Abstract
In contrast to mRNAs, rRNAs are transcribed by RNA polymerase I or III and are not believed to be polyadenylated. Here we show that in Saccharomyces cerevisiae, at least a small fraction of rRNAs do have a poly(A) tail. The levels of polyadenylated rRNAs are dramatically increased in strains lacking the degradation function of Rrp6p, a component of the nuclear exosome. Pap1p, the poly(A) polymerase, is responsible for adenylating the rRNAs despite the fact that the rRNAs do not have a canonical polyadenylation signal. Polyadenylated rRNAs reside mainly within the nucleus and are in turn degraded. For at least one rRNA type, the polyadenylation preferentially occurs on the precursor rather than the mature product. The existence of polyadenylated rRNAs may reflect a quality-control mechanism of rRNA biogenesis.
Polyadenylation is a crucial step in 3′ end processing of mRNA in eukaryotes. Although the precise functions of poly(A) tails remain to be elucidated, they have been proposed to affect mRNA stability, nuclear export, and translation (1). Poly(A) tails have been thought to be a distinctive feature of RNA polymerase II products, the mRNAs, because RNA polymerase II is required for efficient polyadenylation. In addition, the polyadenylation of mRNA is coupled with cleavage and requires a large complex of factors, including cleavage and polydenylation specificity factor (CPSF) and cleavage stimulation factor (CstF), as well as cleavage and polyadenylation elements in the RNA sequence. A poly(A) polymerase, Pap1p, is responsible for adding a poly(A) tail to the 3′ end of mRNA after successful cleavage (2–4). rRNAs, which constitute the vast majority of the nucleic acid content of an organism, are products of RNA polymerase I or III. The 18S, 5.8S, and 25S rRNAs are initially synthesized by RNA polymerase I as a polycistronic precursor and then extensively processed and modified to the mature products. 5S rRNA is independently transcribed by RNA polymerase III (5–8). These RNAs were not believed to have poly(A) tails because they lack a recognizable polyadenylation element and are not synthesized by RNA polymerase II.
The riboexonuclease Rrp6p is located within the nucleus and is a member of the nuclear exosome (9), which plays a central role in the precise formation of the 3′ ends of many RNAs and also in the degradation of excess rRNAs or their precursors and processing intermediates. Rrp6p has been shown to be involved in 5.8S rRNA 3′ end formation. Deletion of Rrp6p causes accumulation of 5.8S rRNA with an ≈30-nt extension at the 3′ end. Rrp6p has also been proposed to degrade mRNAs in competition with Pap1p, the poly(A) polymerase (9, 10). A recent finding shows that Rrp6p and Cbc1p, the large subunit of the mRNA cap-binding complex, are involved in an mRNA degradation pathway in the nucleus. Deletion of RRP6 or CBC1 stabilizes normal mRNAs that are artificially retained in the nucleus (11).
RNA degradation in Saccharomyces cerevisiae was investigated by using Affymetrix Yeast S98 GeneChips to determine genomewide RNA expression in rrp6-Δ and cbc1-Δ strains, as well as the upf1-Δ strain, which is defective in the nonsense mediated decay pathway. The Affymetrix Yeast S98 chip contains 9,336 probe sets representing mRNAs, noncoding RNAs, and many other regions in the yeast genome. The Affymetrix Enzo BioArray HighYield RNA Transcript Labeling Kit, which initiates reverse transcription with the bacteriophage T7-Oligo(dT) promoter primer that contains the T7 promoter and 24 Ts, allowed us to monitor the expression profile of genome-wide polyadenylated RNAs. We were surprised to find that many probe sets on the chip representing rRNAs produced significant fluorescent signals. Strikingly, most of the rRNA signals were significantly enhanced on the chips that were hybridized with rrp6-Δ samples, compared to the normal strain. This prompted us to study the possibility that some rRNAs can actually be polyadenylated and the role of Rrp6p in accumulating polyadenylated rRNAs.
Materials and Methods
The following yeast strains were used in this study: B-9037 (normal), MATa cyc1-512 trp2-1 ura3-52; B-9038 (cbc1-Δ), MATa cyc1-512 cbc1::URA3 trp2-1 ura3-52; B-9848 (upf1-Δ), MATa cyc1-512 upf1::URA3 trp2-1 ura3-52; B-11872 (rrp6-Δ), MATa cyc1-512 rrp6::URA3 trp2-1 (11); BPO2 (normal), MATα ade1 ade2 lys2 gal1 ura1 his7 tyr1; BPO2KAN (rrp6-Δ), MATα ade1 ade2 lys2 gal1 ura1 his7 tyr1 rrp6::KAN; UR31481B (pap1-1), MATα ade1 ade2 lys2 gal1 ura1 his7 tyr1 pap1-1; UR31481BK (pap1-1 rrp6-Δ), MATα ade1 ade2 lys2 gal1 ura1 his7 tyr1 pap1-1 rrp6::KAN (12). Unless stated otherwise, B-9037, B-9038, B-9848, and B-11872 were used as the normal, cbc1-Δ, upf1-Δ, and rrp6-Δ strains, respectively. The following plasmids used in this study have been described by Phillips and Butler (13): pGFP-N-FUS (CEN6); pGFP-RRP6 (CEN6, RRP6); pGFP-RRP6-H3 (CEN6, rrp6-3); and pGFP-RRP6-H10 (CEN6, rrp6-13). Primers and probes used in this study are presented in Table 1.
Table 1. Primers and probes used in this study.
| Name | Sequence | Note |
|---|---|---|
| PT* | 5′-CACTCGAGT- (T) 30-3′ | poly(A) |
| P1† | 5′-GGCGGAAAGGCCTTGGGTGCTTGCTGGCG-3′ | 25S internal |
| P2† | 5′-GCCGCGAAGCTACCATCCGCTGGATTATGGCTG-3′ | 25S internal |
| P3* | 5′-ACAAATCAGACAACAAAGGCTTAATCTCAGCAG-3′ | 25S 3′ end |
| 27RD* | 5′-GGCCAGCAATTTCAAGTTA-3′ | 27S ITS2 |
| 25RD5* | 5′-ACTCCTACCTGATTTGAGGTCAAAC-3′ | 25S 5′ end |
| 25RDM* | 5′-CCTTAGGACATCTGCGTTATCGTTT-3′ | 25S internal |
| 58RD† | 5′-GGATCTCTTGGTTCTCGCATC-3′ | 5.8S 5′ end |
| 58RDN* | 5′-CGCTGCGTTCTTCATCGATGCG-3′ | 5.8S |
| 5RD† | 5′-GGTTGCGGCCATATCTACCAGAAA-3′ | 5S 5′e nd |
| 18RD† | 5′-GTTGATTACGTCCCTGCCCTTTGT-3′ | 18S internal |
| ACT† | 5′-CCGGTGGTACCACCATGTTCCCAGG-3′ | ACT1 internal |
| RDPT-Cy3* | 5′- (T) 21-ACAAATCAGACAACAAAGGCTTAATCTCAGCAGAG/3Cy3ph/-3′ | 25S + poly(A) |
| RD-Cy3* | 5′-----ACAAATCAGACAACAAAGGCTTAATCTCAGCAGAG/3Cy3ph/-3′ | 25S |
Antisense sequence used for a reverse-transcription primer, a PCR 3′ primer, a fluorescent in situ hybridization probe, or a Northern blot probe.
Sense sequence used for a PCR 5′ primer.
Microarray Analysis. High-density oligonucleo arrays (Affymetrix Yeast S98) were used to analyze the expression profiles of normal and mutant strains. Sample preparation, array hybridization, and scanning followed protocols provided by the manufacturer (Affymetrix). Hybridization data were analyzed with genetraffic 2.8 (Iobion, La Jolla, CA) by using Robust Multichip Analysis with the normal strain profile as baseline.
RT-PCR Assay. A total of 1 μg of total RNA was reverse transcribed by Moloney's murine leukemia virus (M-MLV) reverse transcriptase with 50 pmol of antisense primer in a 20-μl reaction volume. A total of 1 μl of reverse transcriptase was then used as PCR template and amplified by Taq polymerase with 2.5 pmol of primers of interest in 25 μl. Cycle numbers were monitored to perform saturated or unsaturated amplification. Five microliters of PCR products was resolved on 2% agarose gel stained with ethidium bromide and quantitated with imagequant 5.0 (Molecular Dynamics). Contrast was inverted for better display. PCR products were subcloned into PCR2.1 (Invitrogen) and sequenced with big dye 3.1 (Applied Biosystems). The primers are described in Table 1.
Fluorescence in Situ Hybridization (FISH) Analysis. FISH of 25S rRNA and 25S+poly(A) rRNA was performed by using Cy3-labeled probes, and DNA was stained with 4′,6-diamidino-2-phenylindole, as described (11). The probes are described in Table 1.
Total RNA isolation, poly(dT) selection, and Northern blot analysis were performed as described (14).
Results
Detection of Polyadenylated 25S rRNAs. Two independent Micro-Array assays were performed by using Affymetrix Yeast S98 chips following the protocol recommended by the manufacturer. A summary of all of the rRNA probe sets included in the Affymetrix S98 GeneChip is listed in Table 2. The fold change of individual rRNA probes varied among the two sets because of the signal fluctuation of the MicroArray analysis. However, the accumulation pattern of the rRNA signals in the rrp6-Δ strain was reproducible in both assays, and the fold changes of the rRNA signal were always the highest of the entire chip. The sample labeling initiates from a reverse transcription with T7-Oligo(dT) primer. Therefore, only RNAs with long poly(A) tracts were expected to be labeled. In contrast to rrp6-Δ, the other two mutant strains tested, cbc1-Δ and upf1-Δ, did not show a significant increase in the rRNA signal. Thus, we believe that the unusual rRNA signal is not due to systematic fluctuation or nonspecific labeling but instead results from reverse transcription of rRNAs with the T7-Oligo(dT) promoter primer. Considering that the longest poly(A) tract within the genome sequence of nascent polycistronic rRNA precursor, 37S rRNA, is only 11 nt long (Saccharomyces Genome Database, www.yeastgenome.org), we reasoned that the poly(dT) primer is most likely priming poly(A) sequences that are added to the rRNAs after transcription.
Table 2. Relative levels of representative transcripts of rRNAs.
|
rrp6-Δ
|
|||||
|---|---|---|---|---|---|
| Probe | Probe number | Experiment 1 | Experiment 2 | cbcI-Δ | upfI-Δ |
| 35S rRNA | 3806_s_at | 2.91 | 2.77 | 0.73 | 0.42 |
| 35S rRNA | 3807_s_at | 35.37 | 35.06 | 10.32 | 1.80 |
| 35S rRNA | 3808_s_at | 3.07 | 7.84 | 1.09 | 0.50 |
| 35S rRNA | 3809_s_at | 2.26 | 16.86 | 0.90 | 0.54 |
| 35S rRNA | 3810_s_at | 28.29 | 113.14 | 1.14 | 0.94 |
| 35S rRNA | 3811_s_at | 169.12 | 21.92 | 1.26 | 0.93 |
| 35S rRNA | 3764_s_at | 138.62 | 3.49 | 7.77 | 0.63 |
| 25S rRNA | 3765_s_at | 1.45 | 2.68 | 1.08 | 0.92 |
| 25S rRNA | 3766_s_at | 76.02 | 1.51 | 18.84 | 0.42 |
| 18S rRNA | 3767_s_at | 301.00 | 123.40 | 1.23 | 0.84 |
| 5S rRNA | 3768_i_at | 372.82 | 70.46 | 2.02 | 2.10 |
| 5S rRNA | 3769_s_at | 409.53 | 163.23 | 0.91 | 0.88 |
| 5S rRNA | 3770_i_at | 28.95 | 8.67 | 1.29 | 1.13 |
| 25S rRNA | AFFX-25srRnaa_at | 1.29 | 6.47 | 1.42 | 1.29 |
| 25S rRNA | AFFX-25srRnab_at | 8.63 | 74.61 | 1.03 | 0.79 |
| 25S rRNA | AFFX-25srRnac_at | 6.74 | 74.30 | 1.26 | 0.87 |
| 25S rRNA | AFFX-25srRnad_at | 218.57 | 71.0 | 1.00 | 0.83 |
| 25S rRNA | AFFX-25srRnae_at | 48.38 | 91.74 | 15.95 | 0.44 |
| 18S rRNA | AFFX-18srRnaa_at | 6.85 | 21.39 | 5.97 | 2.43 |
| 18S rRNA | AFFX-18srRnab_at | 58.27 | 37.13 | 17.72 | 1.97 |
| 18S rRNA | AFFX-18srRnac_at | 170.81 | 82.27 | 1.83 | 0.98 |
| 18S rRNA | AFFX-18srRnad_at | 203.10 | 84.61 | 1.42 | 1.10 |
| 18S rRNA | AFFX-18srRnae_at | 225.74 | 129.53 | 1.41 | 0.88 |
Expression data were normalized by using Robust Multichip Analysis implemented by genetraffic 2.8 (Iobion). The normal strain was used as baseline. Relative levels of all rRNA probe sets on the Affymetrix array were presented as fold changes against the normal strain.
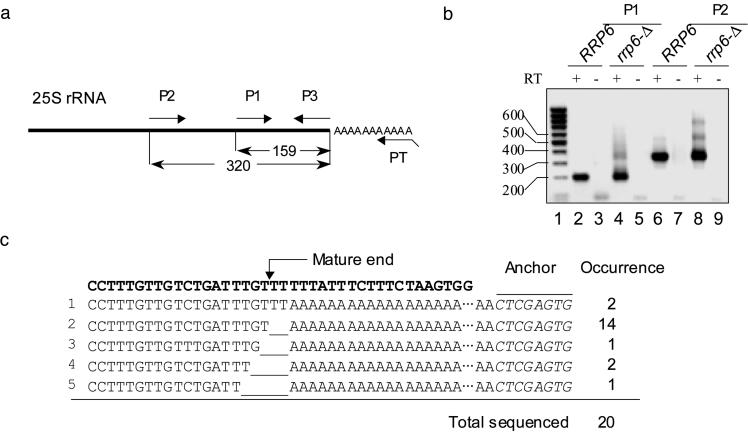
To verify that rRNAs can actually contain 3′ poly(A) tails, we first tested the 3′ end of the 25S rRNA, with RT-PCR (Fig. 1 a and b). Total RNA from a normal strain and rrp6-Δ strain was reverse transcribed with an anchored poly(dT)30 primer (PT) and then PCR amplified with the same PT and either of two specific internal primers, P1 or P2. Both primer sets yielded a distinctive product of the expected length in both the normal and rrp6-Δ strains. The PCR-amplified products were subcloned and sequenced. A total of 14 of 20 randomly selected clones showed a poly(A) sequence at least 30 nt long immediately downstream of the mature 3′ end of 25S rRNA (Fig. 1c). There is no apparent poly(A) tract at this region or cis-acting element reported for polyadenylation (15). Therefore, we believe that the poly(A) detected in this assay was not due to nonspecific priming, and that the poly(A) is added to the 25S rRNA at least after the mature 3′ end has been generated, which is several steps after transcription and Rnt1p cleavage during rRNA biogenesis. [For further review of rRNA biogenesis, see Fatica and Tollervey (7) and Butler (16).] The frequency of rRNA polyadenylation several nucleotides away from the mature end is low, indicating that polyadenylation occurs mainly at the normal 3′ end. Although this assay reveals the position of the 3′ poly(A) tails, it does not reveal information about the identity of the 5′ end(s) of these RNAs. Therefore, this population will be referred to as 25S-related RNAs, which includes the possible variation at the 5′ end.
Fig. 1.
Polyadenylated 25S related rRNAs are detected by RT-PCR. (a) Schematic diagram of the primers (Table 1). (b) Total RNA from the normal (B-9037) and rrp6-Δ (B-11872) strains was reverse-transcribed with the PT primer and then PCR-amplified with the PT primer and 5′ primer P1 or P2. A non-reverse-transcribed control was included. (c) The 3′ terminal sequences of subcloned reverse transcription products. The RDN25-1 genomic sequence is shown in bold. The various lengths of poly(A) were at least 30 nt.
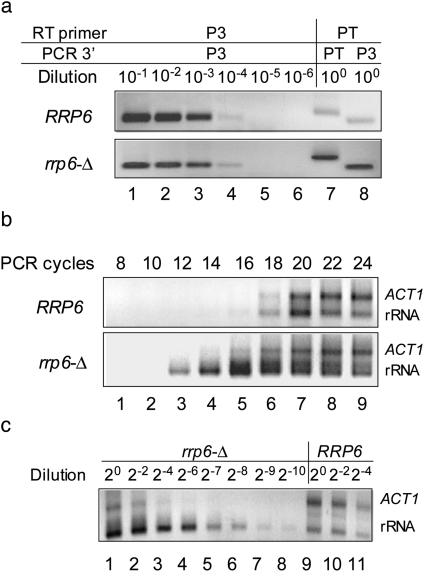
A Small Fraction of 25S-Related RNA Are Polyadenylated. In an effort to estimate relative levels of polyadenylated 25S-related RNA in both the normal and rrp6-Δ strains, the levels of these products were compared with a series of RT-PCR assays performed on diluted samples of the same RNAs (Fig. 2a, lanes 1–6). This analysis showed that the amount of the amplified products from each strain falls within the saturation curve, thus allowing the estimation that the polyadenylated 25S rRNA product is somewhere between 0.01% and 0.1% of total 25S rRNA in normal strain, and that the percentage increases to >1% in the rrp6-Δ strain (Fig. 2a, lanes 7 and 8). A RT-PCR time-course assay (Fig. 2b) indicated that the polyadenylated rRNA in the rrp6-Δ strain was saturated six to eight cycles earlier than that in the normal strain, whereas ACT1 mRNA saturated at about the same number of cycles with RNA from either strain. RT-PCR serial dilution assays (Fig. 2c) again revealed that the amount of amplified polyadenylated 25S-related RNA product in wild type was ≈2–7 of that in rrp6-Δ, assuming that the ACT1 mRNA product was at the same level for both strains. Combining the information described above, we conclude that <0.1% of 25S-related RNA is polyadenylated in normal strain, and this population increases up to 100 times in rrp6-Δ.
Fig. 2.
A small fraction of 25S-related RNA are polyadenylated. (a) Reverse transcripts were made with the P3 or PT primers and RNA from the RRP6 (B-9037) and rrp6-Δ (B-11872) strains. Lane 1–6, serial dilution of P3 transcripts PCR amplified with P1 and P3. Lane 7, PT transcripts amplified with P1 and PT. Lane 8, PT transcripts amplified with P1 and P3. (b) Time course of PCR for PT reverse transcripts. 25S rRNA was amplified with P1 and PT; ACT1 was amplified with ACT and PT in the same reaction. PCR cycle numbers are indicated. (c) PT reverse transcripts were diluted serially, and 18 cycles of PCR were performed. Lanes 1–8, the rrp6-Δ strain; lanes 9–11, the normal strain. Dilution factors are indicated. (See Table 1 for the sequences of the primers.)
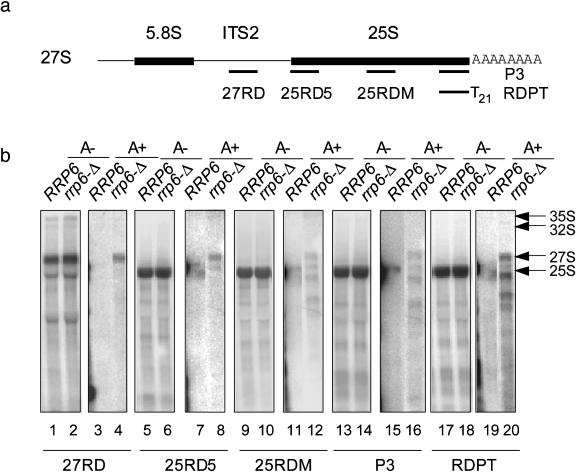
Polyadenylated 25S-Related RNAs Are Degraded from the 5′ End. In the MicroArray assay, the relative signal strength of 25S and 18S probe sets in rrp6-Δ exhibits a polarity pattern. It appears that the probe sets near the 3′ end of the transcript produce higher signals (Table 2; note the probe sets from AFFY-25srRnaa_at through AFFY-25srRnae_at and AFFY-18SrRnaa_at through AFFY-18srRnae_at). Although the reverse transcription used in the Affymetrix sample labeling procedure may also produce a higher signal at the 3′ end, measuring the fold changes against the normal strain compensated for this effect. Furthermore, the cbc1-Δ and upf1-Δ strains did not show this pattern. Therefore, we suggest that there is a 5′ exonuclease activity degrading the polyadenylated 25S related RNAs. In poly(dT) selection and Northern blot analysis, many bands of smaller length were detected by 25S rRNA probes to the 3′ end of the transcript (Fig. 3b, lanes 16 and 20). In contrast, probes directed toward the 5′ end of the 27S prerRNA detected only one (27RD), two (25RD5), or five (25RDM) bands (Fig. 3b, lanes 4, 8, and 12). This pattern is consistent with the hypothesis that the polyadenylated 25S-related RNAs are degraded from the 5′ end. The bands with smaller lengths detected here may actually be fragments of 25S-related RNAs with poly(A) tails, suggesting pausing of the 5′ exonuclease activity at secondary structures within 27S rRNAs. On the other hand, we could not rule out the possibility that 3′ degradation also exists, because this process would remove the poly(A) tail so that the fragments, if any, would escape poly(dT) selection. Indeed, the dependence of the polyadenylated rRNA on rrp6-Δ implies that these molecules may be degraded from the 3′ end. It is likely that the fate of polyadenylated 25S-related RNAs is to be degraded.
Fig. 3.
Polyadenylated 25S related RNAs are degraded from the 5′ end. (a) Schematic diagram of probes used in the Northern blot analysis. (For sequences of the probes, see Table 1.) (b) Poly(dT) selection and Northern blot analysis with 25S-related probes (1% argarose). The probes are indicated under each blot. A – designates the unbound fraction, whereas A+ designates the bound fraction. For each set of probes, the sensitivity of the phosphorimage was adjusted to produce similar intensities for the A– and A+ panels. The following A–/A+ ratios were used with the following probes: 42, 27RD; 11, 25RD5; 53, 25RDM; 160, P3; and 4, RDPT.
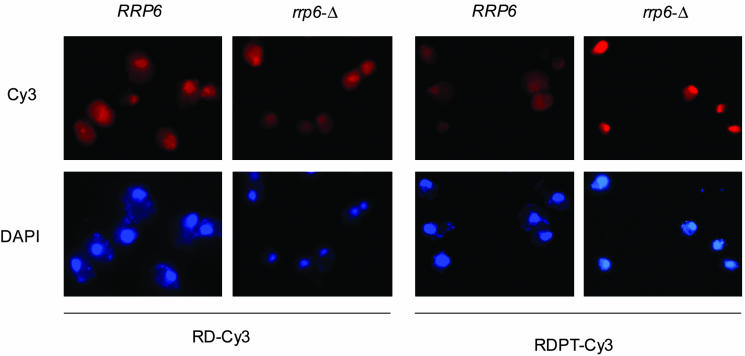
Polyadenylated 25S rRNAs Are Located in the Nucleus. That rRNA or its precursors can be polyadenylated prompted us to localize these polyadenylated species. A Cy3-labeled probe specific to polyadenylated 25S rRNA and normal 25S rRNA was used for fluorescent in situ hybridization (Fig. 4). The diffuse pattern of normal 25S rRNA is similar between normal and rrp6-Δ strains. The location of the center of Cy3 staining differs from the 4′,6-diamidino-2-phenylindole (DAPI) location. However, the polyadenylated 25S-related RNA in rrp6-Δ showed a strong colocalization with the DAPI signal, indicating that polyadenylated 25S-related RNAs are associated with the nucleus and possibly reside within the nucleolus, where the rRNA is processed and assembled into ribosomes. The retention of polyadenylated rRNAs may suggest that these RNAs have a different fate than normal rRNAs and are most probably not assembled in the ribosome but degraded by the cell as defective molecules.
Fig. 4.
Polyadenylated 25S related RNAs are retained in the nucleus. Fluorescence in situ hybridization with the Cy3-labeled 25S rRNA probe (RD-Cy3) or the 25S+poly(A) probe (RDPT-Cy3). 4′,6-Diamidino-2-phenylindole (DAPI) stains DNA and represents the position of the nucleus. Note that polyadenylated rRNA is localized in the nucleus of the rrp6-Δ strain.
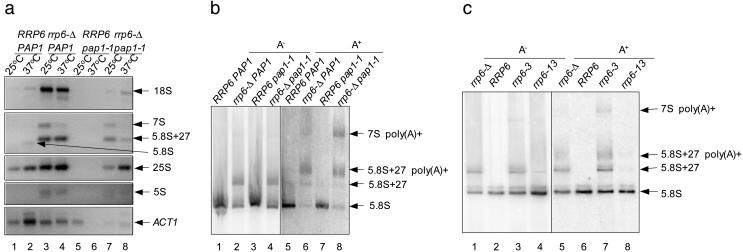
Polyadenylation of Other rRNAs in rrp6Δ Strains. Further study revealed that not only the 25S-related RNA but also the other three types of rRNA can be polyadenylated and accumulated in the rrp6-Δ strain (Fig. 5a, lanes 1–4). 5S rRNA is a product of RNA polymerase III, suggesting that all of the rRNAs can receive a poly(A) tail regardless of their origin, and Rrp6p has a role in affecting the level of a diverse set of polyadenylated rRNAs. Pap1p is the poly(A) polymerase in yeast. The temperature-sensitive mutant pap1-1 abolishes polyadenylation of mRNAs at nonpermissive temperature (12, 14) (Fig. 5a bottom, lanes 5 and 6). Interestingly, the polyadenylated rRNAs are not detectable in the pap1-1 strain even at the permissive temperature (Fig. 5a, lanes 5 and 6) but are partially restored by deleting Rrp6p (Fig. 5a, lanes 7 and 8). A similar result was also seen when RNAs are analyzed by poly(dT) selection and Northern blotting (Fig. 5b, lanes 7 and 8), suggesting that Pap1p plays a role in adenylating the rRNAs despite the fact that they lack a recognizable polyadenylation signal and are synthesized by RNA polymerase I or III. Two point mutants of Rrp6p, rrp6-3 and rrp6-13, were examined to determine the role of Rrp6p. The rrp6-3 lacks both rRNA processing and degradation functions of Rrp6p, whereas rrp6-13 lacks only the processing function (13). Interestingly, the polyadenylated 5.8S rRNA product was not detectable in the rrp6-13 strain but was present in the rrp6-3 strain (Fig. 5c, lanes 7 and 8), suggesting that the Rrp6p may degrade the polyadenylated rRNAs or shorten their poly(A) tail, which is consistent with the proposed role of the Rrp6p in mRNA processing and polyadenylation (12, 16).
Fig. 5.
A diverse set of rRNAs is polyadenylated and degraded. (a) Cells were grown at 25°C and transferred to 37°C for 1 h. Each of the 5S, 18S, 5.8S, 25S rRNAs and the ACT1 mRNA were amplified with PT and the corresponding 5′ primer. (b) Total RNA was extracted from cells grown at 25°C. Poly(dT) selection and Northern blot analysis with 5.8S rRNA probe were performed with total RNAs from strains indicated (8% PAGE) A– designates the unbound fraction, whereas A+ designates the bound fraction. The bound fraction was exposed 50 times longer than the unbound fraction. Strains used in a and b: BPO2 (normal), BPO2KAN (rrp6-Δ), UR31481B (pap1-1), and UR31481BK (pap1-1 rrp6-Δ). (c) Total RNA was extracted from cells grown at 30°C. Poly(dT) selection and Northern blot analysis were performed as described in b. All strains used in the analysis are BPO2KAN containing one of the following plasmids: lanes 1 and 5, pGFP-N-FUS (CEN6); lanes 2 and 6, pGFP-RRP6 (CEN6 RRP6); lanes 3 and 7, pGFP-RRP6-H3 (CEN6 rrp6-3); and lanes 4 and 8, pGFP-RRP6-H10 (CEN6 rrp6-13).
Discussion
In this study, we have determined that in S. cerevisiae, all of the rRNAs can be polyadenylated at a certain level, and the polyadenylated rRNAs accumulate dramatically in an rrp6-Δ strain. Moreover, the poly(A) polymerase, Pap1p, appears responsible for the polyadenylation of rRNAs.
There are at least two possible explanations for this surprising phenomenon. First, the rRNAs, although having no recognizable poly(A) signal, are polyadenylated by Pap1p at a certain level, and Rrp6p may have a role in degrading them or removing their poly(A) tails. However, the rRNAs may not be the preferred substrates of Pap1p, because only a small fraction of rRNA appears to be polyadenylated. We also observed that there is a diffuse set of products running slower than the 25S rRNA RT-PCR product in rrp6-Δ (e.g., Fig. 2b, lanes 3 and 7). These products may correspond to either RNAs with various lengths of the poly(A) tail or to various lengths of 3′ premature species. However, in randomly selected clones, we did not find sequences corresponding to the premature 3′ end of 25S rRNA. Because the polyadenylated 3′ premature 25S rRNA is at a very low level, and because there is no evidence that Rrp6p is directly involved in 25S rRNA 3′ end formation, we suggest it is likely that Rrp6p is responsible for degradation of the polyadenylated rRNAs, or that it competes with polyadenylation machinery to shorten the poly(A) tail. This is consistent with the finding that polyadenylated rRNA did not accumulate in an rrp6-13 strain. Therefore, Rrp6p may play a wide role in removing the poly(A) tail or degrading RNAs that carry a poly(A) tail.
The second possibility is that rRNA precursors or aberrant rRNAs may accumulate in rrp6-Δ and may be subsequently polyadenylated. In other words, the ribosome biogenesis defects caused by rrp6-Δ led to polyadenylation of rRNAs. The evidence supporting this hypothesis came from the behavior of 5.8S rRNA, the direct substrate of Rrp6p's processing function. Deletion of RRP6 causes accumulation of 5.8S rRNA that is extended by ≈30 nt at its 3′ end (9, 17). In our RT-PCR assay, polyadenylated mature 5.8S rRNA is barely detectable in normal strain. However, in the rrp6-Δ strain, we were able to amplify a polyadenylated 27-nt extended 5.8S rRNA at a much higher level (Fig. 5a, second blot, sequence data not shown). Northern blot analysis of poly(dT) selected total RNA from the normal and rrp6-Δ strains also revealed the accumulation of polyadenylated 5.8S + 27 rRNA in the rrp6-Δ strain (Fig. 5b). It is likely that these rRNA precursors are more susceptible to polyadenylation compared to the mature product, given that the abundance of mature 5.8S rRNA, and 5.8S + 27 rRNA is similar in the rrp6-Δ strain (Fig. 5b, lane 2). This phenomenon appears similar to the polyadenylation of stable RNAs in prokaryotes, which also accumulate in the absence of processing exoribonucleases, including the Rrp6p homologue, RNase D. It has been proposed that poly(A) may serve as a degradation signal for deficiently processed noncoding RNAs in prokaryotes (18, 19). In fact, the signals of almost all of the noncoding RNAs included in the Affymetrix S98 chip were increased in the rrp6-Δ strain (data not shown), agreeing with previous findings that several premature small nucleolar RNAs were accumulated in the polyadenylated format in certain exosome mutants, including the RRP6 deletion (17, 20). Therefore, we believe it is reasonable to assume that S. cerevisiae may use a similar strategy to achieve quality control of noncoding RNAs. The deletion of Rrp6p might not only reduce the ability to degrade polyadenylated rRNA but might also cause the accumulation of inappropriately processed RNAs, thus leading to their polyadenylation. Unlike the preferential polyadenylation on 5.8S + 27 rRNA, 25S rRNA was mainly polyadenylated at the mature 3′ end. However, with the length limitation of RT-PCR, we could not rule out the possibility that rrp6-Δ indirectly causes defective processing at the far 5′ end of some 25S-related RNA precursors and in turn causes the population to be a target of polyadenylation. If the hypothesis that poly(A) serves as a marker for aberrant or precursor rRNAs is true, we expect polyadenylation of rRNAs in other rRNA processing mutants. A transient appearance of polyadenylated 25S rRNA was reported to occur in Candida albicans when the cells were stressed by serum treatment (21). This phenomenon could also be explained by the surveillance mechanism, which removes rRNAs that are polyadenylated because of aberrant processing.
The discovery of polyadenylated rRNA changes the traditional view that poly(A) is a distinctive feature of mRNAs. Furthermore, although the relative amount of polyadenylated rRNA is low, the amount is still comparable to mRNAs, given the high preponderance of rRNA, >95% of total RNA in yeast. Our findings imply that one may obtain a high “background” when using oligo(dT) to represent mRNA because of polyadenylated rRNAs. Thus, some of the previous results considering poly(A) RNA as mRNA may need to be revaluated, especially when a mutant that affects rRNA biogenesis was used.
Acknowledgments
We thank Dr. Yisang Yoon (University of Rochester) for assistance in the use of the fluorescence microscope, Ms. Linda Salamone (University of Rochester) for processing MicroArray analysis, and Dr. Biswadip Das (University of Rochester) for useful discussions. This work was supported by National Institutes of Health Grants R01 GM12702 (to F.S.) and R01 CA95913 (to J.S.B.). Affymetrix chips and processing were provided in part by the Nathan Shock Center for Excellence in Aging Research Microarray Core and National Institutes of Health Grant P30 AG18254.
Abbreviation: PT, poly(dT)30 primer.
References
- 1.Colgan, D. F. & Manley, J. L. (1997) Genes Dev. 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 2.Hirose, Y. & Manley, J. L. (1998) Nature 395, 93–96. [DOI] [PubMed] [Google Scholar]
- 3.Proudfoot, N. & O'Sullivan, J. (2002) Curr. Biol. 12, R855–R857. [DOI] [PubMed] [Google Scholar]
- 4.Zhao, J., Hyman, L. & Moore, C. (1999) Microbiol. Mol. Biol. Rev. 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng, W. T., Robinson, M. D., Mnaimneh, S., Krogan, N. J., Cagney, G., Morris, Q., Davierwala, A. P., Grigull, J., Yang, X., Zhang, W., et al. (2003) Cell 113, 919–933. [DOI] [PubMed] [Google Scholar]
- 6.Sherman, F. (2002) Methods Enzymol. 350, 3–41. [DOI] [PubMed] [Google Scholar]
- 7.Fatica, A. & Tollervey, D. (2002) Curr. Opin. Cell Biol. 14, 313–318. [DOI] [PubMed] [Google Scholar]
- 8.Wahle, E. & Ruegsegger, U. (1999) FEMS Microbiol. Rev. 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 9.Briggs, M. W., Burkard, K. T. & Butler, J. S. (1998) J. Biol. Chem. 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- 10.Burkard, K. T. & Butler, J. S. (2000) Mol. Cell. Biol. 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, B., Butler, J. S. & Sherman, F. (2003) Mol. Cell. Biol. 23, 5502–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proweller, A. & Butler, S. (1994) Genes Dev. 8, 2629–2640. [DOI] [PubMed] [Google Scholar]
- 13.Phillips, S. & Butler, J. S. (2003) RNA 9, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel, D. & Butler, J. S. (1992) Mol. Cell. Biol. 12, 3297–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Z. & Sherman, F. (1996) Trends Biochem. Sci. 21, 477–481. [DOI] [PubMed] [Google Scholar]
- 16.Butler, J. S. (2002) Trends Cell Biol. 12, 90–96. [DOI] [PubMed] [Google Scholar]
- 17.Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E. & Tollervey, D. (1999) EMBO J. 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Z., Pandit, S. & Deutscher, M. P. (1998) Proc. Natl. Acad. Sci. USA 95, 12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Z., Reimers, S., Pandit, S. & Deutscher, M. P. (2002) EMBO J. 21, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoof, A., Lennertz, P. & Parker, R. (2000) Mol. Cell. Biol. 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann, J. & Liu, H. (2001) Gene 265, 71–76. [DOI] [PubMed] [Google Scholar]