Abstract
The protein CTCF plays an essential role in the action of a widely distributed class of vertebrate enhancer-blocking insulators, of which the first example was found in a DNA sequence element, HS4, at the 5′ end of the chicken β-globin locus. HS4 contains a binding site for CTCF that is necessary and sufficient for insulator action. Purification of CTCF has revealed that it interacts with proteins involved in subnuclear architecture, notably nucleophosmin, a 38-kDa nucleolar phosphoprotein that is concentrated in nuclear matrix preparations. In this report we show that both CTCF and the HS4 insulator element are incorporated in the matrix; HS4 incorporation depends on the presence of an intact CTCF-binding site. However the DNA sequence in the neighborhood of HS4 is not like that of canonical matrix attachment regions, and its incorporation into the matrix fraction is not sensitive to ribonuclease, suggesting that the insulator is a distinct matrix-associated element.
Insulators are DNA sequence elements that can act either to block the extension of a condensed chromatin domain into a transcriptionally active region (barrier activity), or to prevent the interaction of a distal enhancer with a promoter when placed between the two (1, 2). Elements with the latter property, called enhancer blocking insulators, have been found in Drosophila and in vertebrates. In flies the most studied insulator element is gypsy, which when placed between two enhancers in a series of enhancers found in the yellow locus, blocks the action of all enhancers distal to the insertion but has no effect on those more proximal to the promoter (3). It has been shown that the insulator action of gypsy is mediated by a DNA-binding protein, Suppressor of Hairy wing [Su(Hw)], and a cofactor, Mod(mdg4) (4). Gypsy elements appear to localize to the nuclear envelope, where they cluster and organize the neighboring chromatin into loop domains (5). It is thought that the loop domain structure gives rise to the insulating activity either by preventing regulatory elements on different loops from interacting or by interfering with a “tracking” signal that would ordinarily proceed from enhancer to promoter (6–8). Loop domains can be established by attachment to other fixed sites in the nucleus. For example, a barrier function that prevents heterochromatinization of an active gene can be generated by tethering DNA elements to nuclear pore proteins (9). Loop domains can also arise simply from interactions that cause the insulator-bound proteins to stick to each other.
A different enhancer blocking insulator activity has been described in vertebrates. First found at the 5′ end of the chicken β-globin locus, it is part of a compound element (HS4) at that site that has both barrier and enhancer-blocking action (10). These two activities are separable; the enhancer-blocking insulation arises from a single DNA site that binds the protein CTCF (11). Insulator elements that bind CTCF have also been found at many other loci including the human and mouse β-globin cluster, near the promoter of the Tsix gene at the mouse X inactivation center, and the imprinted control region of the Igf2/H19 locus, where it plays an important role in regulation of imprinted Igf2 expression (12–15).
We have shown recently that CTCF in nuclear extracts forms a stable and well defined complex with nucleophosmin, a protein found at high concentration in the nucleolus (16). Chromatin immunoprecipitation experiments show that nucleophosmin and CTCF are both bound in the neighborhood of HS4. Furthermore, copies of the insulator sequence stably integrated into an erythroid cell line are found by fluorescence in situ hybridization analysis to be localized at the nucleolar periphery. This suggests a model quite similar to the one proposed for gypsy, in which the insulator serves to generate loop domains that isolate enhancer and promoter in separate loops.
There is an extensive literature concerned with the organization of nuclear structural proteins that is based on the isolation of a nuclear matrix fraction and its associated DNA sequences or matrix attachment regions (MARs) (17). The solvent extraction and nuclease digestion procedures used to prepare the matrix are intended to retain only the most tightly bound proteins and their associated DNA. A wide variety of proteins, including nuclear lamins, nucleophosmin, and topoisomerase II, and many regulatory factors are found in the matrix (18). The MARs themselves tend to be quite rich in A-T base pairs (19); they were originally proposed to organize metaphase chromosomes into loops by attachment of MARs (or scaffold attachment regions) to a proteinacious scaffold or backbone (20, 21). Given the strong interaction between CTCF and nucleophosmin, it seemed important to determine whether CTCF was incorporated in the nuclear matrix fraction. We show here that CTCF and the HS4 insulator sequence are highly concentrated in that fraction. Furthermore, ≈1.5 kb of DNA 3′ of the insulator is also protected against nuclease digestion and appears in the matrix fraction. Although this would appear to justify classifying the insulator as an MAR, the properties of the GC-rich-binding site (Fig. 1) and the behavior during purification suggest that CTCF and the insulator it binds may be different from usual components of the nuclear matrix.
Fig. 1.
Schematic representation and sequence of the HS4 core insulator (underlined) and flanking regions. The binding of CTCF and nucleophosmin are based on previous chromatin immunoprecipitation data (16).
Materials and Methods
Matrix Preparation. High salt matrices were prepared essentially as described (22). Cells were incubated for 10 min in RSB buffer (10 mM Tris, pH 7.4/10 mM NaCl/3 mM MgCl2/PMSF) on ice, homogenized 10 times with a Dounce homogenizer by using a “loose” pestle, and centrifuged at 1,000 × g for 5 min at 4°C. Pelleted nuclei were washed twice in RSB and 0.25 M sucrose, resuspended in RSB and 2 M sucrose, and centrifuged for 10 min at 34,000 × g. Pelleted nuclei were washed once in RSB and 0.25 M sucrose, resuspended in RSB, 0.25 M sucrose, 1 mM CaCl2, and PMSF, and digested for 3 h with 100 μg/ml RNase-free DNase I (Roche) at room temperature on a rotating platform. After digestion, an equal volume of 20 mM Tris (pH 7.4), 4 M NaCl, 10 mM EDTA, and PMSF was added, and the nuclei were incubated on ice for 10 min followed by centrifugation at 1,500 × g for 15 min. Pellets were washed twice with 10 mM Tris (pH 7.4), 2 M NaCl, 5 mM EDTA, and 0.25 mg/ml PMSF and stored in RSB plus 0.25 M sucrose, 0.25 mg/ml BSA, and 50% glycerol at –20°C.
For Western analyses of soluble and matrix proteins, digested nuclei were centrifuged at 1,500 × g for 5 min, and the supernatant was removed. Pelleted nuclei were then extracted with extraction buffer (20 mM Tris, pH 7.4/10 mM EDTA) containing an increasing amount of NaCl to 2 M as indicated. For each concentration of salt, the digested nuclei were washed three times with 3-pellet vol of extraction buffer each, followed by centrifugation at 1,500 × g for 5 min. The three washes were combined at each step and mixed with loading buffer, and an equal volume of each was used for Western blotting. The final insoluble nuclear matrix was solubilized directly in loading buffer with a volume equal to the other steps.
In Vitro Matrix Assay. The 1.2-kb insulator fragment was digested from the pNI vector (10) with XbaI and agarose gel purified. The 1.2-kb λ fragment was gel purified from a λ/BstPI DNA ladder (Panvera, Madison, WI). Both fragments were end-labeled with [32]ATP and T4 polynucleotide kinase (NEB, Beverly, MA). Matrices from K562 cells were prepared by the high-salt extraction procedure or by the lithium salt procedure (see below), and the binding assays were performed as described (22). Matrices from ≈1 × 107 cells were mixed with labeled probe in assay buffer (10 mM Tris, pH 7.4/50 mM NaCl/0.25 M sucrose/2mM EDTA/0.25 mg/ml BSA/100 μg/ml sonicated Escherichia coli DNA) for 2 h at room temperature. Matrices were then washed once with assay buffer and once with assay buffer minus carrier DNA with centrifugation of the matrices at 10,000 × g for 60 s. Washed matrices were solubilized in 10 mM Tris/1 mM EDTA (pH 8) (TE) plus 0.5% SDS and 0.4 mg/ml proteinase K, digested overnight at 50°C, phenol:chloroform extracted, and EtOH precipitated with 20 μg of tRNA as carrier. Pelleted DNA was resolved on a 1.2% agarose gel, which was then dried and exposed on a phosphoimager.
In Vivo Matrix Assay. Low ionic strength matrices from 6C2 cells and 10-day chicken RBCs were prepared essentially as described (21). Cells were washed in PBS, incubated for 10 min on ice in isolation buffer [3.75 mM Tris, pH 7.4/20 mM KCl/0.5 mM EDTA (potassium hydroxide)/0.05 mM spermine/0.125 mM spermidine/0.1% digitonin/PMSF] and homogenized 15 times with a “loose” pestle. Nuclei were pelleted by centrifugation at 900 × g for 10 min at 4°C and washed three times with isolation buffer. Nuclei were resuspended in 100 μl of isolation buffer without EDTA and incubated in a 37°C waterbath for 20 min. Extraction buffer (5 mM Hepes/NaOH, pH 7.4/2 mM KCl/2 mM EDTA/0.25 mM spermidine/0.1 mM digitonin/25 mM lithium 3,5-diiodosalicylate) was slowly added to a final volume of 7 ml, and extracted nuclei were incubated for 5 min at room temperature.
After protein extraction, nuclei were digested with 100 μg/ml RNase-free DNase I (Roche) alone or with 200 units/ml of micrococcal nuclease (Worthington) for 3 h at room temperature. Matrices were centrifuged for 5 min at 1,500 × g, washed twice with digestion buffer, and resuspended in TE, 0.1% SDS, and 1 mg/ml proteinase K. Matrices were digested at 50°CO/N, phenol:chloroform extracted, and EtOH precipitated. Quantitative PCRs were carried out by using primers/probes as described (16). Standard PCR of matrix DNA was carried out in 25-μl reactions [2.5 μl of 10X buffer/0.5 μl of 10 mM dNTP mix/0.5 μl each of 10 μM forward and reverse primer/0.25 μlof Taq (Roche)/4 μl of DNA template/16.57 μl of H2O] with the following cycles [94°C for 2 min, 33 times (94°C for 15 s, 70°C for 15 s, and 72°C for 15 s) and 72°C for 3 min]. PCRs were resolved in a 2% agarose gel in 0.5× 40 mM Tris-acetate (pH 8.5)/1 mM EDTA and visualized by ethidium bromide staining. In parallel, PCRs were carried out with 6C2 cell genomic DNA. Primer sequences are available on request.
Results
In our initial experiments, we examined the solubility properties of CTCF protein in the nucleus. Typically, nuclear matrix preparative procedures used to investigate protein extractability have relied on the digestion of isolated nuclei with DNase I, followed by treatment with 2 M NaCl. However, some proteins that are soluble at lower salt concentrations bind more tightly to chromatin at such high salt concentrations (23). To avoid this problem, nuclei from K562 cells, digested with DNase I, were extracted with buffers of increasing ionic strength, in a stepwise manner up to 2 M NaCl. The release of CTCF protein was then compared to the extractability of insoluble lamins and soluble histone H3 (Fig. 2). Although a fraction of CTCF was soluble in buffers of lower ionic strength, the majority of CTCF remained in the insoluble material, even when detergent was added to the buffers. The release of CTCF was not improved by combined digestion with DNase I and micrococcal nuclease (Mnase) or by subsequent digestion with RNase A (data not shown), indicating that CTCF insolubility is not dependent on the presence of intact RNA. In this respect and others (see below), CTCF differs from some other components of the nuclear matrix, which are released upon RNase digestion. Similarly, it has recently been shown that CTCF is not extracted from a human MCF7 cells after Triton X-100 (0.5%) extraction and DNase I digestion (24).
Fig. 2.
Extraction of CTCF protein from nuclei reveals soluble/insoluble fractions. Nuclei were digested with DNase I and then extracted with buffer of increasing ionic strength. Release of CTCF protein was monitored by Western analyses and compared to known insoluble lamins and soluble histone H3. The extraction of CTCF was partially improved with detergent.
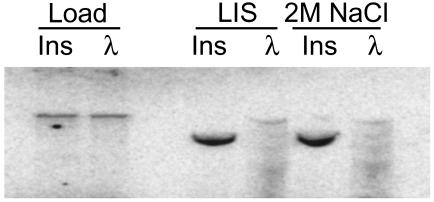
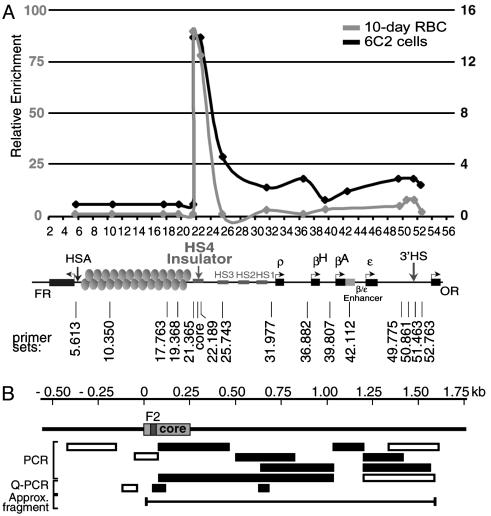
Given that some CTCF protein in the nucleus is insoluble and that CTCF can interact with matrix proteins, it seemed reasonable to ask whether insulators containing CTCF sites could be bound to the nuclear matrix. In a binding assay in vitro, nuclear matrix preparations were incubated with labeled DNA fragments. Matrices prepared by two different methods bound specifically a labeled 1.2-kb HS4 insulator fragment but not a λ DNA control fragment (Fig. 3). It seems likely that residual DNase I present in the matrices from the preparation causes some digestion of the labeled fragments. In the case of the insulator DNA, attachment allows only partial digestion resulting in enrichment of a slightly smaller fragment. The unattached λ control DNA, however, becomes almost completely digested into many smaller fragments. Endogenous DNA in nuclear matrix preparations also can be measured by a method more likely to preserve native attachment sites (21). In this approach, proteins are stripped from the nuclei by mild detergent followed by digestion of the DNA with DNase I. After digestion, the remaining DNA fragments (MARs) are precipitated and used in quantitative PCRs to detect which sequences are present. In both chicken erythroid 6C2 cells and primary 10-d chicken RBCs, after extensive DNase I digestion (Fig. 4A) or DNase I/Mnase digestion (data not shown), the HS4 element of the β-globin locus remained tightly bound, whereas most of the surrounding regions were digested away.
Fig. 3.
The chicken HS4 insulator behaves as a matrix attachment site in vitro. An in vitro MAR assay by using a 32P-labeled 1.2-kb HS4 insulator probe (Ins) or a control λ DNA probe. Prepared matrices (lithium salt and 2 M NaCl) were incubated with the labeled DNA fragments and washed, and bound DNA was precipitated and resolved by agarose gel electrophoresis. The 1.2-kb insulator specifically bound, whereas the control DNA did not.
Fig. 4.
Matrix attachment properties of DNA in the neighborhood of the chicken HS4 insulator. (A) An in vivo MAR assay of the endogenous HS4 insulator. Matrices from pre-erythroid 6C2 cells or primary 10-d RBCs were prepared by using the lithium salt extraction procedure to preserve native attachment sites. After extensive DNase I digestion, residual DNA fragments were precipitated and analyzed by quantitative PCR by using primers across the chicken β-globin locus. Only DNA fragments of the HS4 insulator remained. (B) Representation of PCR analyses of matrices from 6C2 cells centered on the HS4 core and surrounding sequences. All primer sets were tested with genomic DNA. Open bars represent amplified regions for the genomic DNA but were absent from the matrix preparation. Filled bars represent amplified products for both genomic and matrix DNA samples.
To determine precisely the extent of the protected region generated by this method, we carried out a series of PCR experiments with primers extending on both sides of the CTCF site (Fig. 4B). The region of attachment begins at the immediate 5′ end of the 250-bp core and extends ≈1.2 kb beyond the 3′ end of the core (toward the globin genes), resulting in an ≈1.5-kb-protected fragment in 6C2 cells. This very GC-rich fragment was detected even after prolonged nuclease digestion. Known MARs are generally A-T rich, suggesting that the HS4 insulator and surrounding sequences represent an atypical matrix-associated site (19).
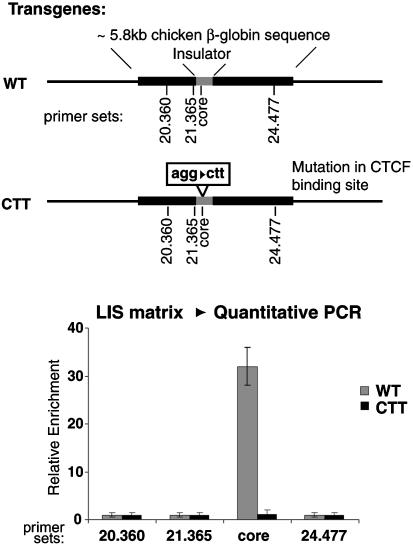
Is CTCF itself responsible for the attachment of the insulator DNA to the matrix? The in vivo MAR-binding assay was repeated by using a human cell line carrying a stably integrated transgene with ≈5.8 kb of the chicken HS4 insulator and its flanking sequences (16). In parallel, a cell line was transformed with a similar transgene containing a mutation in the CTCF-binding site. After matrix preparation, the WT insulator remained with the matrix, whereas a transgene carrying an insulator mutated at the CTCF-binding site did not (Fig. 5). This data indicates that an intact CTCF-binding site is required for the association of the insulator itself with the matrix. We did not determine the size of the protected region in these experiments.
Fig. 5.
The intact CTCF site is responsible for the matrix attachment. Transgenic cell lines containing a fragment of the HS4 insulator and flanking sequences were generated in parallel with lines carrying transgenes with a mutation in the CTCF-binding site. After matrix preparation, only the lines containing the WT CTCF-binding site were found to be associated with the nuclear matrix.
Discussion
In earlier studies, we reported that CTCF forms a strong and well defined complex with nucleophosmin/B23, a protein that is present throughout the nucleus but is largely concentrated on the nucleolar surface (16). Nucleophosmin copurifies with the nuclear matrix (25), and it seems reasonable to suggest that it is the interaction between this protein and CTCF that leads to the appearance of CTCF in the matrix fraction. Chromatin immunoprecipitation experiments have shown that nucleophosmin and CTCF colocalize at insulator sites in vivo, presumably explaining our observation that the HS4 insulator sequence is also concentrated in the matrix fraction. However it is clear from the known abundance of CTCF and nucleophosmin that most molecules of these two proteins are not likely to be attached in vivo to strong DNA-binding sites for CTCF.
We have proposed that the tethering of CTCF-dependent insulators to the nucleolar periphery creates separate “loop” domains that prevent interaction between an enhancer and promoter situated on opposite sides of the insulator (16). Such a structural impediment located between an enhancer and promoter could block the enhancer while having no direct effect on the promoter, consistent with the observation that insulators are neutral with respect to transcriptional activation. The enhancer blocking activity could arise either because elements in different loop domains are physically unable to make direct contact or because some activating signal that normally tracks from enhancer to promoter (e.g., by “facilitated tracking”) (6, 26, 27) is blocked by the insulator at its attachment point. This is topologically equivalent to the model proposed earlier by Corces and Labrador (2) explaining the action of gypsy, an enhancer-blocking insulator in Drosophila. The gypsy element binds the protein Su(Hw) and its associated protein Mod(Mdg4), which form clusters to create discrete loop domains that are also tethered, in this case to the nuclear periphery (5). Subsequent studies from the Corces laboratory have shown that these two proteins also fractionate with the nuclear matrix (28).
Recently it has been shown that although the full gypsy element concentrates at the nuclear envelope, transgenes containing only the Su(Hw)-binding sites, albeit still active insulators, are not localized in this way (29). Tethering to the nuclear envelope is thus likely to depend both on Su(Hw) binding and on the presence of another as yet uncharacterized element. Candidate elements containing A-T-rich MAR-like sequences are interleaved with the Su(Hw)-binding sites (30, 31), but more recent evidence implicates gypsy sequences that lie outside the cluster of Su(Hw) sites (29). It has been suggested that Su(Hw) may also function through local interactions with fixed interior chromatin components. As we have pointed out, these models do not require any particular point of attachment within the nucleus; in another variant of the model, the insulator sites could be attached in clusters only to each other to create equivalent loop domain structures (1, 16).
Although there is good reason to implicate tethering mechanisms in the function of enhancer-blocking insulators, the relationship between tethering and fractionation with the matrix is not a necessary one. The nuclear matrix, as operationally defined here and elsewhere in terms of a set of preparative methods, includes contributions from the nuclear envelope, the nucleolus, and from DNA-binding proteins that may play other structural roles (17, 23). Our results in any case indicate that CTCF does not behave like most of the proteins associated with the matrix fraction. First, it is not released by a combination of high salt and ribonuclease treatments and is thus quite different in behavior from many MAR-associated proteins, including Su(Hw) and Mod(mdg4) (28, 32). Equally important, canonical matrix attachment sites are typically very rich in A-T base pairs, whereas the binding domain of CTCF is not, and the extended DNA region around the HS4 insulator that it brings into the matrix fraction (Fig. 4B and see below) is very rich in G-C base pairs (Fig. 1).
We have focused here on the properties of the element upstream of the chicken β-globin locus that carries the enhancer-blocking insulator activity. We have shown that this element is carried into the matrix fraction through its interaction with CTCF. Of particular interest is the extent of protection of the surrounding DNA afforded by that interaction. As shown in Fig. 4B, a stretch of ≈1.5 kb of DNA is resistant to nuclease digestion and appears in the matrix fraction. This extends unidirectionally 3′ of the CTCF-binding site; PCR probes detect no protected sequence 5′ of the site. It should be noted that the hypersensitive site associated with this region has been located near the 5′ end of the 250-bp core, perhaps helping to define the 5′ end of the protected region. However the procedures used to generate this fragment involve removal, before digestion, of histones and quite possibly other proteins that help to generate the hypersensitive site. We have not yet been able to determine whether this extended protection depends on CTCF binding. It is possible that recruitment of nucleophosmin, which can oligomerize (33), is responsible for this behavior.
As noted above, the preponderance of CTCF is located elsewhere in the nucleus, possibly involved in completely different (noninsulating) functions. Furthermore, we do not know whether all or even most CTCF-binding sites are associated with MAR activity or with nucleophosmin. Possibly because of the somewhat weaker binding of CTCF at the 3′-HS of the chicken β-globin locus 11, 16), we have been unable to detect those sequences in the MAR fraction. It also has been reported that the multiple CTCF-binding sites in the mouse Igf2/H19-imprinted control region are either not associated or only weakly associated with the MAR fraction (34). However we have recently found another CTCF insulator upstream of the mouse DadI locus (35) that is located at a peak of MAR activity. We have not yet determined the cofactors bound at that site, either to CTCF or to adjacent DNA sequences, nor have we shown that the MAR activity in that region depends on CTCF binding.
Although the CTCF-binding site at the 5′ end of the β-globin locus and its associated proteins appear in the matrix-associated fraction, the presence of a DNA sequence in that fraction does not necessarily mean that it will function as an insulator. Some MAR elements may have insulator activity, but whether that activity resides in the A-T-rich sequences themselves or in embedded and undetected binding sites for specific DNA-binding proteins is not yet determined. In the case of the CTCF site in the globin insulator, our results suggest a more specific model in which nucleophosmin, acting in concert with CTCF, provides a high affinity complex, resistant to dissociation and nuclease digestion, which carries the site into the nuclear matrix fraction.
Acknowledgments
We thank M. Gaszner, B. Burgess-Beusse, and V. Mutskov for their insightful comments during the preparation of the manuscript.
Abbreviations: MAR, matrix attachment region; Su(Hw), Suppressor of Hairy wing.
References
- 1.West, A. G., Gaszner, M. & Felsenfeld, G. (2002) Genes Dev. 16, 271–288. [DOI] [PubMed] [Google Scholar]
- 2.Labrador, M. & Corces, V. G. (2002) Cell 111, 151–154. [DOI] [PubMed] [Google Scholar]
- 3.Geyer, P. K. & Corces, V. G. (1992) Genes Dev. 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- 4.Gerasimova, T. I., Gdula, D. A., Gerasimov, D. V., Simonova, O. & Corces, V. G. (1995) Cell 82, 587–597. [DOI] [PubMed] [Google Scholar]
- 5.Gerasimova, T. I., Byrd, K. & Corces, V. G. (2000) Mol. Cell 6, 1025–1035. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood, E. M. & Kadonaga, J. T. (1998) Science 281, 61–63. [DOI] [PubMed] [Google Scholar]
- 7.Liu, Y., Bondarenko, V., Ninfa, A. & Studitsky, V. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F. & Fraser, P. (2002) Nat. Genet. 32, 623–626. [DOI] [PubMed] [Google Scholar]
- 9.Ishii, K., Arib, G., Lin, C., Van Houwe, G. & Laemmli, U. K. (2002) Cell 109, 551–562. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. H., Whiteley, M. & Felsenfeld, G. (1993) Cell 74, 505–514. [DOI] [PubMed] [Google Scholar]
- 11.Recillas-Targa, F., Pikaart, M. J., Burgess-Beusse, B., Bell, A. C., Litt, M. D., West, A. G., Gaszner, M. & Felsenfeld, G. (2002) Proc. Natl. Acad. Sci. USA 99, 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell, A. C. & Felsenfeld, G. (2000) Nature 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 13.Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. (2000) Nature 405, 486–489. [DOI] [PubMed] [Google Scholar]
- 14.Farrell, C. M., West, A. G. & Felsenfeld, G. (2002) Mol. Cell. Biol. 22, 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao, W., Huynh, K. D., Spencer, R. J., Davidow, L. S. & Lee, J. T. (2002) Science 295, 345–347. [DOI] [PubMed] [Google Scholar]
- 16.Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. (2004) Mol. Cell 13, 291–298. [DOI] [PubMed] [Google Scholar]
- 17.Pederson, T. (2000) Mol. Biol. Cell 11, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belgrader, P., Siegel, A. J. & Berezney, R. (1991) J. Cell Sci. 98, 281–291. [DOI] [PubMed] [Google Scholar]
- 19.Singh, G. B., Kramer, J. A. & Krawetz, S. A. (1997) Nucleic Acids Res. 25, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson, J. R. & Laemmli, U. K. (1977) Cell 12, 817–828. [DOI] [PubMed] [Google Scholar]
- 21.Mirkovitch, J., Mirault, M. E. & Laemmli, U. K. (1984) Cell 39, 223–232. [DOI] [PubMed] [Google Scholar]
- 22.Cockerill, P. N. & Garrard, W. T. (1986) Cell 44, 273–282. [DOI] [PubMed] [Google Scholar]
- 23.Hancock, R. (2000) Chromosoma 109, 219–225. [DOI] [PubMed] [Google Scholar]
- 24.Dunn, K. L., Zhao, H. & Davie, J. R. (2003) Exp. Cell Res. 288, 218–223. [DOI] [PubMed] [Google Scholar]
- 25.Mattern, K. A., Humbel, B. M., Muijsers, A, O., de Jong, L. & van Driel, R. (1996) J. Cell. Biochem. 62, 275–289. [DOI] [PubMed] [Google Scholar]
- 26.Hatzis, P. & Talianidis, I. (2002) Mol. Cell 10, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 27.Engel, J. D. & Tanimoto, K. (2000) Cell 100, 499–502. [DOI] [PubMed] [Google Scholar]
- 28.Byrd, K. & Corces, V. G. (2003) J. Cell Biol. 162, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, Q., Li, M., Adams, J. & Cai, H. N. (2004) J. Cell Sci. 117, 1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabirochkin, S., Ossokina, M. & Heidmann, T. (1998) J. Biol. Chem. 273, 2473–2479. [DOI] [PubMed] [Google Scholar]
- 31.Scott, K. C., Taubman, A. D. & Geyer, P. K. (1999) Genetics 153, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fey, E. G., Ornelles, D. A. & Penman, S. (1986) J. Cell Sci. Suppl. 5, 99–119. [DOI] [PubMed] [Google Scholar]
- 33.Yung, B. Y. & Chan, P. K. (1987) Biochim. Biophys. Acta 925, 74–82. [DOI] [PubMed] [Google Scholar]
- 34.Weber, M., Hagege, H., Murrell, A., Brunel, C., Reik, W., Cathala, G. & Forne, T. (2003) Mol. Cell. Biol. 23, 8953–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magdinier, F. M., Yusufzai, T. M. & Felsenfeld, G. (April 13, 2004) J. Biol. Chem., 10.1074/jbc.M403121200. [DOI] [PubMed]