Abstract
Sclerosing epithelioid fibrosarcoma (SEF) is a rare soft tissue tumor exhibiting considerable morphologic overlap with low grade fibromyxoid sarcoma (LGFMS). Moreover, both SEF and LGFMS show MUC4 expression by immunohistochemistry. While the majority of LGFMS cases are characterized by a FUS-CREB3L1 fusion, both FUS-CREB3L2 and EWSR1-CREB3L1 fusions were recently demonstrated in a small number of LGFMS and SEF/LGFMS hybrid tumors. In contrast, recent studies pointed out that SEF harbor frequent EWSR1 rearrangements, with only a minority of cases showing FUS-CREB3L2 fusions. In an effort to further characterize the molecular characteristics of pure SEF and hybrid SEF/LGFMS lesions, we undertook a clinicopathologic, immunohistochemical and genetic analysis of a series of 10 SEF and 8 hybrid SEF/LGFMS tumors. The mortality rate was similar between the two groups, 44% within the pure SEF group and 37% in the hybrid SEF/LGFMS with a mean overall follow-up of 66 months. All but one pure SEF and all hybrid SEF/LGFMS tested cases showed MUC4 immunoreactivity. The majority (90%) of pure SEF cases showed EWSR1 gene rearrangements by FISH with only one case exhibiting FUS rearrangement. Of the 9 EWSR1 positive cases, 6 cases harbored CREB3L1 break-apart, two had CREB3L2 rearrangement (a previously unreported finding) and one lacked evidence of CREB3L1/2 abnormalities. In contrast, all hybrid SEF/LGFMS tumors exhibited FUS and CREB3L2 rearrangements. These results further demarcate a relative cytogenetic dichotomy between pure SEF, often characterized by EWSR1 rearrangements, and hybrid SEF/LGFMS, harboring FUS-CREB3L2 fusion; the latter group recapitulating the genotype of LGFMS.
Keywords: sclerosing epithelioid fibrosarcoma, fibromyxoid sarcoma, EWSR1, FUS, CREB3L1, CREB3L2, fusion
INTRODUCTION
First characterized by Meis-Kindblom in 1995 (Meis-Kindblom 1995), sclerosing epithelioid fibrosarcoma (SEF) is a rare soft tissue tumor exhibiting a characteristic morphologic pattern of epithelioid cells arranged in strands, cords, nests and sheets embedded in a sclerosed and hyalinized stroma. A subset of SEF shares morphologic overlap with low grade fibromyxoid sarcoma (LGFMS), an entity coined by Evans in 1987 (Evans 1987, 1993), that features a deceptively benign histology of bland appearing spindle cells in an alternating fibrous and myxoid stroma. Interestingly, the so-called hyalinizing spindle cell tumor with giant rosettes (HSCT), initially considered its own entity (Lane 1997; Folpe 2000), was subsequently reclassified as a variant of LGFMS, which in some cases appears to have hybrid features between SEF and LGFMS (Reid 2003). Furthermore, hypercellular spindle cell areas resembling fibrosarcoma have been described in both SEF and LGFMS (Evans 1987, 1993; Meis-Kindblom 1995; Antonescu 2001).
SEF tends to have an aggressive clinical course with high rates of local recurrence (50%), distant metastasis (40–80%) and mortality (25–57%) (Meis-Kindblom 1995; Antonescu 2001) whereas LGFMS has been characterized as having a “low-grade” protracted clinical course with local recurrences and late metastasis (Evans 1987, 1993). Nevertheless, in Evans’ most recent comprehensive study of 33 LGFMS cases with long-term follow-up (mean of 14 years), half of the patients developed metastases and 42% died of disease (Evans 2011).
The majority of LGFMS exhibit the characteristic t(7;16)(q33;p11) translocation resulting in FUS-CREB3L2 fusion (Panagopoulos 2004; Matsuyama 2006), with rare reported cases harboring a t(11;16)(p11;p11) secondary to a FUS-CREB3L1 fusion (Mertens 2005; Guillou 2007; Odem 2013; Rubinstein 2014). The same FUS-CREB3L2 fusion was subsequently demonstrated in a small number of SEF/LGFMS hybrid tumors (Rekhi 2011; Doyle 2012). In contrast, in a series of 22 morphologically pure SEFs, FUS gene rearrangements were found in only 2 cases (9%)(Wang 2012), after extensive sampling of the tumors to rule out the presence of a LGFMS component. In addition to the rare FUS-CREB3L1 variant, EWSR1-CRE3BL1 fusions were also described in two LGFMS cases and in a small number of tumors with SEF/LGFMS hybrid morphology (Rekhi 2011; Doyle 2012).
SEF and LGFMS share overexpression of MUC4 by immunohistochemistry. Using global expression analysis, MUC4 was found to be among the top upregulated genes in LGFMS and thus detectable via immunohistochemistry (Moller 2011), subsequent comprehensive studies (Doyle 2011) demonstrated that MUC4 immunohistochemistry represents a sensitive and specific marker for this entity. Interestingly enough, expression of MUC4 is also present in up to 78% of SEFs according to another publication from the same group (Doyle 2012).
As genetic differences are emerging among members of this fibrosarcoma family, we undertook a comparative FISH analysis between pure SEF and hybrid SEF/LGFMS tumors, spanning a wide variety of clinical presentations and anatomic locations, in an attempt to delineate correlations between morphology and gene fusion type.
MATERIALS AND METHODS
The Pathology files of MSKCC and the personal consultations of the corresponding author (CRA) were searched for cases of sclerosing epithelioid fibrosarcoma (SEF) and low grade fibromyxoid sarcoma (LGFMS). Pathologic diagnosis and immunohistochemical stains were re-reviewed in all cases. Cases were included in the study if adequate material was available for MUC4 immunohistochemistry and/or FISH molecular studies. Four cases with classic morphologic features of LGFMS were included as controls. The study was approved by the Institutional Review Board 02-060.
Clinicopathologic Findings
The histologic requirement for inclusion in the study was the presence of classic morphologic appearance of SEF, namely a proliferation of predominantly epithelioid cells arranged in strands, nests and sheets, set in a fibrotic and extensively hyalinized stroma. SEF cases displaying significant nuclear pleomorphism and/or extensive areas of spindling with fascicular growth were excluded. However, focal areas of spindling, as described in the original descriptions under the term ‘fibrosarcoma-like’ areas were tolerated. With regards to the LGFMS areas, the classic morphologic picture of a bland spindle cell proliferation arranged in a swirling pattern in a myxoid and variably fibrotic background was used to define such a component. The selected tumors were then reclassified as into pure SEF and hybrid SEF/LGFMS. The presence of spindle cell areas with either fibroma-like or hypercellular fibrosarcoma-like morphologies was semi-quantitatively assessed in the pure SEF cases (focal <10%; moderate >10–25%), while in the cases with mixed features, the percentage of each SEF and LGFMS components was recorded. In addition, the presence of collagenous rosettes as well as higher-grade areas with small blue round cell tumor phenotype (SBRCT-like) was noted. Mitotic activity per 10 high power fields (HPFs) and the extent of necrosis (focal and extensive) were also recorded. Patient charts were reviewed to document recurrences, metastasis and ultimate outcome.
Immunohistochemistry for MUC4
Immunohistochemical staining for MUC4 (anti-human mouse monoclonal antibody clone 8G7, 1:2000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) was performed on 12 cases and the positive immunoreactivity was recorded taking in consideration the intensity (weak, moderate and strong) and extent (rare 1+, scattered 2+, patchy 3+, diffuse 4+) of the signal.
RNA Sequencing
In one case with frozen tissue available and adequate quality RNA, RNA sequencing was performed using the standard Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from the total RNA (2 µg) and then was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer (Illumina). To reduce the inclusion of artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library because of inefficient A-tailing reactions that lead to self ligation of blunt-ended template molecules (Quail 2008) an additional size-selection step (capturing 350–400 bp) was introduced prior to the adapter ligation step. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA 1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2500 (Illumina).
Analysis of RNA Sequencing Results with FusionSeq
All reads were independently aligned with STAR alignment software against the human genome reference sequence (hg19) and a splice junction library, simultaneously (Dobin 2013). The mapped reads were converted into Mapped Read Format (Habegger 2011) and analyzed with FusionSeq (Sboner 2010) to identify potential fusion transcripts. FusionSeq is a computational method successfully applied to paired-end RNA-seq experiments for the identification of chimeric transcripts (Tanas 2011; Pierron 2012; Mosquera 2013). Briefly, paired-end reads mapped to different genes are first used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcript candidates. Once a confident list of fusion candidates was generated, they were ranked with several statistics to prioritize the experimental validation. In these cases, we used the DASPER score (difference between the observed and analytically calculated expected SPER): a higher DASPER score indicated a greater likelihood that the fusion candidate was authentic and did not occur randomly. See reference 23 for further details about FusionSeq.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified as potential fusion partners in the RNA-seq experiment. BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu), see Supporting Information Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A positive score was interpreted when at least 20% of the nuclei showed a breakapart signal. Nuclei with incomplete set of signals were omitted from the score.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An aliquot of the RNA extracted above from frozen tissue (Trizol Reagent; Invitrogen; Grand Island, NY) was used to confirm the fusion transcript identified by FusionSeq. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). One microgram of total RNA was used for cDNA synthesis by SuperScript VR III First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA). RT-PCR was performed using the Advantage-2 PCR kit (Clontech, Mountain View, CA) for 33 cycles at a 64.5°C annealing temperature, using the following primers: EWSR1 Exon 6 Fwd: 5’–CACCTCCATCCTACCCTCCTAC–3’ and CREB3L2 Exon 7 Rev: 5’–GGACAGCCTGGAGAAAAAAGTGG–3’. Amplified products were purified and sequenced by Sanger method.
Long-range PCR
Genomic DNA was extracted from frozen tissue using the Phenol/Chloroform assay and quality was confirmed by electrophoresis. About 0.5 mg genomic DNA was amplified with the Advantage 2 PCR kit (Clontech, Mountain View, CA) in order to assess the intronic breaks. The following primer sets were used EWSR1 Intron 7 Fwd: 5’– CTCTCAGCAGAACACCTATGGG –3’ and CREB3L2 Exon 6 Rev: 5’–CTGTCAAAATCAGAGGAGAAGGC– 3’.
RESULTS
Clinicopathologic Features
A total of 18 cases were included and classified into two groups: pure SEF (10 cases) and hybrid SEF/LGFMS (8 cases) according to the above mentioned morphologic criteria. Four cases from the pure SEF group (SEF1, 2, 5 and 7) and two cases from the SEF/LGFMS hybrid group (SEF/LGFMS2 and 6) were previously reported in the SEF series by Antonescu et al. (Antonescu 2001). One case from the SEF/LGFMS hybrid group (SEF/LGFMS5) previously reported as LGFMS with giant collagenous rosettes (Woodruff 1999), was found to have areas morphologically consistent with SEF. In addition to additional immunohistochemical and cytogenetics data, updated follow up data for these cases was also included. There were a total of 11 females and 7 males. The pure SEF group showed a striking female preponderance (9 females and 1 male) and the hybrid SEF/LGFMS showing a majority of male patients (6 males and 2 females). Taking in consideration both groups the overall mean age at diagnosis was of 44 years (range 18–78). When considering the separate groups the mean age was 41 years-old for pure SEF and 47 years-old for the hybrid SEF/LGFMS. The tumors were found in deep locations which included: lower extremities, 6 (thigh, 5; buttock, 1); upper extremity, 3 (arm, 2; shoulder, 1); trunk, 4 (back/paraspinal, 2; pelvis/penis, 1; retroperitoneum, 1), and skull 2 cases. Three analyzed cases were metastases, two cases to lung and one case to iliac bone, originating from primaries in soft tissue of the neck and thigh. Both SEF and SEF/LGFMS tumors presented overall as large masses with an average tumor size of 9.2 cm (range 1.0 – 19.5 cm) with seven tumors measuring 10 cm or more.
Pure SEF Group
The tumors composing this group (n = 10) exhibited the classic morphologic features of this entity, namely a vaguely lobular proliferation of relatively uniform small to medium-size epithelioid cells with clear cytoplasm which were arranged in single files, cords, nests, irregular shaped sheets and flanked by deeply eosinophilic columns and bands of hyalinized collagen (Figures 1A–C). Six tumors showed areas of spindling with morphologic features ranging from innocuous appearing, almost fibroma-like (Figure 1D, left) to hypercellular, fibrosarcoma-like zones (Figure 1E). This latter feature recapitulates the description from the original series of this entity (1). A semi-quantitative assessment of the relative amount of this component was carried out and revealed that 40% of the tumors had a moderate degree of spindle cell component, while 20% showed only focal areas of spindling. The mitotic activity ranged from 2–7/10 HPFs (mean: 4/10 HPFs) and focal amount of necrosis was present in 3 cases.
Figure 1. Morphologic spectrum of pure SEF correlated with the fusion type.
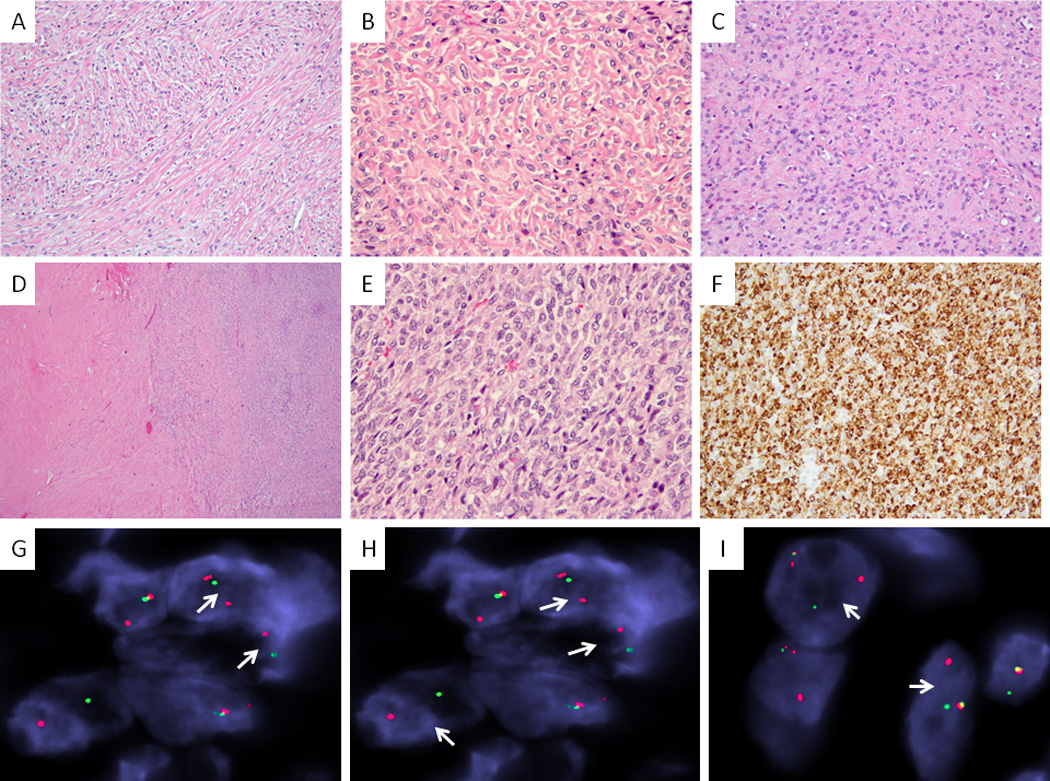
Monotonous proliferation of epithelioid cells arranged in single files and cords, separated by deeply eosinophilic columns and bands of hyalinized collagen (A. EWSR1-CREB3L1 fusion positive SEF2, 100×; B. EWSR1-CREB3L2 positive SEF3, 200×); or by a more confluent fibrotic stroma (C. EWSR1-positive SEF10, 200×). Tumors may additionally show deceptively bland, hypocellular fibroma-like areas (D, SEF8), as well as cellular components of fibrosarcoma-like zones (E, SEF3, left side, 40×). MUC4 immunoexpression is typically diffuse and strong (F, SEF3, 200×). FISH studies show EWSR1 and CREB3L1 break-apart in the majority of pure SEF (G &H, SEF4); however CREB3L2 gene rearrangements (a novel finding) were also identified (I, SEF8)(arrows, red, centromeric; green telomeric).
Follow-up clinical information was available in 9 patients, ranging from 7–141 months (mean 54 months). Three patients suffered local recurrences at a mean of 27 months from diagnosis (range 12 to 46 months). Five patients developed metastases (55%), most frequently to the lungs (4 patients), the mean time from diagnosis to first metastasis was 35 months, with a wide time interval average of detection; ranging from metastases at presentation in one patient, to 10 years in a different patient. Four patients died of disease (44%) at a mean time of 36 months after diagnosis (range 18 to 52 months)(Table 1). Three were alive with residual disease (33%) and 2 patients were disease-free after 7 and 67 months follow-up period, respectively.
Table 1.
Pure SEF Cases: Demographics, Pathologic features, FISH results and Follow-up data
| SEF # |
Age/ Sex |
Location/ Size |
Spindle Cell Component |
Recurrence (R)/ Metastases (M) |
Follow Up | MUC4 | EWSR1/ FUS |
CREB3L1/ CREB3L2 |
|---|---|---|---|---|---|---|---|---|
| 1• | 45/F | Paraspinal/10 cm | None | M Lung (0 mo) | DOD (24 mo) | Strong/+4 | EWSR1 | CREB3L1 |
| 2• | 52/F | Skull/6 cm | Moderate | R (12 mo)/M Ribs (13 mo) | DOD (52 mo) | Strong/+4 | EWSR1 | CREB3L1 |
| 3 | 28//F | Thigh/8.9 cm | Moderate | N/A | NED (7 mo) | Strong/+4 | EWSR1 | CREB3L2* |
| 4 | 28/F | Arm/2.5 cm | Focal | R (24 mo)/M Lung (24 mo) | AWD (64 mo) | Strong/+4 | EWSR1 | CREB3L1 |
| 5• | 37/F | Skull/>15 cm | Focal | R (46 mo) | AWD (65 mo) | ND | EWSR1 | CREB3L1 |
| 6 | 50/F | Met Iliac /1.0 cm | None | M Bone, Lungs (124 mo) | AWD (141 mo) | Strong/+4 | EWSR1 | CREB3L1 |
| 7• | 53/F | Arm/9 cm | None | M Lymph node, Lung (15 mo) | DOD (18 mo) | ND | EWSR1 | CREB3L1 |
| 8 | 25/F | Shoulder/4 cm | Moderate | N/A | NED (67 mo) | Strong/+4 | EWSR1 | CREB3L2 |
| 9 | 78/F | Thigh/2.5 cm | Moderate | ND | ND | ND | FUS | ND |
| 10 | 20/M | RP/13.5 cm | None | Incomplete resection | DOD (52 mo) | Negative | EWSR1 | NEG** |
Abbreviations: M: male, F: female, R: local recurrence, N/A: Not Applicable, mo: months, ND: No Data, DOD: Died Of Disease, AWD: Alive With Disease, NED: No Evidence of Disease
previously reported (Antonescu 2001);
negative by FISH, positive by RNASeq and RT-PCR;
negative also for POU5F1, PBX1, ZNF444 and FLI1 rearrangements by FISH
Hybrid SEF/LGFMS Group
When examined microscopically for relative percentages of SEF or LGFMS components, the hybrid SEF/LGFMS group of tumors (n=8) showed in half of the cases a preponderance of SEF morphology (50% or more), while the other half exhibited a dominant LGFMS component (Figure 2A–C). The mitotic count ranged from 2–7/10 HPFs (mean: 5/10 HPFs), while necrosis was present only focally in half of the cases. Three cases from this group (SEF/LGFMS 3, 5 and 8) showed giant collagenous rosettes, as seen previously described in the Hyalinizing Spindle Cell Tumor (HSCT) with giant rosettes variant of LGFMS (Figures 2D–G)(Folpe 2000; Lane 1997; Reid 2003). Two cases (SEF/LGFMS 6 and 7) exhibited solid sheets of small to intermediate-sized cells with scant cytoplasm and vesicular nuclei, reminiscent of a small blue round cell tumor (SBRCT) appearance (Figure 2C). Similar morphology was previously described in Evans’ LGFMS latest series as “de-differentiated areas” and was found in recurrent and metastatic tumors with deadly outcome (Evans 2011). Indeed in our series both patients harboring tumors with such morphology succumbed of their disease at 10 and 73 months from diagnosis.
Figure 2. Histologic and FISH findings of hybrid SEF/LGFMS.
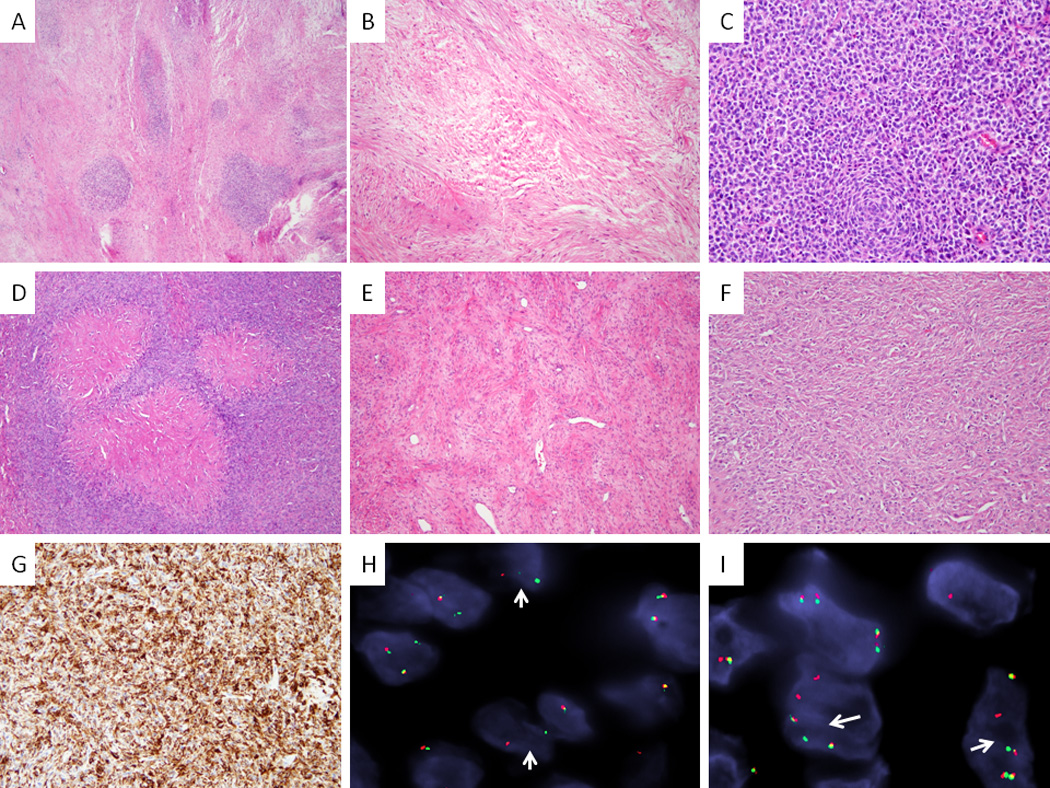
A–C. SEF7 showing predominant LGFMS background with scattered nodular areas of SEF (FUS-CREB3L2 positive, A, 40×); typical LGFMS morphology with bland spindle cells arranged in fascicles (B, 100×) and high power of SEF showing a predominant SBRCT phenotype (C, 200×). D–G. A hybrid SEF/LGFMS (SEF8), with distinctive hyalinizing giant rosettes (FUS-CREB3L2 positive, D, 100×), showing areas of classic LGFMS (E, 100×), as well as solid areas of SEF (F, 100×); MUC4 strong and diffuse immunoreactivity (G, 100×). FISH showing (H, SEF/LGFMS4) and CREB3L2 (I, SEF/LGFMS4) split-apart signals (arrows) in SEF (red, centromeric; green telomeric)
Clinical follow-up data was available for all cases in this group and ranged from 7–271 months (mean: 97 months). Three patients (37%) developed local recurrences at a mean time of 34 months from diagnosis (Table 2). Five patients (62%) exhibited metastases, with lungs being the preferred site and the mean time from diagnosis to detection metastases was 31 months. Three patients died of disease (37%) at an average of 118 months after diagnosis (range 10 to 271 months), two patients were alive with disease and three were free of disease.
Table 2.
Hybrid SEF/LGFMS Cases: Demographics, Pathologic features, FISH results and Follow-up data
| # | Age/ Sex |
Location/ Size |
%SEF vs FMS |
Recurrence (R)/ Metastasis (M) |
Follow-Up | MUC4 | EWSR1/ FUS |
CREB3L1/ CREB3L2 |
|---|---|---|---|---|---|---|---|---|
| 1 | 56/M | Thigh / 16.7 cm | 70%/30% | N/A | NED (21 mo) | Strong/+4 | FUS | CREB3L2 |
| 2◆ | 54/M | Penis / 9.7 cm | 40%/60% | N/A | NED (105 mo) | Strong/+4 | FUS | CREB3L2 |
| 3 | 18/M | Thigh / 7.5 cm | 20%/80%* | N/A | NED (7 mo) | Strong/+4 | FUS | CREB3L2 |
| 4 | 75/M | Gluteus /15 cm | 60%/40% | R (12 mo)/M Lung (12 mo) and Liver (18 mo) | AWD (23 mo) | ND | FUS | CREB3L2 |
| 5• | 41/M | Lung Met / 9 cm | 10%/90%* | M Lung (48 mo) | DOD (271 mo) | Strong/+4 | FUS | CREB3L2 |
| 6◆ | 44/F | Paraspinal/ 15 cm | 80%/20%+ | R (34 mo)/M Lung (35 mo) | DOD (73 mo) | Moderate/ +3 | FUS | CREB3L2 |
| 7 | 54/M | Thigh/19.5 cm | 80%/20%+ | R (8 mo)/M Lung (13 mo) | DOD (10 mo) | Strong/+4 | FUS | CREB3L2 |
| 8 | 35/F | Lung Met /1.3 cm | 20%/80%* | M Lung (48 mo) | AWD (266 mo) | Strong/+4 | FUS | CREB3L2 |
Abbreviations: M: male, F: female, %SEF/LGFMS: Percentage of SEF and LGFMS components, R: Local recurrence, N/A: Not Applicable, mo: months, ND: No Data, DOD: Died of Disease, AWD: Alive with disease, NED: No evidence of disease
previously reported as SEF (Antonescu 2001)
previosuly reported as LGFMS with giant collagenous rosettes (Woodruff 1999);
Cases with collagenous rosettes;
Cases with high-grade SRBCT-like areas
Immunohistochemistry for MUC4
A total of 14 cases (7 pure SEF and 7 hybrid SEF/LGFMS) were analyzed for MUC4 expression by immunohistochemistry. Thirteen of the analyzed tumors were positive with this marker (92%). Only one case from the SEF group (SEF 10) did not express MUC4 (1/7, 14%) and all of the tested SEF/LGFMS hybrid tumors were positive (7/7, 100%). The majority of the MUC4 positive cases showed strong and diffuse (+4) granular cytoplasmic immunoreactivity (Figure 1F, 2G) and only one hybrid SEF/LGFMS case exhibited moderate positivity distributed in a patchy fashion (3+).
EWSR1-CREB3L1 fusion is associated with pure SEF morphology
The presence of either FUS or EWS rearrangements was mutually exclusive. The overwhelming majority of pure SEF cases (9/10, 90%) showed EWSR1 rearrangements. Six out of nine examples of the EWSR1-rearranged lesions exhibited CREB3L1 gene break-apart by FISH (Figures 1G,H), two cases revealed a CREB3L2 rearrangements (one case by FISH, Figure 1I and the other by RNAseq, see below) and the remaining case lacked abnormalities in both genes. Since the latter case was also MUC4 negative, additional loci were interrogated using custom BAC probes (POU5F1, PBX1, ZNF444, FLI1) as previously described (Antonescu 2010) to rule out other entities such as a myoepithelial tumor and Ewing sarcoma, all with negative results. The only SEF case investigated by RNAseq (SEF 3) was selected due to a positive EWSR1 rearrangement result, but negative FISH results for both CREB3L1/2 gene abnormalities. However, the FusionSeq identified an EWSR1-CREB3L2 fusion candidate (Figure 3). The false negative FISH result detected was most likely secondary to a complex / cryptic translocation. Only one case from this group showed a FUS gene rearrangement, however, no further material was available to assess the CREB3L1/2 break-apart status. It was noted that no morphologic feature, including the presence and/or amount of spindle cell component, correlated with the type of rearrangement found in these tumors.
Figure 3.
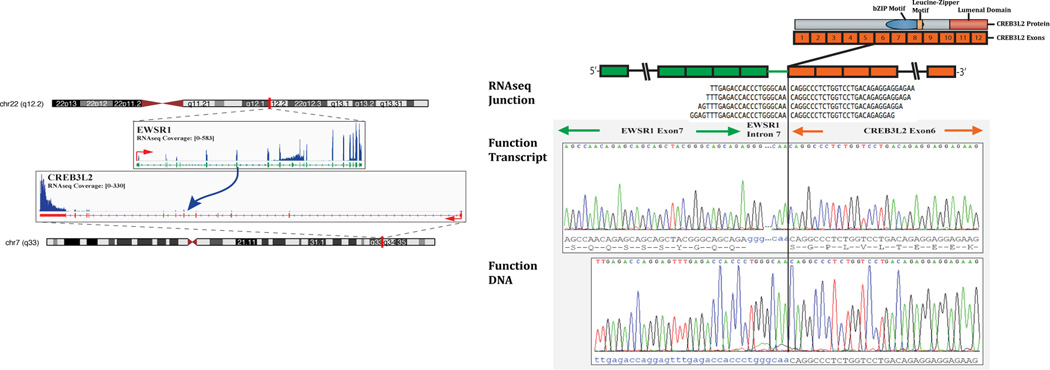
Schematic representation indicating the EWSR1 locus located on chromosome 22 is joined to CREB3L2, situated on chromosome 7, resulting in a t(7;22)(q33;q12.2) translocation, as detected by RNAseq (left panel). RNA reads covering the fusion junction were isolated independent to the FusionSeq analysis work flow, supporting the EWSR1-CREB3L2 fusion candidate (upper right image); RT-PCR experimental validation of the fusion shows the junction sequence between EWSR1 exon 7 and CREB3L2 exon 6 (middle right panel), with an intervening short fragment of EWSR1 intron 7. The retained 3’ portion of CREB3BL2 preserve the functional domains of the protein (basic ZIP, leucine Zipper motif, luminal C terminus domain). DNA PCR further confirms the DNA break between intron 7 of EWSR1 and intron 5 of CREB3L2 (bottom right image).
Hybrid SEF/LGFMS tumors exclusively exhibit FUS and CREB3L2 rearrangements
In contrast to the pure SEF group, the hybrid SEF/LGFMS tumors showed the presence of a FUS-CREB3L2 fusion in all cases tested. Similarly, most cases from the control LGFMS group showed that the most prevalent abnormalities were FUS and CREB3L2 gene rearrangements (Figures 2H,I), present in three of the cases, in keeping with the previously reported literature. Unexpectedly, one of these cases with classic LGFMS morphology, presenting in the kidney of an infant exhibited EWSR1 and CREB3L1 rearrangements. The findings related to this particular case were recently reported elsewhere (Rubinstein 2014), emphasizing the potential diagnostic pitfall of an EWSR1-positive FISH result, due to the large morphologic spectrum of EWSR1-positive tumors in this age group.
DISCUSSION
Herein, we report a series of 18 SEF cases, 10 with pure SEF morphology and 8 exhibiting SEF/LGFMS hybrid features with confirmed genetic abnormalities. As previously described (Meis-Kindblom 1995; Antonescu 2001; Rekhi 2011; Doyle 2012; Arbajian 2014), these tumors arose predominantly in the deep soft tissue locations of the limbs and trunk of young to middle aged individuals (mean 44 years old). Of interest, there was a striking female predisposition noted in the lesions with pure SEF morphology (90% of patients), feature that has not been documented previously in the largest SEF series published to date (Meis-Kindblom 1995; Doyle 2012; Wang 2012), except for a slight female tendency in Antonescu et al. (Antonescu 2001) SEF cohort, in which 62% of the patients were female.
On histologic evaluation, the pure SEF tumors showed areas with spindle cell morphology at least focally in 60% of the cases. Considering both groups, the tumors showed a relatively low mitotic count (mean 4–5 mitoses/10 HPF) and less than half of cases showed areas of necrosis. The overall clinical behavior spanning both cohorts demonstrated a mortality rate of 41%, while an additional 30% of the patients were alive with local recurrences and/or metastases. The mortality rate was slightly higher in patients with pure SEF (44%), compared to the hybrid SEF/LGFMS group (37%). In the hybrid SEF/LGFMS cohort, 2 cases showing the presence of zones of SBRCT-like morphology (referred as ‘dedifferentiated areas’ in Evans’ latest publication on his LGFMS cohort (Evans 2011)), were associated with a fatal outcome (SEF/LGFMS 6 and 7).
Fluorescence in situ hybridization (FISH) analysis using custom break-apart probes interrogating the EWSR1, FUS, CREB3L1 and CREB3L2 loci revealed that a great majority of the pure SEF tumors (83%) exhibited rearrangements of EWSR1, which was accompanied by CREB3L1 break-apart in all except two cases, showing an EWSR1-CREB3L2 fusion instead. To our knowledge, this fusion variant (EWSR1-CREB3L2) was not reported before in any soft tissue tumor. Only one example from the pure SEF group (10%) showed a FUS rearrangement. Although this particular case exhibited histologically a moderate amount of spindling the tumor was extensively sampled and no LGFMS component was found. Since a moderate amount of spindling was also present in 60% of the EWSR1 rearranged tumors, we concluded that probably there is no correlation between the presence of spindle cell component and the type of rearrangement in an otherwise typical SEF. In contrast, the hybrid SEF/LGFMS group showed the presence of FUS-CREB3L2 fusion in all cases tested, which also represents the cytogenetic aberration most frequently found in LGFMS (Panagopoulos 2004; Matsuyama 2006; Doyle 2011).
The presence of EWSR1 rearrangements in SEF was first recognized by Doyle et al. (Doyle 2012) and subsequently highlighted in a short communication (Doyle and Hornick 2013) after the report by Lau et al. (Lau 2013) of two LGFMS cases harboring EWSR1-CREB3L1 fusions. This was followed by recent study by Arbajian et al.(Arbajian 2014) composed of a total of 15 cases of both morphologically “pure” SEFs (10 cases) and SEF/LGFMS hybrid tumors (5 cases) explored the prevalence of EWSR1, FUS, CREB3L1 and CREB3L2 rearrangements in this group of tumors with a combination of RT-PCR and FISH techniques in an effort to better characterize the shared pathogenesis between SEF and LGFMS. Their results show a bona fide recurrent EWSR1-CREB3L1 fusion/rearrangement by either RT-PCR or FISH in four of their SEF cases (4/10), while the remainder SEF cases (6/10) revealed either loss of 3’ EWSR1 locus (3/10), an altogether hemizygous deletion of EWSR1 (2/10) or a negative FISH and RT-PCR result for all loci (EWSR1, FUS and CREB3L1/2) in one case (1/10). Only one of their SEF cases exhibited a hemizygous deletion of FUS with a concomitant EWSR1 hemizygous deletion and loss of 5’ signal of CREB3L1. Additionally, 5/7 SEF interrogated cases were positive for CREB3L1 rearrangements by FISH and two were negative. There were no CREB3L2 gene alterations identified in the 5/10 cases tested, this is in contrast with our findings, where two EWSR1 rearranged SEF exhibited a CREB3L2 rearrangement (2/10, 20%), a previously unreported finding. Interestingly, one of their SEF cases was positive for EWSR1-CREB3L1 only by RT-PCR, while being negative by FISH (FUS, EWSR1 and CREB3L1/L2). This phenomenon is similar to our CREB3L2 false negative FISH result in SEF 3. Overall, our results are quite similar; especially with regards to EWSR1 rearrangements/aberrations predominantly identified in pure SEF cases (9 of the 10 cases), while the hybrid SEF/LGFMS lesions were exclusively characterized by FUS-CREB3L2 fusion transcripts by RT-PCR. These findings led the authors to conclude that pure SEF and LGFMS (either pure or in mixed histologies) are indeed separate entities.
The soft tissue tumors that harbor aberrations involving EWSR1 rearrangements are legion (Antonescu and Dal Cin 2014; Fisher 2014). This is comprehensible since EWSR1 product has a wide range of functions, from controlling microtubules during mitotic and meiotic processes (Azuma 2007; Li 2007; Leemann-Zakaryan 2009) through DNA repair (Li 2007) to a variety of features related to RNA, such as the capacity of RNA to bind proteins as well as regulation of RNA-splicing among other functions (Bertolotti 1996; Bertolotti 1998). EWSR1, FUS (a.k.a. TLS) and TAF15 belong to the TET family of transcription factors, sharing an RNA binding domain (Delattre 1992; Stolow and Haynes 1995; Bertolotti 1996; Riggi 2007). This similarity in function between EWSR1 and FUS is evidenced by the plethora of soft tissue tumors that can interchangeably exhibit fusions involving EWSR1 and FUS, namely angiomatoid fibrous histiocytoma, myxoid liposarcoma and Ewing sarcoma. This fact could also partially explain that SEF and LGFMS can exhibit the same morphology independently from their association with either EWSR1 or FUS rearrangements. On the other hand, the presence of CREB3L1/2 complex in the SEF/LGFMS spectrum of tumors appears to be constant, suggesting a pivotal role in the development of these tumors. CREB3L1 and CREB3L2, also known as OASIS and BBF2H7, respectively, form the OASIS group of transcription factors, which is composed of basic leucine zipper (bZIP) transmembrane transcription factors. Their function includes overcoming failure of the endoplasmic reticulum’s (ER) adaptive capacity, which may result in accumulation of unfolded or malfolded proteins in the ER lumen (ER stress) (Kondo 2007, 2011) and this group was recently implicated in chondrocyte development along with Sox9 (Hino 2014).
MUC4 is a glycoprotein secreted by a variety of glandular epithelia that belongs to the same group as EMA (MUC1) can also be found to be overexpressed in a wide range of adenocarcinomas (Bafna 2008). When up-regulated, the tumorigenic effect of MUC4 is thought to be elicited through interactions with ERBB2 (HER2) to enhance the proliferation, motility, and tumorigenic capacity of epithelial cancer and fibroblastic cells (Bafna 2008, 2010). Interestingly enough, LGFMS are also known to express EMA (MUC1) (Guillou 2007). One could speculate that the acquisition of an epithelioid morphology (in SEF) as well as expression of this epithelial marker by these soft tissue entities is perhaps related to underlying mesenchymal-to-epithelial transition (MET) mechanisms (Yang 2014). Despite their apparent genetic dichotomy, both SEF and LGFMS share their expression of MUC4 by immunohistochemistry, which has been considered to be a quite sensitive and specific marker for these two groups of tumors, particularly for LGFMS (Doyle 2011) while 78% of SEF are MUC4 positive (Doyle 2012). Intriguingly, a rare case report of a translocation positive, MUC4-negative LGFMS was recently published (Linos 2014). Only one EWSR1-rearranged pure SEF was negative for MUC4 staining in our series, while all the SEF/LGFMS hybrid tumors tested expressed MUC4. These results sum further evidence of the validity of this marker as a sensitive and specific tool in the diagnosis of this particular group of soft tissue tumors.
In addition to further delineate the different groups composing the SEF/LGFMS spectrum, we encounter some potential important diagnostic caveats when faced with these neoplasms. Although distinctive, it is recognized that the morphologic features of SEF can overlap with other tumors with epithelioid/round cell morphology, such as poorly differentiated carcinomas, some lymphomas, SBRCTs and myoepithelial tumors. A similar situation applies to LGFMS, which can easily be mistaken for desmoid-type fibromatosis and soft tissue perineurioma among other entities. In selected cases the distinction of SEF from both myoepithelial tumors and SBRCTs is further complicated if one were solely rely on the presence of EWSR1 rearrangement, without interrogating the CREB3L1/2 status and/or immunohistochemistry for MUC4. It appears that one should not always confidently rely on MUC4 expression since according to Doyle et al. series, 1/10 of their tested myoepithelial tumors was strongly MUC4 positive (Doyle 2012). For these reasons, if stringent classification is needed, interrogation of FLI1 and POU5F1 gene rearrangement status might be carried out to further characterize ambiguous cases lacking CREB3L1/2 gene abnormalities. However, due to the rarity of these entities, it is likely that this situation will be only rarely encountered, with the majority of cases needing EWSR1 and/or FUS FISH and MUC4 immunohistochemistry for correct classification. Another caveat could emerge when facing a possible false negative FISH result, which can occur secondary to a cryptic rearrangement, such as seen in one of our pure SEF cases (SEF 3), which despite a classic SEF morphology and MUC4 positivity, lacked any FISH abnormalities in CREB3L1, to be further characterized by the sensitive RNAseq assay as having an EWSR1-CREB3L1 fusion. As discussed above, Arbajian et al. were faced with a similar situation in one of their pure SEF cases (Arbajian 2014). An additional potential pitfall could stem from the highly hyalinized eosinophilic matrix of SEF being potentially mistaken for osteoid. When confronted with problematic cases arising from bone, in addition to MUC4 immunostaining and cytogenetic findings one could resort to immunohistochemistry for SATB2, a recently described marker for osteoblastic differentiation from the Special AT-Binding Protein family (Wojcik 2014).
In summary, our results using gene specific FISH probes for interrogating the genetic abnormalities in the SEF/LGFMS spectrum, confirm results from previous smaller studies mainly using RT-PCR technology (Rekhi 2011; Doyle 2012; Wang 2012; Arbajian 2014) of a cytogenetic demarcation between pure SEF (mostly EWSR1 rearranged) from LGFMS variants (mainly FUS rearranged). Nevertheless, significant morphologic, immunohistochemical and cytogenetic overlap still exists in a minority of cases. One could propose that, similarly to other soft tissue entities, such as dermatofibrosarcoma protuberans and giant cell fibroblastoma (Dal Cin 1996; Pedeutour 1996), as well as myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor (Antonescu 2011), among others, SEF and LGFMS may represent potentially related entities, possibly at opposite ends of the spectrum of the same disease process. This observation will require further research investigation to detail underlying molecular mechanisms that may be driving these tumors.
Supplementary Material
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA), Cycle for Survival (CRA), Kristin Ann Carr Foundation (CRA).
REFERENCES
- Antonescu CR, Dal Cin P. Promiscuous genes involved in recurrent chromosomal translocations in soft tissue tumours. Pathology. 2014;46:105–112. doi: 10.1097/PAT.0000000000000049. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Rosenblum MK, Pereira P, Nascimento AG, Woodruff JM. Sclerosing epithelioid fibrosarcoma: a study of 16 cases and confirmation of a clinicopathologically distinct tumor. Am J Surg Pathol. 2001;25:699–709. doi: 10.1097/00000478-200106000-00001. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Nielsen GP, Rosenberg AE, Dal Cin P, Fletcher CD. Consistent t(1;10) with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. Genes Chromosomes Cancer. 2011;50:757–764. doi: 10.1002/gcc.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbajian E, Puls F, Magnusson L, Thway K, Fisher C, Sumathi VP, Tayebwa J, Nord KH, Kindblom LG, Mertens F. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am J Surg Pathol. 2014;38:801–808. doi: 10.1097/PAS.0000000000000158. [DOI] [PubMed] [Google Scholar]
- Azuma M, Embree LJ, Sabaawy H, Hickstein DD. Ewing sarcoma protein EWSR1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS One. 2007;2:e979. doi: 10.1371/journal.pone.0000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafna S, Singh AP, Moniaux N, Eudy JD, Meza JL, Batra SK. MUC4, a multifunctional transmembrane glycoprotein, induces oncogenic transformation of NIH3T3 mouse fibroblast cells. Cancer Res. 2008;68:9231–9238. doi: 10.1158/0008-5472.CAN-08-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. Embo j. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin P, Sciot R, de Wever I, Brock P, Casteels-Van Daele M, Van Damme B, Van Den Berghe H. Cytogenetic and immunohistochemical evidence that giant cell fibroblastoma is related to dermatofibrosarcoma protuberans. Genes Chromosomes Cancer. 1996;15:73–75. doi: 10.1002/(SICI)1098-2264(199601)15:1<73::AID-GCC10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle LA, Hornick JL. EWSR1 rearrangements in sclerosing epithelioid fibrosarcoma. Am J Surg Pathol. 2013;37:1630–1631. doi: 10.1097/PAS.0b013e3182a05a6b. [DOI] [PubMed] [Google Scholar]
- Doyle LA, Moller E, Dal Cin P, Fletcher CD, Mertens F, Hornick JL. MUC4 is a highly sensitive and specific marker for low-grade fibromyxoid sarcoma. Am J Surg Pathol. 2011;35:733–741. doi: 10.1097/PAS.0b013e318210c268. [DOI] [PubMed] [Google Scholar]
- Doyle LA, Wang WL, Dal Cin P, Lopez-Terrada D, Mertens F, Lazar AJ, Fletcher CD, Hornick JL. MUC4 is a sensitive and extremely useful marker for sclerosing epithelioid fibrosarcoma: association with FUS gene rearrangement. Am J Surg Pathol. 2012;36:1444–1451. doi: 10.1097/PAS.0b013e3182562bf8. [DOI] [PubMed] [Google Scholar]
- Evans HL. Low-grade fibromyxoid sarcoma. A report of two metastasizing neoplasms having a deceptively benign appearance. Am J Clin Pathol. 1987;88:615–619. doi: 10.1093/ajcp/88.5.615. [DOI] [PubMed] [Google Scholar]
- Evans HL. Low-grade fibromyxoid sarcoma. A report of 12 cases. Am J Surg Pathol. 1993;17:595–600. doi: 10.1097/00000478-199306000-00007. [DOI] [PubMed] [Google Scholar]
- Evans HL. Low-grade fibromyxoid sarcoma: a clinicopathologic study of 33 cases with long-term follow-up. Am J Surg Pathol. 2011;35:1450–1462. doi: 10.1097/PAS.0b013e31822b3687. [DOI] [PubMed] [Google Scholar]
- Fisher C. The diversity of soft tissue tumours with EWSR1 gene rearrangements: a review. Histopathology. 2014;64:134–150. doi: 10.1111/his.12269. [DOI] [PubMed] [Google Scholar]
- Folpe AL, Lane KL, Paull G, Weiss SW. Low-grade fibromyxoid sarcoma and hyalinizing spindle cell tumor with giant rosettes: a clinicopathologic study of 73 cases supporting their identity and assessing the impact of high-grade areas. Am J Surg Pathol. 2000;24:1353–1360. doi: 10.1097/00000478-200010000-00004. [DOI] [PubMed] [Google Scholar]
- Guillou L, Benhattar J, Gengler C, Gallagher G, Ranchere-Vince D, Collin F, Terrier P, Terrier-Lacombe MJ, Leroux A, Marques B, Aubain Somerhausen Nde S, Keslair F, Pedeutour F, Coindre JM. Translocation-positive low-grade fibromyxoid sarcoma: clinicopathologic and molecular analysis of a series expanding the morphologic spectrum and suggesting potential relationship to sclerosing epithelioid fibrosarcoma: a study from the French Sarcoma Group. Am J Surg Pathol. 2007;31:1387–1402. doi: 10.1097/PAS.0b013e3180321959. [DOI] [PubMed] [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K, Saito A, Kido M, Kanemoto S, Asada R, Takai T, Cui M, Cui X, Imaizumi K. Master regulator for chondrogenesis, Sox9, Regulates Transcriptional Activation of the ER Stress Transducer BBF2H7/CREB3L2 in Chondrocytes. J Biol Chem. 2014;289:13810–13820. doi: 10.1074/jbc.M113.543322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Saito A, Asada R, Kanemoto S, Imaizumi K. Physiological unfolded protein response regulated by OASIS family members, transmembrane bZIP transcription factors. IUBMB Life. 2011;63:233–239. doi: 10.1002/iub.433. [DOI] [PubMed] [Google Scholar]
- Kondo S, Saito A, Hino S, Murakami T, Ogata M, Kanemoto S, Nara S, Yamashita A, Yoshinaga K, Hara H, Imaizumi K. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol. 2007;27:1716–1729. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane KL, Shannon RJ, Weiss SW. Hyalinizing spindle cell tumor with giant rosettes: a distinctive tumor closely resembling low-grade fibromyxoid sarcoma. Am J Surg Pathol. 1997;21:1481–1488. doi: 10.1097/00000478-199712000-00011. [DOI] [PubMed] [Google Scholar]
- Lau PP, Lui PC, Lau GT, Yau DT, Cheung ET, Chan JK. EWSR1-CREB3L1 gene fusion: a novel alternative molecular aberration of low-grade fibromyxoid sarcoma. Am J Surg Pathol. 2013;37:734–738. doi: 10.1097/PAS.0b013e31827560f8. [DOI] [PubMed] [Google Scholar]
- Leemann-Zakaryan RP, Pahlich S, Sedda MJ, Quero L, Grossenbacher D, Gehring H. Dynamic subcellular localization of the Ewing sarcoma proto-oncoprotein and its association with and stabilization of microtubules. J Mol Biol. 2009;386:1–13. doi: 10.1016/j.jmb.2008.12.039. [DOI] [PubMed] [Google Scholar]
- Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, O'Shea J, Lee SB. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linos K, Bridge JA, Edgar MA. MUC 4-negative FUS-CREB3L2 rearranged low-grade fibromyxoid sarcoma. Histopathology. 2014 doi: 10.1111/his.12422. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Hisaoka M, Shimajiri S, Hayashi T, Imamura T, Ishida T, Fukunaga M, Fukuhara T, Minato H, Nakajima T, Yonezawa S, Kuroda M, Yamasaki F, Toyoshima S, Hashimoto H. Molecular detection of FUS-CREB3L2 fusion transcripts in low-grade fibromyxoid sarcoma using formalin-fixed, paraffin-embedded tissue specimens. Am J Surg Pathol. 2006;30:1077–1084. doi: 10.1097/01.pas.0000209830.24230.1f. [DOI] [PubMed] [Google Scholar]
- Meis-Kindblom JM, Kindblom LG, Enzinger FM. Sclerosing epithelioid fibrosarcoma. A variant of fibrosarcoma simulating carcinoma. Am J Surg Pathol. 1995;19:979–993. doi: 10.1097/00000478-199509000-00001. [DOI] [PubMed] [Google Scholar]
- Mertens F, Fletcher CD, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- Moller E, Hornick JL, Magnusson L, Veerla S, Domanski HA, Mertens F. FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. Clin Cancer Res. 2011;17:2646–2656. doi: 10.1158/1078-0432.CCR-11-0145. [DOI] [PubMed] [Google Scholar]
- Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, Wexler LH, LaQuaglia MP, Edelman M, Sreekantaiah C, Rubin MA, Antonescu CR. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odem JL, Oroszi G, Bernreuter K, Grammatopoulou V, Lauer SR, Greenberg DD, Vogler CA, Batanian JR. Deceptively benign low-grade fibromyxoid sarcoma: array-comparative genomic hybridization decodes the diagnosis. Hum Pathol. 2013;44:145–150. doi: 10.1016/j.humpath.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Storlazzi CT, Fletcher CD, Fletcher JA, Nascimento A, Domanski HA, Wejde J, Brosjo O, Rydholm A, Isaksson M, Mandahl N, Mertens F. The chimeric FUS/CREB3l2 gene is specific for low-grade fibromyxoid sarcoma. Genes Chromosomes Cancer. 2004;40:218–228. doi: 10.1002/gcc.20037. [DOI] [PubMed] [Google Scholar]
- Pedeutour F, Simon MP, Minoletti F, Barcelo G, Terrier-Lacombe MJ, Combemale P, Sozzi G, Ayraud N, Turc-Carel C. Translocation, t(17;22)(q22;q13), in dermatofibrosarcoma protuberans: a new tumor-associated chromosome rearrangement. Cytogenet Cell Genet. 1996;72:171–174. doi: 10.1159/000134178. [DOI] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R, de Silva MV, Paterson L, Ryan E, Fisher C. Low-grade fibromyxoid sarcoma and hyalinizing spindle cell tumor with giant rosettes share a common t(7;16)(q34;p11) translocation. Am J Surg Pathol. 2003;27:1229–1236. doi: 10.1097/00000478-200309000-00006. [DOI] [PubMed] [Google Scholar]
- Rekhi B, Folpe AL, Deshmukh M, Jambhekar NA. Sclerosing epithelioid fibrosarcoma - a report of two cases with cytogenetic analysis of FUS gene rearrangement by FISH technique. Pathol Oncol Res. 2011;17:145–148. doi: 10.1007/s12253-010-9277-3. [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Suva ML, Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins. Part 1: The fellowship of TET. J Pathol. 2007;213:4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- Rubinstein J, Visa A, Zhang L, Antonescu CR, Christison-Lagay ER, Morotti RA. Primary Low Grade Fibromyxoid Sarcoma of the Kidney in a Child, with the Alternative EWSR1-CREB3L1 Gene Fusion. Pediatr Dev Pathol. 2014 doi: 10.2350/14-05-1487-CR.1. [DOI] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolow DT, Haynes SR. Cabeza, a Drosophila gene encoding a novel RNA binding protein, shares homology with EWS and TLS two genes involved in human sarcoma formation. Nucleic Acids Res. 1995;23:835–843. doi: 10.1093/nar/23.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Wang WL, Evans HL, Meis JM, Liegl-Atzwanger B, Bovee JV, Goldblum JR, Billings SD, Rubin BP, Lopez-Terrada D, Lazar AJ. FUS rearrangements are rare in 'pure' sclerosing epithelioid fibrosarcoma. Mod Pathol. 2012;25:846–853. doi: 10.1038/modpathol.2011.214. [DOI] [PubMed] [Google Scholar]
- Wojcik JB, Bellizzi AM, Dal Cin P, Bredella MA, Fletcher CD, Hornicek FJ, Deshpande V, Hornick JL, Nielsen GP. Primary Sclerosing Epithelioid Fibrosarcoma of Bone: Analysis of a Series. Am J Surg Pathol. 2014 doi: 10.1097/PAS.0000000000000265. [DOI] [PubMed] [Google Scholar]
- Woodruff JM, Antonescu CR, Erlandson RA, Boland PJ. Low-grade fibrosarcoma with palisaded granulomalike bodies (giant rosettes): report of a case that metastasized. Am J Surg Pathol. 1999;23:1423–1428. doi: 10.1097/00000478-199911000-00015. [DOI] [PubMed] [Google Scholar]
- Yang J, Du X, Wang G, Sun Y, Chen K, Zhu X, Lazar AJ, Hunt KK, Pollock RE, Zhang W. Mesenchymal to epithelial transition in sarcomas. Eur J Cancer. 2014;50:593–601. doi: 10.1016/j.ejca.2013.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


