Abstract
Single-stranded DNA (ssDNA)-binding (SSB) proteins are uniformly required to bind and protect single-stranded intermediates in DNA metabolic pathways. All bacterial and eukaryotic SSB proteins studied to date oligomerize to assemble four copies of a conserved domain, called an oligonucleotide/oligosaccharide-binding (OB) fold, that cooperate in nonspecific ssDNA binding. The vast majority of bacterial SSB family members function as homotetramers, with each monomer contributing a single OB fold. However, SSB proteins from the Deinococcus-Thermus genera are exceptions to this rule, because they contain two OB folds per monomer. To investigate the structural consequences of this unusual arrangement, we have determined a 1.8-Å-resolution x-ray structure of Deinococcus radiodurans SSB. The structure shows that D. radiodurans SSB comprises two OB domains linked by a β-hairpin motif. The protein assembles a four-OB-fold arrangement by means of symmetric dimerization. In contrast to homotetrameric SSB proteins, asymmetry exists between the two OB folds of D. radiodurans SSB because of sequence differences between the domains. These differences appear to reflect specialized roles that have evolved for each domain. Extensive crystallographic contacts link D. radiodurans SSB dimers in an arrangement that has important implications for higher-order structures of the protein bound to ssDNA. This assembly utilizes the N-terminal OB domain and the β-hairpin structure that is unique to Deinococcus and Thermus species SSB proteins. We hypothesize that differences between D. radiodurans SSB and homotetrameric bacterial SSB proteins may confer a selective advantage to D. radiodurans cells that aids viability in environments that challenge genomic stability.
In all organisms, single-stranded DNA (ssDNA)-binding (SSB) proteins sequester and protect ssDNA intermediates that arise during DNA replication, recombination, and repair (1). Their prominent roles in genome maintenance reactions make SSB proteins a requirement for cellular life (2). Although the sequences of SSB family members are highly variable, two common functional themes have emerged that link this class of proteins across evolution. The first is that SSB proteins use a conserved domain called an oligonucleotide/oligosaccharide-binding (OB) fold to bind ssDNA (3, 4). OB domains bind ssDNA in a cleft formed primarily by β-strands, by using aromatic residues that stack against nucleotide bases and positively charged residues that form ionic interactions with the DNA backbone (5–8). The second common feature of cellular SSB proteins is obligate oligomerization that brings together four DNA-binding OB folds. For example, Escherichia coli SSB contains a single OB fold per monomer, but the active form of the protein is a homotetramer with four OB folds (1). This general arrangement appears to define a structural paradigm for bacterial SSB family proteins because all but three of the >250 currently identifiable bacterial ssb genes encode proteins with a single OB fold.
Different strategies are used to assemble four DNA-binding OB folds in nonbacterial cells (reviewed in ref. 9). In eukaryotes, replication protein A (RPA) acts as the SSB protein. RPA family members are heterotrimeric and contain six OB folds, four of which bind DNA. In terms of domain organization, archaeal SSB proteins are divided into two groups: those that resemble bacterial SSB (found in crenarchaea) and those that resemble RPA (found in euryarchaea). Interestingly, despite the bacterial-like domain organization of the crenarchaeal Sulfolobus solfataricus SSB, its structure more closely resembles that of RPA than bacterial SSB proteins (10). Exceptions to the general four-OB-fold rule exist outside of cellular SSB family members, including bacteriophage and viral SSB proteins (reviewed in ref. 4).
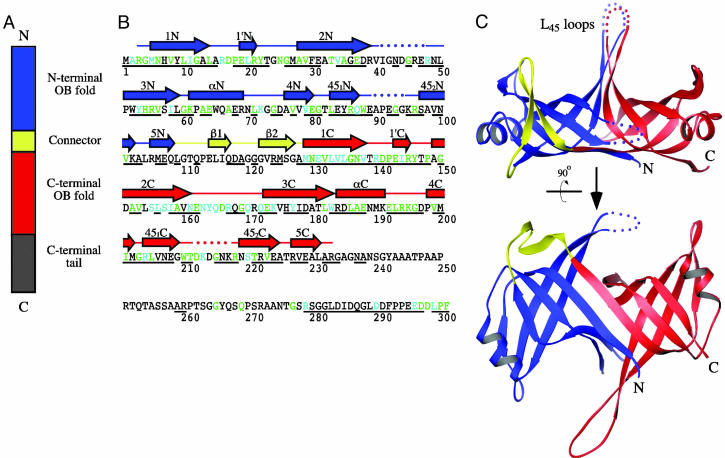
Although virtually all bacterial SSB family members act as homotetramers, recent discoveries have shown that SSB proteins from the Deinococcus-Thermus genera of bacteria adopt a different structural architecture. Bacteria from this group thrive in extreme environments that would kill most cells (desiccation, massive DNA-damaging, and/or high-temperature conditions) by using mechanisms that are presently unclear (11, 12). One distinguishing DNA metabolic feature of Deinococcus-Thermus bacteria is that their SSB proteins are homodimeric, with each SSB monomer encoding two OB folds linked by a conserved spacer sequence (Fig. 1 A and B) (13–15). The impact of this arrangement on SSB function is not yet understood.
Fig. 1.
Structure of D. radiodurans SSB. (A) Schematic diagram of D. radiodurans SSB primary structure. Two OB folds are present in each D. radiodurans SSB monomer: one is N-terminal (blue, residues 1–108), and one is C-terminal (red, residues 129–233). These folds are linked by a connector peptide (yellow, residues 109–128). The C-terminal OB fold is followed by a flexible tail sequence (gray, residues 234–301). (B) Secondary structure of D. radiodurans SSB. The D. radiodurans SSB protein sequence is colored to indicate residues that are highly similar (teal) or identical (green) to E. coli SSB (28). Underlined residues are conserved with SSB proteins from T. aquaticus and T. thermophilus (13). Helices (boxes) and β-strands (arrows) are shown above the sequence (colored as in A) and labeled as for E. coli SSB (28) but with identifiers that indicate whether the element comprises part of the N-terminal (N) or the C-terminal (C) OB fold. β-strands in the connector region are labeled β1or β2 to distinguish them from elements in either OB fold. Dotted lines indicate regions for which electron density was not observed. Electron density was also absent for the C-terminal tail. (C) Orthogonal views of a ribbon diagram of the crystal structure of D. radiodurans SSB. The structure is colored as in A. Dotted lines indicate regions that could not be modeled because of a lack of electron density. The L45 loop region is indicated. Ribbon diagrams were rendered by using ribbons (33).
SSB proteins may have an expanded role in DNA metabolism in extremeophiles. E. coli maintain an estimated 300–2,000 SSB tetramers per cell and do not increase SSB levels significantly in response to DNA damaging conditions (16). In contrast, Deinococcus radiodurans has an estimated 19,500 SSB dimers per cell and increases that level to ≈56,000 in response to treatment with ionizing radiation (S. Saveliev and M.M.C., unpublished data). The D. radiodurans SSB protein has recently been purified and shown to stimulate strand-exchange activity of the D. radiodurans RecA protein (15).
To better understand the structural and functional consequences of the unusual domain arrangement of Deinococcus-Thermus SSB proteins, we have determined a high-resolution x-ray crystal structure of D. radiodurans SSB. The structure shows that D. radiodurans SSB contains two OB folds that are connected by a β-hairpin linker. Sequence differences between the two OB folds impose an asymmetry that is likely to affect the DNA binding properties and other functions of each domain. D. radiodurans SSB assembles a four-OB-fold arrangement through symmetric dimerization. A major crystal contact buries 1,288 Å2 of surface area between adjacent dimers, revealing a potentially important site for higher-order assembly of multiple D. radiodurans SSB dimers on long ssDNA molecules. This surface is formed by the N-terminal OB domain and the conserved β-hairpin that links OB folds in the Deinococcus-Thermus bacterial group and, thus, could provide a unique mechanism for cooperative ssDNA binding by these proteins to facilitate survival in extreme conditions.
Materials and Methods
Crystallization of D. radiodurans SSB. Full-length D. radiodurans SSB protein was purified as described (15). Crystals of D. radiodurans SSB were grown by hanging drop vapor diffusion by mixing 1 μl of protein (10 mg/ml in 10 mM Tris·HCl, pH 8.0) with 1 μl of well solution [25% polyethylene glycol (PEG) 200/3–10% PEG 4000/0.1 M sodium acetate, pH 5.5/0.3 M potassium chloride] and equilibrating at room temperature. Crystals diffracted with C2 symmetry and the unit cell dimensions a = 91.3 Å, b = 63.4 Å, c = 54.9 Å, and β = 106.3°, consistent with one molecule of D. radiodurans SSB per asymmetric unit (17). Crystals with C2 symmetry and similar unit cell parameters were also grown with 2.0 M sodium formate/0.1 M sodium acetate (pH 4.6) (form II; a = 90.9 Å, b = 63.3 Å, c = 54.9 Å, β = 106.0°) and 5% dioxane/0.1 M Mes (pH 6.5)/1.4 M ammonium sulfate (form III; a = 91.3 Å, b = 62.2 Å, c = 51.5 Å, β = 110.6°) well solutions. Selenomethionine-incorporated D. radiodurans SSB was overexpressed as described in ref. 18 and purified as per native protein, except that 0.8 mM tri(2-carboxyethyl) phosphine hydrochloride (TCEP) was included as the reducing agent in place of 1 mM 2-mercaptoethanol. Crystals of selenomethionine-incorporated D. radiodurans SSB were grown by using the same method as the native crystals, but the well solution was modified to 10% PEG 200/0.5% PEG 4000/0.1 M sodium acetate (pH 5.5)/0.5 M potassium chloride/1 mM TCEP. Selenomethionine-incorporated SSB crystals were stabilized by transfer to a cryoprotectant solution (25% PEG 200/0–1% PEG 4000/0.1 M sodium acetate, pH 5.5/0.5 M potassium chloride/1 mM TCEP) and then frozen in liquid nitrogen for data collection.
Multiwavelength Anomalous Dispersion Phasing and Model Refinement. The structure of selenomethionine-incorporated D. radiodurans SSB was solved to a 1.8-Å resolution by using multiwavelength anomalous dispersion phasing (Table 1). Data were indexed and scaled with mosflm (19) and scala (20). Selenium sites were identified by using solve (21) and further refined by using mlphare (22). Solvent flattening with dm (23) resulted in interpretable experimental electron density maps for model building. Ninety percent of the model was built automatically by using arp/warp (24), and the remainder was built manually with the program o (25). The model was improved by rounds of refinement with refmac5/arp (24, 26) and manual rebuilding. The arrangement of D. radiodurans SSB proteins in crystal forms II and III was determined by molecular replacement, with the refined selenomethionine-substituted structure as a search model.
Table 1. Data collection, phasing, and refinement.
| Anomalous peak | Remote | Form II | Form III | Phasing/model statistics | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength, Å | 0.9795 | 0.9568 | 1.5418 | 1.5418 | |
| Resolution (last shell), Å | 34 to 1.8 (1.89 to 1.8) | 32 to 1.8 (1.89 to 1.8) | 50 to 2.56 (2.6 to 2.56) | 50 to 2.8 (2.85 to 2.8) | |
| Reflections measured/unique | 212,761/27,979 | 115,292/27,686 | 35,699/9,674 | 25,562/6,747 | |
| Multiplicity | 7.6 (7.5) | 4.1 (3.6) | 3.65 (1.72) | 3.78 (3.78) | |
| Completeness (last shell), % | 99.9 (99.9) | 99.7 (99.7) | 99.0 (79.4) | 99.8 (100) | |
| Rsym (last shell), %* | 6.1 (41.8) | 7.0 (44.3) | 8.0 (37.8) | 5.4 (36.8) | |
| I/σ (last shell) | 15.1 (5.1) | 10.9 (2.5) | 12.4 (3.6) | 16.6 (3.1) | |
| Phasing statistics | |||||
| Resolution, Å | 20.0 to 1.8 | ||||
| Figure of merit | 0.439 | ||||
| After density modification | 0.788 | ||||
| Refinement | |||||
| Resolution, Å | 20.0 to 1.8 | ||||
| Rwork/Rfree† | 21.7/23.2 | ||||
| rms deviation bond lengths, Å | 0.012 | ||||
| rms deviation bond angles, Å | 0.98 | ||||
| PDB code | 1SE8 |
Rsym = ΣΣj|Ij - <I>|/ΣIj, where Ij is the intensity measurement for reflection j, and <I> is the mean intensity for multiply recorded reflections.
Rwork/Rfree = Σ||Fobs| -|Fcalc||/Fobs|, where the working and free R factors are calculated by using the working and free reflections sets. The free relections (5% of the total) were held aside throughout refinement.
Results
Structure of the D. radiodurans SSB Protein. We have solved the crystal structure of D. radiodurans SSB to 1.8-Å resolution by multiwavelength anomalous dispersion phasing with a selenomethionine-substituted variant of the full-length protein. Our structure contains amino acids 3–233, excluding residues 41–47, 89–94, and 212–216, for which electron density was not observed. The C-terminal tail, residues 234–301, is also absent from our model because of a lack of interpretable electron density. The lack of electron density in these regions likely reflects a high degree of flexibility. The structure was refined with good bond geometry and crystallographic quality statistics (Table 1). No bond angles in the final model fall into disallowed regions of Ramachandran space.
The structure of D. radiodurans SSB showed that the protein consists of a major β-barrel domain from which several β-hairpin loops extend (Fig. 1C). As predicted from the protein sequence (15), the secondary structural elements of D. radiodurans SSB fold together to form two OB folds. The two OB folds are linked by a rigid β-hairpin connector that packs against the N-terminal OB domain of the protein. It is likely that a similar β-hairpin connector is also present in the Thermus species proteins because this sequence is conserved in Thermus aquaticus and Thermus thermophilus SSB proteins (Fig. 1B).
The arrangement of OB folds within the D. radiodurans SSB monomer is quite similar to that seen in the dimeric arrangements of other bacterial SSB proteins that contain a single OB fold per polypeptide (27–29). In both types of proteins, the first β-strands in each OB fold lie adjacent to one another, forming an extended antiparallel β-sheet (Fig. 1C). This configuration creates a large hydrogen bond network within the D. radiodurans SSB monomer and positions the two OB folds in a similar relative orientation to that seen in E. coli SSB (28) (rms deviation = 1.7 Å for 154 Cα atoms in the two proteins). A second interaction between the L45 loops that extend orthogonally from the β-barrel platform is also formed in both D. radiodurans and single-OB-fold bacterial SSB proteins (Fig. 1C). Thus, despite its unusual number of OB domains, the arrangement of the N- and C-terminal OB folds in D. radiodurans SSB strongly resembles that seen in E. coli SSB, the prototypical bacterial SSB.
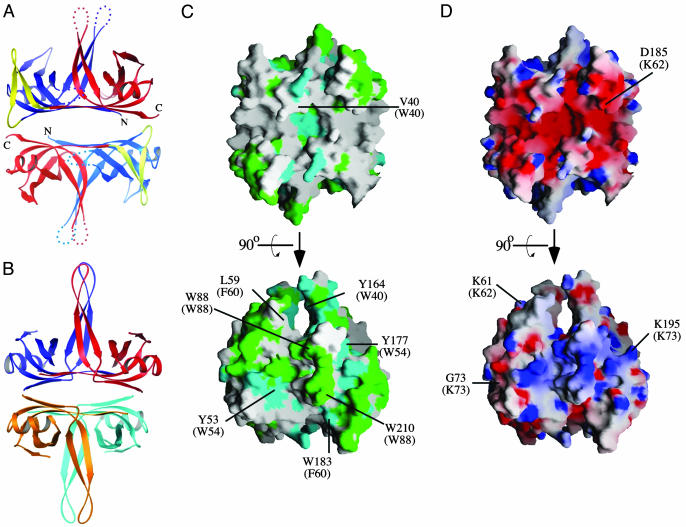
Dimeric Structure of D. radiodurans SSB. With the exception of SSB proteins from Deinococcus-Thermus genera bacteria, all previously studied bacterial SSB family members form homotetramers to bring together four OB domains in their active state. D. radiodurans SSB also forms a four-OB-fold-containing species, but does so by means of dimerization (15). Although the crystal form reported here contains only a single protein per asymmetric unit, a two-fold symmetry axis exists in the crystals that relates two monomers of D. radiodurans SSB through an interface that buries 2,191 Å2 of surface area (Fig. 2A). The orientation of the two monomers in D. radiodurans SSB places its OB folds into different positions from that seen in E. coli SSB (Fig. 2B). To fit the E. coli SSB arrangement, one of the D. radiodurans SSB monomers would need to be rotated ≈40° relative to the other. This rotational difference from E. coli SSB is quite similar to one that has been observed recently in the Mycobacterium tuberculosis SSB structure (29).
Fig. 2.
Dimeric structure of D. radiodurans SSB. (A) Ribbon diagram of the D. radiodurans SSB dimer. Coloring is as in Fig. 1 A for Upper and is shown in paler colors for Lower to aid in distinguishing between the two protomers. (B) Ribbon diagram of the E. coli SSB tetramer (28). Each protomer is colored differently (red, blue, orange, and teal). (C) Orthogonal views of the surface conservation of D. radiodurans SSB. The dimeric protein surface is colored as in Fig. 1B by its conservation with E. coli SSB. Structurally similar residues present at the positions of four key aromatic residues in E. coli SSB are indicated by their single-letter code with the equivalent E. coli residues indicated in parentheses. Upper is the same orientation as in A. (D) Orthogonal views of the surface potential of D. radiodurans SSB. The dimeric protein surface is colored by its electrostatic surface potential at ±6 kBT/e for positive (blue) or negative (red) charge potential by using the program grasp (34). Positions of several residues in D. radiodurans SSB are indicated in single-letter code, and electropositive DNA-binding residues in structurally conserved locations in E. coli SSB are indicated in parentheses. Upper is the same orientation as in A.
Unlike other bacterial SSB proteins, D. radiodurans SSB possesses an inherent structural asymmetry that arises from sequence differences between its N- and C-terminal OB domains. Although the C-terminal OB domain in the protein contains homologous residues to the four aromatic base-stacking residues used for DNA binding in E. coli SSB (7), only two of these residues are conserved in the N-terminal OB domain of the protein (Fig. 2C). The E. coli SSB base-stacking residues are Trp-40, Trp-54, Phe-60, and Trp-88; the structurally related residues in D. radiodurans SSB are Val-40, Tyr-53, Leu-59, and Trp-88 in the N-terminal OB domain and Tyr-164, Tyr-177, Trp-183, and Trp-210 in the C-terminal OB domain. Additional sequence asymmetries lead to differences between charged residues in the two D. radiodurans OB domains that are related to ssDNA-binding residues in E. coli SSB (7) (Fig. 2D). A structural equivalent to the ssDNA-binding E. coli SSB Lys-73 residue is present in the D. radiodurans C-terminal OB domain (Lys-195) but is replaced by a Gly residue in its N-terminal OB domain (Gly-73). A second key ssDNA-binding residue in E. coli SSB (Lys-62) is conserved in the D. radiodurans N-terminal OB domain (Lys-61) but is replaced with an Asp residue (Asp-185) in its C-terminal OB domain. These differences could lead to differential ssDNA binding by the individual OB folds in D. radiodurans SSB.
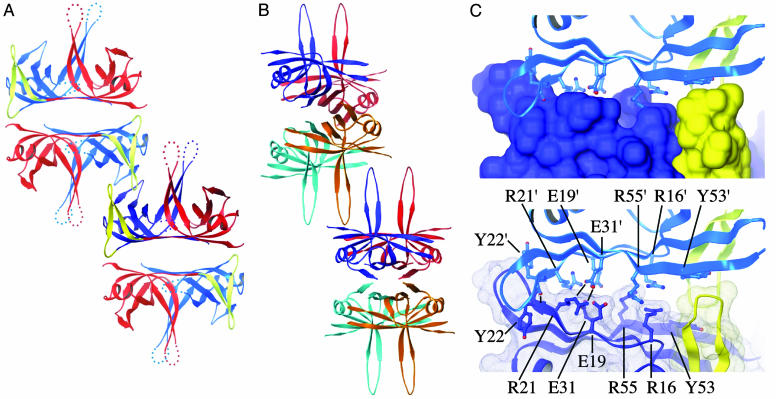
Higher-Order Assembly of D. radiodurans SSB. Inspection of protein–protein interfaces in the crystal structure revealed a large interaction surface that linked neighboring D. radiodurans SSB dimers (Fig. 3). This interface buries 1,288 Å2 of surface area in an extensive interlocking grip that is formed by the β-hairpin connectors and N-terminal OB domains of adjacent molecules. As will be described in Discussion, this interaction is distinct from the interface that mediates contacts between E. coli SSB tetramers (7, 28) (Fig. 3B). Numerous molecular interactions contribute to the D. radiodurans SSB dimer–dimer interface (Fig. 3C). First, extensive van der Waal's contacts link the molecules. The Tyr-22 side chain from each of the molecules packs against a loop (residues 23–26) that connects two β-strands in the adjacent protomer. These loops also pack against each other in the interface. In addition, the β-hairpin connectors of each molecule pack against Tyr-53 of the opposing molecule and against each other. Second, ionic and hydrogen bonding contacts contribute to the interface. The ionic interactions are formed between Arg-21 and Glu-31 side chains from opposing SSB molecules, and intermolecular β-sheet hydrogen bonds are formed between Tyr-22 main-chain atoms. Potentially repulsive residue side chains are also present in the interface, including an acidic residue (Glu-19) and two basic residues (Arg-16 and Arg-55) that are not in ion pairs.
Fig. 3.
Higher-order assembly of D. radiodurans SSB. (A) Ribbon diagram of two crystallographically related D. radiodurans SSB dimers. One monomer is colored as in Fig. 1 A, with the other three shown in paler colors to aid in visualization. The N-terminal OB domains and β-hairpin connectors of the two central monomers interlock to form an extensive interaction surface. (B) Ribbon diagram of two crystallographically related E. coli SSB tetramers (28). Monomers are colored as in Fig. 2B. Interaction between tetramers is mediated by the L45 loops of two monomers. (C) Close-up view of the interaction surface between D. radiodurans SSB dimers. (Upper) The surface of one D. radiodurans SSB dimer is enveloped by its crystallographically related contact partner. Surfaces and ribbons are colored as in Fig. 1C, indicating that the interaction is mediated exclusively by the N-terminal OB domains and β-hairpin connectors of the adjacent protomers. (Lower) Residues that mediate the protein–protein contact (R21, Y22, E31, and Y53) and unpaired charged residues (R16, E19, and R55) are indicated. Residues labeled with a prime symbol are from the upper molecule.
To determine whether this interface could be an artifact of the particular crystallization conditions used, we have collected diffraction data from two other D. radiodurans SSB crystal forms (forms II and III; Table 1). These two crystal forms and the form previously described were produced under conditions that differed substantially in pH, precipitant, and salt identity and concentration (see Materials and Methods). Despite these differences, all of the crystals diffracted with C2 symmetry and with similar unit cell parameters. Not surprisingly, the dimer–dimer interfaces in forms II and III were found to be essentially the same as that observed in the first form (data not shown), indicating that the interface is preserved in a wide variety of chemical conditions. Sequence conservation implies that this interface is likely to be present in Thermus species SSB proteins as well (Fig. 1B).
Discussion
In this article, we describe a high-resolution structure of the D. radiodurans SSB protein, a central factor in the unusually vigorous genome maintenance pathways of the bacterium. With two OB folds per monomer, this is the first structure of any bacterial SSB that differs from the canonical single-OB-fold arrangement. The protein forms a symmetric dimer to bring a total of four OB domains together in its active state. Key sequence differences between the N- and C-terminal OB folds of the protein lead to structural asymmetry that likely reflects specialized roles for each of the two domains. The structure shows that adjacent D. radiodurans SSB dimers bind each other by using an extensive surface area that is formed by their N-terminal OB domains and β-hairpin connectors. This interface appears to be unique to SSB proteins from Deinococcus-Thermus genera bacteria and is hypothesized to be important for the robust survival mechanisms that are common among these organisms.
Effects of the Two-OB-Domain Arrangement in D. radiodurans SSB. The consequences of linking two OB folds together in a single SSB protomer are dramatic. One major advantage of this arrangement is that the two OB domains can evolve separately, allowing the development of specialized functions for each fold. This arrangement is not possible in bacteria, such as E. coli, that use homotetrameric single-OB-fold SSB proteins, because any mutation would be present in all four of the SSB OB domains. The differences between the two OB folds in D. radiodurans SSB indicate that there are distinct roles for each. As described in Results, the C-terminal domain retains nearly all of the residues that would be predicted to bind ssDNA based on the E. coli SSB/ssDNA complex (7), whereas the N-terminal OB domain lacks a number of potentially important ssDNA-binding residues (Fig. 2). These differences are likely to lead to a lower affinity for ssDNA in the N-terminal OB domain relative to the C-terminal OB fold.
Interestingly, asymmetric DNA binding to OB domains has previously been observed for E. coli SSB despite its homotetrameric nature. E. coli SSB can bind ssDNA in two different modes: a (SSB)65 mode in which all four protomers bind DNA and a (SSB)35 mode in which only two of the protomers are DNA-bound (1). Both modes display cooperative DNA-binding features that are likely to be linked to interactions between tetramers. Structural analysis has led to models of the two E. coli SSB binding modes and predicts that, in the (SSB)35 mode, the two DNA-bound monomers are diagonally related within a tetramer (7). This analysis places the DNA-binding protomers into positions that are essentially equivalent to the D. radiodurans SSB dimer C-terminal OB domains. It is therefore possible that an asymmetric ssDNA binding mode similar to the (SSB)35 binding mode of E. coli SSB is conserved in the D. radiodurans protein but that it is directed by sequence differences between the N- and C-terminal OB domains. Such a model is consistent with the proposed specialized roles of the two OB domains in the D. radiodurans SSB structure, in which the C-terminal domain binds DNA and the N-terminal domain mediates the association of neighboring dimers (described below). Given that biochemical experiments that measure DNA binding properties of D. radiodurans SSB have not yet been performed, further data are needed to determine its precise modes of DNA binding.
Another potentially important difference between tetrameric and dimeric SSB proteins is that dimeric proteins contain only two C-terminal tails in their active forms. C-terminal tails in SSB proteins have been implicated as binding sites for other proteins involved in genome maintenance through acidic patches at their extreme C-termini (30, 31). Reducing the number of C-terminal tails by half could dramatically affect the function of two-OB-fold-containing SSB proteins in vivo. There are several ways in which D. radiodurans could overcome this difference. Other acidic areas of the D. radiodurans protein could act in place of the tails to make these interactions (e.g., the dimer interface; see Fig. 2D), or the protein could form the necessary complexes by using only two acidic tails as mediators. It is also possible that proteins recruited by interactions with SSB C-terminal tails in other bacteria are unneeded or are recruited by using other factors in D. radiodurans.
Finally, physical linkage of multiple, nonidentical OB domains in D. radiodurans SSB may represent an evolutionary convergence between homotetrameric bacterial/cenarchaeal SSB and eukaryotic/euryarchaeal RPA family members. Because of their homotetrameic nature, most bacterial SSB proteins are evolutionarily restricted relative to their RPA counterparts, given that their OB folds cannot evolve independently. In contrast, RPA family members have evolved as complex assemblies of OB folds in which some of the domains are key DNA-binding elements and others mediate non-DNA-binding functions. The structure of the D. radiodurans SSB suggests that, although the protein is bacterial in origin, it has distinct functions for its tandem OB domains analogous to those of RPA. A plausible evolutionary explanation of this arrangement is that the D. radiodurans protein arrived at an RPA-like arrangement of multiple OB domains within a single SSB polypeptide through gene duplication.
Assembly of Multiple D. radiodurans SSB Dimers. A surprising feature of the D. radiodurans SSB structure is the presence of an interaction surface that links neighboring dimers (Fig. 3). Several aspects of this interface suggest that it has biological relevance. The interface is large (1,288 Å2), uses a complex mixture of interactions (van der Waal's, hydrogen, and ionic bonds), and is preserved in diverse chemical conditions, consistent with bona fide protein–protein interaction surfaces. This interaction is distinct from the L45-loop-mediated interface that links E. coli SSB tetramers (7, 28) (Fig. 3, compare A with B). The L45-type association places E. coli SSB tetramers into a relative orientation that differs by ≈90° from D. radiodurans SSB dimers, which leaves all of its OB domains available for ssDNA binding. In contrast, extensive portions of the N-terminal D. radiodurans SSB OB domain are buried in its dimer–dimer interface. This interface occupies much of the predicted ssDNA-interaction surface from the N-terminal OB domain and even co-opts a residue that would be predicted to directly bind to ssDNA (Tyr-53). Thus, it appears that the major role of the N-terminal OB domain is to mediate homotypic SSB protein interaction. Given that the β-hairpin peptide that connects the two OB domains in D. radiodurans SSB also forms an important element in the interaction surface, this interface is likely to represent a specialized structure that is present in all Deinococcus-Thermus bacterial SSB proteins.
What possible function could this interaction surface provide for D. radiodurans SSB? The only known ssb genes with the features described in this structure are from bacteria that have evolved to live in extreme environments. Because a number of thermophilic bacterial species use single-OB-fold SSB proteins, it is unlikely that the two-OB-fold arrangement arose as a requirement for thermophilicity. It is possible, however, that the unusual arrangement of OB folds in the D. radiodurans SSB structure evolved to aid in desiccation or radiation-damage survival and that the structure has been retained by species in the Deinococcus-Thermus group of bacteria. Deinococcus species are commonly found in arid environments in which nutrients are scarce, and they can survive long periods of time in these conditions (11). In a long-term static state, it could be advantageous to have an SSB protein that forms stable protective fibers over exposed ssDNA until nutrients become available to drive genome metabolic processes. The extensive interaction surface observed between D. radiodurans SSB dimers in this study is a good candidate for helping build such a superstructure. The unusually high concentrations of SSB dimers present in D. radiodurans cells could aid SSB fiber assembly through mass action. Because genes that aid in desiccation resistance are also important for radioresistance in Deinococcus bacteria (32), this feature of D. radiodurans SSB could also be critical for protecting ssDNA that is exposed by the DNA repair processes after treatment with DNA damaging agents. In summary, the structure presented here provides a new model for SSB protein function in Deinococcus-Thermus group bacteria that could help explain their well noted vigorous survival mechanisms.
Acknowledgments
We thank the Advanced Photon Source Beamline staff (BioCARS Beamline 14-ID-B Station) for assistance in data collection. This work was supported by a grant from the Shaw Foundation for Medical Research (to J.L.K.) and National Institutes of Health Grants GM068061 (to J.L.K.) and GM52725 (to M.M.C.). D.A.B. was supported by National Institutes of Health Training Grant GM08293. M.P.K. is a Cremer Scholar.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ssDNA, single-stranded DNA; SSB, ssDNA-binding; OB, oligonucleotide/oligosaccharide-binding; RPA, replication protein A; PEG; polyethylene glycol.
Data deposition: The model coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PBD ID code 1SE8).
References
- 1.Lohman, T. M. & Ferrari, M. E. (1994) Annu. Rev. Biochem. 63, 527–570. [DOI] [PubMed] [Google Scholar]
- 2.Mushegian, A. R. & Koonin, E. V. (1996) Proc. Natl. Acad. Sci. USA 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murzin, A. G. (1993) EMBO J. 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suck, D. (1997) Nat. Struct. Biol. 4, 161–165. [DOI] [PubMed] [Google Scholar]
- 5.Shamoo, Y., Friedman, A. M., Parsons, M. R., Konigsberg, W. H. & Steitz, T. A. (1995) Nature 376, 362–366. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., Pfuetzner, R. A., Edwards, A. M. & Frappier, L. (1997) Nature 385, 176–181. [DOI] [PubMed] [Google Scholar]
- 7.Raghunathan, S., Kozlov, A. G., Lohman, T. M. & Waksman, G. (2000) Nat. Struct. Biol. 7, 648–652. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto, T., Morimoto, Y., Shibata, N., Kinebuchi, T., Shimamoto, N., Tsukihara, T. & Yasuoka, N. (2000) J. Biochem. (Tokyo) 127, 329–335. [DOI] [PubMed] [Google Scholar]
- 9.White, M. F. (2003) Biochem. Soc. Trans. 31, 690–693. [DOI] [PubMed] [Google Scholar]
- 10.Kerr, I. D., Wadsworth, R. I., Cubeddu, L., Blankenfeldt, W., Naismith, J. H. & White, M. F. (2003) EMBO J. 22, 2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battista, J. R. (1997) Annu. Rev. Microbiol. 51, 203–224. [DOI] [PubMed] [Google Scholar]
- 12.Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V. & Daly, M. J. (2001) Microbiol. Mol. Biol. Rev. 65, 44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabrowski, S., Olszewski, M., Piatek, R., Brillowska-Dabrowska, A., Konopa, G. & Kur, J. (2002) Microbiology 148, 3307–3315. [DOI] [PubMed] [Google Scholar]
- 14.Dabrowski, S., Olszewski, M., Piatek, R. & Kur, J. (2002) Protein Expression Purif. 26, 131–138. [DOI] [PubMed] [Google Scholar]
- 15.Eggington, J. M., Haruta, N., Wood, E. A. & Cox, M. M. (January 12, 2004) BMC Microbiol. 4, www.biomedcentral.com/1471-2180/4/2. [DOI] [PMC free article] [PubMed]
- 16.Meyer, R. R. & Laine, P. S. (1990) Microbiol. Rev. 54, 342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews, B. W. (1968) J. Mol. Biol. 33, 491–497. [DOI] [PubMed] [Google Scholar]
- 18.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105–124. [DOI] [PubMed] [Google Scholar]
- 19.Leslie, A. G. W. (1992) Newsletter on Protein Crystallography, No. 26 (Daresbury Laboratory, Warrington, U.K.).
- 20.Kabsch, W. (1988) J. Appl. Crystallogr. 21, 916–924. [Google Scholar]
- 21.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski, Z. (1991) in Proceedings of the CCP4 Study Weekend, eds. Wolf, W., Evans, P. R. & Leslie, A. G. W. (Daresbury Laboratory, Warrington, U.K.), pp. 80–86.
- 23.Cowtan, K. (1994) Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr. 31, 34–38. [Google Scholar]
- 24.Lamzin, V. S. & Wilson, K. S. (1993) Acta Crystallogr. D 49, 129–147. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 26.Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D. 57, 122–133. [DOI] [PubMed] [Google Scholar]
- 27.Yang, C., Curth, U., Urbanke, C. & Kang, C. (1997) Nat. Struct. Biol. 4, 153–157. [DOI] [PubMed] [Google Scholar]
- 28.Raghunathan, S., Ricard, C. S., Lohman, T. M. & Waksman, G. (1997) Proc. Natl. Acad. Sci. USA 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saikrishnan, K., Jeyakanthan, J., Venkatesh, J., Acharya, N., Sekar, K., Varshney, U. & Vijayan, M. (2003) J. Mol. Biol. 331, 385–393. [DOI] [PubMed] [Google Scholar]
- 30.Curth, U., Genschel, J., Urbanke, C. & Greipel, J. (1996) Nucleic Acids Res. 24, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genschel, J., Curth, U. & Urbanke, C. (2000) Biol. Chem. 381, 183–192. [DOI] [PubMed] [Google Scholar]
- 32.Mattimore, V. & Battista, J. R. (1996) J. Bacteriol. 178, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carson, M. (1997) Methods Enzymol. 277, 493–505. [PubMed] [Google Scholar]
- 34.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]