Abstract
Protein adsorption on nanoparticles is closely associated with the physicochemical properties of particles, in particular, their surface property. We synthesized two batches of polyacrylic acid-coated nanoparticles under almost identical conditions except for heating duration and found differences in the head-group structure of the polyacrylic acid. The structure change was confirmed by NMR and MS. The two batches of particles had varied binding affinities to a selected group of proteins. Computational work confirmed that the head group of the polymer on the surface of a nanoparticle could directly interact with a protein, and small structural changes in the head group were sufficient to result in a significant difference in the free energy of binding. Our results demonstrate that protein adsorption is so sensitive to the surface property of particles that it can reveal even small variations in the structure of a nanoparticle surface ligand, and should be useful for quick assessment of nanoparticle properties.
1. Introduction
Nanoparticles (NPs) have been replacing bulk materials for application in diverse areas. With increasing exposure of humans to NPs, much research attention has been drawn to the interactions between NPs and biomolecules,1–3 which could affect the behaviors of NPs in biosystems. Protein adsorption has been found to be strongly influenced by the surface properties of NPs.4–9 Some adsorption behaviors are governed by the overall surface charge and hydrophobicity, similar to those observed on a flat surface made by bulk materials.10 However, the comparable dimensions of NPs and proteins suggest that protein–NP interactions could mimic protein–protein interactions, and the NP surface ligands may target particular domains on proteins via particular functional groups.11–13 This feature suggests that protein adsorption could be very sensitive to changes in the surface property of NPs.
One possible change in the NP surface property is alteration of ligand structure during particle preparation, which could be induced by slight variations in synthesis conditions or use of alternative methods. For example, high-quality nanocrystals, including the II–VI binary semiconductors and metal oxides, that possess unique optical, electrical, and magnetic properties, are typically prepared by pyrolysis reactions or solvothermal processes. These approaches all involve conditions such as high temperature, high pressure, and/or elongated reaction duration to achieve tight size control and/or high crystallinity.14–18 Nevertheless, these harsh synthetic conditions may lead to structural changes in the capping agents that are used to stabilize the nanocrystals. Additionally, ligand exchange and surface modification are common in preparing water-soluble NPs for biomedical research and applications. Varied ligand exchange efficiency may lead to different surface coverage with the hydrophilic ligands on NPs; and chemical modification often requires multiple treatments that may lead to unexpected structural alteration to the surface ligands as well.19 In many cases changes in ligand density and its structure could be subtle enough to not affect ligand binding to the NP surface and dispersion of NPs in solution. However, they could lead to variation in the surface properties of these ligand-capped nanocrystals, and ultimately affect the performance of NPs as biosensors, drug carriers, and in other targeted applications. Thus, such changes should be detected before the deployment of NPs. Though, detailed study of the surface properties of NPs could be challenging because most of the NP core could strongly interfere with many spectroscopic measurements, such as FT-IR spectroscopy, NMR, and MS.20–23 Commonly, the ligands are removed from the NP surface by exchange reaction or chemical cleavage24,25 and are studied without the presence of NPs. In addition, the NP core could be digested by strong acids and the surface ligands purified extensively after tedious procedures before structure analysis.
In the present study, we investigated whether protein adsorption could reveal small changes in the surface ligand of NPs. Highly stable iron oxide NPs were coated with polyacrylic acid (PAA). Up to 20-fold difference in the dissociation constant of the protein–NP complex was detected on the NPs synthesized under different heating durations. Investigation of the PAA structures by NMR and MS confirmed that the heating duration could affect the head-group of PAA. Computational work also verified that the subtle difference in the head-group structure was sufficient to alter the binding energy to a target protein. Our results support that protein adsorption could be a quick and simple way to evaluate particle surfaces and assess small variation in surface ligands, before detailed characterizations were performed by more sophisticated techniques. Such assessments are highly important for particles intended for biomedical applications or with biosafety concerns.
2. Experimental
2.1 Synthesis of PAA-Fe3O4 NPs
The highly water-soluble superparamagnetic iron oxide (Fe3O4) NPs coated with PAA (average Mw 1.8 kDa) were used in our study. The reagents for synthesis were from Sigma-Aldrich Corporation. A NaOH/diethylene glycol (DEG) stock solution was prepared by dissolving 50 mmol of NaOH in 20 mL of DEG. The synthesis approach used in this study followed the procedure previously reported by the Yin group.15 In brief, a mixture of PAA (4 mmol), FeCl3 (2 mmol), and DEG (15 mL) was heated up to 220 °C under nitrogen atmosphere with stirring. Upon a rapid injection of the NaOH/DEG stock solution (4.5 mL), the mixture was further heated for 10 min or 12 hr at 220 °C to yield the magnetite nanoparticles. The excess PAA or its possible by products and other reagents were removed by centrifugation-assisted washing several times using a mixture of deionized water and ethanol. Finally, the Fe3O4 NPs were suspended in 10 mL of water to form the stock solutions.
2.2 Capillary Electrophoresis
Capillary electrophoresis (CE) separation for KD measurement was reported in our previous work.26 Briefly, a 50 cm fused-silica capillary (75 μm id, 365 μm od; PolymicroTechnologies, AZ, USA) with an effective length of 40 cm was sequentially rinsed at 30 psi with 0.1 M NaOH (2 min), deionized water (1 min), and the separation buffer (6 min) prior to injection. All CE separations were done at 25 kV at room temperature. Capillary zone electrophoresis (CZE) was performed using 10 mM borate buffer at pH 8.3 as the separation buffer, in which the samples were pre-incubated in 17.5 mM phosphate buffer (pH 7.5). In affinity capillary electrophoresis (ACE) experiments, no pre-incubation of protein and NPs was needed, and the separation buffer was 17.5 mM sodium phosphate (pH 7.5).
2.3 Characterization of NPs
A Philips Tecnai 12 transmission electron microscope (TEM) was used to investigate the morphology of NPs. Inductively coupled plasma atomic emission spectroscopy (ICP-AES) tests were performed on a Perkin-Elmer Optima 2000 DV optical emission spectrometer to measure the concentration of Fe element in NPs. With the average diameter of the spherical iron oxide NPs measured under TEM and the element content obtained from ICP-AES, molar concentration of the NPs stock solutions was obtained. Dynamic light scattering (DLS) was performed on a Beckman Coulter Delsa Nano C particle analyser at 25 °C. Zeta potential was calculated from the mobility of NPs in CZE.27
2.4 Characterization of PAA structures
The structure of PAA was analyzed using mass spectroscopy (MS) and NMR spectroscopy. The poly(acrylic acid) (PAA) underwent the same hydrolysis procedure as that in synthesis of NPs but without the addition of FeCl3. Once dissolved in DEG, the PAA solution was heated to 220 °C under N2 followed by injection of NaOH/DEG. The PAA structure obtained at different hydrolysis durations such as 10 min, 1 hr, 2 hr, and 12 hr, were tested by sampling 1 mL of the reaction mixture at the corresponding time point.
MS measurement was done by a Thermo-Fisher Electrospray Ionization LTQ mass spectrometer (ESI-LTQ-MS) in the negative ionization mode. Ten μL of the resulted PAA solution was mixed with 190 μL water and 200 μL HPLC grade acetonitrile. The ESI conditions used were as below: spray voltage = −1.5 kV, capillary temperature = 200 °C, capillary voltage = −38 V, tube lens voltage = −100 V.
A combination of one- and two-dimensional (2D) 1H and 13C nuclear magnetic resonance (NMR) spectroscopy were performed on the Varian Inova 500 NMR instrument (Varian Inc., Palo Alto, CA). The 2D NMR included correlation spectroscopy (COSY) and Heteronuclear single-quantum correlation spectroscopy (HSQC). The COSY experiment correlates bond coupling interactions between protons. The diagonal peaks correspond to the peaks in a 1D-NMR experiment, while the off diagonal cross peaks indicate coupling between pairs of nuclei. The 1H-13C HSQC provides correlation between a proton and the carbon it is directly bonded to.
2.5 Computational details for calculating PAA-protein interaction energy
The 3-dimantional (3D) experimental structure of CaM was extracted from Protein Data Bank with PDB code 1CFD.28 We created the 3D structures of the dimeric acrylic acid fragment with two head groups, head group A (HGA) (γ-lactone) and HGB (–C(OH)(CH3)2), using the VegaZZ program29. The Vconf program30 was used to carry out conformational search to obtain the lowest energy state as the starting molecules for the following docking. Ligand-protein docking was performed by the Autodock31 program with the Lamarckian genetic algorithm, which fixed the protein and allowed the polymer fragment to move around in the docking box. AutoDockTools 1.5.432 was used to assign Gasteiger33 charges to the PAAs. The AutoDock scoring function is a subset of the AMBER force field that treats molecules using the United Atom model. We focused docking simulations on six potential binding sites: spots near Gln8, Asp24, Thr79, Glu84, Glu114 and the center of the CaM. For each binding site, Autogrid version 4.0 was used to create affinity grids with 0.375 Å spacing. The cubic grid box with a dimension of 2.25 nm was assigned centering six different locations on CaM for each docking simulation. The final docking result was obtained by 10 runs of simulation with one million rounds of energy evaluation in each run. Each round of energy evaluation was for one possible orientation of the head group structure at the docking site. Ligand conformations with negative computed binding free energies were further analysed.
3. Results and discussion
3.1 IONPs synthesized under different heating durations carried PAA capped with different head groups
To test whether protein adsorption is sensitive enough to reveal small variations in the NP surface ligands, we prepared two batches of the superparamagnetic Fe3O4 NPs coated with PAA (average Mw 1.8 kDa). We picked this type of NPs because they have been widely used in biomedical research and practice for examining molecule or cell isolation, cancer treatment, drug delivery, and MRI.34–38 Additionally, polyelectrolytes have gained great popularity as surface ligands on NPs owing to their exceptional capability in stabilizing NP suspension, assisting NP assembly, enhancing drug loading capacity, and control of protein resistance or attraction.39–42 In particular, PAA allows easy protein immobilization and promotes high particle stability in aqueous solutions.43–46 The highly simple NP preparation strategy we used involves solution-phase hydrolysis at a high temperature, of 220°C, in DEG.15 The particles prepared by this method are Fe3O4 as confirmed by X-ray Adsorption Spectroscopy in the previous report from the Yin group.15 Hydrolysis at elevated temperature is a common approach in preparation of nanomaterials, and hydrolysis duration is often tuned to improve the formation and crystallinity of NPs. Because PAA can undergo thermal degradation at temperature > 160 °C,47, 48 we postulated that if we prepared the NPs under two very distinct hydrolysis durations, the PAA structure might be altered, resulting in NPs having the same chemical compositions and close physical properties, but carrying surface ligands with small variations.
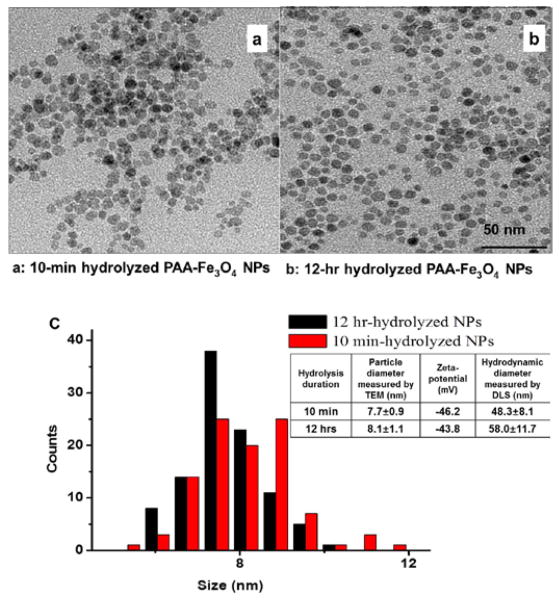
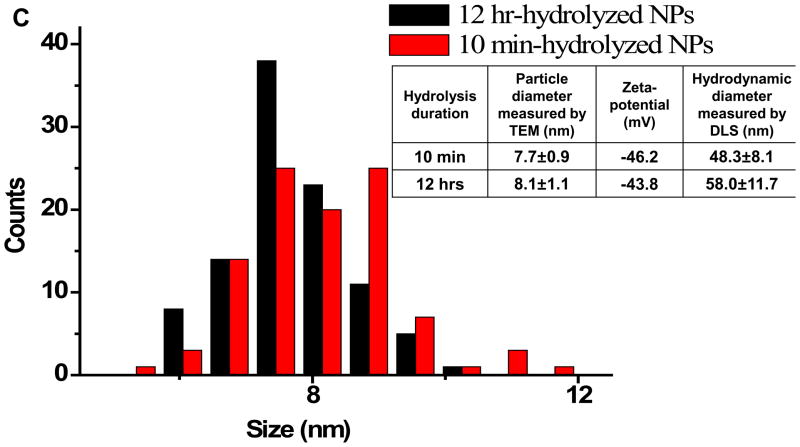
Indeed, NPs prepared by 10-min and 12-hr hydrolysis shared good similarity in particle shape (spherical), diameters measured by TEM (Figure 1) and DLS, as well as zeta potential (inserted Table in Figure 1c). To be more specific, the difference in the particle diameter obtained from the TEM images was only 5%. Although 12-hr hydrolysed NPs had relatively larger (17%) hydrodynamic diameters, the surface charge densities were similar, which was reflected by the 5% difference in zeta potentials. The particles were highly stable in water, with no precipitation even after several months’ storage. Additionally, when analysed by capillary electrophoresis (Electronic Supplementary Information (ESI), Figures S1), both particles gave out sharp and close to symmetrical peak shape. Both facts support that the particles were stably covered by PAA and had quite uniform charge to size ratio.
Figure 1.
TEM images of the PAA-Fe3O4 NPs prepared by 10-min (a) or 12-hr (b) hydrolysis. The two TEM image share the same scale bar. (c) Distribution profiles of the particle diameters measured in the TEM images for 100 particles. The inserted Table showed the average particle diameters measured by TEM and DLS; and zeta-potentials calculated from mobility in capillary electrophoresis (Figure S1).
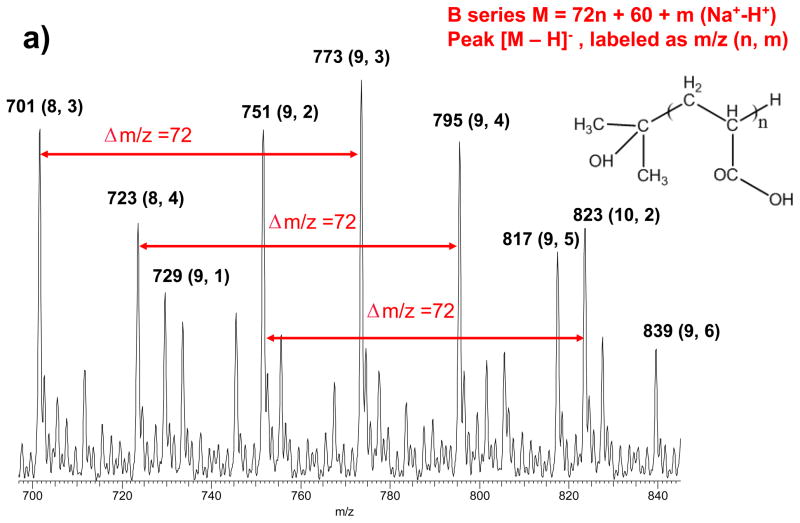
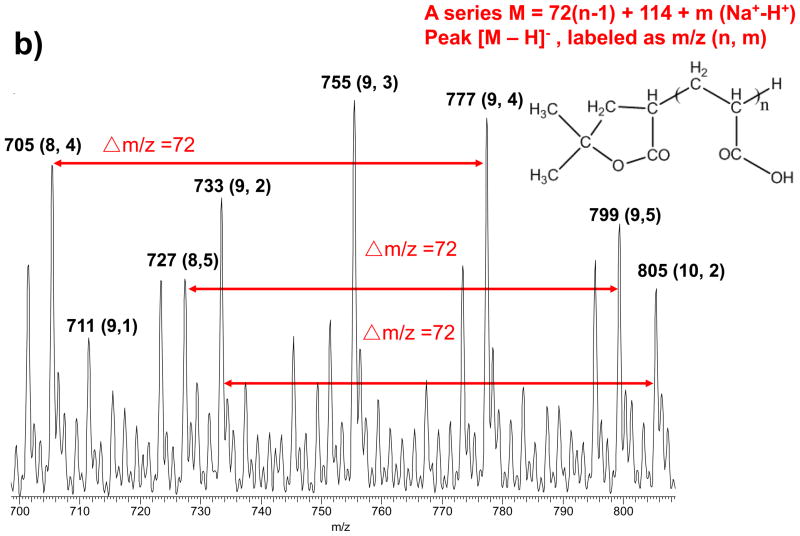
To confirm the PAA structures were different when hydrolyzed for 10 min or 12 hr, we measured the purchased PAA before and after hydrolysis by ESI-MS. Before hydrolysis, the PAA contained two types of the –(CH2CH(COOH))– polymer chains (Figure S2, ESI).48 One type of the PAA chains was capped with a head-group of –C(OH)(CH3)2 (HGB), and with a molecular weight (Mw) pattern of 72n+60. This type was called the B series PAA (bPAA) in the following text. The other type of PAA was capped with a γ-lactone (HGA) and had a Mw pattern of 72(n-1)+114. We referred this type as the A series (aPAA), which should be produced after the bPAA went through an esterification reaction between the –OH group and the –COOH group of the adjacent acrylic monomer. The peak intensities for both types of PAA chains were comparable before hydrolysis. However, after the PAA was hydrolyzed at 220°C in NaOH/DEG for 10 minutes, most of the MS peaks observed belonged to the B series (Figure 2a); while the A series was the dominant species in samples heated for 12 hr (Figure 2b). The intensity of peaks belonging to the A series gradually increased with longer heating duration compared to that of the B series peaks and reached saturation after 4 hr (Figure S3, Supporting Information).
Figure 2.
Mass spectrum of a) B series PAA obtained by 10 min thermal treatment and b) A series PAA by 12 hours heating treatment in the range of m/z 680–850. The quasimolecular ions of (M − H)- were formed in the negative ionization mode, with M losing one H+. Both MS uniformly showed Δm/z = 72, the exact mass of the acrylic acid monomer; and the peaks were all sodium adducts due to the presence of NaOH: among “n” numbers of monomers on each PAA chain, various numbers (dictated as “m” in Figure 2) of monomers carried –COO-Na+ instead of –COO−H+. The molecular weight difference after H+ being replaced by Na+ is +22.
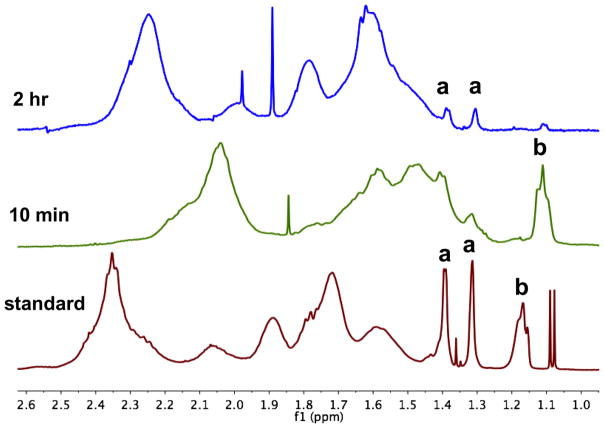
NMR experiments also verified the structure change in the PAA head group at different hydrolysis durations. The result of one-dimensional 1H NMR spectroscopy is displayed in Figure 3, with the 2D 1H and 13C NMR spectroscopy results shown in Figure S4 (ESI). In Figure 3, the peaks labeled as “a” correspond to the two methyl groups in the five-membered ring of HGA in the aPAA series. Due to the different environment above and below the plane of the ring, these two methyl groups have different chemical shifts. The peak labeled as “b” corresponds to the two methyl groups in HGB of the bPAA series. These two methyl groups are chemically identical and have the same chemical shift. Because PAA is a mixture of polymer chains with different lengths, it would be expected to have line broadening and chemical shift effects associated with the different polymer chain lengths and head-group interactions, which can be seen in the asymmetrical peak shapes of all the methyl resonances, and is more pronounced in peak b. The area ratio of peak “a” and “b” also showed gradual change with the hydrolysis duration. In the standard PAA, the ratio of peak “a”:”b” was 1.6:1. In the 10-min hydrolyzed PAA, peak “b” did not change much, while peak “a” decreased in intensity, which indicates that bPAA was the dominant species when hydrolyzed for 10 min. However, after 2 hr of hydrolysis, the situation has reversed, and the ratio of peak “a” to “b” increased to 4.9:1, meaning that more aPAA were present. Unfortunately, neither peak “a” nor “b” were found in the 12-hr hydrolyzed PAA sample, and probably were masked by the debris from PAA fragmentation over the long heating event (Figure S4a, ESI). Still, the trend of change in the relative abundance of the aPAA and bPAA series with hydrolysis duration agreed with that observed by MS.
Figure 3.
1H NMR spectra for standard PAA and PAA heated for 10 min and 2 hr.
3.2 The head-group change in PAA on NP surface could be detected by protein adsorption
Because the hydrolysis durations of 10 min and 12 hr should have yielded the PAA with HGB and HGA, the NPs synthesized at 10-min hydrolysis should be coated with bPAA and that produced at 12-hr hydrolysis should be coated with aPAA. The aPAA seemed to yield a hydrodynamic size of the NPs 17% larger than the bPAA, although the core sizes of the NPs measured by TEM differed by only 5%. To determine whether protein adsorption is sensitive enough to detect such a small difference in the surface PAA structure, despite the highly comparable physical and chemical properties of these NPs, we evaluated the adsorption of a series of proteins on these NPs. We selected human serum albumin (HSA), β-casein, calmodulin (CaM), myoglobin, and cytochrome c (Cyt c) because they possess relatively strong affinity to a large variety of flat surfaces or NPs,49–52 have varied Mw and isoelectric point (pI) values, and differ in structure ridigity. Three are acidic proteins, all with pI < 5: CaM, β-casein, and HSA, with Mw increasing from CaM to HSA (Table 1). CaM, myoglobin, and Cyt c have comparable Mw, but carry negative, zero, and positive charges, respectively, at pH 7. CaM (no Ca2+ bound) and β-casein are considered highly flexible proteins, while HSA, myoglobin and Cyt c are globular proteins with relatively rigid structures.
Table 1.
Affinity measurement and kinetics evaluation on protein-NP interaction
| Protein | Hydrolysis duration of NPs | KD (μM) | n | Dissociation kinetics |
|---|---|---|---|---|
| HSA 66 kDa, pI 4.8 |
10 min | 1.4 ± 0.11 | 1.0 | Fast |
| 12 hr | 5.1 ± 0.18 | 1.1 | Slow | |
| β-casein 24 kDa, pI 4.6 |
10 min | 0.23 ± 0.0085 | 2.7 | Fast |
| 12 hr | 4.6 ± 0.093 | 1.3 | Slow | |
| Calmodulin 17 kDa, pI 3.9 |
10 min | 0.28 ± 0.024 | 0.4 | Fast |
| 12 hr | 0.015 ± 0.00063 | 2.1 | Fast | |
| Myoglobin 17 kDa, pI 6.8 |
10 min | 1.8 ± 0.11 | 1.3 | Fast/Medium |
| 12 hr | 4.0 ± 0.27 | 1.4 | Slow | |
| Cytochrome C 12 kDa, pI 10.7 |
10 min | 18 ± 4.8 | 1.9 | Fast/Medium |
| 12 hr | 2.9 ± 0.091 | 1.1 | Slow |
We measured affinity by a method developed in our group. The f molar fraction of the protein-bound NPs, θ, was obtained with capillary electrophoresis (CE), and the data were fitted to the Hill equation:53
where KD is the microscopic dissociation constant of the NP-protein complex and n is the binding cooperativeness. An “n” < 1 represents strong repulsion between proteins, which prevents adsorption of more than one protein on the same NP, and an “n” > 1 but < 2 often corresponds to a 1:1 binding ratio.53, 54 The KD and n values for interaction of the selected proteins and the two batches of NPs are in Table 1 (CE spectra and the affinity fitting curve shown in Figure S5, ESI).
We detected significant affinity differences, ranging from 2- to 20-fold, between the NPs produced at varied hydrolysis durations in all proteins. The NPs obtained from 10-min hydrolysis interacted more strongly with HSA, β-casein, and myoglobin, with KD differing by 3.6-, 20-, and 2.2-fold, respectively, whereas the NPs prepared by 12-hr hydrolysis formed a stronger interaction with CaM (the Ca2+-free form) and Cyt c, with 20- and 6-fold difference in KD values. The n values for the two highly flexible proteins, β-casein and CaM, were quite different as well. β-casein bound to the NPs heated for 10 min with an n of 2.7 but to the other type with an n of 1.3. A high n suggests high cooperativity during binding and high possibility of having two β-casein molecules on the NPs with 10-min hydrolysis. In contrast, we found an n of 0.4 for CaM binding to the NPs heated for 10 min. Such a small n indicates high repulsion between the protein molecules bound to the same NP and thus low possibility of having more than one protein on the same particle. In agreement with the findings of protein adsorption onto PAA-coated flat surfaces,55 the binding affinity seemed to be irrelevant to the net charge of protein: the positively charged Cyt c did not show higher affinity to the negatively charged PAA-coated NPs than the acidic proteins. Instead, the highly flexible proteins, CaM and β-casein, had the smallest KD values. Easier adaptation to the NP surface curvature may have generated more interaction points to strengthen the binding.
3.3 Calculation of the interaction energy between CaM and PAA head groups
To better understand the high sensitivity of protein adsorption over a tiny structure change on the surface of NPs, we calculated the interaction energy between the protein and the PAA head groups. Moreover, we illustrated atomistic details of the binding modes of a PAA head group and protein to provide insights into how the binding was established. We chose CaM for this calculation because it showed the largest KD difference in the two sets of NPs and its experimental 3D structure was available.28 Our studies showed that the computed trends of binding affinities between different PAA head groups were insensitive to use of a dimer or a trimer of an acrylic acid. From the cavity size of the ligand binding site, we linked the head-group to a dimer of the acrylic acid to mimic the freely movable PAA head on the NP surface to save computation time in our calculation. In addition to exploring inter-molecular interactions, we examined the orientation of the complex structures with the lowest binding free energy and evaluated the difference between the polymer fragments with HGA and HGB.
Our docking results showed that both HGA and HGB could successfully fit into the binding cavity within the docking boxes, and the HGA:CaM complex generally had more negative binding free energy than the HGB:CaM complex (Table 2). This trend agrees well with our experimental observation that the NPs coated by aPAA bound to CaM stronger than that coated by bPAA. The measured affinity difference between these two complexes corresponded to a ΔG difference of 1.8 kcal/mol (difference in ΔG = −RTln(KAaPAA) − (−RTln(KAbPAA) = RTln (KDaPAA)/(KDbPAA) ≈ −1.8 kcal/mol; KA is the association constant, equal to 1/KD), which fell within the calculated energy difference of 0.57–2.75 kcal/mol.
Table 2.
Autodock results for the simulated interaction between CaM and head groups of PAA
| polymer fragment | binding free energy (kcal/mol) | vdw energy (kcal/mol) | electrostatic energy (kcal/mol) |
|---|---|---|---|
| Head group A | −3.79 | −6.34 | +0.76 |
| Head group B | −1.52 | −3.59 | −0.02 |
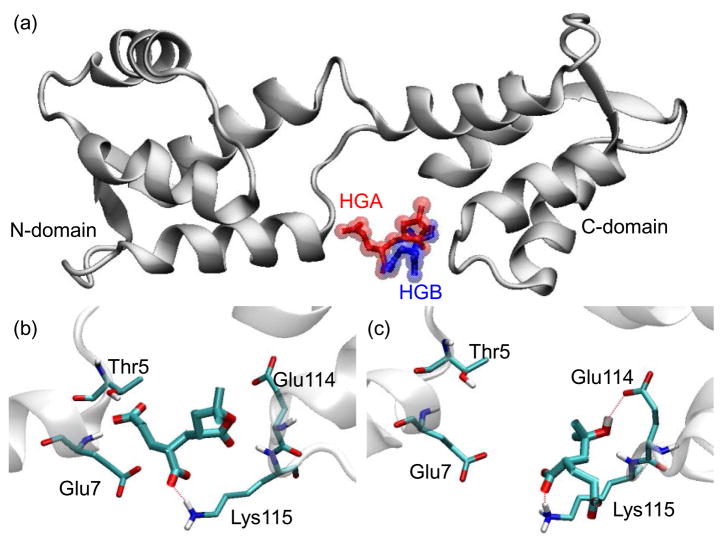
The binding of HGA and HGB to the cavity between the N- and C-terminal domains is shown in Figure 4a as a representative of the simulated complex structure. HGA and HGB face towards the protein and interact with residue Thr5, Glu7, Glu114 and Lys115 (Figure 4b and 4c). The orientation of the head groups agrees with the established concept that the carboxylate groups on the PAA side chains strongly coordinate to the iron cations on NP surface to form a robust coating, whereas the head groups extend into the aqueous solution and play an important role in NP interaction with proteins. Among the overall free energy change, the non-polar van der Waals (vdw) energy between HGA and CaM (−6.34 kcal/mol) is more favorable than that of HGB and CaM (−3.59 kcal/mol) (Table S1, ESI). The binding mode of HGA illustrates that the bulky head group can fit nicely into the binding cavity, as compared with that formed by HGB, which also agrees with the energy calculations. The electrostatic energy described here is a boarder category, which includes conventional electrostatic interaction, hydrogen bonding and salt bridge. HGB shows favorable electrostatic energy (−0.02 kcal/mol) when binding with CaM as compared with HGA (+0.76 kcal/mol) (Table S1) because of the hydrogen bonding formed between the Glu114, Lys115 and the hydroxyl group of HGB (Figure 4c). However, this hydrogen bonding still cannot result in strong attraction for HGB. Taken together, the overall binding free energy between HGA (−3.79 kcal/mol) and CaM was more negative than with HGB (−1.52 kcal/mol), with the vdw attractions contributing more to determine the interaction.
Figure 4.
(a) Overview of docking the head-groups (HGA in red and HGB in blue) to the default center of CaM that locates between the N- and C-domain, in which the head group faced towards CaM and the dimeric acrylic acid part of the structure pointed away from CaM. The zoom-in view of detailed interactions of amino acid side chains with (b) HGA and (c) HGB. The red dotted lines represented the H-bonds.
The calculations did not account for potential ligand-induced protein conformation changes.7,9,56,57 However, the insignificant spatial difference in the two head groups is unlikely to result in considerably different complex conformations in the PAA bound state. The scoring function in Autodock approximated the solvent effect and desolvation, and entropic penalty. Because both aPAA and bPAA have the same acrylic acid backbone, which contributes mainly to solvent effect and PAA flexibility, the backbone may not contribute to the affinity differences.45,46 Additionally, these two types of NPs had comparable size and surface curvature, and CaM has a highly flexible structure without Ca2+ binding. Both features suggest comparable CaM conformational change on binding to these two types of NPs.
The same computer simulation was also performed with the other proteins having an available 3D structure, including HSA (only to drug-binding site 2 because of the large size of HSA; site 2 was identified as the binding site of PAA-NP on HSA13), myoglobin and Cyt c (Table S2, ESI). Like CaM, Cyt c simulation results agreed with the protein-particle affinity trend: HGA showed stronger binding energy than HGB to Cyt c. However, for HSA and myoglobin, HGA showed more binding free energy than did HGB, which was opposite to the KD value trend.
Nevertheless, the simulation results all suggest that, both good fitting into the protein binding cavity by the NP surface ligand, and large overall interaction energy established between the protein amino acid side chains and the ligand are required for stable protein adsorption on NPs. These two requirements determine that protein adsorption would be strongly affected by subtle structural changes on the ligand, especially when the change happens to the portion of the ligand molecule that directly faces the protein.
3.4 Protein adsorption for assessment of NP surface property after ligand exchange
In the above study, variation in the structure of surface ligand was introduced due to small change in the condition of particle synthesis. We also prepared two batches of NPs that were particularly coated with ligands having small structural difference and tested if that could be detected by protein adsorption. The Au NPs originally carried the Ni-nitrolotriacetic acid (NTA) chelate on the surface; and the ligands were two types of polyhistidine (6×His)-peptides, one carrying a free –COOH group (P2) at the C-terminal and the other having the C-terminal blocked by –CONH2 (P1). The 6 × His peptides were conjugated to the Au NPs via their high affinity with Ni-NTA. This is a common approach to couple His-tagged biomolecules on NPs for biomedical applications. During preparation, P1 and P2 were incubated with the Ni-NTA-Au NPs at the ratios of 0:100 or 5: 95. After removal of the free P1 and P2 by the 10-kDa Amicon filter, the obtained Au NPs, named Au-100P2 and Au-95P2, respectively, showed the same color in solution, which indicated no change in particle diameter; and the same mobility in CE, meaning comparable zeta-potential (Figure S6a, ESI). Despite the high similarity in their physicochemical properties, they bound to β-casein quite differently. More protein-NP complex was formed with the Au-95P2 particles than the Au-100P2 ones when incubated with the same concentration of β-casein (Figure S6b). The measured affinity was about 5-fold higher for the Au-95P2, even though only 5% of the surface ligands were different between these two batches of particles. Increase in the neutral P1 percentage may reduce electrostatic repulsion between the –COO− end of 6 × His peptide and acidic β-casein, leading to stronger protein-NP interaction.
4. Conclusions
The PAA ligand has an average molecular weight of 1.8 kDa, thus the head group structure is only a small portion of the entire molecule. The hydrodynamic diameter of the NPs carrying PAA with HGA was 17% larger than that with HGB, although the core sizes of NPs differed by only 5%. More importantly, they induced big variations in the NPs’ affinity to proteins. The high sensitivity of protein adsorption to the head group structure is mainly because, as pointed out by computer simulation, the head group of PAA located at the binding interface with the protein. In order for stable binding to be formed, part of the NP surface ligand should fit well onto certain binding pockets on the protein surface. Even slight structural change in this part could alter the fitting and change the binding energy, ultimately varying the affinity to the protein.
Overall, our results support that screening interaction between NPs and judiciously selected proteins could be an effective way for quick and initial evaluation of the particle surface of NPs. Such screening will be useful for rapid assess of the surface properties of NPs produced in different batches or with varied preparation procedures, which is particular important for quality control of NPs made for biomedical purposes. It is even possible that, by analyzing the interaction change with proteins of distinct properties, such as pI value, shape, hydrodynamic size, and surface hydrophobicity, more information about the kind change being occurred to the NP surface ligand could be revealed.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Dan Borchardtand at the Analytical Chemistry Instrumentation Facility of UCR for his assistance in NMR measurement. We are also grateful to Mr. Le He and Dr. Yadong Yin’s guidance on nanoparticle preparation. This work was supported by National Institute of Environmental Health Sciences Grant No. 1R21ES017870-01A1 to W. Zhong; by National Science Foundation Grant MCB-0919586 and the University of California, Riverside to C. E. A. Chang.
Footnotes
Electronic Supplementary Information (ESI) available: [It includes the electropherograms obtained by CE which were used for calculation of the zeta-potential of the NPs (Figure S1); the mass spectrum for the standard PAA obtained under the same experimental conditions as Fig. 2 (Figure S2); the plot showing gradual increase of the aPAA in the heated PAA sample with longer heating duration and the corresponding mass spectra (Figure S3); comparison of the 1H NMR spectrum from the PAA sample heated for 12 hr with those obtained at other heating durations (Figure S4a); the HSQC and COSY NMR spectra for standard PAA (Figure S4b and S4c); the electropherograms and the affinity fitting curve for KD measurement (Figure S5); the electropherograms of the two types of Au-NPs coated with different ratios of His-tagged peptides on surface and their electropherograms when incubating with β-casein (Figure S6). Two supporting tables (Table S1 and S2) were also included, and they display the calculated binding energy with calmodulin, myoglobin, cytochrome c, and HSA. See DOI: 10.1039/b000000x/
Notes and references
- 1.Grainger DW, Castner DG. Adv Mater. 2008;20:867. [Google Scholar]
- 2.Riehemann K, Schneider SW, Luger TA, Godin B, Ferrari M, Fuchs H. Angew Chem, Int Ed. 2009;48:872. doi: 10.1002/anie.200802585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanhai WR, Sakamoto JH, Canady R, Ferrari M. Nat Nanotechnol. 2008;3:242. doi: 10.1038/nnano.2008.114. [DOI] [PubMed] [Google Scholar]
- 4.De Paoli Lacerda SH, Park J-J, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF. ACS Nano. 2010;4:365. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 5.Lind K, Kresse M, Muller RH. Electrophoresis. 2001;22:3514. doi: 10.1002/1522-2683(200109)22:16<3514::AID-ELPS3514>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc Natl Acad Sci USA. 2008;105:14265. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyano DF, Rotello VM. Langmuir. 2011;27:10376–10385. doi: 10.1021/la2004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayraktar H, You C-C, Rotello VM, Knapp MJ. J Am Chem Soc. 2007;129:2732. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]
- 9.You C-C, Arvizo RR, Rotello VM. Chem Comm. 2006:2905. doi: 10.1039/b605508g. [DOI] [PubMed] [Google Scholar]
- 10.De M, You C-C, Srivastava S, Rotello VM. J Am Chem Soc. 2007;129:10747. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 11.Calzolai L, Franchini F, Gilliland D, Rossi F. Nano Lett. 2010;10:3101. doi: 10.1021/nl101746v. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, Jana S, Guchhait NJ. Phys Chem B. 2012;116:1155. doi: 10.1021/jp2094752. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Zeng S, He L, Zhong W. Anal Chem. 2011;83:6929. doi: 10.1021/ac201889j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casula MF, Jun Y-w, Zaziski DJ, Chan EM, Corrias A, Alivisatos AP. J Am Chem Soc. 2006;128:1675. doi: 10.1021/ja056139x. [DOI] [PubMed] [Google Scholar]
- 15.Ge J, Hu Y, Biasini M, Dong C, Guo J, Beyermann WP, Yin Y. Chem--A Eur J. 2007;13:7153. doi: 10.1002/chem.200700375. [DOI] [PubMed] [Google Scholar]
- 16.Rao CNR, Agrawal VV, Biswas K, Gautam UK, Ghosh M, Govindaraj A, Kulkarni GU, Kalyanikutty KP, Sardar K, Vivekchand SRC. Pure Applied Chem. 2006;78:1619. [Google Scholar]
- 17.Si S, Li C, Wang X, Yu D, Peng Q, Li Y. Cryst Growth Des. 2005;5:391. [Google Scholar]
- 18.Yan Z-G, Yan C-HJ. Mat Chem. 2008;18:5046. [Google Scholar]
- 19.Waddell JN, Mullen DG, Orr BG, Banaszak Holl MM, Sander LM. Phys Rev E. 2010;82:036108/036101-036108/036106. doi: 10.1103/PhysRevE.82.036108. [DOI] [PubMed] [Google Scholar]
- 20.Morasso C, Verderio P, Colombo M, Prosperi D. ACS Sym Ser. 2011;1091:69. [Google Scholar]
- 21.Padmos JD, Zhang PJ. Phys Chem C. 2012;116:23094. [Google Scholar]
- 22.Polito L, Colombo M, Monti D, Melato S, Caneva E, Prosperi D. J Am Chem Soc. 2008;130:12712. doi: 10.1021/ja802479n. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Du F, Li X, Zhang B, Li W, Yan B. J Phys Chem C. 2008;112:19360. [Google Scholar]
- 24.Zhou H, Li X, Lemoff A, Zhang B, Yan B. Analyst. 2010;135:1210. doi: 10.1039/c0an00066c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Chen S, Zhao X, Kuznetsova LV, Wong SS, Ojima I. J Am Chem Soc. 2008;130:16778. doi: 10.1021/ja805570f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Zeng S, Le H, Zhong W. Anal Chem. 2010;82:7460. doi: 10.1021/ac101627p. [DOI] [PubMed] [Google Scholar]
- 27.Schnabel U, Fischer C-H, Kenndler E. J Microcolumn Sep. 1997;9:529. [Google Scholar]
- 28.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Nat Struct Biol. 1995;2:768. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 29.Pedretti A, Villa L, Vistoli GJ. Computer-Aided Mol Design. 2004;18:167. doi: 10.1023/b:jcam.0000035186.90683.f2. [DOI] [PubMed] [Google Scholar]
- 30.Chang C-E, Gilson MKJ. Computational Chem. 2003;24:1987. doi: 10.1002/jcc.10325. [DOI] [PubMed] [Google Scholar]
- 31.Morris GMJ. Comput Chem. 1998;19:1639. [Google Scholar]
- 32.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Computational Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219. [Google Scholar]
- 34.Ai H. Adv Drug Delivery Rev. 2011;63:772. doi: 10.1016/j.addr.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang J-D, Song E. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kievit FM, Zhang M. Acc Chem Res. 2011;44:853. doi: 10.1021/ar2000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F, Laurent S, Fattahi H, Vander Elst L, Muller RN. Nanomedicine. 2011;6:519. doi: 10.2217/nnm.11.16. [DOI] [PubMed] [Google Scholar]
- 38.Tassa C, Shaw SY, Weissleder R. Acc Chem Res. 2011;44:842. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabe M, Verdes D, Seeger S. Adv Colloid Interface Sci. 2011;162:87. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Neoh KG, Kang ET. Polymer Chem. 2011;2:747. [Google Scholar]
- 41.Chen T, Yang M, Wang X, Tan LH, Chen H. J Am Chem Soc. 2008;130:11858. doi: 10.1021/ja8040288. [DOI] [PubMed] [Google Scholar]
- 42.Tan LH, Xing S, Chen T, Chen G, Huang X, Zhang H, Chen H. ACS Nano. 2009;3:3469. doi: 10.1021/nn900917z. [DOI] [PubMed] [Google Scholar]
- 43.Dai J, Bao Z, Sun L, Hong SU, Baker GL, Bruening ML. Langmuir. 2006;22:4274. doi: 10.1021/la0600550. [DOI] [PubMed] [Google Scholar]
- 44.Hollmann O, Czeslik C. Langmuir. 2006;22:3300. doi: 10.1021/la053110y. [DOI] [PubMed] [Google Scholar]
- 45.Wittemann A, Ballauff M. Phys Chem Chem Phys. 2006;8:5269. doi: 10.1039/b609879g. [DOI] [PubMed] [Google Scholar]
- 46.Wittemann A, Haupt B, Ballauff M. Phys Chem Chem Phys. 2003;5:1671. [Google Scholar]
- 47.Roux FX, Audebert R, Quivoron C. Eur Polym J. 1973;9:815. [Google Scholar]
- 48.Lattimer RP. J Anal Appl Pyrolysis. 2003;68–69:3. [Google Scholar]
- 49.Hlady V, Buijs J Curr Opin Biotechnol. 1996;7:72. doi: 10.1016/s0958-1669(96)80098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milthorpe B. Sur Interfaces Biomat. 2005:763. [Google Scholar]
- 51.Schmidt DR, Waldeck H, Kao WJ. In: Biological Interactions on Materials Surfaces. Puleo DA, Bizios R, editors. Springer Science + Business Media, LLC; 2009. p. 1. [Google Scholar]
- 52.Claesson PM, Blomberg E, Froberg JC, Nylander T, Arnebrant T. Adv Colloid Interface Sci. 1995;57:161. [Google Scholar]
- 53.Weiss JN. FASEB J. 1997;11:835. [PubMed] [Google Scholar]
- 54.Abeliovich H. Biophys J. 2005;89:76. doi: 10.1529/biophysj.105.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Czeslik C, Jansen R, Ballauff M, Wittemann A, Royer CA, Gratton E, Hazlett T. Phys Rev E. 2004;69:021401. doi: 10.1103/PhysRevE.69.021401. [DOI] [PubMed] [Google Scholar]
- 56.Lemieux RU. Acc Chem Res. 1996;29:373. [Google Scholar]
- 57.You CC, Agasti SS, Rotello VM. Chem--A Eur J. 2008;14:143. doi: 10.1002/chem.200701234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.