Non-viral gene delivery using synthetic cationic polymeric vectors is widely recognized as an attractive alternative to viral gene delivery which suffers from inherent immunogenicity and various side effects.[1] The transfection efficiency and chemotoxicity of these polymeric vectors are often closely related to their cationic charge density.[2] Materials with low charge density usually show low toxicity but are often poor transfection agents. Polycations with high charge density could mediate effective gene transfer which is however often associated with significant, charge-induced toxicity. [3] When modified with various charge-reducing moieties, including saccharides,[4] hydrocarbons,[5] and poly(ethylene glycol) (PEG),[6] polycations often benefit from improved safety profiles while in the meantime suffer from significantly reduced gene delivery capabilities. In addition to the charge-induced toxicity, excessive positive charges on polycations would also enhance the electrostatic attraction with the nucleic acids to restrict intracellular gene release.[3g,7] Therefore, it would be of great interest to develop a highly charged polycation that possesses full transfection capacity and membrane activity during the course of gene transfer, but can be triggered to transform to a less charged or uncharged material with low membrane activity post-transfection, such that intracellular DNA unpackaging can be facilitated and toxicity can be reduced.[8]
Cell penetrating peptides (CPPs), notable for their excellent membrane activities, have been developed and used in drug and gene delivery.[9] Helical structure is often observed in CPPs or formed in CPPs during membrane transduction, and has been tied to their membrane activity.[10] Mechanistically, the helical CPP presents a rigid amphiphilic structure that interacts with and destabilizes lipid bilayers, creating transient pathways to facilitate the passive diffusion of exogenous materials.[9a] Well known examples of CPPs include oligoarginine, HIV-TAT, and penetratin. Despite their excellent membrane permeability, CPPs are often too short (10-25 peptide residues) and lack adequate cationic charge to efficiently condense and deliver genes by themselves. As such, CPPs often serve as membrane-active ligands incorporated or conjugated to delivery vehicles to improve delivery efficiencies.[11]
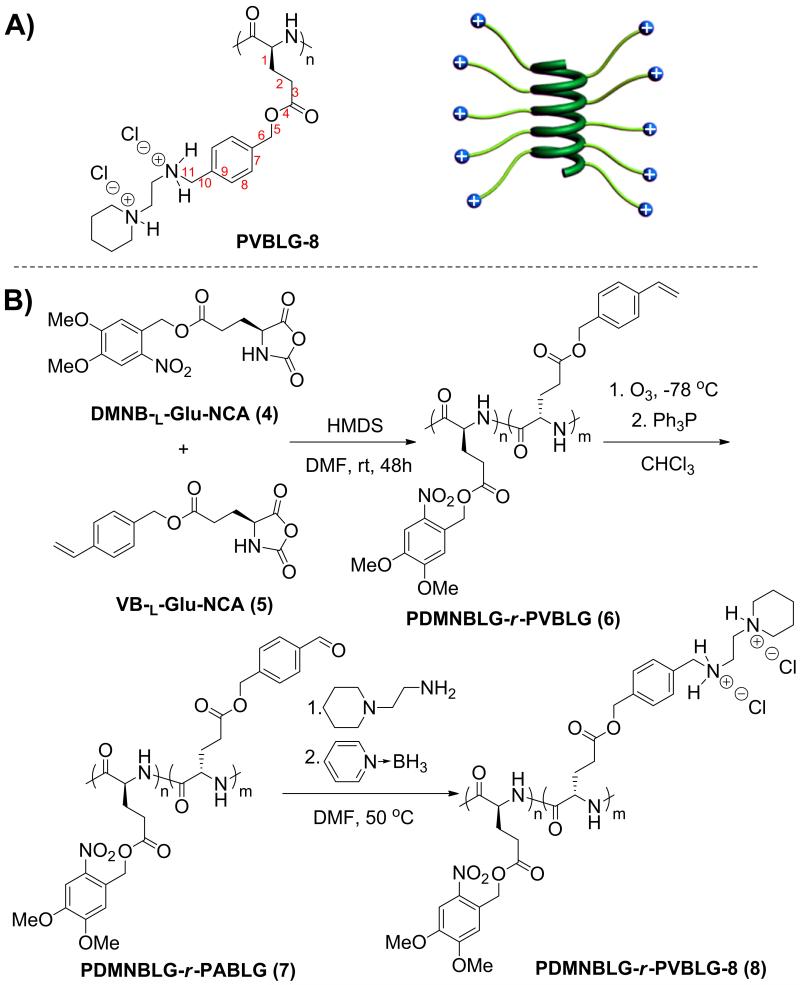
We recently developed high molecular weight (MW), cationic, cell-penetrating, α-helical polypeptides, termed PVBLG-8 (Scheme 1A).[12] By maintaining a minimum separation distance of 11 σ-bonds between the backbone and the side chain charge, the helical structure of PVBLG-8 is stabilized via increased hydrophobic interaction of the side chains and reduced side-chain charge repulsion. Because of the high charge density and higher MW as compared to traditional CPPs, PVBLG-8 can condense and deliver DNA to mammalian cells much more effectively, making it a better gene delivery vector.[12b] However, PVBLG-8 shows notable cytotoxicity at high concentrations and therefore shares the same concerns as many other polycations. In consistence with previous reports on the correlation between cytotoxicity and helicity of CPPs, the helical structure of PVBLG-8 also contributes to its observed toxicity. Moreover, because of its high cationic charge density, PVBLG-8 also suffers from inefficient DNA release. Thus, in attempts to reduce material toxicity as well as facilitate intracellular DNA release, we sought a strategy to reduce both the charge density and the helical content of PVBLG-8 at the post-transfection state. Here, we report the design of cationic α-helical poly(γ-(4,5-dimethoxy-2-nitrobenzyl)-l-glutamate)-r-PVBLG-8 (PDMNBLG-r-PVBLG-8, Scheme 1B) which maintains high membrane activity during the course of transfection due to high charge density and helical contents, while transforms to a toxicity-reduced and DNA-repelling state with distorted helix and diminished cationic charge density post-transfection in response to external triggers.
Scheme 1.
(A) Schematic illustration of cationic helical PVBLG-8. (B) Synthetic route of PDMNBLG-r-PVBLG-8.
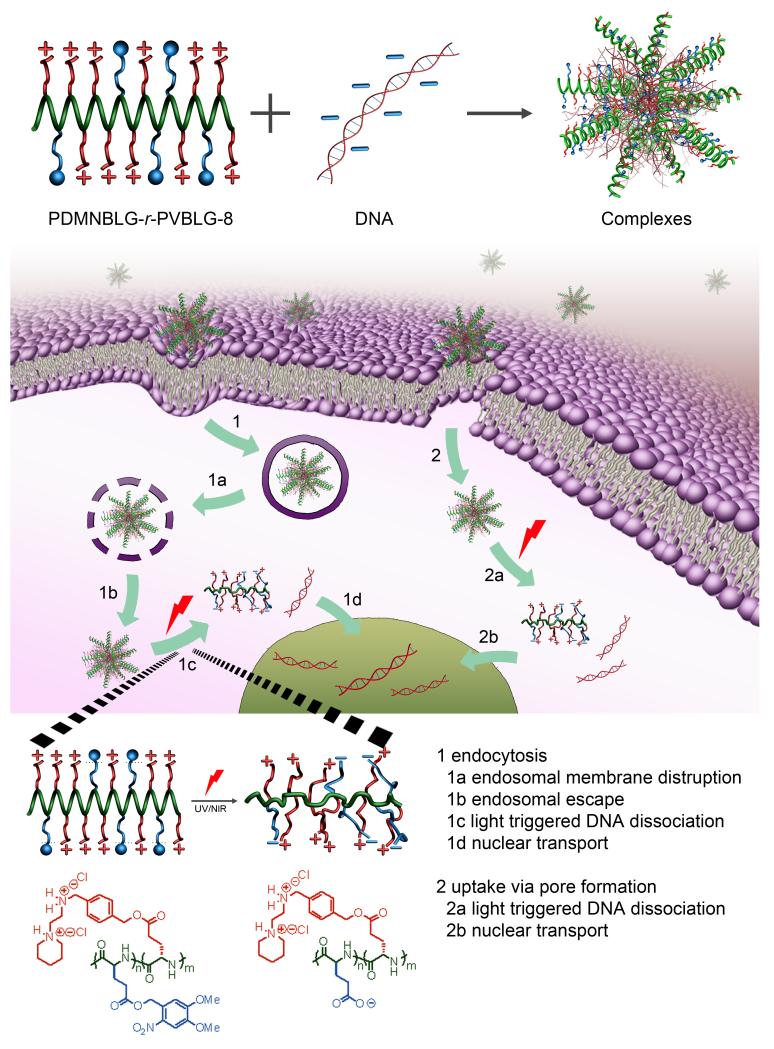
PVBLG-8 contains stable pendant benzyl ester bonds that are difficult to cleave under physiological conditions, thus prohibiting the conversion of the material into the desired non-toxic, negatively charged, random-coiled poly(glutamic acid). By incorporating various amounts of light-sensitive 4,5-dimethoxy-2-nitrobenzyl-glutamate (DMNBLG) into PVBLG-8 to make PDMNBLG-r-PVBLG-8 random copolymers, we could enable the photonic manipulation of the material toxicity and gene release profiles post-transfection. Because the DMNBLG residues are uncharged and hydrophobic, the polycationic nature and helical structure would be well maintained in PDMNBLG-r-PVBLG-8 to exert membrane activity. When a photonic stimulus is applied post-transfection, the PDMNBLG domain would yield pendant carboxylate groups (blue segment of the illustration and chemical structure in Fig. 1) via light-triggered de-esterification, and the polypeptide would thus have much reduced cationic charge density. The intramolecular electrostatic attraction between the negatively charged carboxylate groups and the positively charged amine side groups of the original PVBLG-8 would transform the helical conformation of the parental polypeptide to the helix-disrupted conformation of the light-treated polypeptide. Collectively, the light treatment of PDMNBLG-r-PVBLG-8 would lead to a net effect of reducing the material cytotoxicity and promoting intracellular gene release (Fig. 1). While light-enhanced gene transfection has been reported either via charge-switching multivalency [8a,8b] or supramolecular recognition, [8d] the current study provides a novel strategy to modulate the gene transfection and cytotoxicity by regulating the polypeptide helicity.
Figure 1.
Intracellular kinetics of PDMNBLG-r-PVBLG-8/DNA complexes, including uptake via endocytosis and passive diffusion, endosomal escape via membrane destabilization, light-triggered charge and secondary structure conversion of the polypeptide, subsequently facilitated intracellular DNA unpackaging, and nuclear transport.
To test the above-mentioned design, photo-responsive PDMNBLG-r-PVBLG-8 with a fixed degree of polymerization of 200 and various DMNBLG molar contents (10%, 20%, 30%, and 40%, designated as P10, P20, P30, and P40, respectively) were synthesized via ring-opening copolymerization of DMNB-l-Glu-NCA and VB-l-Glu-NCA. Side chains of the resulting PDMNBLG-r-PABLG were aminated to yield PDMNBLG-r-PVBLG-8 (Scheme 1B). In addition, P0, a non-responsive control polymer containing no DMNBLG (P0 = PVBLG-8) was also prepared.[12b] Hexamethyldisilazane (HMDS) as the initiator ensured well-controlled polymerization, evidenced by the monomodal GPC curves (Supplementary Fig. S2), well-defined MWs, and low polydispersity index (PDI < 1.1, Table 1).[13] All synthesized polypeptides exhibited excellent solubility in aqueous solution at pH < 9 and adopted α-helical conformations (Fig. 2A). The helicities were as high as 90% (Supplementary Fig. S6) and were remarkably stable against pH change between 1 and 9 (Fig. 2B), demonstrating that addition of up to 40 mol% DMNB-l-Glu residues in the random copolymer did not affect the helical conformation of PVBLG-8.
Table 1.
Properties of PDMNBLG-r-PVBLG-8
| Name | M/I a | Mn (Mn*) ×103 b,c | PDI c | Composition d |
|---|---|---|---|---|
| P0 | (200+0)/1 | 47.7 (49.0) | 1.08 | PVBLG-8194 |
| P10 | (180+20)/1 | 47.9 (50.6) | 1.02 | PDMNBLG19-r-PVBLG-8170 |
| P20 | (160+40)/1 | 57.2 (52.2) | 1.02 | PDMNBLG44-r-PVBLG-8175 |
| P30 | (140+60)/1 | 55.6 (53.8) | 1.03 | PDMNBLG62-r-PVBLG-8145 |
| P40 | (120+80)/1 | 52.7 (55.4) | 1.06 | PDMNBLG76-r-PVBLG-8114 |
Feed ratio of (DMNB-l-Glu-NCA+VB-l-Glu-NCA)/HMDS.
Obtained MW for PDMNBLG-r-PVBLG (expected MW*).
Obtained MW and PDI were determined by GPC.
The composition was determined by 1H NMR.
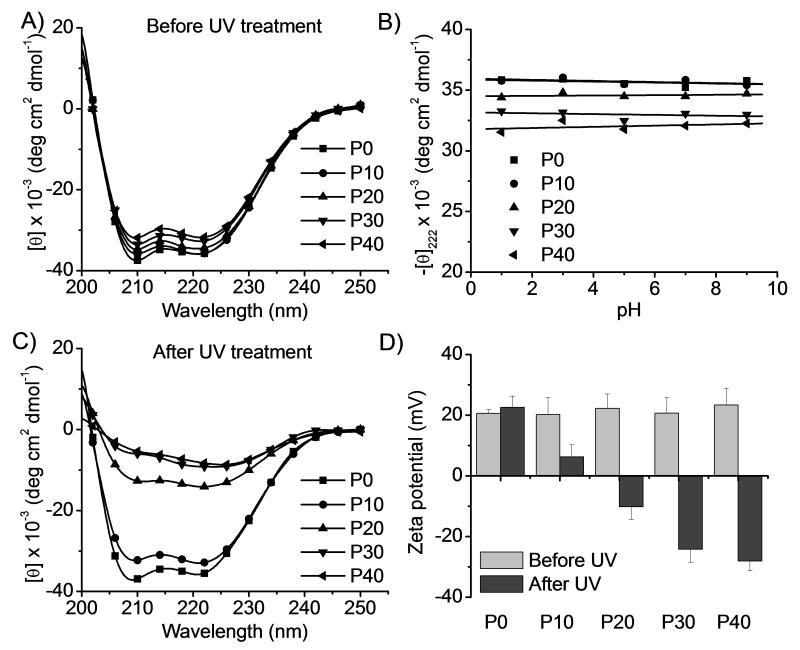
Figure 2.
PDMNBLG-r-PVBLG-8 with reduced helicity and cationic charge upon UV irradiation (20 mW/cm2, 10 min). (A) CD spectra of PDMNBLG-r-PVBLG-8 in water. (B) Helices of PDMNBLG-r-PVBLG-8 at different pH. (C) CD spectra of UV-irradiated PDMNBLG-r-PVBLG-8 in water. (D) Change of the zeta potential of polypeptide/DNA complexes (N/P ratio = 20) upon UV treatment.
The helicities of PDMNBLG-r-PVBLG-8 were demonstrated to be photo-responsive. When an aqueous solution of the polypeptide was irradiated with UV (λ=365 nm, 20 mW/cm2), an efficient model light trigger, the absorption at 346 nm in the UV/Vis spectroscopy decreased whereas the absorption at 400 nm increased (Supplementary Fig. S5), indicating cleavage of the photo-labile ester bond and generation of nitrobenzaldehyde.[14] By plotting OD346 against irradiation time, we determined that the photochemical reaction approached maximum conversion upon 10-min UV-treatment. UV treatment also significantly attenuated the α-helicities of P20, P30, and P40 with significant portions of photo-responsive DMNBLG residues while minimal changes were seen in P0 and P10 (Fig. 2C and Supplementary Fig. S6). The observed helix disruption was attributed to the intramolecular charge attraction between amine groups of PVBLG-8 and carboxylate groups generated after the removal of the DMNB group. The formation of carboxylate groups and positive charge reduction of the materials were also supported by the observation that the zeta potentials of all polypeptide/pCMV-Luc DNA complexes (at the optimal N/P ratio of 20, diameter of 130-170 nm) were decreased upon UV irradiation (Fig. 2D).
P20, having the highest cationic charge density among the three PDMNBLG-r-PVBLG-8 analogues (P20, P30 and P40) susceptible to light-induced helicity reductions (Fig. 1C), was selected to study the light-triggered changes of membrane activity and cytotoxicity. Fluorescein-tris(hydroxymethyl)methanethiourea (FITC-Tris, a membrane-impermeable fluorescent dye in the non-reactive form of FITC after reaction of FITC with Tris, was used as a biomarker for membrane pore formation.[9b] The polypeptides facilitated the uptake of FITC-Tris in HeLa and COS-7 cells by 8-10 fold (Fig. 3A), presumably because P20 induced pore formation on cell membranes.[9b] To validate this hypothesis, FITC-Tris internalization in the presence of UV pre-treated P20 (20 mW/cm2, 10 min) was studied, which showed that light-triggered reduction of cationic charge density and loss of α-helicity indeed reduced the membrane activity and pore formation capability of P20 (P20(UV)+FITC-Tris group vs. P20(non-UV)+FITC-Tris group, p<0.05). It was therefore not surprising to observe that the cytotoxicities of all photo-responsive polypeptides (P10-P40) were notably reduced upon UV treatment (Supplementary Fig. S9) as determined by the MTT assay. In a typical experiment in accordance with the transfection process, polypeptide/DNA complexes (N/P ratio of 20) were first incubated with cells for 4 h followed by UV treatment for 5 min and further culturing for 20 h before viability assessment (Fig. 3B and Supplementary Fig. S10). Consistently, complexes formed from P10-P40 but not the non-responsive P0 showed diminished cytotoxicity in response to UV treatment. These results collectively validated the proposed strategy of eliminating the membrane activity and improving the cell tolerability of polypeptides by post-transfection light exposure. UV is not a biocompatible light source; but cells receiving low intensity UV irradiation for a short period of time (20 mW/cm2, 5 min) showed uncompromised viability (96.8 ± 4.2%, n=3), indicating that UV irradiation in this model, proof-of-the-concept system did not induce appreciable cytotoxicity that would otherwise jeopardize the analysis of trigger-induced toxicity reduction. The cleaved DMNB also showed minimal toxicity to HeLa cells following 24-h incubation at 0.1 μmol/mL that corresponded to P40/DNA complexes (N/P ratio of 20) at the DNA transfection concentration of 5 μg/mL (Supplementary Fig. S11).
Figure 3.
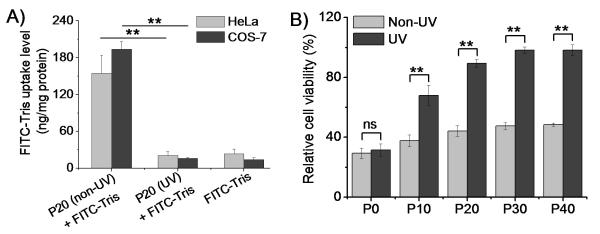
Polypeptides display diminished membrane activity and cytotoxicity upon UV treatment. (A) Uptake of FITC-Tris in HeLa and COS-7 cells in the presence of P20 and UV-treated P20 (n=3). (B) Cytotoxicity of polypeptide/DNA complexes in HeLa cells (N/P ratio of 20, 5 μg DNA/mL) with/without in situ UV treatment (20 mW/cm2, 5 min) as assessed by the MTT assay (n=3); ns denotes no significant difference (p>0.05).
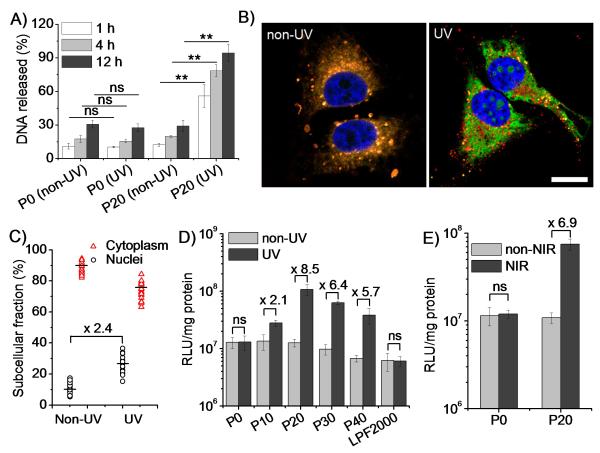
We next investigated whether light-triggered charge and conformational alteration of the polypeptides would promote intracellular DNA unpackaging and gene transfection. As expected, the incorporation of DMNBLG moieties did not compromise the DNA delivery capacity of the polypeptides, leading to notable uptake level of YOYO-1-labeled DNA in both HeLa and COS-7 cells via caveolae-mediated endocytosis and energy-independent non-endocytosis (Supplementary Fig. S12). UV-induced DNA release was monitoed by the heparin replacement assay.[15] As shown in Fig. 4A and Supplementary Fig. 13, UV treatment for 5 min notably facilitated the DNA release from the P20/DNA complexes, leading to almost complete DNA dissociation within 12 h. Comparatively, UV treatment exerted no effect on the P0/DNA complexes, confirming that the helix-distorted and cationic charge-reduced polypeptides promoted DNA unpackaging. Complex size was markedly augmented upon UV treatment, consistently signifying reduced DNA condensation by the polypeptides (Supplementary Fig. S8).
Figure 4.
UV (365 nm, 20 mW/cm2, 5 min)/NIR (750 nm, 3.2 μJ/cm2/pulse, 1.5 h) irradiation improves transfection efficiency by facilitating intracellular DNA unpackaging and nuclear transport. (A) DNA release from non-treated and UV-treated complexes (n=3). (B) CLSM images of HeLa cells incubated with RhB-P20 (red)/YOYO-1-DNA (green) complexes with/without UV treatment (bar = 20 μm). (C) Subcellular distribution of YOYO-1-DNA in HeLa cells following P20/DNA complex treatment and UV irradiation. (D) Transfection efficiency in HeLa cells (N/P = 20, 5 μg DNA/mL) with/without UV irradiation (n=3). (E) Transfection efficiency of P20/DNA complexes in HeLa cells (N/P = 20, 5 μg DNA/mL) with /without NIR irradiation (n=3). ns denotes no significant difference (p>0.05).
Apart from the charge conversion that would facilitate DNA unpackaging, we also examined the impact of secondary structure. PVBLG-8 and PVBDLG-8, two homo-polypeptides possessing the same charge density but different conformation (α-helix for PVBLG-8 prepared with l-Glu and random-coil for PVBDLG-8 prepared with racemic d,l-Glu[12b])—were allowed to form complexes with DNA (N/P ratio of 20). PVBLG-8 showed higher DNA condensation than PVBDLG-8 (Supplementary Fig. S13), suggesting that the reduction in both charge and helicity in UV-treated PDMNBLG-r-PVBLG-8 could synergistically promote the DNA release. By labeling P20 with rhodamine (RhB) and DNA with YOYO-1, we further studied the intracellular DNA unpackaging using confocal laser scanning microscopy (CLSM). Compared to non-treated cells wherein red and green fluorescence were largely overlapped, UV-treated cells exhibited notably enhanced separation of green fluorescence from red fluorescence (Fig. 4B), suggesting trigger-induced intracellular DNA release. The unpackaged DNA spread to the entire cytoplasm and some was localized inside the nuclei (Fig. 4B). Since DNA needs to enter the nuclei before it can be transcribed, we further quantified the nuclear distribution of YOYO-1-DNA.[16] As illustrated in Fig. 4C, following the 4-h complex treatment and subsequent UV treatment, 30% of the internalized DNA was distributed to the nuclei, representing a 2.3-fold increase over non-treated cells. As a result, UV treatment led to up to 8.5- and 5.6-fold increase in luciferase expression in HeLa (Fig. 4D) and COS-7 cells (Supplementary Fig. S16), respectively. Maximal transfection efficiency was noted for UV-treated P20, outperforming commercial reagent Lipofectamine™ 2000 (LPF2000) by 10-18 fold. The promoted gene transfection of P20/DNA complexes was also noted by flow cytometry when plasmid encoding enhanced green fluorescent protein (pEGFP) was used (Supplementary Fig. 17). UV treatment did not alter the transfection efficiency of the non-responsive P0, indicating that light treatment itself did not improve gene expression.
Because UV irradiation often suffers from low penetration and potential genotoxic effects when clinically applied, we went on to evaluate the applicability of near-infrared (NIR) modulation in this system. As shown in Supplementary Fig. S15, NIR irradiation (750 nm, 3.2 μJ/cm2/pulse) eliminated the helicity of P20 which reached the nadir following protracted treatment (1.5 h). In accordance, NIR irradiation for 1.5 h triggered a 6.9-fold increment in the transfection efficiency of P20/DNA complexes in HeLa cells, compared to the unappreciable enhancement of the non-responsive P0/DNA complexes (Fig. 4E). As expected, NIR irradiation did not induce cell death (viability of 95.6 ± 6.2%, n=3). These results validated the potential of regulating the transfection performance of photo-responsive polypeptides using a highly-penetrating and more biocompatible light source. DMNB has negligible absorption in the NIR region, and the slower responsiveness was mainly due to the low two-photon uncaging cross section of the DMNB group.[14] A more NIR-sensitive Glu-protecting ligand (under development in our group) would substantially enhance the applicability of this class of smart, trigger-responsive, non-viral delivery vector.
In summary, we developed a class of cationic helical polypeptides with built-in trigger-responsive domains that control the charge and conformational change of the polypeptides upon external stimuli. The cationic charge reduction and helix disruption subsequently resulted in the reduction of their membrane activities and enhancement of the DNA unpackaging capacities, thus substantially improving gene delivery efficiency with diminished cytotoxicity. As such, one trigger stimulates multiple gene transfection/delivery-relevant changes of material properties, provoking desired intracellular responses to overcome various barriers against non-viral gene delivery.
Supplementary Material
Footnotes
J.C. acknowledges support from the NSF (CHE-1153122) and the NIH (NIH Director’s New Innovator Award 1DP2OD007246, 1R21EB013379).
References
- [1].(a) Salem AK, Searson PC, Leong KW. Nat Mater. 2003;2:668–671. doi: 10.1038/nmat974. [DOI] [PubMed] [Google Scholar]; (b) Roy K, Mao HQ, Huang SK, Leong KW. Nat Med. 1999;5:387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]; (c) Mintzer MA, Simanek EE. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- [2].(a) Lee Y, Miyata K, Oba M, Ishii T, Fukushima S, Han M, Koyama H, Nishiyama N, Kataoka K. Angew Chem Int Edit. 2008;47:5163–5166. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]; (b) Miyata K, Oba M, Nakanishi M, Fukushima S, Yamasaki Y, Koyama H, Nishiyama N, Kataoka K. J Am Chem Soc. 2008;130:16287–16294. doi: 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]; (c) Liu HM, Wang H, Yang WJ, Cheng YY. J Am Chem Soc. 2012;134:17680–17687. doi: 10.1021/ja307290j. [DOI] [PubMed] [Google Scholar]
- [3].(a) McNaughton BR, Cronican JJ, Thompson DB, Liu DR. P Natl Acad Sci USA. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Uchida H, Miyata K, Oba M, Ishii T, Suma T, Itaka K, Nishiyama N, Kataoka K. J Am Chem Soc. 2011;133:15524–15532. doi: 10.1021/ja204466y. [DOI] [PubMed] [Google Scholar]; (c) Srinivasachari S, Fichter KM, Reineke TM. J Am Chem Soc. 2008;130:4618–4627. doi: 10.1021/ja074597v. [DOI] [PubMed] [Google Scholar]; (d) Wei H, Schellinger JG, Chu DSH, Pun SH. J Am Chem Soc. 2012;134:16554–16557. doi: 10.1021/ja3085803. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Li J, Yang C, Li HZ, Wang X, Goh SH, Ding JL, Wang DY, Leong KW. Adv Mater. 2006;18:2969–2974. [Google Scholar]; (f) Sunshine JC, Peng DY, Green JJ. Mol Pharmaceut. 2012;9:3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Shim MS, Kwon YJ. J Control Release. 2009;133:206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]; (h) Zhou JB, Liu J, Cheng CJ, Patel TR, Weller CE, Piepmeier JM, Jiang ZZ, Saltzman WM. Nat Mater. 2012;11:82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McLendon PM, Fichter KM, Reineke TM. Mol Pharmaceut. 2010;7:738–750. doi: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- [5].Zhi DF, Zhang SB, Wang B, Zhao YN, Yang BL, Yu SJ. Bioconjugate Chem. 2010;21:563–577. doi: 10.1021/bc900393r. [DOI] [PubMed] [Google Scholar]
- [6].Takae S, Miyata K, Oba M, Ishii T, Nishiyama N, Itaka K, Yamasaki Y, Koyama H, Kataoka K. J Am Chem Soc. 2008;130:6001–6009. doi: 10.1021/ja800336v. [DOI] [PubMed] [Google Scholar]
- [7].Zhao X, Yin L, Ding J, Tang C, Gu S, Yin C, Mao Y. J Control Release. 2010;144:46–54. doi: 10.1016/j.jconrel.2010.01.022. [DOI] [PubMed] [Google Scholar]
- [8].(a) Kostiainen MA, Smith DK, Ikkala O. Angew Chem Int Edit. 2007;46:7600–7604. doi: 10.1002/anie.200701200. [DOI] [PubMed] [Google Scholar]; (b) Han G, You CC, Kim BJ, Turingan RS, Forbes NS, Martin CT, Rotello VM. Angew Chem Int Edit. 2006;45:3165–3169. doi: 10.1002/anie.200600214. [DOI] [PubMed] [Google Scholar]; (c) Jiang XA, Zheng YR, Chen HH, Leong KW, Wang TH, Mao HQ. Adv Mater. 2010;22:2556–2560. doi: 10.1002/adma.200903933. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nalluri SKM, Voskuhl J, Bultema JB, Boekema EJ, Ravoo BJ. Angew Chem Int Edit. 2011;50:9747–9751. doi: 10.1002/anie.201103707. [DOI] [PubMed] [Google Scholar]; (e) Liu XH, Yang JW, Lynn DM. Biomacromolecules. 2008;9:2063–2071. doi: 10.1021/bm800291v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].(a) Stewart KM, Horton KL, Kelley SO. Organic & Biomolecular Chemistry. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]; (b) Ter-Avetisyan G, Tuennemann G, Nowak D, Nitschke M, Herrmann A, Drab M, Cardoso MC. J Biol Chem. 2009;284:3370–3378. doi: 10.1074/jbc.M805550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].(a) Ruzza P, Calderan A, Guiotto A, Osler A, Borin G. J Pept Sci. 2004;10:423–426. doi: 10.1002/psc.558. [DOI] [PubMed] [Google Scholar]; (b) Daniels DS, Schepartz A. J Am Chem Soc. 2007;129:14578–14579. doi: 10.1021/ja0772445. [DOI] [PubMed] [Google Scholar]
- [11].(a) Johnson RN, Chu DSH, Shi J, Schellinger JG, Carlson PM, Pun SH. J Control Release. 2011;155:303–311. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. J Am Chem Soc. 2008;130:3272–3274. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- [12].(a) Lu H, Wang J, Bai YG, Lang JW, Liu SY, Lin Y, Cheng JJ. Nat. Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]; (b) Gabrielson NP, Lu H, Yin LC, Li D, Wang F, Cheng JJ. Angew Chem Int Edit. 2012;51:1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu H, Cheng JJ. J Am Chem Soc. 2007;129:14114–14115. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- [14].Fomina N, McFearin C, Sermsakdi M, Edigin O, Almutairi A. J Am Chem Soc. 2010;132:9540–9542. doi: 10.1021/ja102595j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Seow WY, Yang YY, George AJT. Nucleic Acids Res. 2009;37:6276–6289. doi: 10.1093/nar/gkp651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhao X, Yin LC, Ding JY, Tang C, Gu SH, Yin CH, Mao YM. J Control Release. 2010;144:46–54. doi: 10.1016/j.jconrel.2010.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.