SUMMARY
In periodontitis, dysbiotic microbial communities exhibit synergistic interactions for enhanced protection from host defenses, nutrient acquisition, and persistence in an inflammatory environment. This Review discusses evidence that periodontitis-associated communities are ‘inflammo-philic’ (= loving or attracted to inflammation) in that they have evolved to not only endure inflammation but also to take advantage of it. In this regard, inflammation can drive the selection and enrichment of these pathogenic communities by providing a source of nutrients in the form of tissue breakdown products (e.g., degraded collagen peptides and heme-containing compounds). In contrast, those species that cannot benefit from the altered ecological conditions of the inflammatory environment, or for which host inflammation is detrimental, are likely to be outcompeted. Consistent with the concept that inflammation fosters the growth of dysbiotic microbial communities, the bacterial biomass of human periodontitis-associated biofilms was shown to increase with increasing periodontal inflammation. Conversely, anti-inflammatory treatments in animal models of periodontitis were shown to diminish the periodontal bacterial load, in addition to protecting from bone loss. The selective flourishing of inflammophilic bacteria can perpetuate inflammatory tissue destruction by setting off a ‘vicious cycle’ for disease progression, in which dysbiosis and inflammation reinforce each other. Therefore, the control of inflammation appears to be central to the treatment of periodontitis, as it is likely to control both dysbiosis and disease progression.
INTRODUCTION
Periodontitis is a biofilm-induced chronic inflammatory disease characterized by loss of bone support of the dentition (Darveau, 2010; Hajishengallis, 2014b). The tooth-associated biofilm (‘dental plaque’) is required but is not sufficient by itself to induce periodontitis. In this regard, it is the host inflammatory response to this microbial challenge that primarily and ultimately causes the degradation of the periodontium, i.e., the tooth-supporting structures such as the gingiva and the underlying alveolar bone (Hajishengallis, 2014b). The precise mechanisms of periodontal pathogenesis are incompletely understood but disease initiation and progression invariably involves the breakdown of periodontal host-microbe homeostasis (Darveau, 2010; Hajishengallis, 2014b).
Periodontal homeostasis can be disrupted by a variety of host- or microbe-related factors. These likely include congenital or acquired host immunodeficiencies, immunoregulatory defects associated with mutations or polymorphisms, old age, systemic diseases such as diabetes, obesity, environmental factors (e.g., smoking, diet, and stress), epigenetic modifications in response to environmental changes, and the presence of keystone pathogens that can transform a symbiotic microbiota into a dysbiotic one (Divaris et al., 2013; Eskan et al., 2012; Hajishengallis, 2014a; Hajishengallis et al., 2012; Laine et al., 2012; Lindroth & Park, 2013; Stabholz et al., 2010; Zhou et al., 2011). These factors could act alone or–more likely– in combination to cause dysbiosis of the periodontal microbiota and progression to periodontitis.
On the basis of evidence from human and animal studies discussed below, this Review proposes that the periodontitis-associated microbiota is inflammophilic (from the Greek suffix philic, meaning attracted to or loving) in that this microbial community has evolved to not only endure inflammation but also to exploit it to promote its adaptive fitness. Specifically, the available evidence is consistent with the notion that the inflamed periodontal pocket is conducive for the creation of a nutritionally rich environment that is permissive for bacterial outgrowth and pathogenesis.
Recent advances in periodontal disease pathogenesis: Keystone pathogens and the PSD model
Recent independent studies employing metagenomic, metatranscriptomic, or mechanistic approaches collectively suggest that periodontitis is not caused by a select few bacteria, historically referred to as ‘periopathogens’ (Abusleme et al., 2013; Dewhirst et al., 2010; Duran-Pinedo et al., 2014; Griffen et al., 2012; Hajishengallis et al., 2011; Jiao et al., 2013; Jorth et al., 2014; Orth et al., 2011). In this respect, it has been proposed that periodontal disease pathogenesis involves polymicrobial synergy and dysbiosis (‘PSD model’) (Hajishengallis & Lamont, 2012, 2014), a notion consistent with the above-cited human meta-genomic/transcriptomic studies, as well as mechanistic studies in animal models and in vitro (Hajishengallis et al., 2011; Kesavalu et al., 2007; Maekawa et al., 2014b; Polak et al., 2009; Ramsey et al., 2011; Settem et al., 2012; Tan et al., 2014). According to the PSD model, periodontitis-associated microbial communities show synergistic interactions for enhanced colonization, nutrient procurement, and persistence in an inflammatory environment. The dysbiosis of the periodontal microbiota signifies an imbalance in the relative abundance or influence of microbial species within the ecosystem (as compared to health), leading to alterations in the host-microbial crosstalk sufficient to mediate destructive inflammation and bone loss (Abusleme et al., 2013; Hajishengallis, 2014b; Hajishengallis & Lamont, 2012). The recent metagenomic studies indicate that the periodontitis-associated microbiota is more heterogeneous and diverse than previously thought (i.e., on the basis of cultural studies) and many of the newly recognized organisms (e.g., Filifactor alocis and other species from the genera Peptostreptococcaceae, Desulfobulbus, and Synergistetes) show as good or better a correlation with disease than the classical red complex bacteria, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia (Abusleme et al., 2013; Curtis et al., 2011; Dewhirst et al., 2010; Griffen et al., 2012; Griffen et al., 2011; Kumar et al., 2006). Moreover, a plethora of virulence factors upregulated in the microbiome of periodontitis patients is primarily derived from species that are not traditionally considered as ‘periopathogens’ (Duran-Pinedo et al., 2014). Furthermore, the presence of the three red complex species in a complex subgingival biofilm model hardly affected the transcriptional profiling of challenged human gingival fibroblasts (Belibasakis et al., 2014). The above should not be interpreted to mean that the red complex bacteria do not play important roles in periodontitis. Simply, their role needs to be re-interpreted in the face of newly emerging evidence.
For instance, P. gingivalis was until recently thought to directly cause periodontitis, a notion consistent with its ability to cause bone loss upon its implantation in the oral cavity of animals, including mice and non-human primates (Baker et al., 1999; Holt et al., 1988). However, it was recently shown that P. gingivalis by itself fails to cause periodontitis in mice (i.e., germ-free mice that lack commensal bacteria). The actual role of P. gingivalis involves its ability to initiate the conversion from a symbiotic community structure to a dysbiotic one capable of causing destructive inflammation (Hajishengallis et al., 2011). In this regard, P. gingivalis expresses a variety of virulence factors (such as gingipains, atypical lipid A structures, and serine phosphatases), which manipulate the host response in ways that create a permissive environment for the growth of both P. gingivalis and bacteria co-habiting the same niche (Hajishengallis & Lamont, 2014; Hajishengallis et al., 2011; Takeuchi et al., 2013; Zenobia et al., 2014). This community-wide impact and the fact that P. gingivalis is a quantitatively minor constituent of the microbiota, has prompted its characterization as a keystone pathogen, by analogy to the crucial role of the literal keystone holding an entire arch together (Darveau et al., 2012; Hajishengallis et al., 2012). It should be noted that certain other periodontal species (e.g., T. forsythia, T. denticola, and Aggregatibacter actinomycetemcomitans) have also been shown to subvert the host response in ways that compromize protective immunity (Jusko et al., 2012; Miller et al., 2012; Potempa & Pike, 2009; Venketaraman et al., 2008). Whether these or other species can also play keystone roles (in similar or different environments or forms of periodontal disease) has not been addressed experimentally yet but it is a plausible hypothesis.
Although originally established in the mouse model, the keystone-pathogen concept in periodontitis is consistent with observations in other animal models and in humans: In rabbits, P. gingivalis causes a shift to a more anaerobic flora and an overall increase in the bacterial load of the dental biofilm (Hasturk et al., 2007; Zenobia et al., 2014). In non-human primates, where P. gingivalis is a natural inhabitant of the periodontal biofilm, a gingipain-based vaccine caused a reduction both in P. gingivalis numbers and in the total subgingival bacterial load (Page et al., 2007), suggesting that the presence of P. gingivalis benefits the entire biofilm. The keystone-pathogen concept is consistent with P. gingivalis also being a quantitatively minor constituent of human periodontitis-associated biofilms, despite its increased prevalence and association with progressive bone loss in periodontal patients (Abusleme et al., 2013; Chaves et al., 2000; Doungudomdacha et al., 2000; Kumar et al., 2006; Moore et al., 1982; Moore et al., 1991). P. gingivalis may additionally be detected, albeit with decreased frequency, in periodontally healthy individuals (Abusleme et al., 2013; Haffajee et al., 1998). The most likely explanations, which are not mutually exclusive, involve alterations in the status of the host or the bacterium, such as strain and virulence diversity within the population structure of P. gingivalis (Darveau et al., 2012). From the host point of view, there may be individuals who can either resist or tolerate the conversion of the periodontal microbiota from a symbiotic to a dysbiotic state, by virtue of their intrinsic immuno-inflammatory status (e.g., hyporesponsive or lack-of-function polymorphisms that attenuate inflammation or microbial immune subversion). This notion is consistent with human genetic variability in innate immunity and susceptibility to periodontitis and with cases of individuals who remain periodontally healthy despite massive accumulation of dental plaque at dentogingival sites (Kinane et al., 2007; Kinane & Hart, 2003; Laine et al., 2012; Socransky & Haffajee, 1994).
It has been proposed that dysbiosis is not dependent so much on the particular microbial roster but rather on the specific gene combinations or collective virulence activity within the microbial community (Hajishengallis & Lamont, 2012). This notion is supported by a recent study that employed gene expression profiling to characterize patient-matched healthy and disease-associated periodontal microbiotas. This investigation showed that disease-associated microbial communities exhibit highly conserved metabolic gene expression profiles, despite high interpatient variability in terms of microbial composition (Jorth et al., 2014). Moreover, this concept and the fact that periodontitis is not uniquely a human disease (Page & Schroeder, 1982) further validates the use of appropriate animal models to study periodontal disease pathogenesis (Graves et al., 2008). The evidence in support of the inflammophilic character of the periodontitis-associated microbiota is based on both animal models and clinical observations in human patients (see below).
Inflammation can drive the selection and enrichment of dysbiotic communities
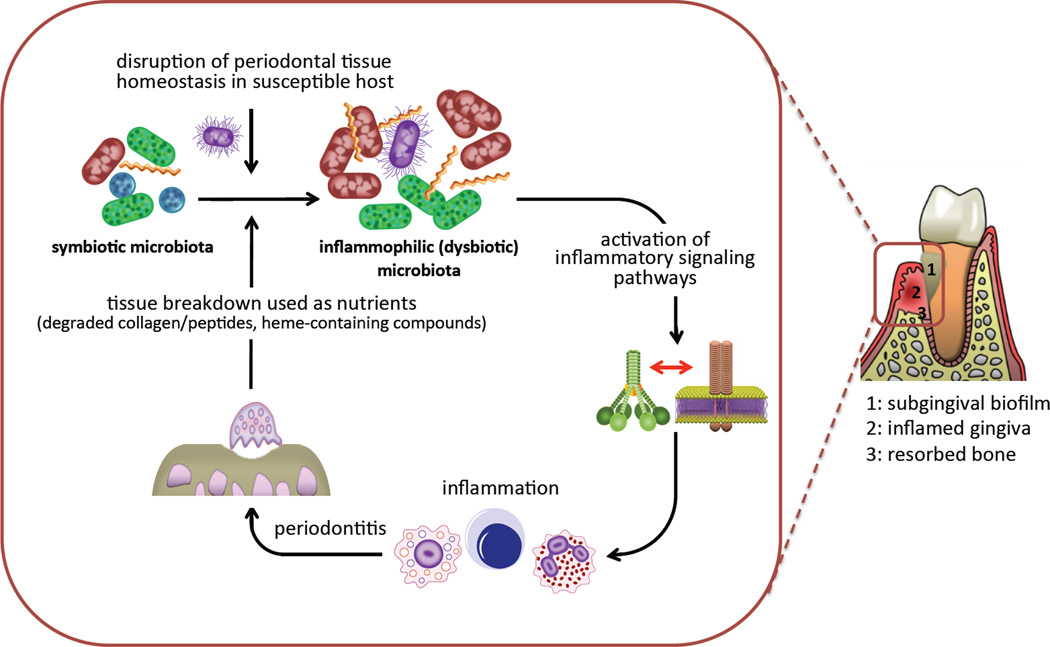
The ecological succession from health to periodontitis reveals the emergence of newly-dominant community members rather than the appearance of novel species (Abusleme et al., 2013). This important recent finding is consistent with the ecologic plaque hypothesis proposed more than ten years ago (Marsh, 2003). According to this hypothesis, ‘periodontal pathogens’ are members of the normal microbiota but at levels too low to cause disease, whereas changes in ecologic conditions could favor the outgrowth of such organisms beyond a threshold sufficient to lead to periodontitis (Marsh, 2003). This threshold could be numerical or physiological, or a combination of both. In other words, this notion views periodontitis as an ecologic catastrophe. It cannot be overstated that periodontal inflammation is not simply a mechanism for host tissue destruction; from a microbe-centric standpoint, inflammation is an important source of nutrients and, therefore, can exert a powerful influence in the composition and numbers of the periodontal microbiota favoring those species that can both endure inflammation and utilize tissue breakdown products (Fig. 1). Stated in lay terms, bacteria do not cause inflammation to destroy the periodontal tissue but to obtain their food. Destructive inflammation generates abundant tissue-breakdown products that serve the nutritional needs of certain types of bacteria; for instance, degraded collagen and heme-containing compounds (haptoglobin, hemopexin, and hemoglobin) can be utilized by proteolytic and asaccharolytic bacteria to obtain essential amino-acids and iron (Fig. 1) (Hajishengallis et al., 2012). Subgingival sites harboring periodontitis-associated bacteria exhibit a low redox potential and are bathed in gingival crevicular fluid (GCF), a nutritionally rich serum-like exudate, the flow of which increases with increasing periodontal inflammation (Marsh, 2003). These conditions support a microbial community with higher proportions of obligately anaerobic bacteria that thrive at the expense of aerobic species or those that cannot take advantage of the inflammatory environment. Indeed, those species that cannot benefit from these environmental changes, or for which host inflammation is detrimental, may have a fitness disadvantage and hence be outcompeted. The selective flourishing of inflammophilic bacteria acting as pathobionts can potentially perpetuate a self-feeding “vicious cycle” for incessant tissue destruction and bacterial overgrowth (Fig. 1) (Hajishengallis et al., 2012).
Figure 1. Inflammation and dysbiosis in periodontitis.
Various factors including defects in the immuno-inflammatory status of the host and/or the presence of keystone pathogens can tip the balance from a symbiotic periodontal microbiota to a dysbiotic state. The inflammation caused by the dysbiotic microbiota depends in great part on crosstalk signaling between complement and pattern-recognition receptors and exerts two major and interrelated effects: it causes inflammatory destruction of periodontal tissue (including bone loss, the hallmark of periodontitis) which in turn provides tissue breakdown-derived nutrients for the bacteria. The latter further promotes dysbiosis and escalates tissue destruction, thereby generating a self-perpetuating pathogenic cycle. Therefore, the periodontitis-associated dysbiotic communities are ‘inflammo-philic’ (= loving or attracted to inflammation) in that they appear to have evolved to not only endure inflammation but also to take advantage of it for enhancing their adaptive fitness.
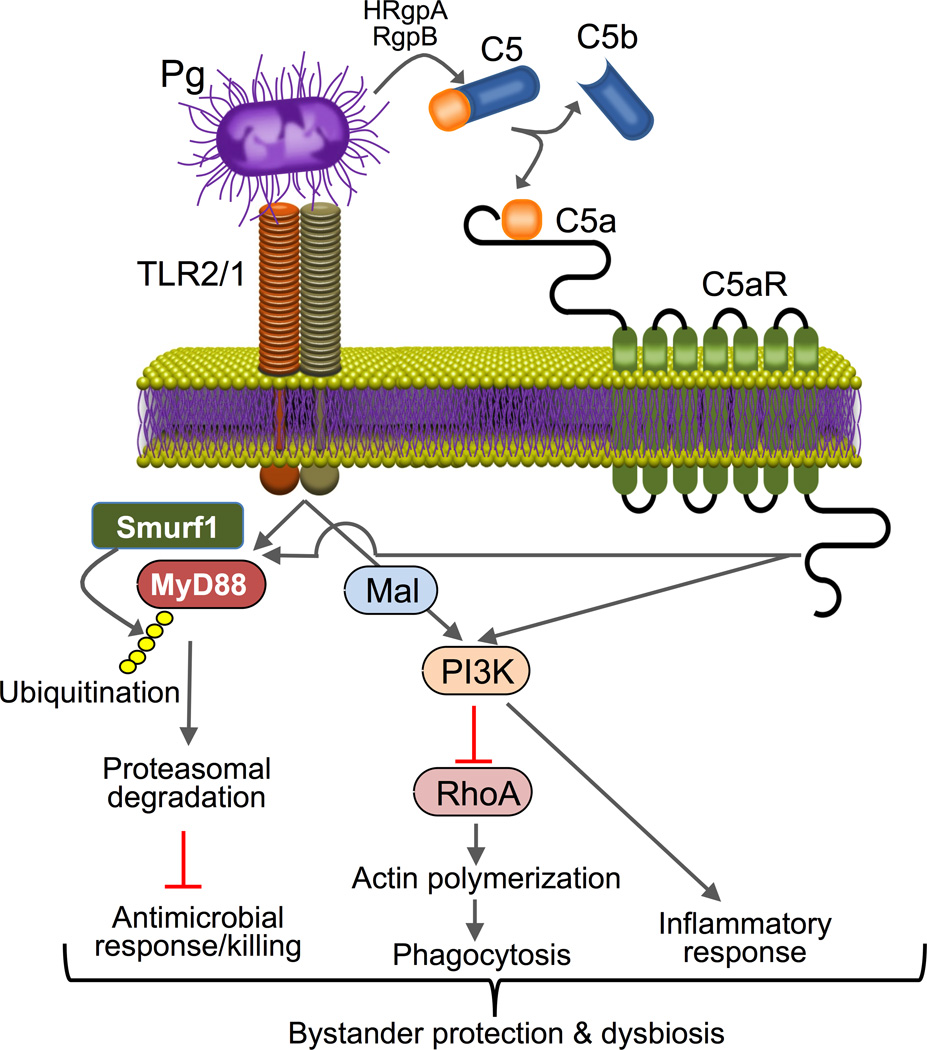
As alluded to above, P. gingivalis is an exemplar of immune subversive activity in the periodontal tissue (Bostanci & Belibasakis, 2012; Hajishengallis & Lambris, 2011; Yilmaz, 2008). This microbe is now thought to orchestrate rather than to directly cause inflammatory bone loss, which is largely mediated by pathobionts, i.e., commensals that under conditions of disrupted homeostasis have the potential to cause disease. P. gingivalis and other immune-subversive organisms, including T. forsythia, T. denticola, and Prevotella intermedia, interact with complement in complex ways including both inhibitory and stimulatory effects (Jusko et al., 2012; Krauss et al., 2010; Miller et al., 2012; Popadiak et al., 2007; Potempa & Pike, 2009). On the one hand, periodontal bacteria need to escape immune elimination, whereas, on the other hand, they have to proactively induce inflammation and thereby stimulate the flow of GCF to obtain essential nutrients (Hajishengallis et al., 2012; Hasturk et al., 2012). In other words, although immunosuppression is an immune evasion strategy used by many microbial pathogens, it is not a viable option for inflammophilic bacteria. In this context, P. gingivalis was shown to block a host-protective TLR2-MyD88 pathway in neutrophils via proteasomal degradation of MyD88, whereas it activates a proinflammatory TLR2MalPI3K pathway that also blocks phagocytosis (Maekawa et al., 2014b). Both subversive pathways require an intimate crosstalk between TLR2 and C5aR instigated by P. gingivalis (Fig. 2). The integrated mechanism mediates ‘bystander’ protection to otherwise susceptible bacteria and promotes dysbiotic inflammation in vivo (Maekawa et al., 2014b). This study has therefore dissected a mechanism by which P. gingivalis can disengage bacterial clearance from inflammation and can thereby contribute to the persistence of microbial communities that drive periodontitis (Fig. 2). This, however, does not imply that all cases of periodontitis are initiated by P. gingivalis, which constitutes one of many factors that can contribute to the disruption of periodontal host-microbe homeostasis (see above).
Figure 2. Model of P. gingivalis subversion of neutrophils leading to dysbiotic inflammation.
P. gingivalis co-activates TLR2 and C5aR in neutrophils and the resulting crosstalk leads to E3 ubiquitin ligase Smurf1-dependent ubiquitination and proteasomal degradation of MyD88, thereby inhibiting a host-protective antimicrobial response. Moreover, the C5aR-TLR2 crosstalk activates PI3K, which prevents phagocytosis through inhibition of RhoA activation and actin polymerization, while stimulating an inflammatory response. In contrast to MyD88, Mal is a component of the subversive pathway acting upstream of PI3K. The integrated mechanism provides ‘bystander’ protection to otherwise susceptible bacterial species and promotes polymicrobial dysbiotic inflammation in vivo. From Maekawa et al., 2014b with permission.
Anti-inflammatory interventions can control the periodontitis-associated microbiota
A clinical study that characterized the periodontal microbiota of progressing initial chronic periodontitis concluded that no species in baseline microbial samples alone were strongly associated with progressing periodontitis (Tanner et al., 2007). Interestingly, as an alternative interpretation, the authors suggested that periodontal inflammation may determine the composition of the microbiota rather than vice-versa, in other words “the organisms are there because of the disease, which raises the possibility that periodontal pathogens may not predict future disease” (Tanner et al., 2007). The notion that inflammation fosters the growth of a pathogenic microbiota (Fig. 1) is consistent with experimental data showing that anti-inflammatory treatments diminish the periodontal bacterial burden. In a rabbit model of P. gingivalis-induced periodontitis, treatment with resolvin E1, a proresolving mediator of inflammation, not only resolved periodontal inflammation but also diminished the numbers of P. gingivalis and other Gram-negative periodontal bacteria (Hasturk et al., 2007). Similarly, treatment of P. gingivalis-induced periodontitis in mice with an antagonist of the complement C5a receptor (C5aR; CD88) reversed both inflammation and the outgrowth of the commensal microbiota caused by P. gingivalis colonization; P. gingivalis itself was essentially eradicated by the C5aR antagonist treatment (Abe et al., 2012; Hajishengallis et al., 2011). Moreover, treatment of mice undergoing ligature-induced periodontitis with meloxicam (a selective inhibitor of cyclooxygenase-2) caused a significant reduction of the periodontal bacterial load (Eskan et al., 2012). A recent study has identified a periodontal pathobiont (designated NI1060) that selectively accumulates in damaged periodontal tissue and thrives under inflammatory conditions, thereby becoming particularly pathogenic in causing bone loss (Jiao et al., 2013). More recently, topical treatment with a probiotic preparation (Lactobacillus brevis CD2) in the oral mucosa of mice undergoing ligature-induced periodontitis not only inhibited inflammation and bone loss but also exerted differential effects on the aerobic and anaerobic microbiotas (Maekawa & Hajishengallis, 2014). Specifically, the probiotic treatment with L. brevis CD2 resulted in significantly higher counts of aerobic bacteria and, conversely, significantly lower counts of anaerobic bacteria, as compared to the placebo-treated control group. This finding is consistent with the notion that periodontitis-associated bacteria are predominantly (if not exclusively) anaerobic and inflammophilic; therefore, their growth will be limited once inflammation is under control (tissue breakdown is minimal and hence the source of nutrients diminishes). However, an alternative (or additional) interpretation is that L. brevis CD2 might have a direct inhibitory effect on periodontal anaerobic bacteria and, conversely, a stimulatory effect on aerobic bacteria.
As discussed above, the capacity of P. gingivalis to colonize the murine periodontium and cause elevation of the total bacterial counts requires intact C5aR signaling (Abe et al., 2012; Hajishengallis et al., 2011; Maekawa et al., 2014b). P. gingivalis can activate C5aR by releasing the C5a fragment from complement component C5 through the action of its Arg-specific gingipains (Liang et al., 2011; Wang et al., 2010; Wingrove et al., 1992) (Fig. 2). Consistent with its ability to activate C5aR independently of the immunological activation of complement, P. gingivalis retains its capacity to colonize the periodontium of C3-deficient (C3–/–) mice (Maekawa et al., 2014a), in which complement cannot be activated by immunological mechanisms (i.e., via the classic, lectin, or alternative pathway) (Ricklin et al., 2010). In C3–/– mice, P. gingivalis could additionally elevate the total microbiota counts, as determined one week post-inoculation. Intriguingly, however, six weeks post-inoculation, the total microbiota counts in P. gingivalis-colonized C3–/– mice were diminished relative to those of P. gingivalis-colonized WT mice and approached the microbiota counts of sham-inoculated C3–/– mice (Maekawa et al., 2014a). P. gingivalis-colonized C3–/– mice also experienced significantly less periodontal inflammation and bone loss than P. gingivalis-colonized wild-type mice (Maekawa et al., 2014a). Therefore, although C3 is not required for P. gingivalis colonization and the initial elevation of the total microbiota counts, C3 is essential for the long-term sustenance of the dysbiotic microbiota and for maximal inflammatory bone loss. The reason why P. gingivalis-induced dysbiosis cannot be sustained in C3–/– mice is likely related to the diminished periodontal inflammation in these mice. This study also provides a model where the presence of a keystone pathogen such as P. gingivalis does not necessarily lead to disease in the absence of critical host signaling pathways. This could explain, at least in part, why the presence of P. gingivalis in the human periodontium is not always associated with disease (Abusleme et al., 2013; Haffajee et al., 1998); a possible scenario could be that hyporesponsive or lack-of-function polymorphisms attenuate inflammation and prevent disease development despite the presence of P. gingivalis or other organisms capable to act as keystone pathogens.
Similar conclusions about the role of inflammation in periodontal dysbiosis can be drawn from the investigation of aggressive forms of periodontitis, as in leukocyte adhesion deficiency. Mice deficient in the LFA-1 integrin (a model of leukocyte adhesion deficiency type I; LAD-I) experience overproduction of periodontal IL-17 and severe bone loss early in life, as do human LAD-I patients (Moutsopoulos et al., 2014). Interestingly, anti-IL-17 treatment not only inhibits inflammation and bone loss but also diminishes the periodontal bacterial burden (Moutsopoulos et al., 2014). Consistent with the animal model studies, a recent metagenomic study showed that the bacterial biomass of human periodontitis-associated biofilms increases with increasing periodontal inflammation (Abusleme et al., 2013).
These studies collectively suggest that the established pathogenic biofilm in periodontitis is inflammophilic. Apparently, inflammation generates an environment that is conducive for the selection and overgrowth of dysbiotic microbial species. Once such environment is established, the periodontitis-associated bacteria provoke further inflammation (to secure nutrients) which collaterally causes even more tissue damage (Fig. 1). Consistent with the notion that inflammation is exploited by the bacteria as a means that serves their nutritional needs, a study investigating the in situ community-wide transcriptome of the subgingival periodontitis-associated microbiota demonstrated increased expression of proteolysis-related genes and genes for iron acquisition and lipopolysaccharide synthesis (Duran-Pinedo et al., 2014).
Therapeutic implications and conclusion
The above-discussed concepts could be summarized as follows. Nutrients derived from inflammatory tissue breakdown select for community members that are inflammophilic. Subsequently, the emerging community proactively induces inflammation to sustain itself. Bacteria-induced inflammation can fuel further changes to the biofilm and stabilize the transition to a disease-provoking microbiota. An obvious implication is that periodontitis can be treated by approaches distinct from conventional treatment, which involves mechanical removal or physical disruption of the tooth-associated biofilm. In this context, it should be noted that conventional periodontal treatment is often not sufficient by itself to control destructive inflammation and many patients develop recurrent disease (Armitage, 2002; Colombo et al., 2012). From an ecological point of view, promising approaches could be those interfering with the environmental factors that drive the selection and enrichment of disease-causing bacteria. For instance, therapeutic interventions could be implemented aiming to alter the local environment by reducing the flow of the GCF or the site could be rendered less anaerobic through the use of oxygenating or redox agents (Marsh, 2003). The GCF flow could be readily reduced by anti-inflammatory approaches. Moreover, a recent study showed that the ratio of receptor activator of NF-κB ligand to osteoprotegerin (RANKL/OPG), which reflects osteoclastic activity, was not reduced following conventional periodontal treatment despite improved clinical parameters (Bostanci et al., 2011). This important finding implies that subclinical inflammatory events may be unaffected by standard modes of treatment, thus highlighting the need for adjunctive anti-inflammatory treatments. Several anti-inflammatory interventions have been tested in preclinical animal models of periodontitis, including non-human primates, which share key clinical, microbiological, and immunohistological features with the human disease (Assuma et al., 1998; Brecx et al., 1985; Holt et al., 1988; Kornman et al., 1981). Such efforts have targeted diverse inflammatory pathways, ranging from upstream events associated with inflammatory cell recruitment, to signaling pathways that amplify and propagate inflammation, as well as promoting the resolution of periodontal inflammation through the use of specific pro-resolution agonists (Abe et al., 2012; Assuma et al., 1998; Eskan et al., 2012; Graves, 2008; Hasturk et al., 2012; Kirkwood et al., 2007; Maekawa et al., 2014b). In conclusion, inflammation is not only the driving force for the destruction of periodontal tissues; from a microbial perspective, the importance of inflammation lies in its providing a source of nutrients that drive the selection and persistence of a disease-provoking microbiota. Taken together, it can be concluded that anti-inflammatory agents are central to the effective treatment of periodontitis as they are likely to control both dysbiosis and disease progression.
ACKNOWLEDGMENTS
The author is supported by grants from the National Institutes of Health, NIDCR (DE015254, DE017138, and DE021685).
REFERENCES
- Abe T, Hosur KB, Hajishengallis E, Reis ES, Ricklin D, Lambris JD, Hajishengallis G. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol. 2012;189:5442–5448. doi: 10.4049/jimmunol.1202339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage GC. Classifying periodontal diseases--a long-standing dilemma. Periodontol 2000. 2002;30:9–23. doi: 10.1034/j.1600-0757.2002.03002.x. [DOI] [PubMed] [Google Scholar]
- Assuma R, Oates T, Cochran D, Amar S, Graves DT. IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J Immunol. 1998;160:403–409. [PubMed] [Google Scholar]
- Baker PJ, Carter S, Dixon M, Evans RT, Roopenian DC. Serum antibody response to oral infection precedes but does not prevent Porphyromonas gingivalis-induced alveolar bone loss in mice. Oral Microbiol Immunol. 1999;14:194–196. doi: 10.1034/j.1399-302x.1999.140309.x. [DOI] [PubMed] [Google Scholar]
- Belibasakis GN, Bao K, Bostanci N. Transcriptional profiling of human gingival fibroblasts in response to multi-species in vitro subgingival biofilms. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12053. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Saygan B, Emingil G, Atilla G, Belibasakis GN. Effect of periodontal treatment on receptor activator of NF-κB ligand and osteoprotegerin levels and relative ratio in gingival crevicular fluid. J Clin Periodontol. 2011;38:428–433. doi: 10.1111/j.1600-051X.2011.01701.x. [DOI] [PubMed] [Google Scholar]
- Brecx MC, Nalbandian J, Ooya K, Kornman KS, Robertson PB. Morphological studies on periodontal disease in the cynomolgus monkey. II. Light microscopic observations on ligature-induced periodontitis. J Periodontal Res. 1985;20:165–175. doi: 10.1111/j.1600-0765.1985.tb00423.x. [DOI] [PubMed] [Google Scholar]
- Chaves ES, Jeffcoat MK, Ryerson CC, Snyder B. Persistent bacterial colonization of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans, and in periodontitis and its association with alveolar bone loss after 6 months of therapy. J Clin Periodontol. 2000;27:897–903. doi: 10.1034/j.1600-051x.2000.027012897.x. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst FE, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, Lange EM, Moss K, Barros SP, Weyant RJ, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 2013;22:2312–2324. doi: 10.1093/hmg/ddt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doungudomdacha S, Rawlinson A, Douglas CW. Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method. J Med Microbiol. 2000;49:861–874. doi: 10.1099/0022-1317-49-10-861. [DOI] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014 doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 2012;13:465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves D. Cytokines that promote periodontal tissue destruction. J Periodontol. 2008;79:1585–1591. doi: 10.1902/jop.2008.080183. [DOI] [PubMed] [Google Scholar]
- Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol. 2008;35:89–105. doi: 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, Vriesendorp B, Faust RA, Janies DA, Leys EJ. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, Socransky SS. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Aging and its impact on innate immunity and inflammation: Implications for periodontitis. J Oral Biosci. 2014a;56:30–37. doi: 10.1016/j.job.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014b;35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis . Eur J Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, Van Dyke TE. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J Immunol. 2007;179:7021–7029. doi: 10.4049/jimmunol.179.10.7021. [DOI] [PubMed] [Google Scholar]
- Hasturk H, Kantarci A, Van Dyke TE. Paradigm shift in the pharmacological management of periodontal diseases. Front Oral Biol. 2012;15:160–176. doi: 10.1159/000329678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Nunez G, Giannobile WV, Raes J, et al. Induction of bone loss by pathobiont-mediated nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5:e01012–e01014. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusko M, Potempa J, Karim AY, Ksiazek M, Riesbeck K, Garred P, Eick S, Blom AM. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J Immunol. 2012;188:2338–2349. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M, Ebersole JL. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane DF, Demuth DR, Gorr SU, Hajishengallis GN, Martin MH. Human variability in innate immunity. Periodontol 2000. 2007;45:14–34. doi: 10.1111/j.1600-0757.2007.00220.x. [DOI] [PubMed] [Google Scholar]
- Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14:430–449. doi: 10.1177/154411130301400605. [DOI] [PubMed] [Google Scholar]
- Kirkwood KL, Cirelli JA, Rogers JE, Giannobile WV. Novel host response therapeutic approaches to treat periodontal diseases. Periodontol 2000. 2007;43:294–315. doi: 10.1111/j.1600-0757.2006.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornman KS, Holt SC, Robertson PB. The microbiology of ligature-induced periodontitis in the cynomolgus monkey. J Periodontal Res. 1981;16:363–371. doi: 10.1111/j.1600-0765.1981.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Krauss JL, Potempa J, Lambris JD, Hajishengallis G. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol 2000. 2010;52:141–162. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000. 2012;58:37–68. doi: 10.1111/j.1600-0757.2011.00415.x. [DOI] [PubMed] [Google Scholar]
- Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186:869–877. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Park YJ. Epigenetic biomarkers: a step forward for understanding periodontitis. J Periodontal Implant Sci. 2013;43:111–120. doi: 10.5051/jpis.2013.43.3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D, Lambris JD, Hajishengallis G. Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. J Immunol. 2014a;192 doi: 10.4049/jimmunol.1400569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Hajishengallis G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. J Periodontal Res. 2014 doi: 10.1111/jre.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou K, Triantafilou M, Hashim A, Hoch S, Curtis MA, Nussbaum JD, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014b doi: 10.1016/j.chom.2014.05.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- Miller DP, Bell JK, McDowell JV, Conrad DH, Burgner JW, Heroux A, Marconi RT. Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease. J Biol Chem. 2012;287:12715–12722. doi: 10.1074/jbc.M112.339721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982;38:1137–1148. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–739. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos NM, Konkel J, Sarmadi M, Eskan MA, Wild T, Dutzan N, Abusleme L, Zenobia C, Hosur KB, Abe T, et al. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17–driven inflammatory bone loss. Sci Transl Med. 2014;6:229ra240. doi: 10.1126/scitranslmed.3007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth RK, O'Brien-Simpson NM, Dashper SG, Reynolds EC. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011;26:229–240. doi: 10.1111/j.2041-1014.2011.00612.x. [DOI] [PubMed] [Google Scholar]
- Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L, Braham P, Persson GR. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis . Oral Microbiol Immunol. 2007;22:162–168. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Page RC, Schroeder HE. Periodontitis in man and other animals- A comparative review. Basel, Switzerland: Karger; 1982. [Google Scholar]
- Polak D, Wilensky A, Shapira L, Halabi A, Goldstein D, Weiss EI, Houri-Haddad Y. Mouse model of experimental periodontitis induced by P. gingivalis/F. nucleatum infection: Bone loss and host response. J Clin Periodontol. 2009;36:406–410. doi: 10.1111/j.1600-051X.2009.01393.x. [DOI] [PubMed] [Google Scholar]
- Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect Immun. 2012;80:2436–2443. doi: 10.1128/IAI.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Evidence of bacterial etiology: a historical perspective. Periodontol 2000. 1994;5:7–25. doi: 10.1111/j.1600-0757.1994.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:138–153. doi: 10.1111/j.1600-0757.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Hirano T, Whitmore SE, Morisaki I, Amano A, Lamont RJ. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog. 2013;9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KH, Seers CA, Dashper SG, Mitchell HL, Pyke JS, Meuric V, Slakeski N, Cleal SM, Chambers JL, McConville MJ, et al. Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses. PLoS Pathog. 2014;10:e1003955. doi: 10.1371/journal.ppat.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner AC, Kent R, Jr, Kanasi E, Lu SC, Paster BJ, Sonis ST, Murray LA, Van Dyke TE. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 2007;34:917–930. doi: 10.1111/j.1600-051X.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Venketaraman V, Lin AK, Le A, Kachlany SC, Connell ND, Kaplan JB. Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microb Pathog. 2008;45:173–180. doi: 10.1016/j.micpath.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis . J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, Darveau RP. Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infect Immun. 2014;82:650–659. doi: 10.1128/IAI.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Leeman SE, Amar S. Signaling mechanisms in the restoration of impaired immune function due to diet-induced obesity. Proc Natl Acad Sci U S A. 2011;108:2867–2872. doi: 10.1073/pnas.1019270108. [DOI] [PMC free article] [PubMed] [Google Scholar]