Abstract
Agonists of mouse STING (TMEM173) shrink and even cure solid tumor by activating innate immunity; human STING agonists are needed to test this therapeutic hypothesis in man. The endogenous STING agonist is 2′3′-cGAMP, a 2nd messenger that signals the presence of cytosolic dsDNA. We report activity-guided partial purification and identification of ENPP1 as the dominant 2′3′-cGAMP hydrolyzing activity in cultured cells. The hydrolysis activity of ENPP1 was confirmed using recombinant protein and was depleted in tissue extracts and plasma from Enpp1-/- mice. We synthesized a hydrolysis-resistant bis-phosphothioate analog of 2′3′-cGAMP (2′3′-cGsAsMP) with similar affinity for human STING in vitro and 10 times more potent at inducing IFN-β secretion from human THP1 monocytes. Studies in mouse Enpp1-/- lung fibroblasts indicate that resistance to hydrolysis contributes significantly to its higher potency. 2′3′-cGsAsMP is therefore improved over natural 2′3′-cGAMP as a model agonist, and has potential as a vaccine adjuvant and cancer therapeutic.
Introduction
Activation of innate immune signaling is a proven therapeutic strategy in vaccination1,2, viral infection3, and cancer (e.g. BCG for bladder cancer and imiquimod for skin cancer). Vaccine adjuvants and immune-activating anti-cancer agents have traditionally depended on relatively broad or non-specific stimuli, such as attenuated or killed bacteria or alum crystals. An improved understanding of innate immune signaling should make it possible to design more precise immunostimulants for treating or preventing specific diseases. The discovery of the STING pathway, a central pathway in anti-viral innate immunity4,5, and the second messenger cyclic dinucleotides (CDNs) that activate it6 opened up several new possibilities in this area. STING agonists would be candidates for clinical testing as adjuvants, and as stimulants for anti-cancer immune activity. We and others recently found that DMXAA, which dramatically shrinks cancer in mouse model systems by activating the innate immune response, is a STING agonist that activates mouse STING (mSTING) but not human STING (hSTING)7-9. To test the therapeutic hypothesis that STING agonists will be effective for cancer treatment and/or as vaccine adjuvants we need human-active molecules. The most potent natural STING agonist in humans is 2′3′-cGAMP (2). We set out to identify a stable agonist for hSTING. To do this, we first investigated the major degradation mechanism of 2′3′-cGAMP and then constructed 2′3′-cGAMP analogs that are resistant to its activity.
STING is an ER transmembrane protein. The cytoplasmic domain of STING forms dimers and CDNs bind at the dimer interface10. Upon ligand binding, the cytoplasmic tail of STING serves as an adapter for TBK-1 (a kinase) and IRF-3 (a transcription factor), resulting in their phosphorylation11. Phosphorylated IRF-3 translocates into the nucleus to induce a panel of host response genes including interferon α/β. In bacterial infection, STING is activated by conserved bacterial second-messengers, cyclic dinucleotides linked through two 3′-5′ phosphodiester linkages (3′3′-CDNs), which can contain two guanosines, two adenosines, or one of each12. A second, more powerful activation signal results from the presence of viral or self dsDNA in the cytoplasm, leading to the synthesis of 2′3′-cGAMP, which is a heterodimer linked by one standard 3′-5′ phosphodiester, and one rare 2′-5′ phosphodiester13-15. The affinity of 2′3′-cGAMP for hSTING is very high, with a dissociation constant of 4.59 nM compared to >1 μM for bacterial 3′3′-CDNs.
CDNs do not resemble typical small molecule drug candidates. Their molecular weight is ∼700; they have two negative charges; and they are built from potentially labile phosphodiester linkages. Nevertheless, they are able to activate the STING pathway, presumably after entering the cell by unknown mechanisms9. Moreover, 2′3′-cGAMP has shown potential as an immune adjuvant in mouse studies16. These observations and its high affinity to hSTING suggest that molecules directly based on 2′3′-cGAMP have therapeutic potential. However, important questions remain, including the hydrolytic stability of CDN derivatives in the circulation and tissues.
Other 2nd messengers containing phosphodiester linkages are rapidly hydrolyzed by specific enzymes, and in some cases the hydrolytic enzymes are important drug targets. For example, cAMP and cGMP are degraded by 11 classes of PDEs, and PDE5 is the target of Viagra17,18. Intracellular cAMP and cGMP are also exported from the cell by ATPase pumps: ABCC4 (MRP4)19, ABCC5 (MRP5)20, and ABCC11 (MRP8)21 have all been implicated in cyclic nucleotide export. No degradation or export pathways for 2′3′-cGAMP are known. Here, we describe the identification of ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP1) as the dominant 2′3′-cGAMP hydrolase in cells, tissue extracts, and in blood. We also report hydrolysis resistant CDN analogs that retain strong hSTING agonist activity.
Results
PDE12 does not hydrolyze 2′3′-cGAMP
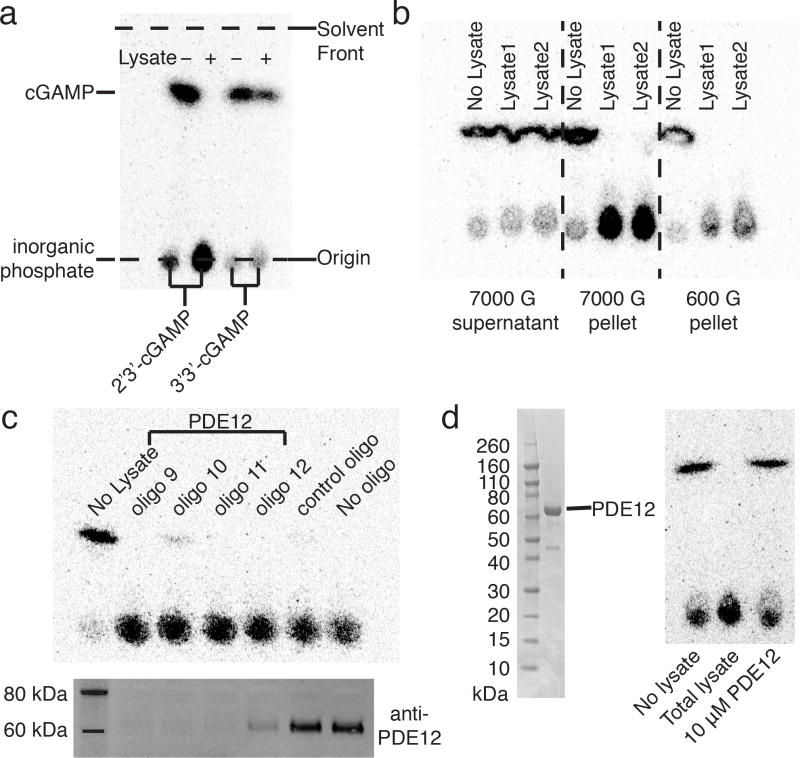
We enzymatically synthesized 2′3′-cGAMP from GTP and ATP using a mouse cGAS construct22,23 and 3′3′-cGAMP (1) using DncV from V. cholerae12. 32P versions were synthesized by including trace [α-32P] ATP. Synthesis and hydrolysis of radio-labeled CDNs were assayed using a simple thin-layer chromatography (TLC) assay (Fig. 1a). The TLC assay separates small molecules by their polarity with less polar compounds migrating higher on the plate. We chose a solvent condition, which moves CDN sup while leaving ATP/GTP and the final hydrolysis product, inorganic phosphate, at the baseline. Using this assay, we first searched for hydrolase activities in cell lines. We used mouse L929 cells and human THP-1 cells, both of which are model cell lines for the STING pathway and are capable of synthesizing 2′3′-cGAMP in response to dsDNA23. By fractioning cells by differential centrifugation, we discovered that the cytosol from these cells contained negligible activity. Detergent (NP-40) extracts of the cells had much higher activity, suggesting that the activity is located in an organelle or on the plasma membrane (Supplementary Results, Supplementary Fig. 1a). We invested considerable effort investigating buffer conditions, and found that Ca2+ was required for hydrolase activity, and both EDTA and EGTA abolished activity (Supplementary Fig. 1b). Resting cytosolic Ca2+ level is in the nano-molar range and can only go up to micromolar range transiently, so this observation again suggested that the activity is unlikely to reside in the cytosol. 2′3′-cGAMP was degraded much faster than 3′3′-cGAMP, suggesting that the predominant, Ca2+-requiring hydrolase may prefer the 2′-5′ phosphodiester bond (Fig. 1a).
Figure 1. There is one dominant hydrolase activity in the plasma membrane/heavy organelles and it is not due to PDE12.
(a) 2′3′-cGAMP and 3′3′-cGAMP hydrolysis reactions. THP-1 cells were lysed using 1% NP-40. The 32P-labeled CDNs were incubated with the cell lysate for 20 hours and the reaction was monitored using a TLC assay and visualized by autoradiography. (b) 2′3′-cGAMP hydrolase activity in differential centrifugation fractions of MDA-MB231 cells. Cells were suspended in isotonic buffer and lysed with a Dounce homogenizer. Supernatant and pellets from spins at the indicated speeds were solubilized and assayed in 1% NP-40, 20 mM Tris-HCl, pH 7.5, 1 mM Ca2+. (c) Knockdown of PDE12 does not decrease hydrolase activity. MDA-MB231 cells were transfected with four siRNA oligos against PDE12 and a control oligo. After 4 days, cells were lysed in the same buffer as (b) and assayed for activity. Upper panel: hydrolase activity in cells treated with different siRNA oligos. Lower panel: PDE12 levels in these cells. (d) Activity of purified PDE12 compared with that of MDA-MB231 whole cell lysate.
Although there are 11 classes of phosphodiesterases (PDE1-PDE11) that degrade the 3′-5′ phosphodiester bond in cAMP and cGMP, these PDEs are not thought to degrade 2′-5′ phosphodiesters. Only PDE12 is known to hydrolyze a 2′-5′ phosphodiester bond24. In addition, PDE12 resides in the matrix of mitochondria, consistent with the organelle/membrane-bound hydrolase activity we observed25. We therefore tested the hypothesis that PDE12 might account for the hydrolase activity in detergent extracts of MDA-MB231 cells, a metastatic breast cancer cell line that expresses high levels of PDE12. We performed differential centrifugation on MDA-MB231 cells and again observed no activity in the cytosol, but in the 7000 G pellet, indicative of heavy membranes or heavy organelles (Fig. 1b). We dissolved the pellet in NP-40 and performed column fractionation. The activity of the fractions indicates that there is one dominant hydrolase (Supplementary Fig. 2 upper panel). To test whether this activity is due to PDE12, we blotted these fractions using a PDE12 antibody (Supplementary Fig. 2 lower panel). Unambiguously, active fractions and fractions containing PDE12 do not overlap, suggesting that PDE12 does not account for the major hydrolase activity. We further confirmed this conclusion by knocking down PDE12 in these cells. All four siRNA sequences effectively knocked down PDE12 on the protein level, but had no effect on the hydrolase activity in the cell lysate (Fig. 1c). Using purified PDE12 protein, we showed that it does not have activity towards 2′3′-cGAMP at concentrations much higher than its physiologically concentration (Fig. 1d). Together, our results demonstrated that even though the hydrolase activity localizes in heavy membranes or organelles, it is not from PDE12.
ENPP1 is the dominant 2′3′-cGAMP hydrolase
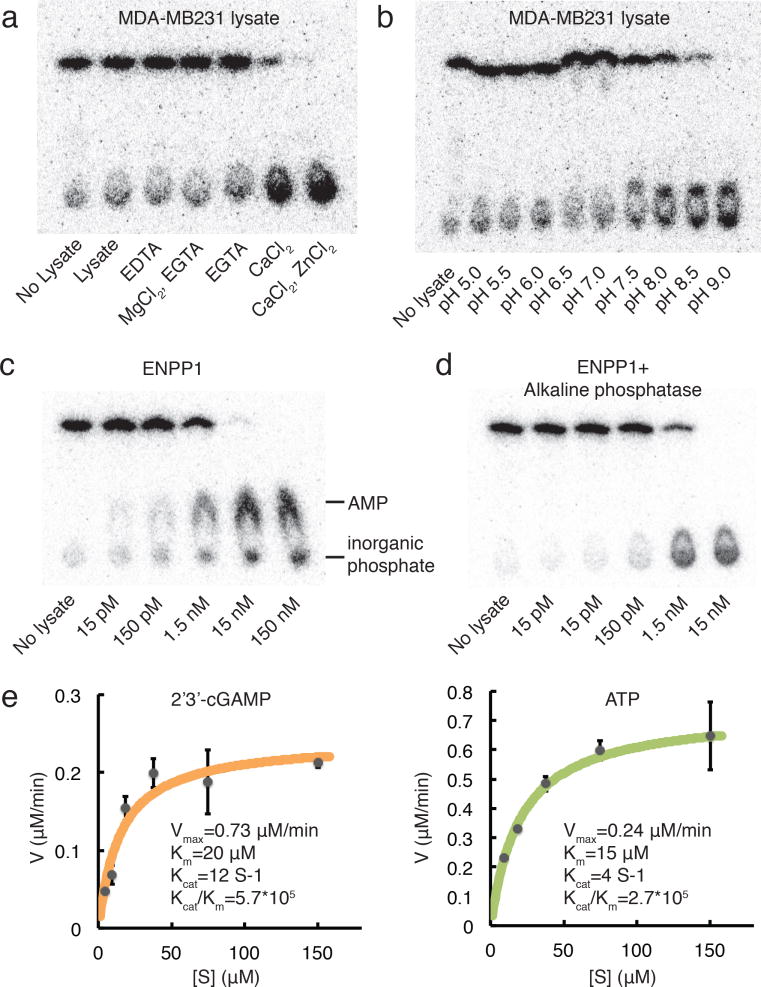
We next sought a bulk tissue source to purify and identify the hydrolase. Liver is a classic source for organelle and enzyme purification. We detected high activity in mouse livers, again was in the 7000 G pellet (Supplementary Fig. 3a). Because a calf liver provides more starting material, we performed differential centrifugation followed by detergent solubilization, anion exchange fractionation, and size exclusion fractionation on calf liver extract (Supplementary Fig. 3b-h). Fraction 26 from the last purification step has low protein concentration undetectable by SDS-PAGE gel, but high hydrolytic activity. Mass spectrometry analysis of this fraction revealed 377 proteins and a top ranking protein annotated as ecto-nucleotide pyrophosphatase/phosphodiesterase (ENPP1) appeared to be the most plausible candidate (Supplementary Dataset)26,27. ENPP1 is a plasma membrane and ER lumen protein28, which agrees with our cell fractionation results. In addition, the structure of ENPP1 revealed that it has a Ca2+ binding domain and chelates two Zn2+ ions in the catalytic site29. Consistent with the ion dependency of ENPP1, addition of Ca2+and Zn2+ both boosted the hydrolase activity in the active fractions (Fig. 2a and Supplementary Fig. 4a). Moreover, we found the optimal pH for the liver hydrolase activity to be 9.0, which agrees with that of ENPP1 (Fig. 2b and Supplementary Fig. 4b). We noticed that 2′3′-cGAMP migrates differently at pH 6.5 and 7.0 compared to other pH conditions. This is due to changed polarity of 2′3′-cGAMP at these conditions rather than its hydrolysis state because the same shift can be observed without lysate (Supplementary Fig. 5a). Nevertheless, 2′3′-cGAMP and the hydrolysis product at the baseline are well separated in all conditions. Finally, recombinant ENPP1 at nano-molar concentrations efficiently degraded 2′3′-cGAMP (Fig. 2c). The degradation product co-migrates with AMP on TLC. Addition of alkaline phosphatase converted the product to inorganic phosphate, showing that no phosphodiester bonds remained after ENPP1 action (Fig. 2d). Together, our results suggest that ENPP1 accounts for the observed hydrolase activity.
Figure 2. ENPP1 is an efficient hydrolase for 2′3′-cGAMP.
(a) Ion dependency and (b) pH preference of the dominant hydrolase activity in MDA-MB231 cells. (c) Activity of recombinant ENPP1 alone or (d) coupled to alkaline phosphatase in buffer condition: 0.2% NP-40, 20 mM Tris-HCl, pH 9.0, 2 mM Ca2+, 200 μM Zn2+. (e) Kinetics of 2′3′-cGAMP and ATP hydrolysis by recombinant ENPP1. 1 nM ENPP1 was tested in the same buffer condition as (f) and (g). Data are presented as mean and standard error.
ENPP1 has high 2′-3′ cGAMP hydrolase activity
The best-characterized substrate of ENPP1 is ATP, which is hydrolyzed to AMP and PPi29. We compared the activity of ENPP1 towards 2′3′-cGAMP and ATP. Kinetic analysis showed that ENPP1 efficiently hydrolyzes both ATP (Kcat= 12 s-1, Km= 20 μM) and 2′3′-cGAMP (Kcat= 4 s-1, Km= 15 μM) (Fig. 2e). Thus it is almost as active on 2′3′-cGAMP as on its known substrate ATP.
ENPP1 is necessary for 2′3′-cGAMP hydrolysis
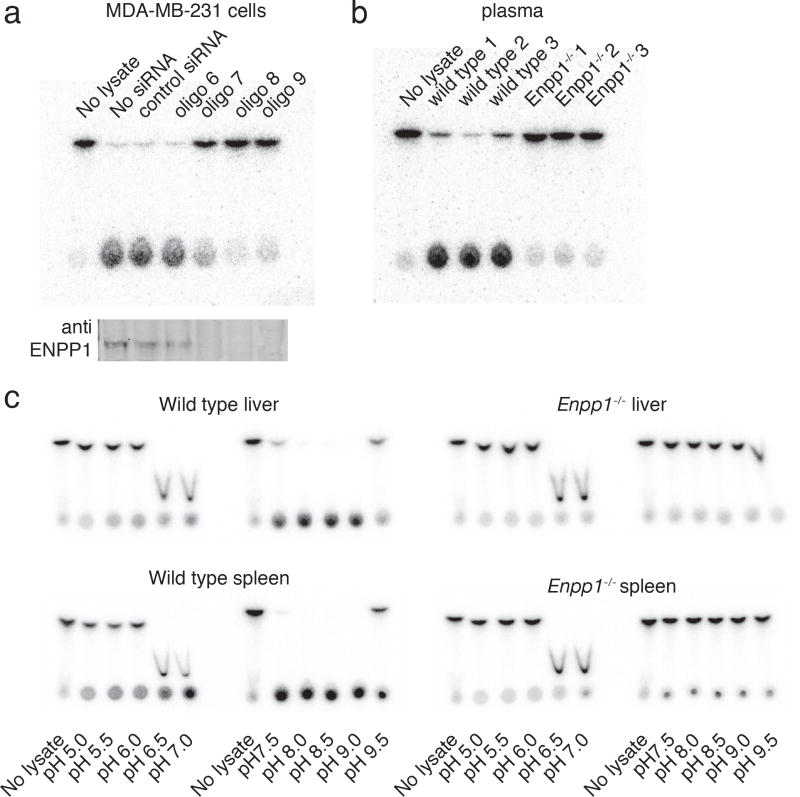
To test whether ENPP1 accounts for the dominant hydrolase activity in cells, we performed knockdown experiments in MDA-MB231 cells. siRNA oligos 7-9 against ENPP1 efficiently knocked down ENPP1 protein level and also significantly reduced the hydrolase activity in the whole cell lysate. Oligo 6 and control siRNA against PDE12 did not affect ENPP1 protein level and also did not change the hydrolase activity. These results indicate that the dominant 2′3′-cGAMP hydrolase in this cell line is ENPP1 (Fig. 3a).
Figure 3. ENPP1 is the dominant hydrolase activity for 2′3′-cGAMP.
(a) Knockdown of ENPP1 in MDA-MB231 cells diminished hydrolase activity. Four siRNA oligos against ENPP1 with a control siRNA sequence against PDE12 were used. 4 days after siRNA transfection, cells were lysed and assayed for activity (upper panel) and blotted for ENPP1 level (lower level). (b) Hydrolase activity in plasma from Enpp1-/- mice and their littermates. (c) Western blot characterization of Enpp1-/- mice. (d) Hydrolase activity in livers and spleens from Enpp1-/- mice and their littermates. Livers and spleens were minced and Dounce homogenized in lysis buffer: 1% NP-40, 20 mM Tris-HCl, pH 7.5, protease inhibitor cocktail. The assay was conducted in 0.2% NP-40, 20 mM Tris-HCl, 150 mM KCl, 2 mM Ca2+, 2 mM Mg2+, 200 μM Zn2+ at the indicated pH. NaOAc buffer was used for pH 5.0-6.0; PIPES buffer was used for pH 6.5 and 7.0 and 2′3′-cGAMP runs at a lower Rf in this buffer; Tris-HCl buffer was used for pH 7.5-9.0, and Borate buffer was used pH 9.5. These buffer conditions were also used in liver and spleen extract studies.
ENPP1 is an ecto-enzyme that can be shed/secreted into the serum30. Indeed, we detected high hydrolase activity in fetal bovine serum and human serum (Supplementary Fig. 6). Like the hydrolase activity in MDA-MB231 cells and in calf liver fractions, bovine and human serum hydrolase activity was also pH dependent, with peak activity at pH ∼9.0, agreeing with the profile of ENPP1. To test whether ENPP1 accounts for most of the serum hydrolase activity, we tested plasma from Enpp1-/- mice. While plasma from their wild-type littermates exhibited hydrolase activity, plasma from Enpp1-/- mice did not (Fig. 3b). Therefore, ENPP1 is the dominant hydrolase for 2′3′-cGAMP in mouse plasma. Given the pH profile and similar activity, we suspect the same is true in human serum.
We next tested whether ENPP1 is the dominant 2′3′-cGAMP hydrolase in the liver and spleen. We chose to study the liver because it is the major site for drug metabolism and the spleen because of its important role in the immune system. We compared the hydrolase activity of livers and spleens from Enpp1-/- mice and their wild type littermates. Since we could have missed other hydrolases during our purification by using conditions not optimized for them, we surveyed a wide range of pH values and divalent ion concentrations. The migration profile of pure 2′3′-cGAMP at different pH conditions was generated to serve as the starting point (Supplementary Fig. 5b). At a point when extracts from wild-type counterparts had completely degraded 2′3′-cGAMP, extracts from ENPP1-/-livers and spleens showed undetectable levels of degradation (Fig. 3c). Together, our results demonstrate that ENPP1 is the dominant 2′3′-cGAMP hydrolase in vivo, at least in mice under our assay conditions.
Phosphothioate analogs are resistant to hydrolysis
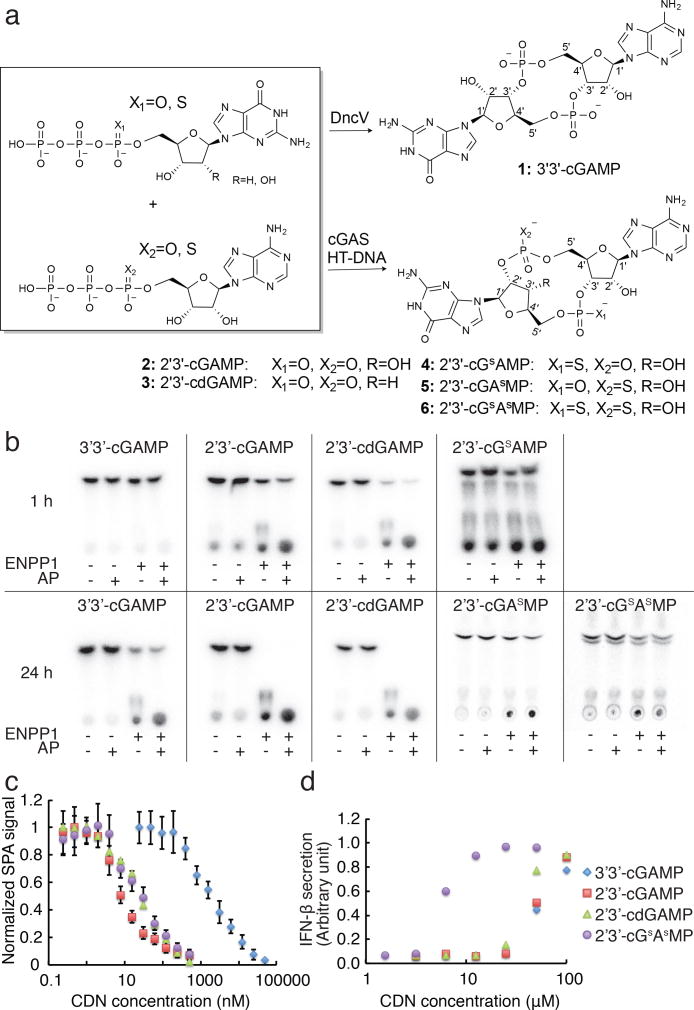
Phosphothioate diester linkages are often resistant to hydrolysis by phosphodiesterases and nucleases, and have been used to build important non-hydrolyzable ATP and GTP analogs. We enzymatically synthesized 2′3′-cGAMP analogs that used phosphothioate linkages in place of either the 2′-5′ (5, 2′3′-cGAsMP) or the 3′-5′ (4, 2′3′-cGsAMP) phosphodiester linkage, or both (6, 2′3′-cGsAsMP) (Fig. 4a and Supplementary Fig. 7-9). A higher enzyme concentration was required for 2′3′-cGsAsMP synthesis and 2′3′-cGsAMP synthesis was incomplete, indicating that cGAS is less efficient with GTPαS as a starting material. Nevertheless, sufficient 2′3′-cGsAMP was made to test its resistance to ENPP1 hydrolysis. We did not attempt to separate the diastereomers of these phosphothioate analogs, but rather used them as mixtures, which are sufficient for proof of concept. We also synthesized a 3′-deoxy analog (3, 2′3′-cdGAMP) to evaluate the role of the 3′-hydroxyl group adjacent to the 2′-5′ phosphodiester bond.
Figure 4. Development of hydrolysis resistant hSTING agonists.
(a) Scheme of enzymatic synthesis of 3′3′-cGAMP and 2′3′-cGAMP analogs. For 3′3′-cGAMP, 50 nM of DncV was incubated with 1 mM ATP and 1 mM GTP in 1 mL buffer containing 20 mM Tris-HCl, pH 8.0, 20 mM MgCl2 for 3 h at room temperature. For 2′3′-cGAMP analogs, 1-10 μM mouse cGAS (amino acid residues 147-507) was incubated with 1 mM ATP, 1 mM GTP, and 0.1 mg/mL HT-DNA in 1 mL of the same buffer for 12 h at room temperature. (b) Hydrolysis reactions of ENPP1 (1 nM) with the analogs (10 μM). (c) Scintillation proximity assay to measure the binding affinity of the analogs towards hSTING (amino acid residues 139-379). Biotinylated hSTING (100 nM) was immobilized onto 96-well streptavidin coated SPA plates. Neat 35S-labeled 2′3′-cGAsMP (500 pM) was used as the probe. N=3 samples. Data are presented as mean and standard error. (d) IFN-β production in THP-1 cells stimulated with the analogs. THP-1 cells were incubated with the analogs at the indicated concentrations for 24 h. IFN-β in the media was measured using a HEK-SEAP cell line. Representative data from a biological triplicate is depicted.
We first tested the stability of these analogs in THP-1 cell lysates. Their relative stability ranks as 2′3′-cGsAsMP>3′3′-cGAMP≫2′3′-cGAMP (Supplementary Fig. 10a). We then tested their stability towards 1 nM recombinant ENPP1, the concentration used in the kinetic analysis, either by itself or coupled with alkaline phosphatase. 2′3′-cGAsMP and 2′3′-cGsAsMP were stable for at least 1 day, while all the other 2′3′-cGAMP analogs exhibited half-lives of around an hour in the presence of ENPP1 (Fig. 4b). 2′3′-cGsAsMP was able to inhibit 2′3′-cGAMP hydrolysis suggesting it is capable of binding to ENPP1 and acting as a competitive inhibitor (Supplementary Fig. 10b). The increased stability of 2′-5′ phosphothioate analogs (at least 40 times more stable than 2′3′-cGAMP) suggest that cleavage of the 2′-5′ phosphodiester linkage is the first step and also the rate-limiting step catalyzed by ENPP1. This is consistent with our early observation that 3′3′-cGAMP, the bacterial CDN, is stable in the presence of mammalian cell lysate. Testing with pure ENPP1 confirmed that 3′3′-cGAMP is not a substrate (Fig. 4b). Since 2′3′-cdGAMP is an ENPP substrate, the 3′-hydroxyl group is dispensable in the catalytic reaction (Fig. 4b). The relative stability of the analogs towards recombinant ENPP1 shares the same trend as in whole cell lysates. This structure-activity relationship study suggests that ENPP1 is the direct hydrolase of 2′3′-cGAMP rather than indirectly involved in regulation/activation of another hydrolase in the cell lysate.
2′3′-cGsAsMP has excellent STING agonist activity
We next tested the hSTING binding activity of 2′3′-cGsAsMP. We previously used fluorescence polarization and a fluorescent derivative of cyclic-di-GMP to measure affinity of drugs for mSTING7, but this approach failed with hSTING. Instead, we developed a scintillation proximity assay (SPA) based competition binding assay (Supplementary Fig. 11). hSTING was biotinylated and immobilized onto streptavidin coated SPA plates. 35S-labeled 2′3′-cGsAsMP was used as a probe. The SPA signal increased with increasing amount of probe and saturated at 10 nM, with a Kd of ∼ 5 nM. We used 500 pM of the probe in the competition assay, a concentration that yielded a high SPA signal, but is well below the estimated Kd of the probe (Kdprobe). We then titrated in the analogs to measure their IC50s. According to the Cheng-Prusoff equation Kd = IC50/(1 + [probe]/Kdprobe). Because [probe]≪Kdprobe, the Kds of the analogs are roughly equal to the measured IC50s in this assay (Fig. 4c). We tested four CDN analogs using this assay. Consistent with previous reports that used calorimetric readouts13, 3′3′-cGAMP is a much weaker ligand for hSTING ligand binding domain in vitro than 2′3′-cGAMP. The affinities of 2′3′-cGsAsMP and 2′3′-cdGAMP were comparable to that of 2′3′-cGAMP. Thus the non-hydrolyzable double phosphothioate analog is a good STING ligand.
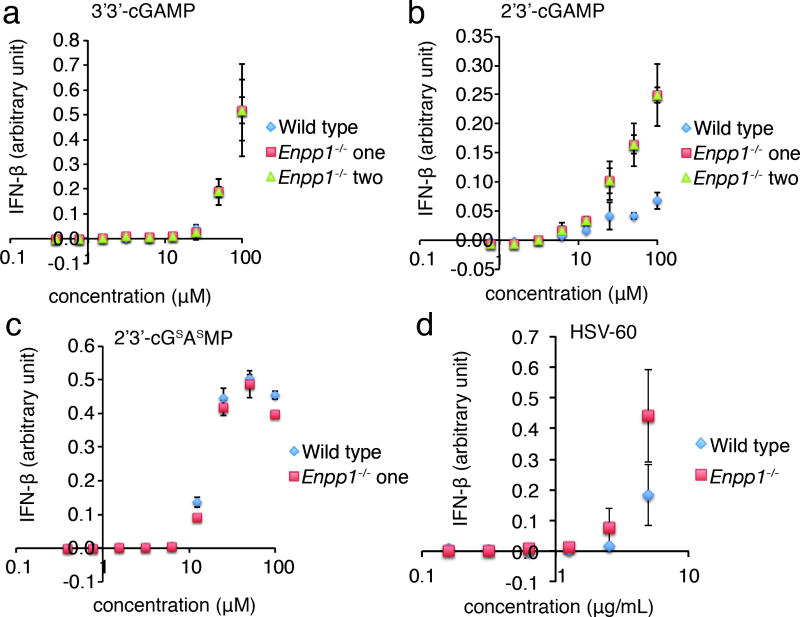
We next tested whether the analogs can activate the STING pathway. All four analogs induced TBK1 phosphorylation within 2 hours of drug addition in HEK 293 cells that express STING, suggesting direct activation (Supplementary Fig. 12a). We also tested HEK 293T cells, a sub-clone of the HEK 293 cells that do not express STING. 2′3′-cGsAsMP activated the TBK1/IRF3 axis in HEK 293, but not in HEK 293T cells, suggesting STING dependent signaling (Supplementary Fig. 12b). To test the performances of these analogs in inducing IFN-β, monocytic human THP-1 cells were treated with the analogs for 24 hours and secreted IFN-β was measured using HEK-Blue™ IFN-α/β Cells, a reporter cell line that responds to IFN-β by secreting alkaline phosphatase. The non-hydrolyzeable 2′3′-cGsAsMP was the most potent analog at activating IFN-β (Fig. 4d), scoring with an EC50 ∼10-fold lower than natural 2′3′-cGAMP. To test whether its higher activity is due to increased biostability, we tested lung fibroblast cells from Enpp1-/- mice and their wild type littermates. The hydrolysis resistant 3′3′-cGAMP and 2′3′-cGsAsMP activated IFN-β to similar levels regardless of ENPP1 expression. On the contrary, 2′3′-cGAMP is more active in Enpp1-/- cells, presumably due to prolonged half-life (Fig. 5a-c for female mice and Supplementary Fig. 13 for male mice). The advantage of 2′3′-cGsAsMP over 2′3′-cGAMP is much smaller in Enpp1-/- cells suggesting that biostability contributes significantly to its improved cellular activity.
Figure 5. ENPP1 dampens 2′3′-cGAMP signaling.
Lung fibroblast cells from wild type and Enpp1-/- female mice were incubated with 2′3′-cGAMP analogs and HSV-60 at the indicated concentrations for 24 h. IFN-β in the media was measured using a B16-SEAP cell line. N=3 samples. Data are presented as mean and standard error.
Finally, to determine whether ENPP1 is important in shutting down the endogenous 2′3′-cGAMP pathway, we used HSV-60 (a 60 bp oligonucleotide containing viral DNA motifs) to activate this pathway. Enpp1-/- cells expressed significantly more IFN-β in response to HSV-60 than wild type cells, demonstrating its role as a negative regulator of the endogenous 2′3′-cGAMP pathway (Fig. 5d).
Discussion
We identified a candidate 2′3′-cGAMP hydrolase by partially purify this activity from a calf liver. Our purification procedures were remarkably short and the final fraction is rather crude. However, mass spectrometry analysis of this crude fraction was sufficient to generate the hypothesis that ENPP1 degrades 2′3′-cGAMP. We then used genetics to show that the dominant hydrolase activity for 2′3′-cGAMP in cells, tissues, and blood indeed is ENPP1. While we cannot rule out the possibility that other enzymes escaped our search because we did not find the optimum assay conditions for them, we showed that MDA-MB231 cells with ENPP1 knocked down and plasma, livers, and spleens from Enpp1-/- mice all have negligible 2′3′-cGAMP hydrolase activity. The modern methods used in this study (early proteomic analysis of fractions and testing of mouse knockout tissues) show how proteins can be identified by activity-guided fractionation much more easily and more reliably now than in the past.
ENPP1 inactivating mutations and overexpression are known to cause a variety of disease phenotypes in man and mice. This enzyme has rather broad substrate specificity, including ATP and NAD+, and we have now shown that 2′3′-cGAMP is almost as good a substrate for recombinant ENPP1 as is ATP. This opens the question of which substrate(s) are involved in the ENPP1-related disease phenotypes. For example, ENPP1 is known to play important roles in bone mineralization. ENPP1 inactivating mutations are the cause of generalized arterial calcification of infancy (GACI), a deadly disease31. The current understanding is that ENPP1 hydrolyzes extracellular ATP to AMP and PPi, and PPi is a potent inhibitor of bone mineralization and calcification32. ENPP1 is also highly expressed in some breast tumors33, and ENPP1 expression has been correlated with tamoxifen resistance34. Finally, a three-allelle haplotype of ENPP1 is associated with obesity and higher risk of type-II diabetes35, although this linkage has not been found in all ethnic groups. The putative molecular mechanism for ENPP1's involvement in insulin signaling is that ENPP1 interacts with the insulin receptor directly to inhibit its phosphorylation and activation36. Studies of ENPP1 in mice also showed that high levels of ENPP1 suppress insulin signaling37. The enzymatic activity of ENPP1 is required for its inhibitory effect on insulin receptor signaling38. Our findings raise the question of whether 2′3′-cGAMP hydrolysis is responsible for any of these ENPP1 related diseases.
Our discovery of ENPP1 as 2′3′-cGAMP's major hydrolase sheds light on the regulation of the dsDNA-cGAS-STING innate immune pathway. This pathway has been shown to be downregulated by ULK-1 activation39 and autophagy-mediated degradation of cGAS40. Loss of cytosolic 2′3′-cGAMP, either through hydrolysis or export, could provide another negative regulation mechanism, but we found no evidence for a 2′3′-cGAMP hydrolase activity in the cytosol from tissue culture cells, liver, or spleen. Our data indicate that this novel 2nd messenger must be longer lived than cAMP and cGMP in the cytosol, which are rapidly hydrolyzed by PDEs in the cytosol. This hypothesis is consistent with the observation that 2′3′-cGAMP can travel long distances through gap junctions41.
ENPP1 activity is not found in the cytosol. Instead it is found both on the basal lateral surface of the plasma membrane in hepatocytes and in the rough endoplasmic reticulum (ER) fraction from the liver. In the ER, its catalytic domain resides in the lumen28, where there is high Ca2+ concentration necessary for its activity. Therefore, 2′3′-cGAMP must be transported across a membrane to be degraded, and may have undiscovered activities once there. We do not know whether the ER lumen or the extracellular space is the major site for 2′3′-cGAMP degradation. There is no information on how permeable the ER membrane is to 2′3′-cGAMP. It is likely that there is an export mechanism for 2′3′-cGAMP, since many other nucleotide derivatives (e.g. ATP, NAD, cAMP, and cGMP) have specific efflux pumps and have interesting extracellular biology. We (Fig. 4d) and others9 have shown that this double negatively charged small molecule functions when added to the media without transfecting reagents, suggesting that a specific import mechanism may also exist. Given these results, we hypothesize that 2′3′-cGAMP may have interesting extracellular biology that is waiting to be explored (Supplementary Fig. 14).
Finally, our findings have major implications for the development of CDN based hSTING agonist drugs. Since 2′3′-cGAMP is the best ligand for hSTING (Fig. 4c)13, it is at least in principle a good starting point for drug development. 2′3′-cGAMP is active when added to the outside of cells, and has shown adjuvant activity in mice when injected intramuscularly16, so it is apparently sufficiently stable to show some drug-like activity. Consistent with this, we find that it has a half-life in the range of minutes to 1 hour in various biofluids and tissue lysates. Our bis-phosphothioate analog 2′3′-cGsAsMP, which is ∼40 X more resistant to ENPP1 hydrolysis, is expected to last much longer in biofluids, and showed ∼10 X higher cell-based activity in culture (Fig. 4d). This higher activity was not due to tighter STING binding (Fig. 4c), and is likely to be due to increased biostability (Fig. 5). We propose that 2′3′-cGsAsMP will be a useful tool compound in further exploring the biology and metabolism of 2′3′-cGAMP, and for testing whether CDN analogs deserve further development as clinical candidates.
Online methods
Reagents, antibodies, and cell lines
ATP, [α-32P]- 3000 Ci/mmol 10 mCi/ml, and ATPαS, [35S]-1250 Ci/mmol, 12.5 mCi/ml, 250 μCi were purchased from Perkin Elmer. Adenosine-5′-O-(1-Thiotriphosphate), Guanosine-5′-O-(1-Thiotriphosphate), and 3′-Deoxyguanosine-5′-Triphosphate were purchased from Trilink Biotechnologies. ATP, GTP, and herring testes DNA were purchased from Sigma-Aldrich. EZ-Link Sulfo-NHS-LC-Biotin was purchased from Pierce (21335). Set of 4: ON-TARGETplus PDE12 siRNA (LQ-017946-01-0002) and Set of 4: ON-TARGETplus ENPP1 siRNA (LQ-003809-00-0002) were purchased from Dharmacon. Anti-PDE12 antibody (ab87738) was from Abcam. Anti-ENPP1 Antibody (#2061) was from Cell Signaling. Rabbit polyclonal antibodies against TBK, pTBK (S172), pIRF3 (S396), STING were purchased from Cell Signaling Technology. HSV-60/LyoVec™, HEK-Blue™ IFN-α/β cells and B16-Blue™ IFN-α/β cells were purchased from Invivogen. Recombinant ENPP1 was purchased from R&D systems (6136-EN-010). Collagenase D was from Roche and Trpsin was from Sigma.
Cell culture and siRNA transfection
L929 and HEK-Blue™ IFN-α/β Cells were maintained in DMEM (Cellgro) supplemented with 10% FBS (GIBCO) (v/v) and 1% of penicillin-streptomycin (Cellgro). THP-1 cells were grown in RPMI (Cellgro) supplemented with 10% FBS, 0.1% β-mercaptoethanol (GIBCO), and 1%of penicillin-streptomycin. MDA-MB231 cells were grown in RPMI supplemented with 10% FBS and 1% of penicillin-streptomycin.
Expression and purification of recombinant proteins
All procedures involving proteins and cell lysates were conducted at 4 °C unless stated otherwise.
DncV expression plasmid was a generous gift from Dr. John J. Mekalanos and the protein was produced using the established protocol12.
DNA sequence encoding mouse cGAS (147-507) was amplified from the cDNA library generated from L929 cells using the primer pair: mcGAS-147-FWD TATTGAGGCTCACAGAGAACAGATTGGTGGT CCGGACAAGCTAAAGAAGGTGCT and mcGAS-REV GGATCCCCTTCCTGCAGTCACCCGGGCTCGAG TCAAAGCTTGTCAAAAATTGGAAA. The PCR product was inserted into the SapI and XhoI sites of pTB146 (a generous gift from Dr. Thomas Bernhard) using isothermal assembly. His-SUMO tagged mcGAS (147-507) was expressed in Rosetta™ Competent Cells. Cells were grown in LB media with Ampicilin (100 ng/mL) and were induced with 0.5 mM IPTG when OD reached 1 and were allowed to grow overnight at 16 °C. Cells were pelleted and lysed. The cell extract was then cleared by ultracentrifugation at 45,000 rpm for 1 h. The cleared supernatant was incubated with Ni-NTA beads (4 mL of beads per liter of bacteria culture). Ni beads were washed with 20 mM imidazole in PBS and protein was eluted with step concentrations of 50 mM to 500 mM imidazole in PBS. Fractions containing his-SUMO-mcGAS were pooled, concentrated and dialyzed against PBS. The protein was further purified by size-exclusion chromatography in running buffer: 20 mM Tris HCl (pH 8.0), 150 mM NaCl, and 2 mM Mg2+. Fractions containing mcGAS (147-507) was pooled, concentrated, and snap frozen for future use.
DNA encoding full-length human PDE12 was cloned into a modified version of pDB-His-MBP vector. The protein was expressed in E. coli BL21 DE3 RIPL Codon-Plus cells. E. coli cell was induced by 0.4 mM IPTG when cell density reached 0.6 and grew at 20°C overnight. Cells were pelleted and lysed. After centrifugation and removal of cell debris, supernatant was incubated with Ni-NTA beads. Ni beads were washed extensively and protein was eluted in lysis buffer with 300 mM imidazole. After cleavage by PreScission protease, the N-terminal His6-MBP was removed by incubation with Ni-NTA beads. PDE12 protein in the flow-through fractions was further purified by size-exclusion chromatography.
Enzymatic synthesis of 3′3′-cGAMP and 2′2′-cGAMP analogs
For 3′3′-cGAMP, 50 nM of DncV was incubated with 1 mM ATP and 1 mM GTP in 1 mL buffer containing 20 mM Tris-HCl, pH 8.0, 20 mM MgCl2 at room temperature for 3 h. For 2′3′-cGAMP analogs, 1-10 μM mouse cGAS (amino acid residues 147-507) was incubated with 1 mM ATP, 1 mM GTP, and 0.1 mg/mL HT-DNA in 1 mL of the same buffer for 12 h at room temperature. The reaction mixtures were then heated at 95 °C for 3 min and filtered through a 3 KDa filter. The product was purified using a silica plug with 5 mM NH4HCO3: 15% H2O: 85% EtOH as the mobile phase. To make the 32P and 35S-labeled CDNs for degradation assays, 10 μCi [α-32P] ATP or [35S] ATPαS were mixed in with the cold starting materials. The reaction mixture was filtered through a 3 KDa filter and used without further purification. To synthesize neat 35S-labeled 2′3′-cGAsMP for the SPA assay, 250 μCi [35S] ATPαS and 100 μM cold GTP were used as the starting material in 100 μL buffer. The product was purified using a silica plug with 5 mM NH4HCO3: 15% H2O: 85% EtOH as the mobile phase.
Mice
Generation and characterization has been reported previously for Enpp1-/- mice42,43. Genotyping was performed with PCR protocols on genomic DNA. Anesthesia was given intraperitonal with Avertin. Blood was collected by cardiac puncture into lithium heparin tubes; tissues were frozen immediately in liquid nitrogen. The Institutional Animal Care and Use Committee (IACUC) approved all animal procedures.
Preparation of cell and tissue extracts
To determine whether THP-1 cells, L929 cells, and MDA-MB231 possess hydrolase activity for 2′3′-cGAMP, the whole cell lysate was generated using lysis buffer:10 mM Tris-HCl, pH 7.5,150 mM NaCl, 1% NP-40, 1.5 mM MgCl, 2 mM DTT, protease inhibitor tablet (Roche) at 1 mL for 100 million cells. To determine the subcellular location of the hydrolase(s), cells were lysed by douncing in either a mitochondria friendly buffer: 10 mM Tris-MOPS (pH 7.4), 10 mM EDTA/Tris, 200 mM sucrose or a hypotonic buffer: 10 mM Tris-HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 2 mM DTT, protease inhibitor tablet. The homogenate was then subjected to centrifugation at 600 G and 7000 G.
To prepare whole cell lysate, liver, and spleen extracts to test the role of ENPP1, the cells and tissue samples were minced and Douce homogenized in detergent containing buffer: 20 mM Tris-HCl, pH 7.5, 1% NP-40, protease inhibitor tablet. The lysate were tested without centrifugation clearance at different pHs in the presence of 150 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 200 μM ZnCl2. 100 mM NaOAc buffer was used for pH 5.0-6.0; 100 mM PIPES buffer was used for pH 6.5 and 7.0; 100 mM Tris-HCl buffer was used for pH 7.5-9.0; and 100 mM Borate buffer was used pH 9.5.
Thin-Layer Chromatography Analysis of CDN synthesis and degradation
This protocol was modified from established protocol 22. Reaction solution (1.5 μL) was spotted onto TLC plates (HPTLC silica gel, 60 Å pores, F254, 1055480001, MERCK Millipore) and the nucleotides were separated with 5 mM NH4HCO3: 15% H2O: 85% EtOH as the mobile phase at 25 °C for 30 min. The plates were visualized by UV (254 nm) or by autoradiography. Images were processed using Adobe Photoshop and Illustrator CS6.
Partial purification of ENPP1 from a calf liver
The calf liver was homogenized in buffer A: 10 mM Tris-MOPS (pH 7.4), 10 mM EDTA/Tris, 200 mM sucrose. For subcellular fractionation, the homogenate was centrifuged at 600 G for 20 min to remove cell debris and nuclei. The supernatant was centrifuged at 7000 G for 20 min to precipitate mitochondria and other heavy organelles. The pellet was re-suspended in 200 mL buffer A and centrifuged at 7000 G for 20 min. Both the supernatant and the pellet were analyzed for activity in reaction buffer: 20 mM Tris-HCl, pH 7.5, 1%NP-40, 1 mM CaCl2. The supernatant was where the activity resided, so was then centrifuged at 45000 G for 1 h and all the activity was precipitated. The pellet was extracted using the detergent-containing reaction buffer and cleared by centrifuging at 45000 G for 1h. The supernatant was subjected to anion exchange fractionation using a 5 mL HiTrap Q column followed by size exclusion fractionation using a superose 6 column. Fraction 26 contained high activity, but undetectable amount of protein by SDS-PAGE gel and coomassie staining. The entire lane was subjected to mass spectrometry analysis at the Taplin Mass Spectrometry Facility at Harvard Medical School as a standard service, which identified 377 proteins (Supplementary Dataset). ENPP1, which ranked #17 on the list, was the top candidate because it is the only phosphodiesterase on the list judging by their annotations.
Scintillation proximity assay
Wild type hSTING (230G/232R, amino acid residue 139-379) was prepared as described before7. The protein was dialyzed in PBS for 3 h to remove trace amount of Tris-HCl that would react with the labeling reagent. The protein was labeled at 300 μM with 3 equivalent of EZ-Link Sulfo-NHS-LC-Biotin at room temperature for 30 min. Another 3 equivalent of the labeling reagent was added and allowed to react for another 30 min. Bovine serum albumin was labeled using the same protocol as a control. The proteins were then dialyzed in PBS to remove free biotin. To immobilize the proteins onto 96-well streptavidin coated SPA plates (Perkin Elmer 1450-551), biotinylated hSTING and the BSA control (100 nM) were incubated on the plates for 30 min. The plates were then washed 3 times with PBS. To decide the Kd of the probe, serial dilutions of neat 35S-labeled 2′3′-cGAsMP (0.01 nM to 20 nM) were incubated with immobilized protein for 30 min. The probe was then aspirated and read on a 1450 MicroBeta TriLux for 3 min. For the competition assay, 500 pM 35S-labeled 2′3′-cGAsMP in PBS was used to make the serial dilutions of the CDNs. The cold CDN and probe mixtures were incubated with the protein plates for 30 min. The wells were aspirated dry and read on the MicroBeta plate reader.
SEAP assay
HEK-Blue™ IFN-α/β Cells were used to measure IFN-α/β produced by THP-1 cells. We followed the manufacturer recommended protocols. Briefly, 24 hours after drug treatment, conditioned media were collected and added to the HEK-Blue™ reporter cells. After 18 hours, SEAP activity was measured using a Victor plate reader (Perkin Elmer). B16-Blue™ IFN-α/β cells were used to measure IFN-α/β produced by mouse lung fibroblast cells. The assay protocol is the same as that for HEK-Blue™ IFN-α/β cells.
Isolation of lung fibroblast cells
Lungs from each mouse were minced in a Petri dish and digested in 5 mL of DMEM with 0.1% collagenase D and 0.2% trypsin at 37 °C for 1 h. The mixture was pipetted up and down every 10 min. Disassociated cells were washed twice in DMEM and pelleted at 1200 G, 4°C for 10 min to remove enzymes. Cells were re-suspended in complete media (DMEM, 10% FBS, Penstrep and Fungizone) and seeded in a 10-cm dish. The fibroblast cells attached in one hour and the rest of the cells were washed away by replacing the media.
Supplementary Material
Acknowledgments
We thank Peter Koch, Dr. Paul Choi, Dr. Kristin Krukenberg, Dr. Aaron Groen, Dr. Martin Loose, Dr. Sujeong Kim, and Dr. Scott Gruver for helpful discussions. We thank Dr. Ruomu Jiang for help with data analysis. We thank Dr. Kiki Chu for providing mouse livers. We thank Dr. Suzanne Walker for sharing the MicroBeta plate reader and Dr. Chenguang Fan for technical assistance. We thank Dr. Greg Heffron and Charles Sheahan for assistance with the NMR data collection and analysis. We thank Dr. Rebecca Ward for help with manuscript preparation. This research was supported by the National Cancer Institute (CA139980, AR53102, AI050872, and 1K99AI108793-01). Lingyin Li thanks the Jane Coffin Childs Fund for her postdoctoral fellowship.
Footnotes
Author Contributions: L.L. and T. J. M. developed the hypothesis and designed the study. L. L., Q. Y., P. K., and Z. M. conducted the experiments. All authors interpreted and discussed the results. L.L. and T. J. M. wrote the manuscript
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Duthie MS, Windish HP, Fox CB, Reed SG. Use of defined TLR ligands as adjuvants within human vaccines. Immunol Rev. 2011;239:178–96. doi: 10.1111/j.1600-065X.2010.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–50. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, et al. Anticancer Flavonoids Are Mouse-Selective STING Agonists. ACS Chem Biol. 2013;8:1396–401. doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon J, et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–25. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–62. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–8. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–70. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–35. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–4. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–61. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XD, et al. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 18.Maurice DH, et al. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13:290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wielinga PR, et al. Characterization of the MRP4- and MRP5-mediated transport of cyclic nucleotides from intact cells. J Biol Chem. 2003;278:17664–71. doi: 10.1074/jbc.M212723200. [DOI] [PubMed] [Google Scholar]
- 20.Jedlitschky G, Burchell B, Keppler D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol Chem. 2000;275:30069–74. doi: 10.1074/jbc.M005463200. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, et al. MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2′,3′-dideoxycytidine and 9′-(2′-phosphonylmethoxyethyl) adenine. J Biol Chem. 2003;278:29509–14. doi: 10.1074/jbc.M304059200. [DOI] [PubMed] [Google Scholar]
- 22.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota K, et al. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J Biol Chem. 2004;279:37832–41. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 25.Poulsen JB, et al. Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 2011;39:3754–70. doi: 10.1093/nar/gkq1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goding JW, et al. Ecto-phosphodiesterase/pyrophosphatase of lymphocytes and non-lymphoid cells: structure and function of the PC-1 family. Immunol Rev. 1998;161:11–26. doi: 10.1111/j.1600-065x.1998.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 27.Bollen M, Gijsbers R, Ceulemans H, Stalmans W, Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit Rev Biochem Mol Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
- 28.Bischoff E, Tran-Thi TA, Decker KF. Nucleotide pyrophosphatase of rat liver. A comparative study on the enzymes solubilized and purified from plasma membrane and endoplasmic reticulum. Eur J Biochem. 1975;51:353–61. doi: 10.1111/j.1432-1033.1975.tb03935.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato K, et al. Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16876–16881. doi: 10.1073/pnas.1208017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belli SI, van Driel IR, Goding JW. Identification and characterization of a soluble form of the plasma cell membrane glycoprotein PC-1 (5′-nucleotide phosphodiesterase) Eur J Biochem. 1993;217:421–8. doi: 10.1111/j.1432-1033.1993.tb18261.x. [DOI] [PubMed] [Google Scholar]
- 31.Rutsch F, et al. PC-1 nucleoside triphosphate pyrophosphohydrolase deficiency in idiopathic infantile arterial calcification. Am J Pathol. 2001;158:543–54. doi: 10.1016/S0002-9440(10)63996-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hessle L, et al. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc Natl Acad Sci U S A. 2002;99:9445–9. doi: 10.1073/pnas.142063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau WM, et al. Enpp1: A Potential Facilitator of Breast Cancer Bone Metastasis. Plos One. 2013;8:5. doi: 10.1371/journal.pone.0066752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umar A, et al. Identification of a Putative Protein Profile Associated with Tamoxifen Therapy Resistance in Breast Cancer. Molecular & Cellular Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyre D, et al. Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet. 2005;37:863–7. doi: 10.1038/ng1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rey D, et al. Amerindians show no association of PC-1 gene Gln121 allele and obesity: a thrifty gene population genetics. Mol Biol Rep. 2012;39:7687–93. doi: 10.1007/s11033-012-1604-1. [DOI] [PubMed] [Google Scholar]
- 37.Maddux BA, et al. Membrane glycoprotein PC-1 and insulin resistance in non-insulin-dependent diabetes mellitus. Nature. 1995;373:448–51. doi: 10.1038/373448a0. [DOI] [PubMed] [Google Scholar]
- 38.Chin CN, et al. Evidence that inhibition of insulin receptor signaling activity by PC-1/ENPP1 is dependent on its enzyme activity. Eur J Pharmacol. 2009;606:17–24. doi: 10.1016/j.ejphar.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–98. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Q, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15:228–38. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–4. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sali A, F JM, Terkeltaub R, Goding JW. Germline deletion of the nucleoside triphosphosphate (NTPPPH) plasma cell membrane glycoprotein (PC-1) produces abnormal calcification of periarticular tissues. In: Vanduffel L, Lemmens R, editors. Ecto-ATPases and Related Ectonucleotidases. Shaker Publishing BV; Maastricht, The Netherlands: 1999. pp. 267–282. [Google Scholar]
- 43.Narisawa S, et al. Novel inhibitors of alkaline phosphatase suppress vascular smooth muscle cell calcification. J Bone Miner Res. 2007;22:1700–10. doi: 10.1359/jbmr.070714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.