Abstract
A primitive genetic code is thought to have encoded statistical, ambiguous proteins in which more than one amino acid was inserted at a given codon. The relative vitality of organisms bearing ambiguous proteins and the kinds of pressures that forced development of the highly specific modern genetic code are unknown. Previous work demonstrated that, in the absence of selective pressure, enforced ambiguity in cells leads to death or to sequence reversion to eliminate the ambiguous phenotype. Here, we report the creation of a nonreverting strain of bacteria that produced statistical proteins. Ablating the editing activity of isoleucyl-tRNA synthetase resulted in an ambiguous code in which, through supplementation of a limited supply of isoleucine with an alternative amino acid that was noncoding, the mutant generating statistical proteins was favored over the wild-type isogenic strain. Such organisms harboring statistical proteins could have had an enhanced adaptive capacity and could have played an important role in the early development of living systems.
The modern genetic code appeared >3 billion years ago (1, 2). This code relates each of the 20 canonical amino acids to specific nucleotide triplets found in the coding sequences of genes and mRNAs (3). These coding sequences are translated into polypeptides with only one type of amino acid at each position in the sequence, so that each protein is a specific and homogeneous chemical entity. The code itself is thought to have started in a primitive form, perhaps with codons composed of two rather than three nucleotides and with different amino acids not precisely assigned to specific codons (4–6). The result was a system of coded peptide synthesis in which a given nucleotide sequence was translated into a related group of statistical, heterogeneous polypeptides, so that a given location in the peptide chain could have one of several related amino acids (7, 8). (For example, hydrophobic amino acids of similar size might be treated as roughly equivalent and be interchangeably inserted at a given codon.) Because they could opportunistically use whatever amino acids were available to complete a protein sequence, organisms harboring statistical proteins could have had a selective advantage in a primitive environment. Also, by having many closely related versions of the same basic sequence, variants with a particular catalytic activity could be produced.
These microvariants might have special adaptive advantages, much in the way that one or more mutations in an enzyme can enhance its activity or broaden its specificity. The selective advantages of more complex organisms that were able to produce their own amino acids and are dependent on higher specificity eventually forced replacement of the ancient statistical systems. Remains of ambiguous codes are still observed in nature as in the Leu/Ser ambiguity in Candida sp. (9).
The genetic code is established in reactions catalyzed by aminoacyl-tRNA synthetases, in which each amino acid is covalently joined to its cognate tRNA. The tRNA bears the complementary nucleotide triplet of the code corresponding to the attached amino acid (10). For hydrophobic amino acids like isoleucine, the catalytic site of the enzyme has limited ability to discriminate between isoleucine and smaller, canonical amino acids such as valine (11) or cysteine, or noncanonical amino acids such as norvaline, O-methyl threonine, or O-methyl serine (12, 13). As a consequence, these alternative amino acids can be joined to tRNAIle and erroneously be incorporated into proteins at positions normally occupied by isoleucine. Certain synthetases, including isoleucyl-tRNA synthetase (IleRS), have a second active site that clears mischarged amino acids and thereby removes errors of aminoacylation (14–21). For example, the ATP-dependent aminoacylation reaction with isoleucine-specific tRNAIle can result in production of Ile-tRNAIle or Val-tRNAIle.
 |
However, the second active site specifically clears Val-tRNAIle so that valine is not incorporated into proteins at positions reserved for isoleucine (22).
 |
The editing reactions of tRNA synthetases are essential for ensuring the production of accurate homogeneous proteins and not statistical mixtures of closely related sequences. Mischarging reactions could, however, have advantages for early cells growing under conditions where the supply of a particular amino acid is limited, because natural or nonnatural alternative amino acids could be recruited to “fill in” for the limiting amino acid in a growing polypeptide.
Materials and Methods
Strains and Growth Conditions. Strain construction steps are described below, and the strains used are listed in Table 1. Bacteria were routinely grown in minimal medium (23) containing 2 g of glucose per liter in LB (24) or in Mueller-Hington (BD Diagnostic Systems, Sparks, MD). Growth media were solidified with 15 g/liter agar (BD Diagnostic Systems) for the preparation of plates. Transformations and P1 transductions were performed following standard procedures (24, 25).
Table 1. Strains.
| Strain | Genotype | Source |
|---|---|---|
| MG1655 | E. coli Genetic Stock Center | |
| PS2306 | ΔileS203::KmR pBADileSWT | Ref. 27 |
| PS2449 | ΔileS203::KmR pBADileSAla | Ref. 27 |
| PS7066 | ileSAla | This work |
| PS2066 | Δtdk::KmR | This work |
| PS6231 | Δtdk::KmRileSAla | This work |
| PS7096 | ΔthyA::KmR pthyAcys146AUA | This work |
| PS7108 | ΔthyA::KmR ileSAla pthyAcys146AUA | This work |
| PS7079 | ΔilvGE pAlaXp | This work |
| PS7081 | ΔilvGE ileSAlapAlaXp | This work |
| PS7068 | ΔilvGE | This work |
| β1412 | ΔilvGE ileSAla | This work |
To test for the reversion phenotype, a strain bearing ileSAla was inoculated in a continuous culture apparatus (26) and grown in minimal medium supplemented with glucose at 30°C. After 30 days in the apparatus, the strain was still sensitive to norvaline and therefore had kept the editing-deficient allele. Growth curves were conducted in an ELX808 Microplate Reader (Bio-Tek, Burlington, VT) as described (27).
Construction of the ΔthyA::KmRand Δtdk::KmR Alleles. The KmR gene from pUC4K (Amersham Pharmacia Biosciences) was amplified by PCR by using the following oligonucleotide pairs tdk.ol1 (5′-ATGATA A ACTCCAGCCA ACT T TAT T TCATATCAT TGAGGGCCTGTGGCTGCCAGTCACGACGT TGTA A A ACGA) and tdk.ol2 (5′-ATCGTGGCGATGCCTTTCCTGAATAGCCGTTAATGAGTCGACTTGTAACAAACAGCTATGACCATGATTAC) amplified the KmR gene with 50-bp overhangs covering regions upstream and downstream of the Escherichia coli tdk gene. thyA.ol1 (5′-TGGTCTGGGCATATCGTCGCAGCCCACAGCAACACGT T TCCTGAGGA ACCCCAGTCACGACGT TGTAAAACGA) and thyA.ol2 (5′-ATGCCCGGATGCGGATCGTAGCCTTCAATCTCAAAGTCTTCGAAACGGTAGAAACAGCTATGACCATGATTAC) amplified the KmR gene with 50-bp overhangs covering regions upstream and downstream of the E. coli thyA gene. The resulting 1.5-kb fragments were transformed by electroporation into strain JC8679 (28) and plated on LB Km (50 μg/ml)/thymidine (dT) (300 μM). Recombinants were tested for resistance to 3′-azido-3′-deoxythymidine (AZT) and dT prototrophy, respectively, and the markers were transduced into MG1655 to give strain PS2260 (Δtdk::KmR) and strain PS2271 (ΔthyA::KmR).
Construction of the ileSAla Allele. The ileSAla mutation was introduced into ileS by ligation of a synthetic oligonucleotide duplex containing 10 alanine-encoding triplet GCG in place of the amino acid codons for 241 to 250, into the SpeI and ClaI sites of pVDC433. The resulting plasmid was designated pTLH33. The following 5′-phosphorylated oligonucleotides (Invitrogen) were used: 5′-pCTAGTA ATCGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGGCGCGCGCAATAT and 5′-pCGATAT TGCGCGCGCCGCCGCCGCCGCCGCCGCCGCCGCCGCCGCGATTA. We sequenced the resulting plasmid, pTLH33, and verified the insertion of 10 alanine codons. The ΔileS203::KmR allele was transduced from the strain IQ839(ΔileS203::KmR λpKS-46) (29) into MG1655 containing plasmid pTLH33 to give strain PS2449. The transductants were selected as described when constructing the isogenic strain overexpressing the WT ileS gene, PS2306 (27).
The 2.6-kb EcoRI and XmaI fragment carrying the ileSAla gene was subcloned from pTLH33 into the corresponding sites in pSH93 to give pSH102. Plasmid pSH93 was constructed as follows: The tdk gene and the region covering the valS promoter region were amplified by PCR from Haemophilus influenzae genomic DNA by using the oligonucleotides tdk1.3 (5′-CCCGTCGACTTAAATTTGACCGCACTTTTCTTTA) and tdk15 (5′-ATGGCGAAACTCTACTTCTATTAC) in one case and ValS1.5 (5′-CCCCTCGAGGATAAAGGTTATCATGTTG) and ValS1.3 (5′-TAGAGTTT-CGCCATTTATTTTTTCTCTTTATTATAA) in the other. A PvalSHItdkHI cassette was then generated by PCR by using the two previous PCR products and the outside oligonucleotides ValS1.5 and tdk1.3 and cloned into the XhoI site of pSW23 after digestion with XhoI and SalI. Plasmid pSW23 is a vector carrying the chloramphenicol (CmR) gene and the R6K replicon that can be maintained only in a strain that expresses the Pi protein (BW19610) (both vector and strains were gifts of D. Mazel, Institut Pasteur, Paris). Strain PS2260 (Δtdk::KmR) was transformed with pSH102, and transformants were selected on Mueller-Hington plates supplemented with Cm (20 μg/ml). Because PS2260 does not express the Pi protein, CmR clones result from single crossover events integrating the CmR gene and the PvalSHItdkHI at the ileS locus. Double crossover recombinants leading to the replacement of the WT ileS gene by the ileSAla allele were then selected by growing cultures of the CmR clones on Mueller-Hington AZT (30 μM) plates and than screening the AZTR colonies for CmS. The AZTR CmS clones were screened for norvaline sensitivity as a test for the presence of the ileSAla allele. NorS clones then were checked by PCR by using the ileSAla-specific oligonucleotide (5′-CGCCGCCGCCGCCGCCGCCGCCGC) and the oligonucleotide (5′-GATTGCGCGTGATGAATTAACC). Four independent Δtdk::KmR ileSAla clones were obtained; one was named PS6231 and used herein. The Δtdk::KmR was eliminated from PS6231 by P1 transduction and selection for growth on Trimethoprim (100 μM) and dT (300 μM). The resulting strain ileSAla was named β1210 and used herein.
Construction of ileSAla Derivatives. The ΔilvGE allele from CU925 (30) was introduced into β1210 by co-linkage with the metE::Tn10 (TetR) allele from strain CAG18491 (31). The WT met+ allele was transduced back in to eliminate the methionine requirement. The final strain, ΔilvGE ileSAla, was named β1412. An isogenic set was derived from β1412 by transducing in the WT ileS allele [selection on MS plates supplemented with Ile, Leu, Val (100 μM), norvaline (5 mM) and 0.2% glucose] to give strain PS7068 (ΔilvGE) or the WT ilv allele [selection on MS glucose to give PS7066 (ileSAla)]. The presence of the ileSAla allele was checked by PCR. After sample preparation using Chelex (Sigma) to prepare DNA from colonies, PCR was performed with the oligonucleotides TLHseq7 (ggttgacggtcaacgc) and ile-Sol7 (5′-ATCCAGGGTAACGTGATCGCCG) to amplify a 500-bp fragment around the mutated region of ileS. An AvaII site present in the WT allele but absent in the ileSAla allele was used as a marker for the presence or absence of the allele through analysis of the PCR products digested by AvaII. The isogenic set β1412 and PS7068 was used in the yield experiments. The plasmid pAlaXp (32) expressing the His-tagged tester protein was transformed into PS7066 and PS7068 to give the strains PS7079 (ΔilvGE pAlaXp) and PS7081(ΔilvGE ileSAla pAlaXp) that were used to purify the AlaXp protein for MS analysis. The ΔthyA::KmR allele was transduced in the isogenic set to give strains PS2271 (ΔthyA::KmR) and PS7087 (ΔthyA::KmR ileSAla). The plasmid pSH20 (pthyACys146UAU) (32) was transformed into the isogenic ΔthyA set to give PS7096 (ΔthyA::KmR pthyAcys146AUA) and PS7108 (ΔthyA::KmR ileSAla pthyAcys146AUA). Strains PS7096 and PS7108 were used in the suppression test.
Protein Purification and in Vitro Assays. AlaXp was purified with Ni-NTA agarose (Qiagen, Hilden, Germany), cut out of an SDS/PAGE preparative gel, and analyzed by matrix-assisted laser desorption ionization and μ-liquid chromatography tandem MS as described (32). WT IleRS and IleRSAla mutant proteins were purified from strains PS2306 and PS2449 as described (27). Transfer RNAIle was isolated from E. coli strain MV1184 containing the plasmid pES300, which allows for the lac-inducible overexpression of tRNAIle, and purified as described (33). The total assay for editing (based on the measurement of ATP hydrolysis) was performed as described (17). Each enzyme was assayed for the ability to catalyze the formation of the misacylated species (27, 34). Val-tRNAIle was produced by using T242P IleRS, as described (27).
Results and Discussion
Nonreverting Disruption of Site for Editing. To investigate the feasibility of sustained growth for cells with an ambiguous genetic code, the active site for editing of E. coli IleRS was ablated in a way that prevented genetic reversion. Earlier work with valyl-tRNA synthetase and isoleucyl-tRNA synthetase demonstrated that a point mutation in the site for editing resulted in uncorrected misacylations and the transient production of error-containing proteins. However, the strains bearing the mutant alleles had a strong propensity for reversion (32, 34, 35). Rather than create a deletion that could destabilize the protein, a critical region of ileS encoding 10 aa (T241-N250) was converted by iterative site-specific recombination to one encoding an oligoalanine decapeptide segment to give the chromosomal ileSAla allele in strain PS7066. This sequence replacement spans T242, a residue shown to be susceptible to mutations that severely diminish the editing capacity of IleRS (34). Because of the large region that was replaced, this construction was genetically stable to reversion.
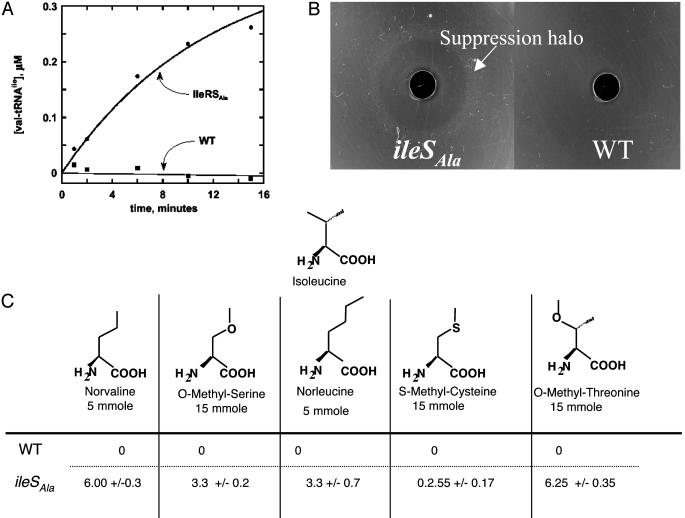
A plasmid-encoded version of the ileSAla allele also was constructed under control of the arabinose-sensitive PBAD promoter (36). This construction enabled us to express sufficient IleRSAla to purify and perform assays for aminoacylation (with Ile as substrate) and misacylation (with Val as substrate). As expected from this allele's maintenance of bacterial growth, the aminoacylation activity was retained. Two separate assays confirmed that the editing activity of IleRS was ablated by the introduction of the polyalanine mutations. First, IleRSAla, but not WT IleRS, produced Val-tRNAIle (Fig. 1A). Second, editing of misactivated valine by WT IleRS resulted in the tRNAIle-dependent hydrolysis of ATP over time. This ATPase activity was absent in IleRSAla (data not shown).
Fig. 1.
Misacylation in vitro and in vivo. (A) Production of Val-tRNAIle by IleRSAla. (B) The cysteine precursor S-carbamoyl-cysteine was loaded in a central well after dilution of overnight cultures of strains PS7108 (Left) and PS7096 (Right) were spread and dried on minimal glucose plates containing thymidine (0.3 mM). The halo of growth around the central well is marked with an arrow for the strain containing ileSAla. (C) Isoleucine analogs were loaded in a central well on plates spread with strains PS2066 and PS6231. The diameter (in cm) of the toxicity halos was measured after 24 h. The standard deviation of three independent experiments is given.
Misacylation in Vivo and Production of Statistical Proteins. Initially, to detect misacylation in vivo, a suppression test was designed to demonstrate the misincorporation of Cys at Ile codons. The ileSAla allele was combined with a ΔthyA::KmR allele by P1 transduction. The resulting strain PS7087 was auxotrophic for dT. Strain PS7087 was transformed with a plasmid carrying the thyA gene in which the essential Cys-146 UGC codon was replaced with the Ile AUA codon (32) to give strain PS7108. Thus, if misacylation of tRNAIle with cysteine occurs in vivo, then the suppression efficiency of strain PS7108 with the ileSAla allele should be increased relative to suppression in the isogenic strain PS7096 harboring the WT ileS allele. Indeed, addition to media of the cysteine analog S-carbamoyl-cysteine (this cysteine analog is hydrolyzed to cysteine over time, thereby assuring a stable supply of nonoxidized cysteine) suppressed the requirement for dT in strain PS7108 but not in the isogenic strain PS7096 harboring the WT ileS allele (Fig. 1B).
In vivo incorporation of other amino acids also was investigated. We tested for toxicity of isoleucine hyposteres in the different backgrounds. In the absence of editing, the ileSAla-containing PS6231 should be more sensitive to excess amounts of a hypostere. Valine is the obvious starting point because of its structural similarity to isoleucine. However, the effects of valine are difficult to measure because of its general toxicity in minimal media, caused by feedback inhibition of the isoleucine biosynthetic pathway (37). For this reason, several other isoleucine hyposteres were tested (norvaline, O-methyl-serine, norleucine, S-methyl-cysteine, and O-methylthreonine) (Fig. 1C). These analogs are nontoxic to the WT strain but exhibited varying levels of toxicity with strain PS6231 containing IleRSAla. Norvaline was the most toxic. In WT background, norvaline concentrations as high as 4 mM had no effect on cellular growth [minimal inhibitory concentration (MIC) > 4 mM]. In contrast, the MIC of norvaline in strain PS7066 dropped to 20 μM. Importantly, the presence of 100 μM Ile suppressed the toxic effect of norvaline in PS6231, consistent with the role of IleRSAla in conferring toxicity (data not shown).
To verify that toxicity was because of misincorporation of norvaline at Ile codons, the yeast protein AlaXp (SwissProt# P53960) was overexpressed and purified from the editing-deficient strain PS7081 and, separately, from the isogenic WT strain, with both strains grown in the presence of norvaline. Protein samples were digested with trypsin and analyzed by MS. Matrix-assisted laser desorption ionization analysis showed that, when AlaXp was produced in strain PS7081, derived peptides contained a mixture of isoleucine and misincorporated norvaline (Fig. 2). The levels of misincorporation ranged from 10% to 15% per codon. Direct sequencing of a norvaline-containing peptide (corresponding to Ile-203–Lys-215) by μ-liquid chromatography tandem MS confirmed that norvaline was specifically misincorporated into position 203 designated by an Ile codon (data not shown).
Fig. 2.
Norvaline misincorporation at Ile codons. The His-tagged protein AlaXp was expressed in Δilv strains containing the WT ileS or the mutant ileSAla allele, in minimal medium containing limiting Ile (0.02 mM) and excess Nor (0.5 mM). AlaXp was purified and analyzed by matrix-assisted laser desorption ionization and μ-liquid chromatography tandem MS (32). The spectrum for peptide Gln-313–Arg-320 containing an Ile residue at the second position is shown in Upper (WT cells, peptide mass = 1027.61 g/mol). It is resolved into two components when isolated from cells bearing the ileSAla allele (Lower). The second component has a mass of 1,013.59, exactly 14 mass units less than the “WT peptide.” Multiple peaks correspond to 13C isotopic forms.
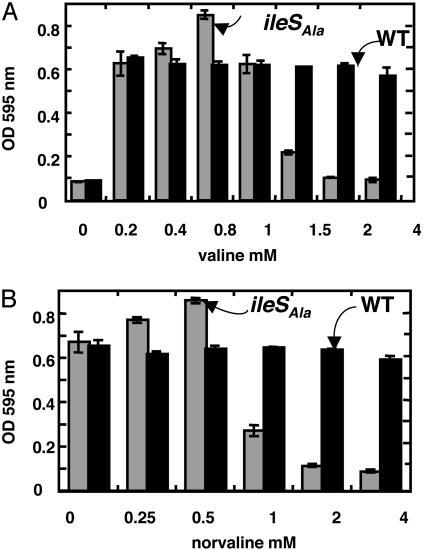
Statistical Proteins Can Give a Growth Yield Advantage. To investigate whether an ambiguous lineage can have an advantage in certain environments, the tolerance of cells to the substitution of isoleucine by hyposteres was investigated. By limiting the concentrations of isoleucine available for growth, hypostere misincorporation into proteins was enhanced. To this end, the ΔilvGE allele conferring auxotrophy for isoleucine, valine, and leucine was combined with the ileSAla allele to create strain β1412. In media containing the required leucine, valine, and isoleucine and supplemented with excess valine (0.6–0.9 mM) or norvaline (0.25–0.5 mM), the growth yield of the editing-defective β1412 ileSAla allele was increased up to 20% compared with the yield obtained with the editing-proficient isogenic PS7068 (Fig. 3). Thus, in the editing-deficient β1412, norvaline, and valine can overcome the effects of a limited concentration of isoleucine and be recruited into statistical polypeptides. As expected, high concentrations of valine or norvaline ultimately became toxic in the editing-deficient strain but not in its WT counterpart. Plausibly, the concentration of catalytically active microspecies in the intracellular statistical population is too low to sustain cell growth of the editing-deficient strain, when the mutant IleRS is confronted with too many amino acid possibilities. To investigate this phenomenon in more detail, we conducted further experiments with norvaline to compare the growth of strains β1412 and PS7068 over a period of hours in the presence and absence of 200 μM norvaline. Although strain PS7068 had an initial advantage in the early stages of growth in the absence of norvaline, both strains were equivalent after 10 h (Fig. 4A). However, in the presence of 200 μM norvaline, by 15 h, strain β1412 had outgrown strain PS7068. This superiority of β1412 in the presence of 200 μM norvaline was seen whether cell mass was measured by turbidity (Fig. 4A) or by actual viable cell count analysis (Fig. 4B).
Fig. 3.
Growth yields in different conditions. Strains β1412 (ΔilvGE ileSAla) and PS7068 (ΔilvGE) were grown overnight in minimal glucose supplemented with Ile (100 μM), Leu (100 μM), and various concentrations of valine (A) or norvaline (B) in an ELX808 Microplate Reader as described (27). The final OD at 595 nm was measured after 20 h to quantitate yield. Experiments were performed in triplicate. Error bars correspond to the standard deviations.
Fig. 4.
Increases in growth yield at a given norvaline concentration. Growth and viable counts of strains β1412 (ΔilvGE ileSAla) and PS7068 (ΔilvGE) in minimal glucose supplemented with Ile (100 μM), Leu (300 μM), or Val (300 μM) with or without Nor (200 μM). Experiments were performed with 12 independent cultures for each condition (error bars correspond to the standard deviations). (A) A representative growth curve for each condition is shown. (B) At stationary phase, viable counts were measured by making two independent dilutions for each culture and spotting, three times, 10 μl of a 10–5 dilution on an LB plate.
Concluding Remarks. In summary, a stable and robust strain with an ambiguous code, and thus harboring statistical proteins, was created by irreversible ablation of the editing activity of a single tRNA synthetase. The WT strain, with its full complement of editing activities, has the decided advantage of being more resistant to the potential toxicity of elevated concentrations of noncoding amino acids (for example, norvaline) (Figs. 3 and 4). However, the editing-deficient strain with its statistical proteins has the capacity to use noncanonical amino acids to fill in at codons specifying (but starved for) particular amino acids such as isoleucine. This capacity is advantageous in circumstances when the organism is confronted with modest concentrations of various amino acids that might have been the only available building blocks for proteins in an early environment. The lack of both specific resources and competing species may have favored early organisms that could maximize yield and therefore maximize the chances of spreading to new resource patches that would otherwise go unused. Thus, organisms with the capacity to generate statistical proteins could plausibly have served as intermediates in the evolution of early living systems.
Acknowledgments
We thank The Scripps Research Institute MS facility for performing the analysis, Dr. M. Lovato (The Scripps Research Institute) for the clone of AlaXp, Dr. M. Berlyn of the E. coli Genetic Stock Center, and Dr. D. Mazel (Institut Pasteur, Paris) for gift of strains. This work was supported by National Institutes of Health Grant GM23562, National Science Foundation Grant MCB-0128901, and a fellowship from the National Foundation for Cancer Research. T.L.H. was a National Institutes of Health postdoctoral fellow.
Abbreviations: Cm, chloramphenicol; dT, thymidine; IleRS, isoleucyl-tRNA synthetase; AZT, 3′-azido-3′-deoxythymidine.
References
- 1.Woese, C. R. (1973) Naturwissenschaften 60, 447–459. [DOI] [PubMed] [Google Scholar]
- 2.Ribas de Pouplana, L. & Schimmel, P. (2000) Cell Mol. Life Sci. 57, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick, F. H. (1968) J. Mol. Biol. 38, 367–379. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez-Sanchez, A. (1995) J. Mol. Evol. 41, 712–716. [DOI] [PubMed] [Google Scholar]
- 5.Di Giulio, M. (1997) Trends Biochem. Sci. 22, 49–50. [DOI] [PubMed] [Google Scholar]
- 6.Ribas de Pouplana, L. & Schimmel, P. (2001) J. Biol. Chem. 276, 6881–6884. [DOI] [PubMed] [Google Scholar]
- 7.Lacey, J. C., Jr., Wickramasinghe, N. S. & Cook, G. W. (1992) Origins Life Evol. Biosphere 22, 243–275. [DOI] [PubMed] [Google Scholar]
- 8.Hartman, H. (1995) J. Mol. Evol. 40, 541–544. [DOI] [PubMed] [Google Scholar]
- 9.Santos, M. A., Moura, G., Massey, S. E. & Tuite, M. F. (2004) Trends Genet. 20, 95–102. [DOI] [PubMed] [Google Scholar]
- 10.Ibba, M. & Söll, D. (2000) Annu. Rev. Biochem. 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 11.Pauling, L. (1958) in Festschrift fuer Prof. Dr. Arthur Stoll Siebzigsten (Birkhauser, Basel), pp. 597–602.
- 12.Loftfield, R. B. & Vanderjagt, D. (1972) Biochem. J. 128, 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smulson, M. E. & Rabinovitz, M. (1968) Arch. Biochem. Biophys. 124, 306–313. [DOI] [PubMed] [Google Scholar]
- 14.Nureki, O., Vassylyev, D. G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T. L., Schimmel, P. & Yokoyama, S. (1998) Science 280, 578–582. [DOI] [PubMed] [Google Scholar]
- 15.Lin, L. & Schimmel, P. (1996) Biochemistry 35, 5596–5601. [DOI] [PubMed] [Google Scholar]
- 16.Schreier, A. A. & Schimmel, P. R. (1972) Biochemistry 11, 1582–1589. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, E. & Schimmel, P. (1994) Science 264, 265–267. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt, E. & Schimmel, P. (1995) Biochemistry 34, 11204–11210. [DOI] [PubMed] [Google Scholar]
- 19.Fersht, A. R. (1977) Biochemistry 16, 1025–1030. [DOI] [PubMed] [Google Scholar]
- 20.Fersht, A. R. (1985) Enzyme Structure and Mechanism (Freeman, New York), pp. 347–368.
- 21.Eldred, E. W. & Schimmel, P. R. (1972) Biochemistry 11, 17–23. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin, A. N. & Berg, P. (1966) J. Biol. Chem. 241, 839–845. [PubMed] [Google Scholar]
- 23.Richaud, C., Mengin-Lecreulx, D., Pochet, S., Johnson, E. J., Cohen, G. N. & Marlière, P. (1993) J. Biol. Chem. 268, 26827–26835. [PubMed] [Google Scholar]
- 24.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 100–108.
- 25.Sambrook, J. E., Fritsch, F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 26.de Crécy-Lagard, V. A., Bellalou, J., Mutzel, R. & Marlière, P. (2001) BMC Biotechnol. 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrickson, T. L., Nomanbhoy, T. K., de Crécy-Lagard, V., Fukai, S., Nureki, O., Yokoyama, S. & Schimmel, P. (2002) Mol. Cell 9, 353–362. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, Y., Muyrers, J. P., Testa, G. & Stewart, A. F. (2000) Nat. Biotechnol. 18, 1314–1317. [DOI] [PubMed] [Google Scholar]
- 29.Shiba, K. & Schimmel, P. (1992) Proc. Natl. Acad. Sci. USA 89, 1880–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh, N. J. & Duggan, D. E. (1972) J. Bacteriol. 109, 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer, M., Baker, T. A., Schnitzler, G., Deischel, S. M., Goel, M., Dove, W., Jaacks, K. J., Grossman, A. D., Erickson, J. W. & Gross, C. A. (1989) Microbiol. Rev. 53, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doring, V., Mootz, H. D., Nangle, L. A., Hendrickson, T. L., de Crécy-Lagard, V., Schimmel, P. & Marlière, P. (2001) Science 292, 501–504. [DOI] [PubMed] [Google Scholar]
- 33.Glasfeld, E., Landro, J. A. & Schimmel, P. (1996) Biochemistry 35, 4139–4145. [DOI] [PubMed] [Google Scholar]
- 34.Hendrickson, T. L., Nomanbhoy, T. K. & Schimmel, P. (2000) Biochemistry 39, 8180–8186. [DOI] [PubMed] [Google Scholar]
- 35.Nangle, L. A., de Crécy Lagard, V., Döring, V. & Schimmel, P. (2002) J. Biol. Chem. 277, 45729–45733. [DOI] [PubMed] [Google Scholar]
- 36.Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995) J. Bacteriol. 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Felice, M., Squires, C., Levinthal, M., Guardiola, J., Lamberti, A. & Iaccarino, M. (1977) J. Mol. Biol. 156, 1–7. [DOI] [PubMed] [Google Scholar]