Abstract
We have examined the issue whether axial rotation of an intracellular DNA segment several thousand base pairs in length is associated with a large friction barrier against the merge of oppositely supercoiled DNA domains. The induction of a site-specific recombinase was used to form intracellular DNA rings bearing different numbers of transcription units, and it was found that DNA rings with a single tetA gene and no other transcription units does not become excessively negatively supercoiled in Escherichia coli cells lacking DNA topoisomerase I. Thus, whereas oppositely supercoiled domains are generated in a tetA-bearing DNA ring through anchoring of the tetA transcripts to cell membrane, these domains appear to readily merge by means of axial rotation of the DNA segment connecting them. The diffusional merge of these oppositely supercoiled domains is not significantly affected by the presence of bent sequences in the intervening DNA segment. Examination of the effects of adding more transcription units to the tetA-bearing ring suggests, however, that DNA bends stabilized by bound protein molecules may significantly impede this process inside E. coli, as suggested by previous in vitro studies [Leng, F. & McMacken, R. (2002) Proc. Natl. Acad. Sci. USA 99, 9139–9144].
Regulation of DNA supercoiling in bacteria is often thought to involve the opposing actions of DNA gyrase, which catalyzes the ATP-dependent negative supercoiling of DNA (1), and DNA topoisomerase I, which specifically relaxes negatively supercoiled DNA (2). The wide acceptance of this scheme lies in its simplicity; it attributes the regulation to two well characterized enzymes that affect DNA supercoiling in opposite ways in vitro. This regulatory scheme failed to explain, however, two intriguing findings in the early 1980s. First, intracellular pBR322, a plasmid widely used in DNA cloning, was found to be in a highly positively supercoiled form upon inhibition of gyrase by novobiocin (3). How could inhibition of an enzyme that catalyzes DNA negative supercoiling lead to positive supercoiling? Second, a large increase in the degree of negative supercoiling upon inactivation of DNA topoisomerase I, termed “hypernegative supercoiling,” was seen in pBR322 but not its derivative, pUC8 (4). Further analysis showed that hypernegative supercoiling of pBR322 in topA– cells was critically dependent on the expression of its tetA gene, which is absent in pUC8 (5). How could this tetA dependence be explained in terms of a regulatory mechanism involving diametric actions of different DNA topoisomerases?
The above questions led to the proposal of the twin-supercoiled-domain model of transcription-induced supercoiling (6). The model starts with the notion that translocation of a transcription ensemble R along a helical DNA requires R, which includes the RNA polymerase, its associated proteins, and the nascent transcript attached to it, as well as proteins associated with the nascent transcript, to rotate around the DNA helical axis (7–9). It then follows that interfering with this rotational movement would force overwinding or positive supercoiling of the DNA template ahead of R, and underwinding or negative supercoiling of the DNA template behind R. Pairs of oppositely supercoiled domains generated by the transcription ensembles could at the same time be modulated by the DNA topoisomerases, or merge by means of axial rotation of the DNA segments connecting them (6).
The model readily explains the questions raised earlier. First, it postulates that both positive and negative supercoils are generated in vivo, by transcription as well as plausibly other processes involving translocation of macromolecular assemblies along intracellular DNA. In Escherichia coli and many other bacteria, positively and negatively supercoiled domains appear to be relaxed by different DNA topoisomerases: gyrase, and to a lesser extent, DNA topoisomerase IV, are responsible for the removal of positive supercoils (10, 11); DNA topoisomerase I, on the other hand, is the predominant activity in negative supercoil removal (12, 13). Thus, the addition of novobiocin, which inhibits both gyrase and DNA topoisomerase IV, but does not affect DNA topoisomerase I, would result in a net accumulation of positive supercoils. The large difference in the degrees of supercoiling of pBR322 and pUC8 in topA– cells (4) could also be accounted for: among the various possibilities that could interfere with the rotation of a transcription ensemble R around intracellular DNA (6), cotranscriptional synthesis of membrane proteins, or proteins designated for export across the cytoplasmic membrane, appears to be the most important (14–17). Membrane association of nascent polypeptides attached to an elongating transcript can effectively anchor R to the membrane and thus prevent its rotation around the DNA helical axis. In the case of pBR322, the nascent tetA polypeptides serve as effective membrane anchors for the transcriptional ensemble of the tetA gene, RtetA, whereas the other nascent polypeptides of pBR322 transcripts are ineffective.
The least studied aspect of the twin-supercoiled-domain model of transcriptional supercoiling is the cancellation of oppositely supercoiled domains by axial rotation of the intervening DNA segments. When the model was first proposed, it was suggested that a connecting DNA segment, even when decorated with bound proteins, was unlikely to provide a significant friction barrier; the axial rotational friction coefficient of DNA, estimated for a cylinder with the same volume as a solvated DNA (18), seemed too small to significantly retard the merge of oppositely supercoiled domains (6). More recently, however, it was suggested that the “plumber's snake” model of DNA used in the earlier estimates might have grossly underestimated this friction coefficient (19). A theoretical analysis indicated that randomly ordered and randomly oriented axial deflections along an average DNA, owing to the nonuniform nature of base pair geometries, might greatly increase the effective hydrodynamic volume of DNA, hence the viscous drag against its axial rotation (19). Several in vitro and in vivo studies also raised questions on the notion that an unanchored connecting DNA segment does not pose a significant friction barrier to the merge of oppositely supercoiled domains (see references cited in ref. 19). Of particular interest was the finding that in topA– cells the presence of an additional strong promoter on a tetA-bearing plasmid greatly increases hypernegative supercoiling, even when transcripts from the added promoters are no more than a few dozen nucleotides long and are thus highly unlikely to serve as DNA anchors (20). These theoretical and experimental findings led us to revisit this particular issue. We conclude that the diffusion barrier posed by a tethering DNA several thousand base pairs in length is too small to significantly impede the merge of negative and positive supercoils. We have also examined the effects of introducing into the tethering DNA segment sequence elements that form static bends, or promoter–terminator combinations that directs the synthesis of very short RNAs (20). These results are presented and discussed below.
Materials and Methods
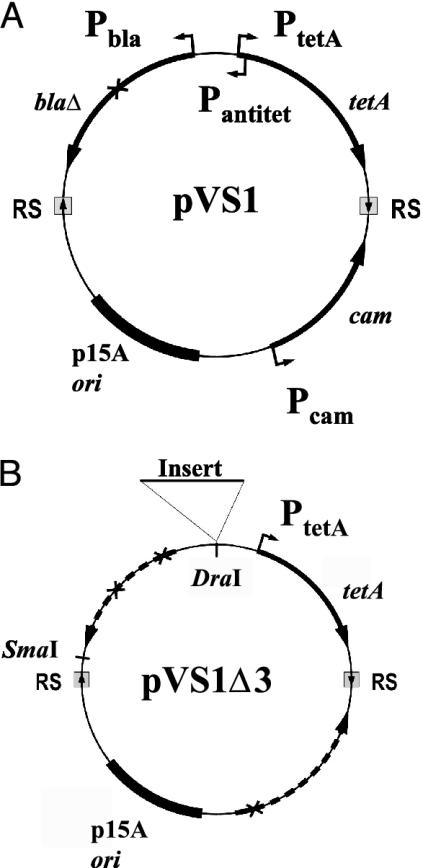
E. coli Strains and Plasmid Construction. Strain DM800 [F– Δ(topA cysB)204 arcA13 gyrB225] was described in DiNardo et al. (21,21); the gyrB225 mutation lowers gyrase activity by a factor of ≈10 (22). Various plasmids were constructed by conventional methods or obtained from commercial sources. The parent of many of the plasmids used in this work, pVS1, was derived from pASLS1 (23) by first inserting a PCR-amplified fragment of the tetA-bla region of pBR322 (base pairs 3296–1268) in between the SmaI and XhoI sites of pASLS1; a 126-bp segment of the bla gene was then deleted to inactivate bla, so that cells harboring both pVS1 and a second plasmid with an ampicilin resistance marker could be selected in media containing ampicilin and chloramphenicol. pVS1Δ1 was derived from pVS1 by replacing the EcoNI–EcoRI DNA segment containing Pantitet, a promoter transcribing away from tetA, with the corresponding fragment from pASLS6 (23), in which Pantitet had been deleted from the tetA regulatory region. Sequential deletion of a 129-bp segment between the EarI and AatII site of pVS1Δ1, which contains the bla promoter Pbla, and a 610-bp BsaAI–NcoI fragment, which contains the promoter region of the cam gene, yielded pVS1Δ2 and pVS1Δ3, respectively. A set of plasmids were also constructed by inserting individual DNA fragments, either chemically synthesized or isolated from DNA samples containing them, into the DraI site of pVS1Δ3 (which is 332 bp upstream of the start of tetA transcription). The plasmid pASLR2 that expresses a site-specific recombinase from an inducible tac promoter had been described (23).
DNA Preparation and 2D Agarose Gel Electrophoresis. To avoid topoisomerase-mediated changes of plasmid DNA linking number during sample preparations, a rapid lysis procedure described (24) was used. 2D agarose gel electrophoresis of the DNA samples was carried out in 0.7% agarose gel slabs as described (25). The chloroquine concentrations during 1D and 2D electrophoresis are specified in the appropriate figure legends. Upon completion of electrophoresis, the DNA bands were transferred to a Zeta-probe membrane (Bio-Rad), and were probed with 32P-labeled DNA prepared by random priming, using a fragment within the tetA region as the template.
Results
Expression of tetA Is Insufficient for Hypernegative Supercoiling of a DNA Ring in ΔtopA Cells. The parent plasmid pVS1 used in this study is depicted in Fig. 1A. It contains three genes bla, tetA, and cam, under the control of the promoters Pbla,PtetA, and Pcam and encoding respectively ampicilin, tetracycline, and chloramphenicol resistance. There is also a fourth promoter, Pantitet, in pVS1 that directs transcription from the PtetA region into bla. The plasmid is divided into two arcs by two RS sequences, the target sites of a site-specific recombinase. Induction of the recombinase gene, located on a second plasmid pASLR2 under the control of a tac promoter (23), would lead to recombination between the RS sequences and thus the partition of pVS1 into two separate rings. Plasmids pVS1Δ1, pVS1Δ2, and pVS1Δ3 were derived from pVS1 by the deletion of Pantitet; Pantitet and Pbla; and Pantitet, Pbla, and Pcam, respectively; a schematic of pVS1Δ3 is depicted in Fig. 1B.
Fig. 1.
A schematic drawing of pVS1, the parent plasmid used in this work. (A) Recombination between the two RS sites, upon induction of a site-specific recombinase gene located on a separate plasmid, divides the two arcs into two separate DNA rings. The other elements on pVS1 are described in the text. The X sign within the bla gene denotes a deletion that rendered the protein product inactive (see Materials and Methods). (B) A schematic of pVS1Δ3, which is derived from pVS1 and bears tetA as its only transcription unit. The X signs mark positions of deletions that inactivate the bla and cam promoters as well as the bla gene protein product. Derivatives of pVS1Δ3 containing various inserts at the DraI site, which is located 332 bp upstream of the start of tetA transcription, were used in studying the effects of these inserts on plasmid supercoiling.
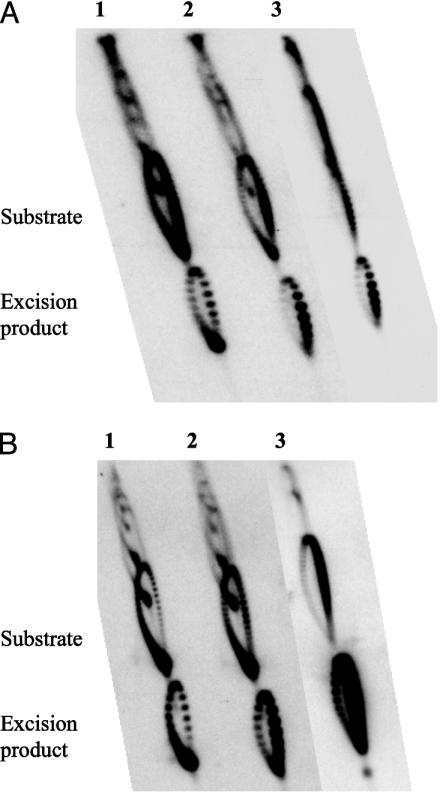
E. coli DM800 ΔtopA cells harboring pASLR2 and pVS1 or one of the pVS1 derivatives were grown to an early logarithmic phase. DNA samples were isolated from the cells by a rapid lysis procedure and were examined by 2D agarose gel electrophoresis. Fig. 2 depicts the topoisomer distributions of various plasmids after induction of the recombinase. In each case, topoisomers of tetA-containing DNA rings were specifically detected by blot hybridization. In Fig. 2, the monomeric form of the tetA-containing plasmid is denoted the “substrate” and its recombinase excision product containing the tetA region the “excision product.” For pVS1, hypernegative supercoiling is evident (Fig. 2 A, lane 1); the topoisomers were concentrated at the tip of the arc where highly negatively supercoiled topoisomers are expected. The excision product bearing tetA and bla but no ori showed a topoisomer distribution similar to pVS1.
Fig. 2.
Distributions of plasmids supercoiled to different degrees. (A and B) Plasmid samples were prepared from E. coli DM800 ΔtopA cells by a rapid lysis procedure, and were analyzed by 2D electrophoresis in 0.7% agarose gel slabs. The electrophoresis buffer contained 0.089 M Tris·borate, 2.5 mM EDTA, and 20 and 80 μg/ml of chloroquine diphosphate for 1D and 2D electrophoresis, respectively. Lanes 1 and 2 in A and B contained pVS1 and pVS1Δ2, respectively. Lane 3 in A contained pVS1Δ3, and lane 3 in B contained a control plasmid pUC18. The radiolabeled probe for detection of the DNA rings, other than the sample analyzed in lane 3 of B, was prepared from a fragment within the tetA region; thus, DNA molecules devoid of the tetA region, including the plasmid pASLR2 that carries ampicilin resistance and was used to introduce an inducible site-specific recombinase gene into strain DM800, were not seen in the autoradiograms shown. For detection of pUC18 (B, lane 3), a radiolabeled probe for the bla region was used. In each lane, the “substrate” refers to the monomeric form of the plasmid, and the “excision product” refers to the product of site-specific recombination that contains the tetA region.
Elimination of both Pantitet and Pbla from pVS1, which leaves two divergently transcribed genes cam and tetA, does not significantly alter the topoisomer distribution of the resulting plasmid pVS1Δ2 (the substrate DNA in lane 2 of Fig. 2 A). The excision product of pVS1Δ2 containing tetA showed, however, a very different topoisomer distribution (compare the excision product in lanes 1 and 2 of Fig. 2 A). The topoisomer distribution of the tetA-containing excision product of pVS1Δ2 is similar to that of a control plasmid pUC18 (Fig. 2B), which, like its pBR322-derived parent pUC8 that carries bla but not tetA, does not exhibit hypernegative supercoiling in ΔtopA cells (4). This result shows that the presence of the tetA transcription unit on the excised DNA ring is insufficient for the hyper-negative supercoiling of the DNA in the absence of DNA topoisomerase I. The same lack of hypernegative supercoiling was also seen in the cases of pVS1Δ3 and its tetA-containing excision product (Fig. 2 A, lane 3). It should be emphasized, however, that the lack of a significant difference between tetA-bearing DNA rings with and without ori should not be construed to imply that the presence of a replication bubble in a DNA segment has little effect on its rotational diffusion; in a steady-state population of an origin-bearing plasmid, only a small fraction of the rings are actually undergoing replication at a particular time.
Bent DNA as a Plausible Friction Barrier in the Rotational Diffusion of DNA. The above results indicate that for a typical DNA fragment, represented here by the 3.66-kb-long sequences constituting the bulk of pVS1Δ3, can be spun around its helical axis without a frictional drag large enough to impede the merge of the oppositely supercoiled domains. Nevertheless, it seems plausible that the presence of a stable macroscopic bend in a connecting DNA segment might pose a significant rotational barrier. Several DNA segments known to contain stable bends (Table 1) were therefore inserted into pVS1Δ3 to test their effects on the topoisomer distribution in ΔtopA cells. We found that none of these bent DNA fragments led to significant hypernegative supercoiling of the plasmid when inserted into pVS1Δ3 (results not shown).
Table 1. List of bent DNA fragments inserted into the DraI site of pVS1Δ3.
| Insert | Description | Ref. |
|---|---|---|
| H1 | A 30-bp synthetic bent DNA fragment (GGAAATTTCC)3 | 32 |
| LtA1 | A 410-bp Sau3A1 fragment of Leishmania tarentolae kinetoplast DNA | 33 |
| KD1 | A 57-bp fragment spanning the first bent region of above | 33 |
| KD6 | Tandem dimer of KD1 | — |
| CF3 | A 458-bp HincII—StuI fragment of Crithidia fasciculata kinetoplast DNA | 34 |
| Y1 | A 117-bp fragment of the regulatory region of Saccharomyces cerevisiae STE3 | 35 |
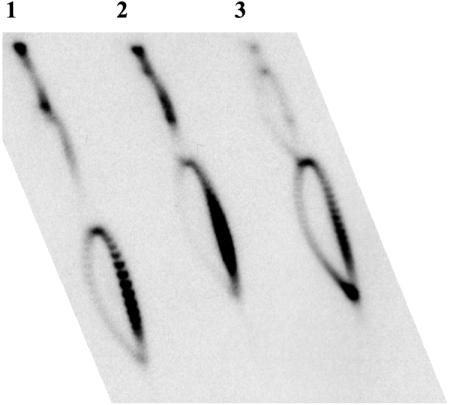
Effect of a Second Transcription Unit on the Topoisomer Distribution of pVS1Δ3. The absence of hypernegative supercoiling in pVS1Δ3 makes it a convenient platform for examining the effects of additional transcription units on its supercoiling. In agreement with previous results (12, 15), insertion of a cam transcription unit into pVS1Δ3 results in a significant shift in topoisomer distribution if cam is transcribed in the opposite direction as tetA, but not if the two are transcribed in the same direction (Fig. 3). Even the presence of two tetA with the same orientation results in little hypernegative supercoiling, as can be seen in the topoisomer distribution of dimeric pVS1Δ3 (arc of topoisomers above that of the monomeric substrate in lane 3, Fig. 2A).
Fig. 3.
Insertion of the cam gene into pVSΔ3 (see Fig. 1B) significantly shifts its topoisomer distribution in DM800 ΔtopA cells toward a higher degree of negative supercoiling, but only if the direction of cam transcription opposes that of tetA transcription. The cam insert, obtained from pACYC184 as a 1,410-bp BsaA1 restriction fragment, was inserted into the DraI site of pVSΔ3 (see Fig. 1B), in two different orientations, to yield the plasmids used in lanes 2 and 3. Lane 1, linking number distribution of pVSΔ3; lanes 2 and 3, derivatives of pVSΔ3 in which the cam and tetA are transcribed in the same (lane 2) or opposite direction (lane 3).
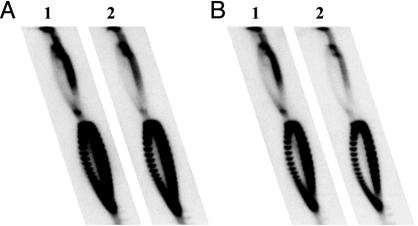
The orientation effect noted above is in sharp contrast to the lack of it when a promoter directing the synthesis of a very short transcript is inserted. As first noted by Chen and Lilley (20), the insertion of a promoter–terminator cassette directing the synthesis of a short transcript significantly enhances hypernegative supercoiling of a plasmid that expresses tetA, independent of the orientation of the cassette relative to tetA. For the pair of plasmids pVS1Δ3-(PtacT)o and pVS1Δ3-(PtacT)s, the tac promoter–terminator cassette PtacT directs the synthesis of a 45-nt transcript; the cassette is located 332 bp upstream of the start of tetA in pVS1Δ3, and the tac promoter in the cassette is either in the opposite or same direction of tetA transcription (denoted respectively by the subscript o or s). Relative to the topoisomer pattern seen with pVS1Δ3, insertion of the PtacT cassette causes a significant shift of the linking number distribution toward lower values of Lk, independent of the orientation of the cassette (Fig. 4A). The same results were seen if a tyrT promoter replaces Ptac (Fig. 4B). Thus, our results are in complete agreement with those of Chen and Lilley (20). Our interpretation of these results differs from theirs, however, and will be covered below (see Discussion).
Fig. 4.
Insertion of a fragment directing the synthesis of a short transcript into pVSΔ3 significantly shifts the topoisomer distribution of the plasmid in DM800 ΔtopA cells toward a higher degree of negative supercoiling, whether the short transcript is transcribed in the same (lane 1) or opposite (lane 2) direction of tetA. The promoter directing the synthesis of the short transcript was Ptac in A and Ptyr in B; oligonucleotide cassettes based on the sequences reported in Chen and Lilley (20) were synthesized and inserted into the DraI site of pVSΔ3. See the text for further details.
Discussion
Whereas the twin-supercoiled-domain model provides an adequate framework for understanding the supercoiling of intracellular DNA, the interdependent nature of the various enzymatic and diffusional pathways that affect this process often makes the supercoiling of even a simple topological domain a rather complicated subject. For clarity, we first discuss several general issues before addressing topics that are more germane to the present study.
Supercoiling of a Small Plasmid with No Anchored Transcript in E. coli. Because of the limited experimental data available, numerical estimates are made here only for small plasmids several kb in length. In the absence of anchored transcripts, the steady level of supercoiling is largely determined by the actions of gyrase and the DNA topoisomerases I and IV, and the specific linking difference σ of a plasmid is typically in the range of –0.05 to –0.09 (13). At this steady-state level of supercoiling, the rates of removing negative supercoils by DNA topoisomerase I and IV were estimated by Zechiedrich et al. (13) to be ≈5 and 0.8 min–1 per plasmid, respectively. Because DNA topoisomerase III apparently has a negligible role in modulating supercoiling, at steady-state gyrase must be introducing negative supercoils at a rate of ≈5 plus 0.8 or ≈6 min–1 per plasmid. For larger plasmids, these estimated rates presumably go up in proportion to their sizes, owing to the higher average numbers of topoisomerase molecules acting on each DNA ring. In general, the steady-state σ varies but little with plasmid size (26).
Effect of Anchoring a Nascent Transcript on Plasmid Supercoiling. We now consider the effect of a plasmid-borne tetA-like gene on the supercoiling of the plasmid. To highlight the effect of an anchored transcript, we assume here that the plasmid is also anchored at a second point. It has been shown that initiation of transcription at a tetA-like gene leads to the anchoring of the nascent transcript in <30 s (15), dividing the plasmid into two topological domains. The anchoring of the nascent transcript is thus followed by the generation of positive supercoils ahead of the translocating RNA polymerase and negative supercoils behind it (6). Assuming a rate of messenger RNA synthesis of 40 nt s–1, then four positive and four negative supercoils s–1 per plasmid, or 240 of each min–1 per plasmid, are generated by a transcription ensemble. These rates are nearly two orders of magnitude higher than the estimated rates for various topoisomerases (see above paragraph). Thus, after the initiation of a tetA-like transcript, large changes in the states of supercoiling ahead and behind the moving polymerase might be expected.
It should be emphasized, however, that the rates of the DNA topoisomerases are highly dependent on the extents of supercoiling of their substrates. When transcriptional supercoiling makes the DNA segment behind the translocating polymerase more negatively supercoiled, the rate of supercoil removal by DNA topoisomerase I is expected to steeply increase (8). Similarly, when transcriptional supercoiling makes the DNA segment in front of a moving polymerase more positively supercoiled, the rates of supercoil removal by gyrase and DNA topoisomerase IV would increase, because both enzymes are known to act more efficiently on a positively supercoiled DNA than on a negatively supercoiled one (11). By simultaneous induction of a pair of divergent transcripts that rapidly anchor to the cell membrane, Cook et al. (15) were able to show that in E. coli ΔtopA cells the combined actions of gyrase and DNA topoisomerase IV were at least comparable in rate to the generation of positive supercoils ahead of the pair of transcribing RNA polymerases.
It is uncertain, however, whether the large differences in the rates estimated by Cook et al. (15) and Zechiedrich et al. (13) can be entirely attributed to the dependence of the DNA topoisomerase rates on supercoiling. The rather slow rates estimated in Zechiedrich et al. (13) probably reflect rate-limiting steps that are not directly related to the intrinsic rates of the DNA topoisomerases. The steady-state negative supercoils, for example, might be stabilized by the binding of proteins. Thus, the rates of removal of such protein-stabilized supercoils might depend on a complex interplay between the actions of the DNA topoisomerases and the dissociation of the bound protein molecules. It is plausible that the transcription process might facilitate the dissociation of such proteins, and consequently alter the rates of topoisomerase-mediated linking number changes. The finding that both gyrase and DNA topoisomerase IV promote replication fork progression (10), the rate of which is much faster than the rate of transcription, also suggests that the DNA topoisomerases are intrinsically capable of much faster rates than those implicated by the data of Zechiedrich et al. (13).
If the intrinsic rates of gyrase and DNA topoisomerases I and IV are much higher than the rates of supercoil generation by translocation processes, including transcription, than in wild-type E. coli cells, these processes might not significantly perturb the steady-state level of supercoiling. Nevertheless, it is likely that the supercoiling of the DNA at a particular location is strongly dependent on the accessibility of the location to various enzymes that modulate supercoiling. It is interesting to note that one or more DNA topoisomerases may be associated with macromolecular assemblies translocating along DNA. E. coli DNA topoisomerase I, for example, has been reported to interact directly with RNA polymerase (27).
Dissipation of Oppositely Supercoiled Domains Through Axial Rotational Diffusion of the Connecting DNA. Our results do not support the idea that a typical DNA segment several kb in length can be an efficient barrier to the cancellation of oppositely supercoiled domains by diffusion. For a DNA ring with a single tetA transcript, hypernegative supercoiling in E. coli ΔtopA cells was not observed. Thus, the positive and negative supercoils on the two sides of an anchored nascent tetA transcript must merge, by spinning the connecting DNA antipodal to the anchored point, at a rate faster than the net rate of positive supercoil removal by gyrase and DNA topoisomerase IV. In other words, the frictional barrier to rotational diffusion of a tethering DNA segment must be relatively low, and no steep gradient in supercoil distribution can build up along the DNA. This conclusion is consistent with the magnitude of the frictional drag estimated from single-molecule measurements of DNA rotating at a very rapid rate of 2,000 turns per s (28).
The presence of a stable macroscopic bend in the tethering DNA also appears to lack a strong effect on hypernegative supercoiling in ΔtopA cells. Insertion of known bent DNA segments into pVS1Δ3 resulted in no significant shift of its topoisomer distribution. It is plausible, however, that a DNA bend more stable than the ones tested in this study may show stronger effects. Of special interest are DNA bends stabilized by protein binding, as discussed below.
Effects of Transcription Units That Do Not Result in Anchoring of Nascent Transcripts. As presented in Results, we have confirmed the observation of Chen and Lilley (20) that insertion of a promoter directing the synthesis of a very short transcript can significantly enhance hypernegative supercoiling of a DNA ring expressing tetA, irrespective of its orientation. Chen and Lilley (20) suggested that repeated helical opening at the inserted promoter, owing to repetitive initiation events, could generate compensatory positive supercoils for conversion to negative supercoils by gyrase, resulting in hypernegative supercoiling of the DNA ring in the absence of DNA topoisomerase I. This interpretation does not explain, however, why tetA expression is necessary for the observed enhancement. Furthermore, it seems rather unlikely that positive supercoils that are locally generated by repetitive polymerase binding could persist long enough, within a region of negatively supercoiled DNA, for gyrase to act on.
An alternative interpretation is that the observed effects are due to a protein-stabilized DNA bend that greatly increases the frictional barrier against rotating the DNA along its helical axis. In a recent in vitro study of plasmid transcription in the presence of gyrase (29), it was found that the binding of a number of sequence-specific DNA-binding proteins, including E. coli RNA polymerase itself, could greatly stimulate the negative supercoiling of the plasmid. It was suggested that nucleoprotein complexes containing sharply bent DNA might serve as a frictional barrier. In their experiments, as well as earlier in vitro experiments (see, for example, ref. 30), severe hindrance to the rotation of the transcribing RNA polymerase around DNA is probably a result of intermolecular interactions between nascent RNA molecules, or between nascent RNA and the reaction vessel. Whereas similar interactions between nucleoproteins could also lead to anchoring of the DNA in vitro, the strong correlation between the presence of a DNA bend in a nucleoprotein and the effectiveness of the nucleoprotein in enhancing plasmid supercoiling supports the idea that a protein-stabilized bend can serve as a significant barrier to DNA rotational diffusion (29).
In both the initiation and elongation complexes of RNA polymerase, the path of the DNA is not straight but bent (for a recent review, see ref. 31). Thus, the presence of a promoter–terminator combination in a DNA segment is likely to cause periodic and sequential formation of the initiation complex, the elongation complex, and finally the termination complex. The stronger the promoter, the shorter the duration between successive transcription cycles and the larger the fractional time the DNA is kept in a stably bent form. If a polymerase-stabilized bend leads to a large increase in the frictional barrier to axial rotation of DNA, then in topA– cells the effect of a promoter–terminator combination on hypernegative supercoiling of a plasmid that also bears a tetA-like transcription unit is expected to increase with the strength of the promoter in the promoter–terminator cassette, as observed by Chen and Lilley (20). In their experiments, even a cassette with a weak promoter Pleu500 exhibits a readily detectable effect, but cassettes containing the much stronger promoters Ptac and PtyrT show much greater effects.
The above interpretation raises an interesting question. On one hand, the addition of even a weak promoter like Pleu500 shows a substantial effect on the hypernegative supercoiling of a tetA-bearing plasmid, irrespective of the direction of the promoter, if it directs the synthesis of a very short transcript. On the other hand, the presence of a long transcription unit like the cam gene elicits hypernegative supercoiling of a similar plasmid, only if the gene is transcribed in the opposite direction of tetA (Fig. 3). Why doesn't a cam transcript going in the same direction as tetA effect hypernegative supercoiling of pVS1Δ3? One might expect that relative to a short transcript directed by a promoter–terminator combination, a much longer message would provide an even higher frictional barrier, irrespective of the direction of transcription.
We believe that the answer to the above perplexity lies in the relative rotational movements of the anchored transcriptional ensemble RtetA and the DNA bend associated with the second transcriptional ensemble, R2nd. During much of a transcription cycle of duration t, the polymerase-associated DNA segment most likely remains in a bent form while the polymerase cycles through the stages of initiation, elongation, and termination. If R2nd makes a very short transcript, the RNA polymerase is in the elongation mode only for a small fraction of t. In such a case, during much of t the DNA bend at R2nd is stationary relative to RtetA. For a gene encoding a long transcript like cam, however, an RNA polymerase transcribing the gene is in the elongation mode most of the time. Thus, if R2nd is moving in the same direction as RtetA, during a large part of t R2nd is turning under its own power in the same direction and at a similar rate as RtetA, and thus provides little resistance to the RtetA-mediated rotation of the DNA template.
Acknowledgments
This work was supported by National Institutes of Health Grant GM24544.
References
- 1.Gellert, M., Mizuuchi, K., O'Dea, M. H. & Nash, H. A. (1976) Proc. Natl. Acad. Sci. USA 73, 3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, J. C. (1971) J. Mol. Biol. 55, 523–533. [DOI] [PubMed] [Google Scholar]
- 3.Lockshon, D. & Morris, D. R. (1983) Nucleic Acids Res. 11, 2999–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruss, G. J. (1985) J. Mol. Biol. 185, 51–63. [DOI] [PubMed] [Google Scholar]
- 5.Pruss, G. J. & Drlica, K. (1986) Proc. Natl. Acad. Sci. USA 83, 8952–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, L. F. & Wang, J. C. (1987) Proc. Natl. Acad. Sci. USA 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maaloe, O. & Kjeldgard, N. O. (1966) Control of Macromolecular Syntheses (Benjamin, New York).
- 8.Wang, J. C. (1973). in DNA Synthesis in Vitro, eds. Wells R. D. Inman, R. B. (University Park Press, Baltimore), pp. 163–174.
- 9.Gamper, H. B. & Hearst, J. E. (1982) Cell 29, 81–90. [DOI] [PubMed] [Google Scholar]
- 10.Khodursky, A. B., Peter, B. J., Schmid, M. B., DeRisi, J., Botstein, D., Brown, P. O. & Cozzarelli, N. R. (2000) Proc. Natl. Acad. Sci. USA 97, 9419–9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crisona, N. J., Strick, T. R., Bensimon, D., Croquette, V. & Cozzarelli, N. R. (2000) Genes Dev. 14, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu, H. Y., Shyy, S. H., Wang, J. C. & Liu, L. F. (1988) Cell 53, 433–440. [DOI] [PubMed] [Google Scholar]
- 13.Zechiedrich, E. L., Khodursky, A. B., Bachellier, S., Schneider, R., Chen, D., Lilley, D. M. & Cozzarelli, N. R. (2000) J. Biol. Chem. 275, 8103–8113. [DOI] [PubMed] [Google Scholar]
- 14.Lodge, J. K., Kazic, T. & Berg, D. E. (1989) J. Bacteriol. 171, 2181–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook, D. N., Ma, D., Pon, N. G. & Hearst, J. E. (1992) Proc. Natl. Acad. Sci. USA 89, 10603–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch, A. S. & Wang, J. C. (1993) J. Bacteriol. 175, 1645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, D., Cook, D. N., Pon, N. G. & Hearst, J. E. (1994) J. Biol. Chem. 269, 15362–15370. [PubMed] [Google Scholar]
- 19.Nelson, P. (1999) Proc. Natl. Acad. Sci. USA 96, 14342–14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, D. & Lilley, D. M. (1999) J. Mol. Biol. 285, 443–448. [DOI] [PubMed] [Google Scholar]
- 21.DiNardo, S., Voelkel, K. A., Sternglanz, R., Reynolds, A. E. & Wright, A. (1982) Cell 31, 43–51. [DOI] [PubMed] [Google Scholar]
- 21.McEachern, F. & Fisher, L. M. (1989) FEBS Lett. 253, 67–70. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, A. S. & Wang, J. C. (1994) J. Mol. Biol. 236, 679–684. [DOI] [PubMed] [Google Scholar]
- 24.Bjornsti, M. A. & Wang, J. C. (1987) Proc. Natl. Acad. Sci. USA 84, 8971–8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peck, L. J. & Wang, J. C. (1983) Proc. Natl. Acad. Sci. USA 80, 6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, J. C. (1969) J. Mol. Biol. 43, 263–272. [DOI] [PubMed] [Google Scholar]
- 27.Cheng, B., Zhu, C. X., Ji, C., Ahumada, A. & Tse-Dinh, Y. C. (2003) J. Biol. Chem. 278, 30705–30710. [DOI] [PubMed] [Google Scholar]
- 28.Thomen, P., Bockelmann, U. & Heslot, F. (June 3, 2004). Phys. Rev. Lett. 10.1103/PhysRevLett.88.248102. [DOI] [PubMed]
- 29.Leng, F. & McMacken, R. (2002) Proc. Natl. Acad. Sci. USA 99, 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsao, Y. P., Wu, H. Y. & Liu, L. F. (1989) Cell 56, 111–118. [DOI] [PubMed] [Google Scholar]
- 31.Cramer, P. (2002) Curr. Opin. Struct. Biol. 12, 89–97. [DOI] [PubMed] [Google Scholar]
- 32.Hagerman, P. J. (1985) Biochemistry 24, 7033–7037. [DOI] [PubMed] [Google Scholar]
- 33.Marini, J. C., Levene, S. D., Crothers, D. M. & Englund, P. T. (1982) Proc. Natl. Acad. Sci. USA 79, 7664–7668, and correction (1982) 80, 7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitchin, P. A., Klein, V. A., Ryan, K. A., Gann, K. L., Rauch, C. A., Kang, D. S., Wells, R. D. & Englund, P. T. (1988) J. Biol. Chem. 261, 11302–11309. [PubMed] [Google Scholar]
- 35.Inokuchi, K., Nakayama, A. & Hishinoma, F. (1988) Nucleic Acids Res. 16, 6693–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]