transfected cells were selected with 300 μg/ml hygromycin. Single cell colonies were isolated to generate cell lines highly and stably expressing Myc-tagged SUMO and Myc-tagged Luc. For immunof luorescence, HEK 293 Tet-On cells harboring pTRE2hyg2-Myc-SUMO were seeded onto eight-well Lab-Tek chamber slide (Nalge Nuc International) and induced with 2 μg/ml Dox for 48 h. Cells were fixed in 3.7% formaldehyde and permeablized with 0.1% Triton X-100. After incubation with anti-Myc primary antibody, the slide was rinsed in 1× PBS (6.5 mM phosphate, pH 7.2/2.7 mM KCl/137 mM NaCl) and incubated with FITC conjugated goat anti-mouse IgG secondary antibody followed by a 5-min incubation in DAPI. After five washes with 1× PBS, images were captured on a Zeiss LSM-5 confocal microscope.
Immunoprecipitation and Immunoblotting. For immunoprecipitation, pTRE2hyg2-Myc-SUMO transfected HEK 293 Tet-on cells were lysed in lysis buffer A (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40/5 mM EDTA/20 mM N-ethylmaleimide/1 mM PMSF/10 μg/ml pepstatin/20 μg/ml leupeptin/10 μg/ml aprotinin). A total of 200 μg of clarified whole cell extracts were incubated with 10 μg of primary antibody for 1 h and then precipitated with 100 μl of 50% protein A-agarose in lysis buffer A. For anti-Myc antibody immunoprecipitation, the agarose conjugated with anti-Myc antibody was used and no protein A-agarose was added subsequently. After being washed five times in lysis buffer A, the agarose beads were boiled in the 4× NuPage loading buffer and resolved by 4–12% NuPage gels (Invitrogen). For immunoblotting, proteins from NuPage gels were transferred onto poly(vinylidene difluoride) (PVDF) membranes and probed with primary antibodies, and the corresponding HRP conjugated secondary antibodies. After five washings, membranes were incubated with Super Signal West Dura (Pierce) substrates and exposed to Kodak BioMax XAR films.
In Vitro Sumoylation and Purification of SUMO-Conjugated Proteins. Biotin–SUMO was purified as described (22), and 6× His-Ubc9 was purified on Ni-NTA column (Qiagen, Valencia, CA). One liter equivalent of HeLa cell nuclei extract (Biovest) was resespended in lysis buffer B [50 mM Tris·HCl, pH 7.5/50 mM NaCl/1 mM EDTA/0.5 mM EGTA/0.25% Nonidet P-40/0.05% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/0.125% sodium deoxycholate/2.5 mM DTT/1 mM PMSF/10 μg/ml pepstatin/20 μg/ml leupeptin/10 μg/ml aprotinin]. The suspension was sonicated and clarified by centrifugation at 14,000 × g for 15 min. For in vitro SUMO-conjugation assay, 40 μg of biotin–SUMO and 2 μg of 6× His-UBC9 were incubated in 2 ml of the HeLa nuclei extract at 37°C for 45 min in 50 mM Hepes (pH 7.5) buffer containing 5 mM MgCl2, 1 mM DTT, 2 mM ATP, and 10 units of inorganic pyrophosphatase. At the end of the reaction, N-ethylmaleimide was added to a final concentration of 10 mM. The reaction was applied directly to a 1-ml Softlink avidin column (Promega) equilibrated with 50 mM Hepes (pH 7.5) containing 5 mM MgCl2 and 1 mM DTT. The column was washed with 10 volumes of 50 mM Hepes (pH 7.5) containing 0.5 M NaCl and 1 mM DTT followed by 10 volumes of 50 mM Hepes (pH 7.5) containing 0.1 M NaCl and 1 mM DTT. SUMO-conjugated proteins were eluted off the column by using 5 mM biotin in 50 mM Hepes (pH 7.5), 0.1 M NaCl, and 1 mM DTT. Eluted fractions were pooled together and stored at 4°C until further use.
Two-Dimensional Gel Electrophoresis. Two-dimensional gel electrophoresis (2-DE) was performed by using either 7- or 18-cm IPG gel strips (pH 3–10, linear; Amersham Pharmacia). Ten and 50 μg of proteins were used for 7- and 18-cm IPG gel strips, respectively. IPG gel strips were focused by ramping up the voltage slowly over a 16- to 20-h period for a total of ≈30,000 and 50,000 Vh for 7- and 18-cm gel strips, respectively. After equilibration, the proteins on gel strips were resolved by SDS/10% PAGE. The 7-cm 2D gels were transferred onto poly(vinylidene difluoride) membrane and probed with anti-Myc antibody; the 18-cm 2D gels were silver stained.
Mass Spectrometric Peptide Sequencing and Protein Identification. Affinity-purified biotin–SUMO-conjugated proteins were precipitated with acetone and redissolved in 100 μl of 6 M urea in 0.1 M Tris·HCl (pH 7.5). Proteins were reduced with 10 mM DTT at room temperature for 1 h and alkylated with 20 mM iodoacetamide for 1 h. The protein mixture was diluted with nine volumes of 25 mM NH4HCO3, and 20 μg of sequencing-grade trypsin were added. Immunoprecipitated Myc-SUMO conjugated proteins were directly digested with 20 μg of sequencing-grade trypsin. The digestion mixtures were incubated at 30°C overnight. The reaction was stopped by adding formic acid to pH 5.
One-dimensional liquid chromatography (LC)/MS analyses of the tryptic digest were carried out by using a modified configuration of the ProteomeX 2D LC/MS workstation (Thermo Finnigan, San Jose, CA). The strong cation exchange (SCX) column was removed from the system, and the tubings were reconnected with a union. The two reversed-phase columns were replaced by two Zorbax 300SB-C18 peptide traps (Agilent Technologies, Wilmington, DE), whereas the ESI source was replaced by a nanospray ionization source and a reversed-phase PicoFrit column (BioBasic C18, 75 μm × 10 cm, tip = 15 μm, New Objective, Woburn, MA). The PicoFrit column was placed directly in front of the ion transfer tube of LCQ DecaXP plus mass spectrometer. The peptides were loaded onto one of the traps by using an autosampler. After washing with solvent A (0.1% formic acid), the peptides were eluted by 0–60% solvent B (acetonitrile) in solvent A for 30 min at a flow rate of ≈200 nl/min using a splitter (75 μl/min before the splitting). The mass/charge (m/z) ratios of peptides and their fragmented ions were recorded by using a data acquisition method that allows the acquisition of three MS2 scans (i.e., top three most intensive peaks were fragmented) after each full MS scan. The raw data files were searched against the human database by using bioworks software (ThermoFinnigan, San Jose, CA), a modified version of sequest based on the algorithm developed by Eng et al. (23).
Results
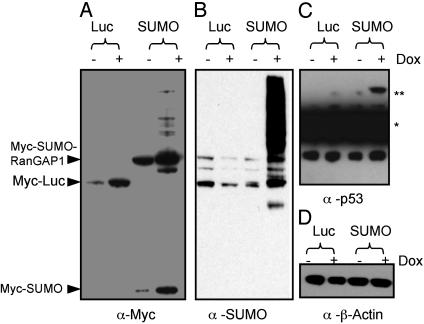
Overexpression of SUMO Increased in Vivo Sumoylation Levels. To overcome the difficulty of identifying low abundance of sumoylated proteins in cells, we set up a stable Myc-SUMO overexpression cell line that generated dramatically higher overall sumoylated protein level than that of untransfected cells. Moreover, the expression of Myc-SUMO is controllable by Dox induction. As shown in Fig. 1A, when induced by Dox, the Myc-SUMO transfected cells expressed at least 10-fold higher free Myc-SUMO, compared to uninduced cells, and some previously undetectable sumoylated proteins also became apparent. The SUMO-RanGAP1 is the most abundant sumoylated protein in cells (24), which was highly sumoylated even in uninduced cells. The Myc-Luc was also found to increase at least 10-fold after induction by Dox (Fig. 1 A). Compared to uninduced cells, the overall cellular sumoylated proteins increased significantly in Myc-SUMO overexpressed cells (Fig. 1B). p53, a known substrate for sumoylation (25), was also found to increase its level of sumoylation in cells overexpressing Myc-SUMO (Fig. 1C). β-actin was used to show equal loading of whole cell extracts (Fig. 1D).
Fig. 1.
Stable cell line expresses elevated SUMO and sumoylated proteins. HEK 293 Tet-On cells were transfected with either pTRE2hyg2-Myc-SUMO or pTRE2hyg2-Myc-Luc vector. Cells stably expressing Myc-SUMO and Myc-Luc were selected with 300 μg/ml hygromycin. After a 48-h incubation with or without 2 μg/ml Dox, whole-cell extracts were resolved by using NuPage gels and probed with anti-Myc (A), anti-SUMO (B), anti-p53 (C), or anti-β-actin (D) antibody. The p53 is marked with a single asterisk, and the sumoylated p53 is marked with double asterisks.
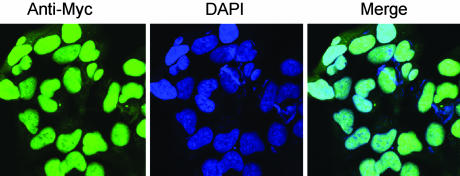
Immunofluorescence showed that Myc-SUMO and its ligated proteins are mainly localized in the nucleus (Fig. 2), which is in agreement with the previous finding (7). In addition, all of the cells contained Myc-tagged sumoylated proteins, indicating that the Myc-SUMO cell lines were successfully established to stably overexpressing Myc-SUMO, and the Myc-tag did not appear to alter the sumoylation capacity of Myc-SUMO.
Fig. 2.
Myc-tagged SUMO and sumoylated proteins mainly localized in the nucleus. After Myc-SUMO stably expressed HEK 293 Tet-On cells were induced with 2 μg/ml Dox for 48 h, cells were fixed with 3.7% formaldehyde and stained with anti-Myc primary antibody and FITC-conjugated secondary antibody. DAPI was used to stain nuclei.
Together, these results indicate that the overexpressed Myc-SUMO functions as the endogenous SUMO and the elevation of the cellular sumoylated protein level corresponds to the overexpressed Myc-SUMO. Therefore, with this system, it is possible to identify low abundant SUMO target proteins that are difficult to find by usual methods.
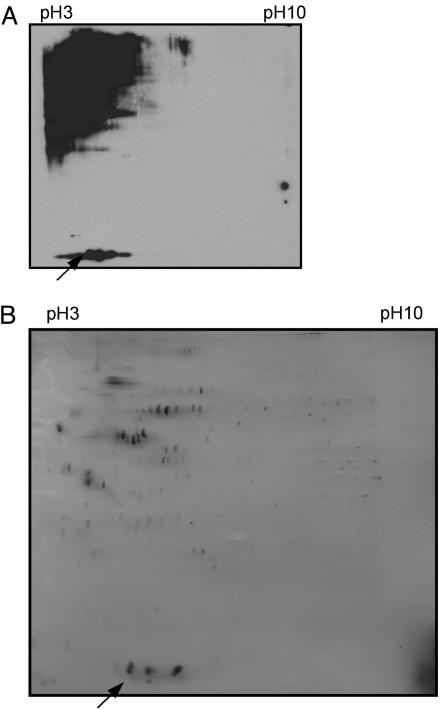
SUMO-Conjugated Proteins Were Isolated in Vivo and in Vitro. For the isolation of sumoylated proteins from Myc-SUMO stably expressing cells, 5 mg of whole cell lysates were used for immunoprecipitation with agarose conjugated with anti-Myc antibody. The immunoprecipitates bound to beads were directly digested by trypsin. Another cell line stably expressing Myc-Luc was used as a control. The SUMO target proteins were also purified in vitro. To do this, biotin–SUMO and enzymatically nonactive biotin–SUMO-ΔGG (biotin–SUMO with C-terminal GG deleted) were used for the in vitro sumoylation reaction. HeLa nuclear extracts were incubated with excess biotin–SUMO and Ubc9. To preserve the integrity of the SUMO conjugates during purification, N-ethylmaleimide was added at the end of sumoylation reaction. The SUMO-modified proteins were affinity purified by column chromatography with monomeric avidin. The choice of monomeric avidin for purification of SUMO conjugates had a considerable advantage over other types of affinity chromatography because this resin allowed a specific elution of biotin SUMO and its conjugated proteins under native conditions. Two-dimensional electrophoresis followed by silver staining of proteins revealed numerous sumoylated proteins (Fig. 3B). The majority of these proteins were acidic, with pI ranging from 3.5 to 6.0 and molecular masses >50 kDa. The overall 2D pattern of the in vitro sumoylated proteins is similar to that of in vivo sumoylation (Fig. 3A). These data indicate that in vitro sumoylation may target similar proteins as those revealed in vivo.
Fig. 3.
Cellular sumoylated proteins show a similar pattern to in vitro isolated sumoylated proteins in 2D gel electrophoresis. Whole cell extracts from HEK 293 Tet-On cells expressing Myc-SUMO were resolved by using 7-cm 2D gel and probed with anti-Myc antibody (A). The Myc-SUMO is marked with an arrow. The in vitro isolated sumoylated proteins were resolved by using 18-cm 2D gel and silver stained (B). Biotin–SUMO is also marked with an arrow.
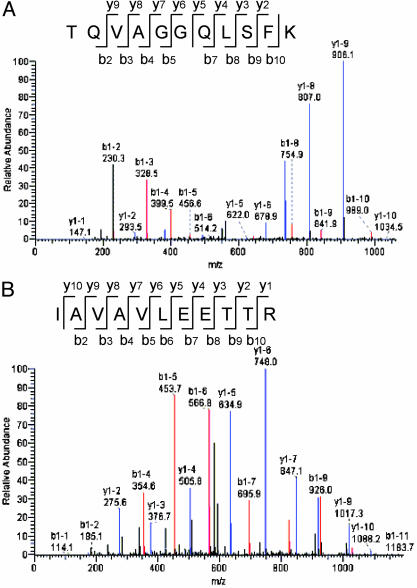
SUMO Substrate Proteins Were Identified by LC/Tandem MS (MS/MS). The in vivo and in vitro isolated sumoylated proteins were trypsinized and identified by LC/MS/MS. Representative MS/MS spectra for one of the peptides detected for RanGAP1 and RanBP2, respectively, are shown in Fig. 4. In both cases, >85% of the (+1) charged y and b ions are covered in the spectra. Putative SUMO target proteins identified from in vivo and in vitro methods are shown in Table 1. The LC/MS/MS sequence analyses of the SUMO-conjugated proteins identified four known SUMO substrates: RanGAP1, RanBP2, DNA topoisomerase II, and sterol regulatory element-binding transcription factor, as well as a number of previously undescribed SUMO substrates (Table 1). The identification of known sumoylated proteins confirms the validity of our method and demonstrates how a proteomic approach can be used to effectively identify SUMO targeted proteins. Proteins appearing in control samples were excluded from our list. All identified proteins contained at least one sumoylation consensus sequence ψKX(E/D) (19). Interestingly, six hnRNPs, including hnRNP A1, hnRNP F, and hnRNP K, and five zinc finger proteins were found to be sumoylated.
Fig. 4.
The MS/MS spectra of RanGAP1 peptide (A), TQVAGGQLSFK, and RanBP2 peptide (B), IAVAVLEETTR, are detected from the LC/MS analyses. Only the (+1) charged ions were indicated in the spectra. Blue vertical lines indicated matches to the theoretical y ions, and red lines indicate matches to the theoretical b ions.
Table 1. Identified SUMO substrates.
| Protein name | GenBank accession number | Number of peptides identified* | Consensus sequence† | sumoplot score† |
|---|---|---|---|---|
| hnRNPs | ||||
| hnRNP A1 | X12671 | 2 | IKED | 0.9444 |
| hnRNP A3 | P51991 | 1 | IKED | 0.9444 |
| hnRNP F | NM_004966 | 1 | LKKD | 0.9056 |
| hnRNP H1 | NM_005520 | 7 | LKKD | 0.9056 |
| hnRNP K | NM_025279 | 2 | IKID | 0.9444 |
| hnRNP U | X65488 | 1 | LKEE | 0.9056 |
| Zinc finger proteins | ||||
| Zinc finger protein 226 | AF228418 | 3 | IKNE | 0.9444 |
| Zinc finger protein 6 | X56465 | 2 | IKTE | 0.9444 |
| Zinc finger protein 221 | AF187987 | 2 | FKCE | 0.8500 |
| Zinc finger protein 409 | AB028979 | 1 | IKEE | 0.9444 |
| Zinc finger protein 291 | AF242528 | 1 | IKKE | 0.9056 |
| Nuclear pore complex proteins | ||||
| Ran GTPase-activating protein 1 (RanGAP1)‡ | X82260 | 7 | LKSE | 0.9056 |
| Ran-binding protein 2 (RanBP2)‡ | S66431 | 19 | IKSE | 0.9444 |
| Ran-binding protein 16 (exportin 7) | AF064729 | 1 | VKVE | 0.9278 |
| Nuclear pore complex protein | AF071076 | 2 | FKAE | 0.8500 |
| Nup98-Nup96 | ||||
| Others | ||||
| Human DNA topoisomerase II, β* | X68060 | 1 | VKVE | 0.9278 |
| Sterol regulatory element-binding transcription factor* | NM_004599 | 1 | IKTD | 0.9444 |
| MAPK/ERK kinase kinase 1 | Q13233 | 2 | IKDE | 0.9444 |
| FUSE binding protein (FBP) | A53184 | 14 | FKPD | 0.8500 |
| FUSE binding protein 2 (FBP2) | U69126 | 2 | FKQD | 0.8500 |
MAPK/ERK, mitogen-activated protein kinase/extracellular signal-related kinase; FUSE, far upstream element.
The number of amino acid residues in identified peptides ranges from 10 to 25.
Sumoylation consensus sequence and theoretical score were generated by sumoplot software.
Previously known SUMO substrates.
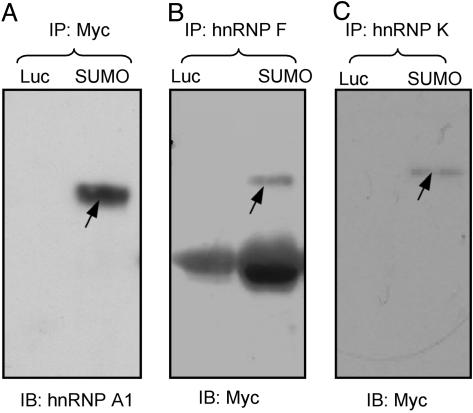
The Sumoylation of hnRNP A1, hnRNP F, and hnRNP K Was Confirmed by Coimmunoprecipitation. The above analysis showed that six hnRNPs were putative SUMO target proteins. To further confirm that hnRNPs are sumoylated in vivo, coimmunoprecipitation method was used. As shown in Fig. 5A, immunoprecipitation by anti-Myc antibody followed by Western analysis with anti-hnRNP A1 antibody identified a protein band of the size of sumoylated hnRNP A1 in the Myc-SUMO transfected cells but not in the control cells, indicating that hnRNP A1 was indeed sumoylated in vivo. Similarly, immunoprecipitation by anti-hnRNP F and anti-hnRNP K, respectively, followed by Western analysis with anti-Myc antibody revealed protein bands of the size of sumoylated hnRNP F (Fig. 5B) and sumoylated hnRNP K, respectively (Fig. 5C) in the Myc-SUMO transfected cells but not in the control cells. These results strongly indicate that both hnRNP F and hnRNP K were sumoylated in vivo.
Fig. 5.
hnRNP A1, hnRNP F, and hnRNP K are sumoylated. The whole-cell extracts from HEK 293 Tet-On cells expressing Myc-SUMO or Myc-Luc were immunoprecipitated with either anti-Myc (A), anti-hnRNP F (B), or hnRNP K (C) antibody. The immunoprecipitates were resolved by NuPage gels and probed with anti-hnRNP A1 (A) or anti-Myc (B and C) antibody. Sumoylated hnRNP proteins are marked with arrows. The lower bands in B are caused by IgG heavy chain cross-reacting with the secondary antibody.
Discussion
Posttranslational modification of proteins by SUMO plays an important role in many cellular processes (7), and SUMO-targeted proteins are being identified with an accelerating rate (8). Because the sumoylation is highly dynamic and only a limited fraction of certain targeted proteins can be sumoylated in cells, it is difficult to identify the relatively low abundant sumoylated proteins or proteins that are sumoylated only under certain conditions. To overcome this problem, we developed both in vivo and in vitro methods to enrich the amounts of sumoylated proteins and affinity purify them to facilitate the MS identification of SUMO target proteins. These two methods are complementary, and each has its own strength. Sumoylated proteins isolated from cell lines stably expressing SUMO support the notion that sumoylation occurs in intact cells. Moreover, the elevated level of SUMO facilitates the identification of low abundant sumoylated proteins. It is not surprising that Ran-GAP1 and RanBP2 were found to be the dominant sumoylated proteins identified by LC/MS/MS, because they are the predominant SUMO-targeted proteins. To minimize suppressive effects caused by ions derived from the overwhelmingly abundant RanGAP1 and RanBP2 during LC/MS analysis, cell extracts were pretreated with antibodies against these two proteins followed by precipitation with protein A agarose to deplete them, and the resulting cell extracts were used for sumoylated protein identification. The MS/MS data obtained by this pretreatment improved significantly, and it allowed us to identify a few more sumoylated proteins. The in vitro sumoylation can also obtain elevated sumoylated proteins. Furthermore, in vitro addition of biotin–SUMO may identify some SUMO substrates that are highly sumoylated under certain stress conditions, but rarely modified in normal biological conditions. However, there was no RanGAP1 identified with this in vitro method. One possible explanation is that the nuclear extracts were used to carry out the in vitro sumoylation by using biotin–SUMO as modifier, and the RanGAP1 bound to the nucleus (26) had already been sumoylated with endogenous SUMO; therefore, no biotin–sumoylated RanGAP1 was isolated. Combining the strengths of in vivo and in vitro methods provided us with the reassurance for our identified sumoylated proteins.
Interestingly, six hnRNPs were among the SUMO substrates identified. The hnRNPs can rapidly bind to nascent preRNA and are involved in subsequent RNA maturation, stabilization, and transport (20). Most hnRNPs contain one or more RNA-binding motifs, such as RNA-binding domain (RBD), K homology (KH) domain, and RGG (Arg–Gly–Gly) boxes (20). The putative sumoylation consensus sequences in hnRNP A1, hnRNP A3, hnRNP F, and hnRNP H are located in their RBD sequences, and in hnRNP K is located in its KH domain. This raises a possibility that sumoylation of these hnRNPs may regulate their RNA-binding activity. The proteins hnRNP A1 and hnRNP K are among the hnRNPs that shuttle between nucleus and cytoplasm (20); however, the mechanism of these shuttling remains to be elucidated. It is of great interest to know whether sumoylation is involved in hnRNP shuttling between cytoplasm and nucleus. Recently, hnRNP F has been found coimmunoprecipitating with RNA polymerase II and TATA-binding protein (27). This suggests that hnRNP F is associated with transcriptional initiation apparatus. Sumoylation of hnRNP F may, thus, modulate transcription processes as well.
FBP (FUSE-binding protein) and FBP2 are known to bind the far upstream element (FUSE) that regulates c-myc gene expression. FBP and FBP2 also bind to RNA and participate in RNA processing and transport through four KH domains (28). The putative sumoylation consensus sequence is located at the end of the KH3 domain in the β-strand. Interestingly, a backbone superposition of KH3 and KH4 domains in FBP exhibits similar 3D structures to the KH3 and KH4 domains of hnRNP K (29). This finding suggests that FBPs RNA-binding activity may also be regulated in the same manner as hnRNP K.
Nuclear pore complex (NPC) plays an important role in nucleocytoplasmic trafficking (30). Two enzymes involved in SUMO conjugation, Ubc9 (E2) and RanBP2 (E3), and one desumoylating enzyme, SENP2, are found localized at the NPC (31). This observation suggests that NPC plays an important role in regulating the state of sumoylation of a number of SUMO-targeted proteins and thus determines their translocation between nucleus and cytoplasm. In addition to the two known sumoylated NPC proteins, RanGAP1 and RanBP2, we identified two previously uncharacterized NPC proteins, Nup98-Nup96 and RanBP16, as SUMO target proteins. The physiological significance of the sumoylation of these NPC proteins has yet to be investigated.
Zinc finger proteins are known to mediate specific protein to protein as well as protein–DNA and protein–RNA interactions. They are widely used for DNA and RNA recognition (32). Five zinc finger proteins were found to be targets for sumoylation. The functions of these five proteins are not known; however, recent data indicate that sumoylation plays an important role in transcription regulation (33). It would be interesting to find out whether sumoylation of these zinc finger proteins is involved in transcriptional regulation.
In conclusion, a stable cell line highly expressing SUMO and in vitro sumoylation were used as complementary ways to isolate SUMO substrates. Together with the power of LC/MS/MS, we identified 20 SUMO target proteins, which included four previously known and 16 previously unknown SUMO substrates. The finding of six hnRNPs as SUMO substrates indicates that sumoylation may play a role in the regulation of RNA metabolism at the transcriptional as well as posttranscriptional levels. The functional effects of sumoylation on these identified substrates await further investigation.
Acknowledgments
We thank Dr. K. Bomsztyk (University of Washington Medical Center, Seattle) and Dr. D. Black (University of California, Los Angeles) for generous gifts of anti-hnRNP K and anti-hnRNP F antibodies, respectively.
Abbreviations: SUMO, small ubiquitin-related modifier; MS/MS, tandem MS; hnRNP, heterogeneous nuclear ribonucleoprotein; Dox, doxycycline; DAPI, 4′-6-diamidino-2-phenylindole hydrochloride; KH, K homology; LC, liquid chromatography.
References
- 1.Stadtman, E. R. & Chock, P. B. (1978) Curr. Top. Cell. Regul. 13, 53–95. [DOI] [PubMed] [Google Scholar]
- 2.Chock, P. B. & Stadtman, E. R. (1995) in Principles of Medical Biology, eds. Bittard, E. E. & Bittar, N. (JAI Press, Greenwich, CT), Vol. 5, pp. 199–218. [Google Scholar]
- 3.Pickart, C. M. (2001) Mol. Cell 8, 499–504. [DOI] [PubMed] [Google Scholar]
- 4.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- 5.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 6.Verger, A., Perdomo, J. & Crossley, M. (2003) EMBO Rep. 4, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichler, A. & Melchior, F. (2002) Traffic 3, 381–387. [DOI] [PubMed] [Google Scholar]
- 8.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 690–699. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz, D. C. & Hochstrasser, M. (2003) Trends Biochem. Sci. 28, 321–328. [DOI] [PubMed] [Google Scholar]
- 10.Melchior, F., Schergaut, M. & Pichler, A. (2003) Trends Biochem. Sci. 28, 612–618. [DOI] [PubMed] [Google Scholar]
- 11.Melchior, F. (2000) Annu. Rev. Cell Dev. Biol. 16, 591–626. [DOI] [PubMed] [Google Scholar]
- 12.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichler, A., Gast, A., Seeler, J. S., Dejean, A. & Melchior, F. (2002) Cell 108, 109–120. [DOI] [PubMed] [Google Scholar]
- 14.Azuma, Y. & Dasso, M. (2002) Dev. Cell 2, 130–131. [DOI] [PubMed] [Google Scholar]
- 15.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- 16.Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N. & Elledge, S. J. (2002) Mol. Cell 9, 1169–1182. [DOI] [PubMed] [Google Scholar]
- 17.Bylebyl, G. R., Belichenko, I. & Johnson, E. S. (2003) J. Biol. Chem. 278, 44113–44120. [DOI] [PubMed] [Google Scholar]
- 18.Li, S. J. & Hochstrasser, M. (2003) J. Cell Biol. 160, 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmstrom, S., Van Antwerp, M. E. & Iniguez-Lluhi, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 15758–15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 21.Matunis, M. J., Xing, J. & Dreyfuss, G. (1994) Nucleic Acids Res. 22, 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, T., Evdokimov, E., Yiadom, K., Yan, Z., Chock, P. B. & Yang, D. C. (2003) Protein Expr. Purif. 30, 140–149. [DOI] [PubMed] [Google Scholar]
- 23.Eng, J. K., McCormack, A. L. & Yates J. R., III (1994) J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 24.Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997) Cell 88, 97–107. [DOI] [PubMed] [Google Scholar]
- 25.Kwek, S. S., Derry, J., Tyner, A. L., Shen, Z. & Gudkov, A. V. (2001) Oncogene 20, 2587–2599. [DOI] [PubMed] [Google Scholar]
- 26.Matunis, M. J., Coutavas, E. & Blobel, G. (1996) J. Cell Biol. 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida, T., Makino, Y. & Tamura, T. (1999) FEBS Lett. 457, 251–254. [DOI] [PubMed] [Google Scholar]
- 28.Duncan, R., Bazar, L., Michelotti, G., Tomonaga, T., Krutzsch, H., Avigan, M. & Levens, D. (1994) Genes Dev. 8, 465–480. [DOI] [PubMed] [Google Scholar]
- 29.Braddock, D. T., Louis, J. M., Baber, J., Levens, D. & Clore, G. M. (2002) Nature 415, 1051–1056. [DOI] [PubMed] [Google Scholar]
- 30.Fahrenkrog, B. & Aebi, U. (2003) Nat. Rev. Mol. Cell Biol. 4, 757–766. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, H., Saitoh, H. & Matunis, M. J. (2002) Mol. Cell. Biol. 22, 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews, J. M. & Sunde, M. (2002) IUBMB Life 54, 351–355. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, D. & Muller, S. (2003) Cell Mol. Life Sci. 60, 2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]