Abstract
Neocortical interactions with dorsal striatum support many motor and executive functions, and such underlying functional networks are particularly vulnerable to a variety of developmental, neurological, and psychiatric brain disorders, including autism spectrum disorders, Parkinson’s disease, and Huntington’s disease. Relatively little is known about the development of functional corticostriatal interactions, and in particular, virtually nothing is known of molecular mechanisms that control generation of prefrontal cortex-striatal circuits. Here, we used regional and cellular in situ hybridization techniques coupled with neuronal tract tracing to show that Cadherin 8 (Cdh8), a homophilic adhesion protein encoded by a gene associated with autism spectrum disorders and learning disability susceptibility, is enriched within striatal projection neurons in medial prefrontal cortex and in striatal medium spiny neurons forming the direct- or indirect-pathways. Developmental analysis of quantitative RTPCR and Western blot data show that Cdh8 expression peaks in prefrontal cortex and striatum at P10, when cortical projections start to form synapses in the striatum. High-resolution immunoelectron-microscopy shows Cdh8 is concentrated at excitatory synapses in dorsal striatum, and Cdh8 knockdown in cortical neurons impairs dendritic arborization and dendrite self-avoidance. Taken together our findings indicate that Cdh8 delineates developing corticostriatal circuits where it is a strong candidate for regulating the generation of normal cortical projections, neuronal morphology, and corticostriatal synapses.
Indexing terms: cadherin, prefrontal cortex, striatum, cell adhesion molecules, synapse, immunogold
Introduction
Neocortical projections to the striatum underlie a host of motor and executive functions. The dorsolateral striatum receives topographically organized projections from sensorimotor cortex and mediates habitual decision processes and learning (Yin et al., 2004). In contrast, the dorsomedial striatum receives projections from the medial prefrontal cortex (mPFC) that control complex goal-directed actions (Berendse et al., 1992, Corbit and Balleine, 2003, Reep et al., 2003, Gabbott et al., 2005, Yin et al., 2005), and other higher-order executive functions by integrating cognitive, sensorimotor, and limbic information (Corbit and Balleine, 2003, Gabbott et al., 2005).
The cortical neurons that furnish projections to the striatum (corticostriatal neurons) are differentially distributed across layers depending on cortical area. In primary motor cortex (M1), the vast majority of corticostriatal neurons reside in layer 5a (L5a) (Wise and Jones, 1977, Sohur et al., 2014), while in the adjacent secondary motor cortex (M2), corticostriatal neurons also become prominent in superficial layers (L2/3) (Wall et al., 2013). In prefrontal cortex (PFC), there is a further shift in distribution, becoming evenly distributed across L2/3, L5a and L5b (Wall et al., 2013).
Corticostriatal projections from all cortical areas are received by two subpopulations of striatal medium spiny neurons (MSNs), which integrate excitatory inputs from cortex and thalamus and form functionally opposing output circuits, the so-called direct- and indirect-pathways of the basal ganglia (Gerfen and Surmeier, 2011, Shepherd, 2013). Imbalances in excitatory drive onto direct- and indirect-pathway MSNs disturb movement and executive function, and the latter is thought to contribute to some neuropsychiatric disorders (Andre et al., 2011, Kravitz et al., 2012).
Excitatory synaptic inputs to the striatum are established in early postnatal life in rodents (Tepper et al., 1998, Christensen et al., 1999, Sohur et al., 2014). Despite the critical importance of thalamostriatal and corticostriatal inputs for balanced information flow through striatal output circuits, there is little known about the molecules that govern their development and targeting. Recent studies indicate that perturbing interactions between Sema3E and PlexinD1 disrupts normal establishment of thalamostriatal input onto direct-pathway MSNs, but has no effect on corticostriatal inputs (Ding et al., 2012). The identity of molecules that regulate formation, targeting, and stability of corticostriatal neurons and their synapses onto direct- and indirect pathway MSNs is unknown.
In other systems, the classic cadherin family of adhesion molecules plays an important role in establishing precise synaptic connections within and between brain regions (Inoue and Sanes, 1997, Benson and Tanaka, 1998, Huntley and Benson, 1999, Poskanzer et al., 2003, Bekirov et al., 2008, Nern et al., 2008, Williams et al., 2011). Consistent with such a role, many cadherins, particularly type II cadherins, exhibit highly restricted expression patterns that are shared between interconnected regions (Suzuki et al., 1997, Miskevich et al., 1998, Bekirov et al., 2002, Gil et al., 2002, Patel et al., 2006), and through homophilic interactions, are thought to participate in a recognition code for establishing connectivity (Williams et al, 2011; Bekirov et al, 2008). In line with this idea, previous studies have shown that Cadherin-8 (Cdh8), which is a type II cadherin, is expressed broadly in cortex, striatum, and other structures during early postnatal development (Korematsu and Redies, 1997, Korematsu et al., 1998, Hertel et al., 2008). Based on such broad distribution patterns, it has been suggested that Cdh8 might function in corticostriatal synaptic development. However, it is not known from these regional surveys which particular cell types in cortex or striatum express Cdh8, whether striatally-localized Cdh8 is synaptic, or what the functional consequences of perturbing Cdh8 expression are to the neurons that compose the corticostriatal projections. Accordingly, we address these and related questions using a combination of high-resolution cellular Cdh8 mRNA localization methods applied to identified corticostriatal neurons and direct- or indirect-pathway MSNs, immunoelectron-microscopy, and functional analysis following Cdh8 knockdown in cortical neurons.
Materials and Methods
Animals
Sprague-Dawley rats of either sex (Charles River Laboraties, Wilmington, MA; RRID:RGD_737891), ranging in age from embryonic day 18.5 (E18.5) to postnatal day 12 (P12), and wild-type (WT) C57BL/6 male mice ranging in age from P0.5 to P60 (Jackson Labs, RRID:IMSR_JAX:000664), were used for this study. In all cases, the use and treatment of animals was in strict accordance with guidelines of the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai and those of the National Institutes of Health.
Isotopic in situ hybridization histochemistry
Slide-mounted brain sections from perfused rats were processed for Cdh8 localization using a 35S-labeled complimentary RNA (cRNA) probe directed against Cdh8, as previously described (Gil et al., 2002). Briefly, frozen sections were pre-treated with Proteinase K (1µg/ml), followed by treatment with 0.25% acetic anhydride in triethanolamine (0.1M). cRNA probe was diluted in hybridization buffer (50% deionized formamide, 50x Denhardt’s solution, 10% dextran sulfate, 0.15 mg/ml yeast tRNA, 0.33 mg/ml denatured salmon sperm DNA, and 40 mM dithiothreital) and hybridized overnight at 50°C in a humidified chamber. Following multiple washes in saline sodium citrate (SSC) of increasing stringencies, slides were exposed to autoradiographic film for 7 – 24 days. Controls consisted of sections hybridized to the sense-strand probe, which showed no specific hybridization signal as expected (Gil et al., 2002).
RNA isolation and cDNA reverse transcription
Prefrontal cortex (PFC) – including medial and lateral regions of the PFC – and striatum from P0.5, P10, P20, and P60 mice (n = 6 mice per age) were quickly dissected on dry ice and homogenized in Trizol reagent (Invitrogen). Total RNA was extracted and diluted in Nuclease-free water, and cDNA was synthesized by incubating 750 ng total RNA in a 20 µl reaction with random primers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. cDNA was amplified by polymerase chain reaction (PCR) at an optimized annealing temperature (55°C) using the following primers to amplify Cdh8: 5’-TTCCAGAAATGGTCAACAACC-3’ (forward) and 5’-TTGCTACAGCCACAGACTCG-3’ (reverse). The amplification products were electrophoresed on a 1.5% agarose gel containing 0.5% ethidium bromide and a single band was detected for Cdh8 at the expected size (196 bp).
Quantitative RT-PCR
Quantitative real-time PCR (qRT-PCR) for Cdh8 and a reference gene, 18s ribosomal RNA (RNA18s), was carried out on an ABI Prism 7900HT thermal cycler (Applied Biosystems) by Mount Sinai's Quantitative PCR Shared Resource Facility using triplicate 10 µl reactions with Hotstart Taq Polymerase (KAPA Biosystem) and SYBR green detection. The following primers were used for RNA18s: 5’-GACTCAACACGGGAAACCTCAC-3’ (forward) and 5’-TCGCTCCACCAACTAAGAACG-3’ (reverse). No significant changes were detected in RNA18s cycle threshold (CT) values between age groups. Cdh8 gene expression for each animal was calculated by averaging triplicate values, and using the ΔΔCT method as described previously (Aujla and Huntley, 2014).
Antibody characterization
Details for antibodies used in this study are summarized in Table 1.
Table 1.
Primary Antibodies
| Antibody | Host, Isotype |
Immunogen | Source | Cat# | Clone #/RRIDs |
|---|---|---|---|---|---|
| Cdh8 | Ms, IgG1 | Extracellular domain, Ms | DSHB | CAD8-1 | N/A/RRID:AB_2078272 |
| Cdh8 | Gt, IgG | C-terminal domain, Hu | Santa Cruz | SC-6461 | C-18/ RRID:AB 2078271 |
| N-cadherin | Ms, IgG1 | aa 802-819 from Ms | BD Transduction Labs | 610920 | 32/Ncadherin RRID:AB_2077527 |
| actin | Ms, IgG2bk | purified from Chk gizzrd | Millipore | MAB1501 | C4/RRID:AB_2223041 |
| vGlutl | GP, IgG | aa 541-560 from rat | Millipore | AB5905 | N/A/ RRID:AB 2301751 |
| vGlut2 | GP, IgG | aa 565-586 from rat | Millipore | AB2251 | N/A/ RRID:AB 1587626 |
| vGlut3 | GP, IgG | aa 569-588 from rat | Millipore | AB5421 | N/A/ RRID:AB 2187832 |
| synGAP | Rb, IgG | aa 947-1167 from rat | Pierce | PA1-046 | N/A/ RRID:AB 2287113 |
| c-myc | Ms, IgG1 | aa 410-419 from Hu | Cell Signaling | 2276 | 9B11/RRID:N/A |
| MAP2 | Rb | purified from cow brain | Abcam | ab24640 | N/A/RRID:AB_44825 |
| GFP | Chk | recombinant GFP | Millipore | AB16901 | N/A/RRID:AB 90890 |
Abbreviations: Cdh8, cadherin-8; vGlut, vesicular glutamate transporter; synGAP, synaptic GTPase-activating protein; MAP2, microtubule-associated protein 2; GFP, green fluorescent proteins; Ms, mouse; Gt, goat; GP, guinea pig; Rb, rabbit; Chk, chicken; aa, amino acid;
Cadherin-8 antibodies
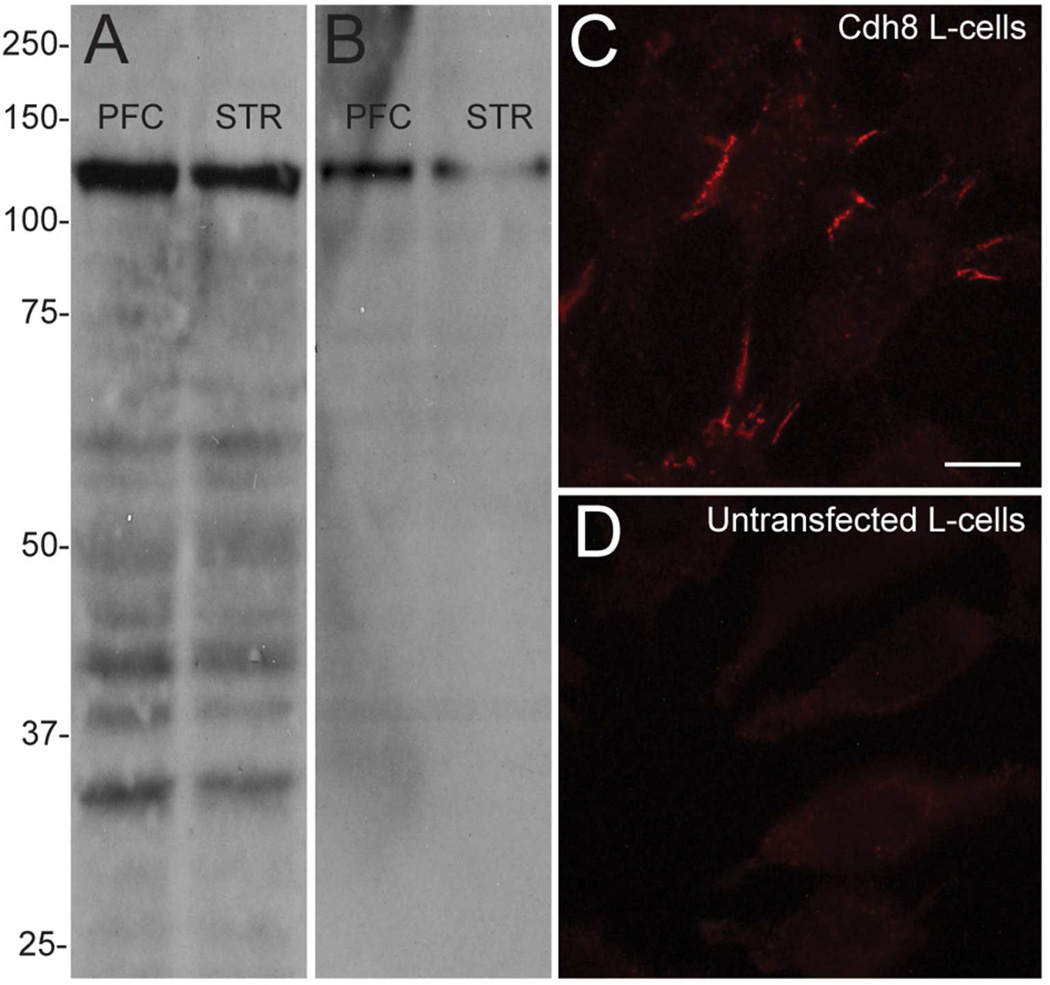
Polyclonal goat anti-Cdh8 antibodies (Santa Cruz Biotechnology, RRID:AB_2078271), which were raised against an 18 amino acid sequence of the C-terminal region of human cadherin-8, detect a prominent band at the expected molecular mass of ~135 kDa in Western analysis of mouse hippocampus (Huntley et al., 2012), and in adult mouse PFC and striatum (Fig 1A). The Cdh8 mouse monoclonal antibody supernatant (CAD8-1 mAb; Develomental Studies Hybridoma Bank, RRID:AB_2078272), which was made from a fusion protein consisting of the extracellular domain of Cdh8 and the Fc region of human immunoglobulin G1 (Suzuki et al., 2007) and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, also detects a single band at the expected size of ~135 kDa in adult mouse PCF and striatum (Fig 1B). To confirm antibody specificity, WT mouse L-cell fibroblasts, which do not endogenously express cadherins (Nose et al., 1988, Shan et al., 2000) and L-cells stably transfected with Cdh8 (gift from Dr. S.T. Suzuki) were immunolabeled with CAD8-1 mAb. As expected, Cdh8-transfected L-cells displayed a characteristic pattern of immunolocalization at cellular junctions formed along apposed cell-membranes (Fig. 1C), while untransfected L-cells were devoid of any immunolabeling (Fig 1D).
Figure 1. Monoclonal Cdh8 antibody specificity.
(A, B) Western blots of adult mouse prefrontal cortex (PFC) and striatum (STR) using (A) polyclonal goat Cdh8 antibodies or (B) CAD8-1 mAb, both of which show a single prominent band at the predicted size of 135 kDa. (C) CAD8-1 mAb immunolabeling in L-cells stably transfected with Cdh8. (D) No immunolabeling is evident in untransfected WT L-cells. Scale bar, 5 µm.
N-cadherin antibody
Mouse monoclonal N-cadherin antibody (BD Transduction, RRID:AB_2077527) was previously shown by us to label N-cadherin-overexpressing L-cells, but not E-cadherin-transfected L-cells (Brock et al., 2004). This antibody recognizes a single band of the appropriate molecular mass (~125 kDa) in immunoblot analysis of brain lysates (Brock et al., 2004, Nikitczuk et al., 2014). Additionally, immunolabeling of hippocampal excitatory synapses is abolished in N-cadherin conditional knockout mice (Nikitczuk et al., 2014). Finally, human embryonic kidney (HEK) 293 cells immunolabeled with anti-N-cadherin antibody displayed the expected localization pattern at cellular junctions (Fig 2C).
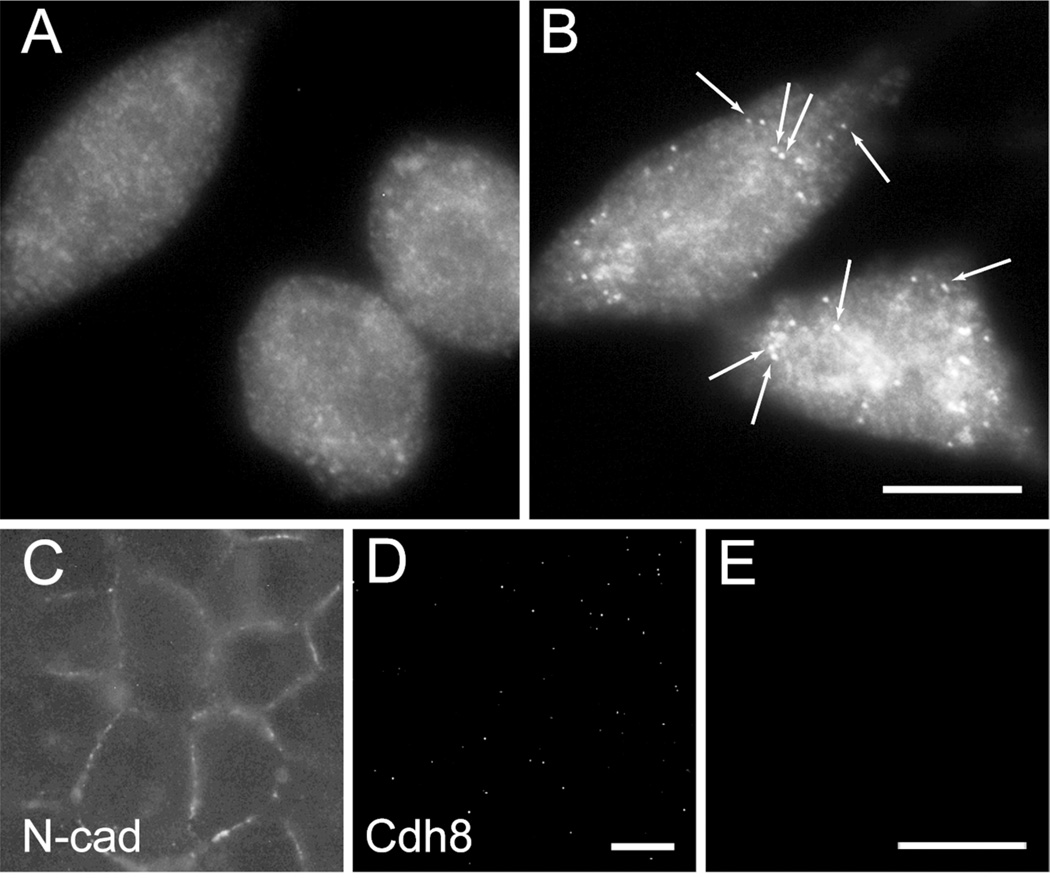
Figure 2. Single-molecule fluorescence in situ hybridization (smFISH) Cdh8 probe specificity.
(A, B) Hybridization of Cdh8 mRNA probes in mouse L-cell fibroblasts. (A) untransfected WT L-cells, which lack endogenous cadherins, do not hybridize Cdh8 probes. (B) L-cells stably transfected with Cdh8 show probe hybridization, revealed by multiple, discrete fluorescent puncta (arrows) (C) HEK293 cells, which endogenously express N-cadherin, show typical N-cadherin immunolocalization pattern of labeled cell-cell membrane appositions. (D-E) HEK293 cells, which do not express endogenous Cdh8, show neither Cdh8 immunolocalization (D) nor Cdh8 smFISH (E). Scale bars, 10 µm.
Actin antibody
Mouse monoclonal actin antibody (Millipore, RRID:AB_2223041) is well-characterized and recognizes a single band at the expected molecular weight of 42 kDa, as described previously by us and others using similar immunoblotting conditions (Nikitczuk et al., 2014).
vGlut antibodies
Guinea-pig polyclonal vesicular glutamate transporter (vGlut) 1, 2, and 3 antibodies (Millipore, RRID:AB_2301751; RRID:AB_1587626; RRID:AB_2187832) were used as an antibody cocktail to label glutamatergic presynaptic sites. Each of these antibodies has been vetted by numerous labs (JCN Antibody Table), and has been used previously by our lab (and others) to identify excitatory presynaptic terminals using preparations similar to those described here (Anderson et al., 2004, Hsiao et al., 2014).
synGAP antibody
Rabbit monclonal synaptic GTPase-activating protein (synGAP) antibody (Pierce, RRID:AB_2287113) recognizes a single band of the appropriate size by Western blot (Oh et al., 2004, Delint-Ramirez et al., 2010), and in our hands immunostaining produced the predicted punctate labeling pattern that colocalized with vGlut labeling as described by others in similar preparations (Harms and Craig, 2005).
c-myc antibody
Mouse monoclonal c-myc antibody (Cell Signaling, RRID: N/A) has been vetted by many other labs (JCN antibody database), and detects myc-tagged full-length Cdh8 at the expected molecular mass of ~135 kDa in Western analysis of transfected HEK293 cells, but is not detected in Western analysis of mouse striatal lysate (data not shown).
GFP antibody
Chicken polyclonal GFP antibodies (Millipore, RRID:AB_90890) are well-characterized and can be used to immunolabel transfected neurons as described previously by us (Mintz et al., 2008) and others (JCN antibody database). No immunolabeling is detected in untransfected cells.
MAP2 antibody
Rabbit polyclonal microtubule-associated protein 2 (MAP2) antibodies (Millipore, RRID:AB_44825) produced the expected somatodendritic labeling pattern (and was excluded from axons), as described previously by us and others using the same immunolabeling conditions (Sepulveda et al., 2013). (Sepulveda et al., 2013)
Western Blot analysis
PFC and striatum from P0.5, P10, P20, and P60 mice (n = 4 mice per age) were dissected on dry ice and homogenized in RIPA buffer (150 mM NaCl, 1.0% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, protease inhibitor cocktail (Roche)). Homogenates were centrifuged at 14,000 rpm for 15 minutes and the resulting supernatant was collected. Protein concentrations were determined by the Bradford method following the manufacturer’s protocol (Bio-Rad Laboratories). Equal amounts of protein were subjected to gel electrophoresis on an 8% polyacrylamide gradient gel and transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked in 10% newborn calf serum and incubated in goat anti-Cdh8 antibody (1:200) or mouse anti-actin antibody (1:2000) overnight. Following incubation with fluorophore-conjugated secondary antibodies (DyLight 800, Cell Signaling and DyLight 680, Pierce), membranes were visualized using the Li-Cor Odessey Clx detection system (Li-Cor Biosciences). In all cases, Cdh8 levels were normalized to actin loading control.
Single-molecule fluorescent in situ hybridization (smFISH) probes in cells
Multiple short (20-nucleotide) probes conjugated to a single fluorophore at the 3’ end were used to identify Cdh8 mRNA at the cellular level. A set of 48 different probes targeted to complimentary regions along the full-length Cdh8 mRNA transcript (GenBank; RRID:nif-0000-02873, accession NM_007667.2) were designed by the Stellaris Probe Designer Version 2.0 (Table 2; https://www.biosearchtech.com/stellarisdesigner) and purchased commercially (Biosearch Technologies). smFISH probes collectively bind along the length of an mRNA transcript and positive signal is only identified from the combined localized fluorescence of multiple probes (> 20 probes), thus providing higher specificity and limiting the detection of off-target transcripts (Raj et al., 2008).
Table 2.
Sequences for single-molecule fluorescent in situ hybridization (smFISH) probes directed against Cdh8
| Probe # | Probe (5’→3’) | Probe position* |
|---|---|---|
| 1 | gccgtgtatacacaagaagg | 70 |
| 2 | aaaacgtgagcctgattcat | 94 |
| 3 | gcttagttccaaaggggatc | 122 |
| 4 | cggttcaggtccagaaaatt | 218 |
| 5 | aggatccagatctgtgtgta | 254 |
| 6 | cctctcggtcaagtcttttg | 363 |
| 7 | tttgtctcaaagtccactgc | 412 |
| 8 | caggtgcattgtcgttgata | 477 |
| 9 | tggacatctctggaacagta | 522 |
| 10 | gtttccataaactgggtcgt | 584 |
| 11 | tgtttcaggctcaatggaaa | 647 |
| 12 | ggcttctctgtccatgttag | 689 |
| 13 | cacccatatctttggcttgt | 732 |
| 14 | taacatcagtcagggtcact | 789 |
| 15 | accacatcttctggtactga | 850 |
| 16 | ggcatcagaagtgatttcga | 971 |
| 17 | cttataacaccatcctgggc | 994 |
| 18 | ctcaaagtccagaggctttc | 1019 |
| 19 | atgtggatattggctgcttc | 1066 |
| 20 | gctgtatccttaaagggtcc | 1108 |
| 21 | acagagttcaaagcagcatt | 1207 |
| 22 | aagtgatatcagggtcacga | 1245 |
| 23 | gttgaactgtctttccaggt | 1301 |
| 24 | gccagtgttatcttcccatc | 1333 |
| 25 | tgttgtgccaaacacttagt | 1371 |
| 26 | tgtggttcctgatctcagta | 1404 |
| 27 | aacaggcactcgagatatct | 1427 |
| 28 | gtcattgacatccagcactt | 1454 |
| 29 | tattcggatgcgaattcagg | 1480 |
| 30 | ctttgtccatggcacttact | 1548 |
| 31 | ttgggttgttgaccatttct | 1611 |
| 32 | ctggcggttgaatccattat | 1682 |
| 33 | gcagaaggtagacttcttgc | 1704 |
| 34 | ggtttccactgtcactgatc | 1731 |
| 35 | taagcttcaacattgcacga | 1819 |
| 36 | actgagtccaataggaagga | 1841 |
| 37 | atatagcaattaaggcgccc | 1863 |
| 38 | agggtaacaaacagaaccac | 1912 |
| 39 | gtggttcatttttgtgcctc | 1935 |
| 40 | cgaacgtcttcgtcatcttt | 1963 |
| 41 | cgtcgtagcgaatgatgttt | 1986 |
| 42 | gttgcaatgtcaaaagcctc | 2032 |
| 43 | aacaccatttggaactggag | 2138 |
| 44 | gggtcattatctgcctcatg | 2188 |
| 45 | cggccttcatagccataaat | 2236 |
| 46 | ttctggtctgagtctgatgt | 2296 |
| 47 | gttcgcccagtctcttaaag | 2346 |
| 48 | gtcactttcaccaacagagt | 2369 |
Position of first nucleotide of probe corresponding to target sequence
To test the specificity of the Cdh8 probes, we hybridized Cdh8 probe-sets with untransfected L-cells, Cdh8-transfected L-cells, and HEK293 cells, which express N-cadherin, but not Cdh8 (Fig 2C, D)(Nollet et al., 2000). Cells were plated onto 25 mm round coverslips that were coated with poly-L-lysine (100 mg/ml), and maintained in DMEM and 10% fetal bovine serum (FBS). Cells were fixed in 4% formaldehyde in PBS for 10 minutes and permeabilized with 70% ethanol for 30 minutes at 4°C. Following equilibration in wash buffer (10% formamide, 2x SSC), probes were diluted in hybridization buffer (10% dextran, 1 mg/ml E. coli tRNA, 200 µg/ml BSA, 2 mM vanadyl ribonucleosides, 10% formamide, 2x SSC) and hybridized overnight in a humidified chamber at 37°C. Following six washes in wash buffer over 2 hours, coverslips were mounted in oxygen-depleted mounting medium (Raj et al., 2008) to minimize photobleaching. All smFISH images were acquired on an Axioplan2 (Zeiss) with a 100×/1.4 N.A. oil-immersion objective. smFISH in L-cells confirms the specificity of the probe set for Cdh8 (Fig 2A, B). Additionally, smFISH in HEK293 cells confirms that Cdh8 smFISH probes do not detect other (off-target) cadherins that share significant sequence homology with Cdh8 (Fig 2E).
smFISH analysis in PFC and striatum
For in vivo smFISH studies, mice aged P10 and P30 were anesthetized with ketamine (100 mg/kg) and xylazine (20 mg/kg) and sacrificed by cervical dislocation (n = 3 mice per age). Brains were dissected, quickly frozen in O.C.T. (Tissue Tek) on dry ice, and stored at −80°C until sectioning. Frozen sections (14 µm) were cut in the coronal plane through PFC and striatum on a cryostat and thaw mounted onto 25 mm round coverslips. Tissue-mounted coverslips were fixed and processed for smFISH as described above.
Retrograde tract tracing
Mice aged P30 were anesthetized with continuous delivery of isoflurane and placed in a stereotaxic frame (David Kopf Instruments). A 0.1% solution of Alexa 488-cholera toxin subunit B (CTB-488; Molecular Probes) diluted in 0.1 M phosphate buffered saline (PBS) was injected bilaterally into the dorsomedial striatum (AP +1.0 mm, ML ±1.2 mm, DV −2.3 mm), globus pallidus external segment (GPe; AP −0.7 mm, ML ±2.0 mm, DV −3.3 mm), or substantia nigra pars reticulata (SNr; AP −3.0 mm, ML ±1.4 mm, DV −4.3 mm) with a Neuros 32 gauge needle syringe (Hamilton Company) (n = 3 mice per injection site). The needle was left in place for 10 minutes. After recovery, mice were returned to their home cages for 7 days before intracardiac perfusion with 4% paraformaldehyde. Images of CTB-488 injection sites and target regions were acquired on an Axioplan 2 (Zeiss) with a 10× objective. In some cases, we combined CTB-488 retrograde labeling with Cdh8 smFISH. For this, brain tissue was processed for smFISH as described above. Images were acquired on an Axioplan 2 at 0.5 µm intervals with a 100×/1.4 N.A. oil-immersion objective and a maximum intensity projection was generated from a 10-image z-stack using ImageJ (NIH).
Dissociated cortical neuron culture
Cortical neurons were cultured from E18.5 Sprague-Dawley rats as described previously (Mintz et al., 2008). Neurons were plated onto 18 mm coverslips coated with 1 mg/ml poly-L-lysine (6.0×104 cells per coverslip; 2.1×104 cells/cm2) and maintained in Neurobasal Media (Invitrogen) containing NS21 supplement (Chen et al., 2008).
Immunocytochemistry in fixed cultured neurons
Cultured neurons were fixed in 4% parafomaldehyde/4% sucrose for 15 minutes and permeabilized with 0.2% Triton X-100 for 5 min. Neurons were blocked in PBS supplemented with 10% bovine serum albumin (BSA), and then incubated overnight with one or more of the following primary antibodies diluted in 1% BSA: SynGAP (1:1000), vGlut antibody cocktail (1:2500), GFP (1:5000), and MAP2 (1:5000). Neurons were incubated in species-specific fluorophore-conjugated secondary antibodies and images were acquired on an LSM510 Meta confocal microscope (Zeiss).
Live-cell Cdh8 immunofluorescence
CAD8-1 mAb supernatant (1:10) was diluted in the culture media of live cultured cortical neurons for 15 minutes at 37°C, washed in PBS, then fixed in 4% parafomaldehyde/4% sucrose for 15 minutes. Neurons were incubated in species-specific fluorophore-conjugated secondary antibody and imaged as described above. To control for the possibility that antibody bound to surface proteins can induce internalization, the distribution of Cdh8 labeling in neurons labeled at 37°C was compared to that labeled at 12°C—a temperature at which little to no internalization takes place. No differences in localization pattern were observed between these two conditions (data not shown).
Immunogold labeling and electron microscopy
Sections through P30 mouse striatum were prepared for electron microscopy as previously described (Mortillo et al., 2012). Briefly, aldehyde-perfused brains were vibratome-sectioned through the dorsal striatum at 200 µm. Trimmed sections were rapidly frozen in liquid propane and embedded and polymerized in Lowicryl (Electron Microscopy Sciences) by freeze-substitution. Ultrathin sections (70 nm) were collected on formvar-coated nickel grids, blocked with donkey serum, incubated overnight in CAD8-1 mAb supernatant (1:10) or goat polyclonal Cdh8 (1:100), and then incubated with colloidal gold-conjugated secondary antibody. Images were acquired on a Hitachi H7000 transmission electron microscope.
Cdh8 cDNA and shRNA constructs
Myc-tagged full-length mouse Cdh8 cDNA was subcloned from a mouse brain library (gift from Dr. Joseph Buxbaum) into a mammalian expression vector driven by the pCAGGs promoter that is also a lentiviral expression vector (assembled by K. Hsiao using pLenti CMVtight; Addgene). The expression vector also contains a Tet-ON system whereby expression is low without co-transfection of a tTA transactivator plasmid. Western blotting with goat polyclonal Cdh8 (1:200) antibodies confirmed that the full-length cDNA was correctly cloned and generated a protein of the expected size (~135kDa). SureSilencing shRNA plasmids (SABiosciences) directed against mouse Cdh8 were tested for Cdh8 knockdown. The set included four shRNA and one scrambled negative control sequences under the control of a U1 promoter and GFP driven by a CMV promoter. HEK293 cells were transiently transfected with myc-tagged full-length mouse Cdh8, a tTa transactivator plasmid (Addgene), and one of the four shRNAs or scrambled negative control plasmids using Lipofectamine 2000 (Invitrogen). After 48 hours, cells were lysed in RIPA buffer and analyzed for Cdh8 expression by Western blotting with mouse monoclonal c-myc antibody (1:2000) using ECL Chemiluminescence detection (Pierce). Dissociated rat neurons were nucleofected before plating with shRNA constructs and eGFP driven by pCAGGs (a mammalian expression vector under the control of chicken β-actin promoter; gift of Jun-ichi Miyazaki, Osaka University Medical School) at 0 days in vitro (DIV) using the Rat Neuron Nucleofection Kit (Lonza). Transfected neurons were identified by GFP immunolabeling.
Image analysis
For smFISH puncta quantifications in direct- and indirect-pathway CTB-488-labeled neurons and co-localization studies in cultured cortical neurons, puncta fluorescence was thresholded using MetaMorph (Molecular Devices, RRID:SciRes_000136), and co-localization was determined from binary images that displayed overlapping fluorescence. Cdh8 knockdown in neurons was measured by average Cdh8 immunofluorescence intensity. Line intensity scans from 3 segments (100 µm) were acquired by MetaMorph and averaged for each neuron. Dendritic processes of transfected neurons were traced in Neurolucida (MBF Bioscience, RRID:nif-0000-10294) and analyzed using the NeuroExplorer module (MBF Bioscience).
Statistics
All data are presented as mean ± SEM and p-value for significance was set at p < 0.05. Significance between groups was determined using one-way ANOVA and post-hoc Dunnett’s multiple comparison test. Significance for Cdh8 fluorescence intensity and dendritic self-crossing was determined by Mann-Whitney test. All other comparisons were determined by unpaired Student’s t test, with Welch’s correction where indicated. Levene's test for homogeneity of variance was used for inclusion in parametric tests (p > 0.05 for Levene's tests). Statistical analyses and graphing were performed with GraphPad Prism v5.0 (GraphPad Software, RRID:rid_000081).
Results
Cdh8 is highly enriched in rodent mPFC and striatum when corticostriatal circuits are forming
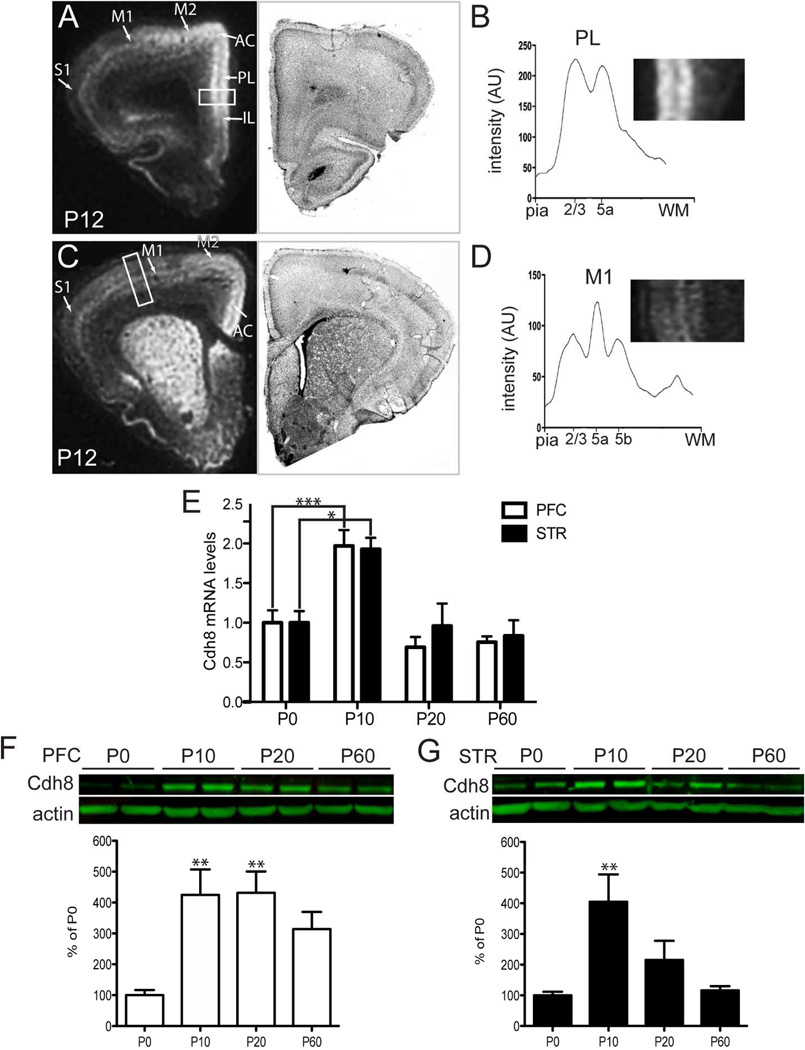
Corticostriatal projections first reach the striatum at P3-4 (Sohur et al., 2012). Between P10 and P21, MSNs undergo a major wave of excitatory synaptogenesis (Sharpe and Tepper, 1998, Tepper et al., 1998). To more precisely define Cdh8 expression in relationship to developing corticostriatal circuitry, we performed in situ hybridization using Cdh8-specific probes in rat brain. Cdh8 mRNA was highly enriched in the PFC at P12 (Fig 3A), and was most abundantly expressed in two heavily labeled bands corresponding to L2/3 and L5 in anterior midline cortex, which includes anterior cingulate (AC), prelimbic (PL), and infralimibic (IL) areas of the mPFC (Fig 3A, B). The two bands of intense Cdh8 probe hybridization correspond to the laminar location of corticostriatal projection neurons in these mPFC regions (Krettek and Price, 1977, Wall et al., 2013). Probe hybridization also extended laterally, through secondary motor area M2, but diminished in intensity in comparison with midline prefrontal cortical areas (Fig 3A). Within primary motor (M1) and somatosensory cortex (S1), Cdh8 probe hybridization was concentrated in L5a, as reported previously (Gil et al., 2002), where the vast majority of corticostriatal projection neurons are localized in these regions (Wise and Jones, 1977, Wall et al., 2013, Sohur et al., 2014)(Fig 3A, D). A similar expression pattern persisted in more caudal AC and sensorimotor cortex (Fig 3C). Cdh8 mRNA was also highly expressed throughout striatum, including the dorsomedial striatum (Fig. 3C), which receives input from the mPFC (Berendse et al., 1992, Gabbott et al., 2005). Within the striatum, Cdh8 mRNA labeling was somewhat heterogenous, consistent with previous work in rodents supporting an enrichment of Cdh8 within the matrix compartment (Fig 3C) (Gerfen, 1992, Heyers et al., 2003, Hertel et al., 2008).
Figure 3. Developmental changes in Cdh8 mRNA in prefrontal cortex (PFC) and striatum.
Film autoradiograms showing isotopic in situ hybridization and semi-adjacent Nissl-stained sections in P12 rat brain in (A) PFC and (C) striatum. Cdh8 mRNA labeling is shown as white signal. Line scans of film autoradiograms taken perpendicular through PL (B) and M1 (D) illustrate discrete intensity peaks in cortical layers projecting to striatum. The white boxes in the film autoradiograms indicate the approximate positions of the line-scans through PL and M1, and correspond to the higher-power insets shown in B, D. Orientation of both insets: pia is to the right, WM is to the left. (E) Relative transcript levels of Cdh8 normalized to housekeeping gene, RNA18s, in mouse PFC and STR during postnatal development (n = 6 per group). (F, G) Western analysis of Cdh8 protein levels in mouse PFC and striatum during postnatal development using polyclonal goat Cdh8 antibodies. Actin was used as a loading control. The relative levels of Cdh8 normalized to actin are shown in the graphs below (n = 4 per group). Abbreviations: AC, anterior cingulate; PL, prelimbic; IL, infralimbic; M1, primary motor area; M2, secondary motor area. Error bars represent S.E.M. *p < 0.05, ** p < 0.001, *** p < 0.0001
We next examined the time course of Cdh8 gene expression in PFC and striatum during early postnatal development. Our qRT-PCR data revealed that in PFC, Cdh8 gene expression peaked between P10 and P20 (one-way ANOVA, n = 6, p < 0.0001; Fig 3E), declining thereafter by P60. Similarly, in striatum Cdh8 expression peaked at P10 (one-way ANOVA, n = 6, p = 0.0072; Fig 3E), when corticostriatal projection neurons are first starting to form synaptic connections with MSNs, but declined by P20, when most MSNs have formed synapses with excitatory cortical afferents, and remained at low levels into early adulthood (P60). Likewise, western blot analyses showed that Cdh8 protein levels, normalized to actin loading control, peaked at P10 in both PFC (one-way ANOVA, n = 4, p = 0.0076; Fig 3F) and striatum (one-way ANOVA, n = 4, p = 0.0074; Fig 3G). While Cdh8 protein levels in the PFC remained high at P20, striatal Cdh8 protein levels decreased at P20 and P60. The matched mRNA and protein expression profiles of these two interconnected regions suggest that Cdh8 is transcriptionally controlled and expressed in regions and layers contributing to corticostriatal pathways when such pathways are forming and refining.
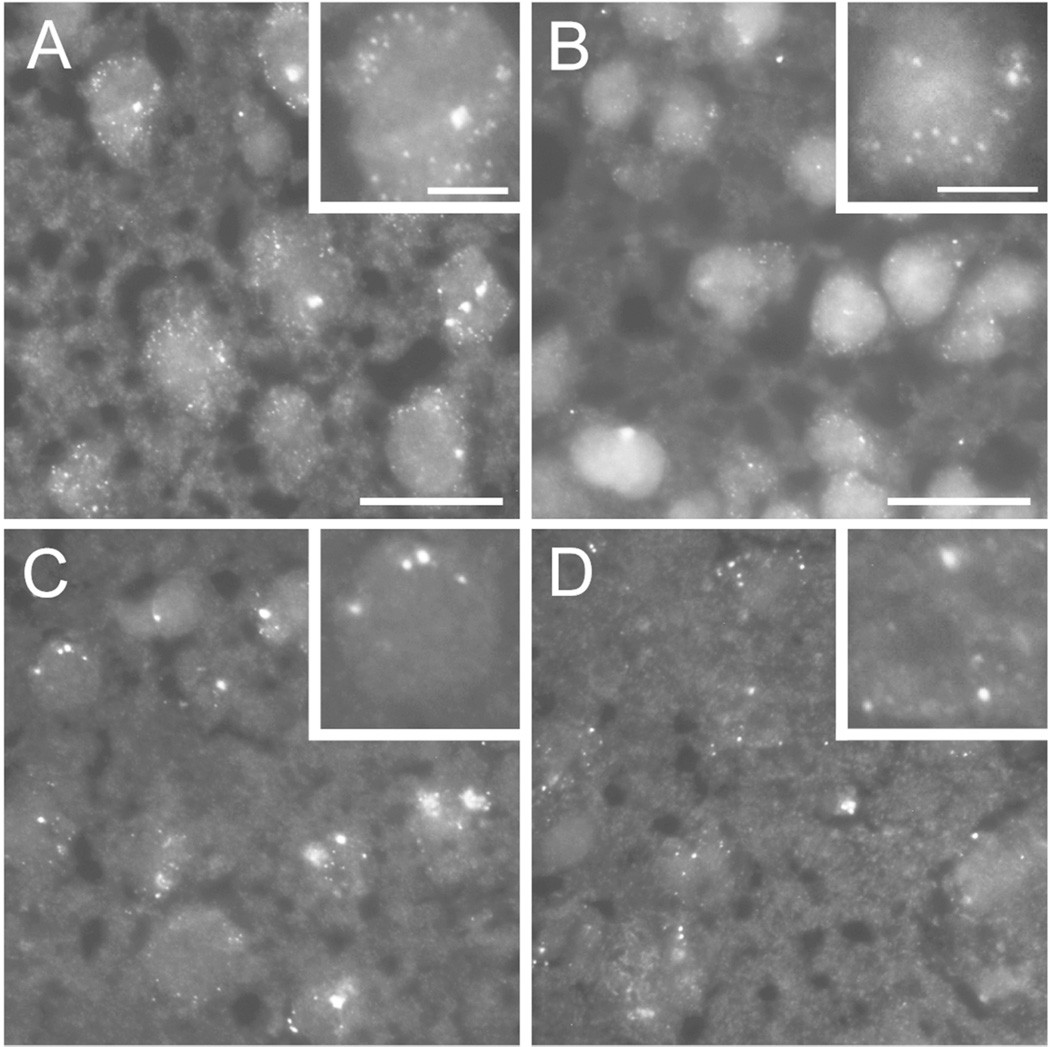
To assess the cellular distribution pattern of Cdh8, we used a single-molecule fluorescent in situ hybridization (smFISH) approach (Raj et al., 2008). At P10, during peak mRNA and protein expression, multiple small discrete puncta, corresponding to individual Cdh8 transcripts, were detected in the mPFC (Fig 4A) and dorsal striatum (Fig 4B). A smaller number of large puncta was also observed. All puncta were restricted to cell bodies, and we saw little evidence for extrasomal transcripts. As expected, smFISH revealed fewer fluorescent puncta at P30 (Fig 4C, D). However, puncta were typically larger in size (Fig 4A, B vs. C, D). Whether these large puncta represent translationally dormant RNA-containing granules (Kiebler and Bassell, 2006) has yet to be determined.
Figure 4. Cdh8 smFISH in neurons during postnatal development.
Single molecule FISH (smFISH) showing Cdh8 mRNA expression in (A, C) medial PFC (mPFC) and (B, D) dorsomedial striatum. High magnification images are shown in inset. Scale bar, 10 µm or 5 µm (inset). (A, B) Top panels show multiple discreet punctate structures indicating high levels of Cdh8 mRNA in mPFC (A) and STR (B) at postnatal day 10 (P10). (C, D) mRNA fluorescent puncta decrease in numbers, but increase in size at P30.
Cdh8 is expressed in corticostriatal neurons
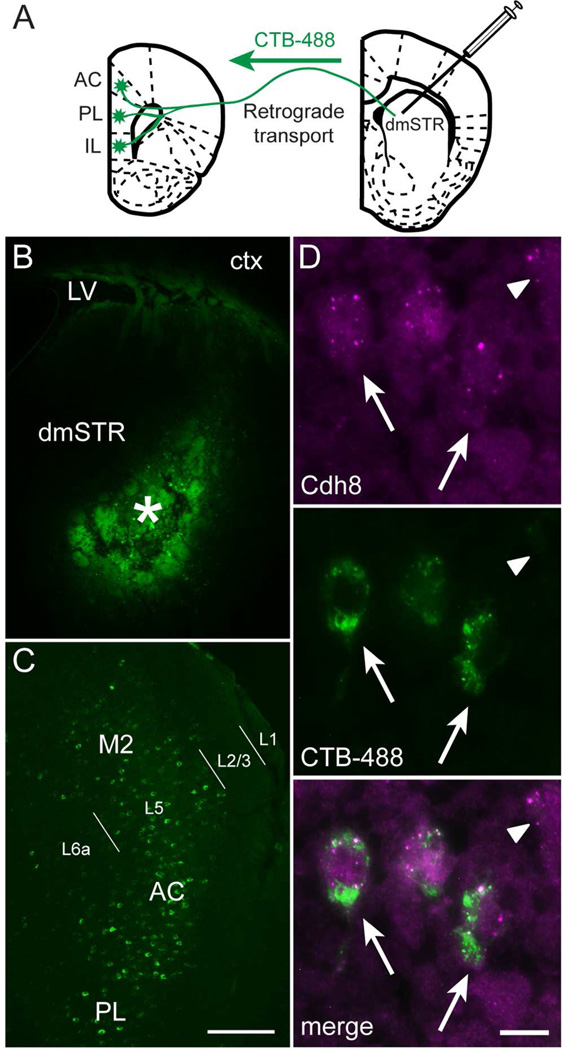
While our data above show that Cdh8 is strongly expressed in mPFC layers that furnish corticostriatal connections, these cortical layers contain multiple populations of projection neurons that target different structures in addition to striatum. To identify if corticostriatal neurons in particular express Cdh8, we used smFISH in combination with tract tracing methods. The retrograde tracer CTB-488 was injected into the dorsomedial striatum (dmSTR) of P30 mice (Fig 5A, B), which resulted in back-labeled corticostriatal projection neurons (Fig. 5C). In combination with smFISH, we found that Cdh8 mRNA was expressed by the vast majority of CTB-488-labeled corticostriatal projecting neurons (arrow, Fig 5D), although, there were other neurons that were Cdh8-positive, but were not back-filled from the striatal injection (arrowhead, Fig. 5D).
Figure 5. PFC-striatal projection neurons express Cdh8.
(A) Schematic of retrograde tracer, Alexa 488 cholera toxin-B (CTB-488), injected into dorsomedial striatum (dmSTR) and retrogradely transported to mPFC. (B) CTB-488 injection site (asterisk) in the dmSTR. LV = lateral ventricle, ctx = cortex. (C) CTB-488 retrogradely labeled L5 cell bodies in AC, PL, and M2 regions of the mPFC (abbreviations as in Figure 3). L1, L2/3, L5, and L6a labels delineate cortical layers. Scale bar, 200 µm. (D) Cdh8 mRNA (magenta) detected in CTB-488-labeled corticostriatal projection neurons (green) in the mPFC (arrows). Arrowhead indicates unlabeled neuron expressing Cdh8. Scale bar, 10 µm.
Cdh8 expression in direct- and indirect-pathway MSNs
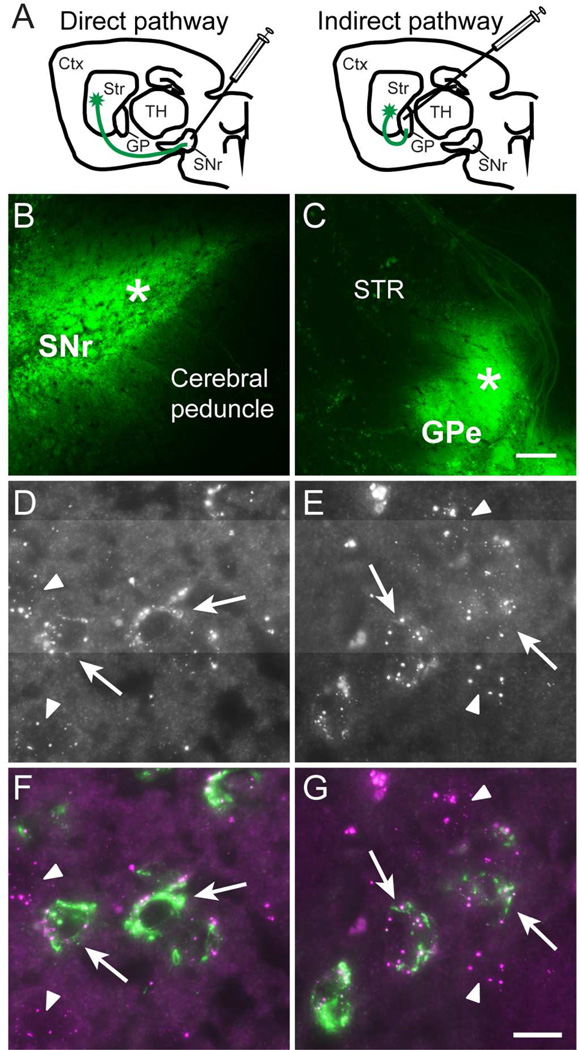
MSNs composing the direct (striatonigral) or indirect (striatopallidal) pathways have distinct molecular profiles, different electrophysiological and synaptic properties (Gerfen and Surmeier, 2011, Shepherd, 2013), and play different roles in information flow through the basal ganglia (Alexander and Crutcher, 1990) Therefore, we next asked if Cdh8 is selectively expressed by MSNs forming one or the other output pathways. To determine this, we combined Cdh8 smFISH with retrograde labeling of direct- or indirect-pathway MSNs following CTB-488 injection into the SNr or GPe, which are the target structures of the direct- or indirect-pathways, respectively (Fig 6A-C). We found that Cdh8 mRNA was expressed in nearly all of the direct-and indirect-pathway MSNs (Fig 6D-G). Quantification of the average number of mRNA puncta per CTB-488-labeled neuron confirmed that direct- and indirect-pathway MSNs expressed similar levels of Cdh8 (puncta per direct MSN: 10.47 ± 1.43, n = 44; puncta per indirect MSN: 10.40 ± 0.99, n = 74; t-test, p = 0.9). These results indicate that Cdh8 is expressed equally by both classes of MSNs.
Figure 6. Similar expression levels of Cdh8 mRNA in direct- and indirect-pathway MSNs.
(A) Schematic of CTB-488 injections at target sites that label direct- and indirect-pathway MSNs. Ctx = cerebral cortex, Str = striatum, TH = thalamus, GP = globus pallidus, SNr = substantia nigra pars reticulata. (B) CTB-488 injected into SNr (asterisk) and (C) globus pallidus external segment (GPe; asterisk). Scale bar, 200 µm. (D-G) High-resolution images of Cdh8 mRNA in CTB-488-labeled MSNs. Cdh8 mRNA in (D) direct pathway MSNs and (E) indirect pathway MSNs. (F, G) Merged images show Cdh8 mRNA (magenta) in CTB-488-labeled (F) direct- and (G) indirect-labeled MSNs (green) in striatum (indicated by arrows). Arrowheads indicate unlabeled neurons expressing Cdh8. Scale bar, 10 µm.
Cdh8 is enriched at glutamatergic synapses on cortical neurons and in striatum
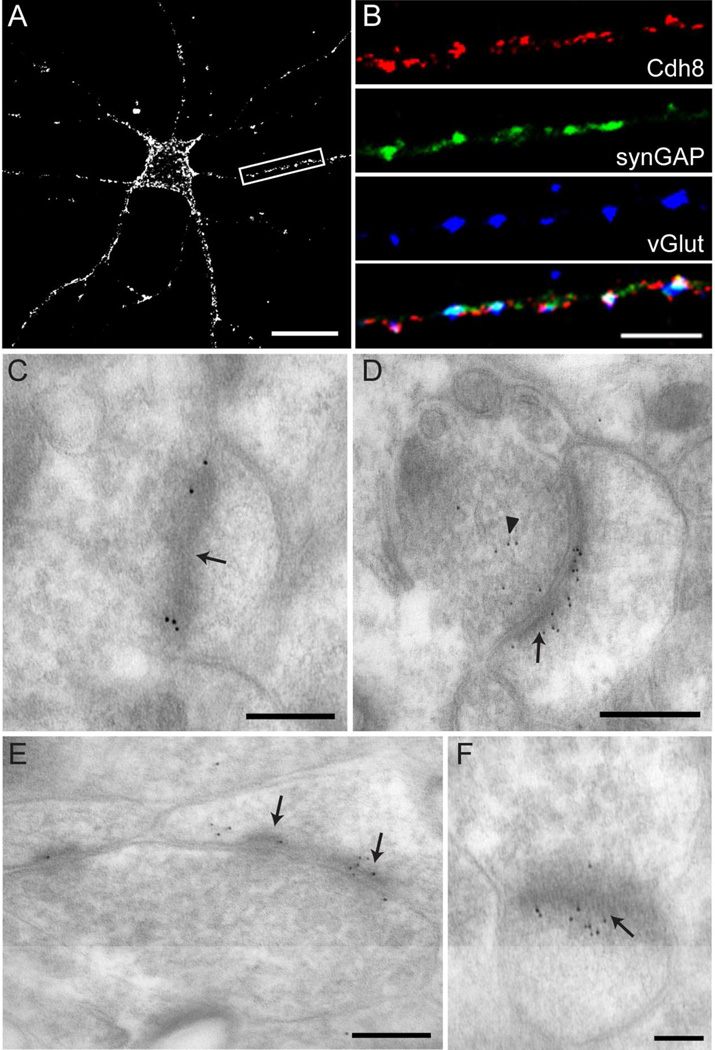
We next investigated the subcellular localization of Cdh8 by first examining Cdh8 immunolocalization in primary cortical neurons at 14 DIV, which is the peak period of synaptogenesis (de Lima et al., 1997). Cdh8 immunofluorescence was visualized using a live staining protocol (see Materials and Methods), which proved to be much more sensitive than light-level immunolocalization following aldehyde fixation. Live neurons were incubated with CAD8-1 mAb, and subsequently fixed and stained with fluorescent secondary antibody. With this method, we found that Cdh8 immunolocalization was punctate and distributed along the surface of cell bodies and dendrites (Fig 7A). To assess whether such Cdh8 puncta corresponded to synapses, we co-labeled neurons with a cocktail of vGlut antibodies (vGlut1, vGlut2, vGlut3) to mark excitatory presynaptic terminals, or synGAP to label glutamatergic postsynaptic sites.
Figure 7. Cdh8 is synaptically localized in cultured cortical neurons and striatum.
(A) Immunofluorescence labeling with monoclonal Cdh8 antibody in primary cortical neurons at 14 DIV visualized using a live staining protocol. Scale bar, 20 µm. (B) High magnification image of dendrite (boxed area in (A)) shows immunofluorescence labeling of Cdh8, glutamatergic presynaptic marker vGluts (green), and glutamatergic postsynaptic marker synGAP (blue). Merged image shown in bottom panel. Scale bar, 5 µm. (C-F) Electron micrographs of sections through the dorsal striatum at P30. (C) Immunogold labeling with mAb CAD8-1 antibody. Scale bar, 200 nm. (D-F) Immunogold labeling with goat polyclonal anti-Cdh8 antibodies. (D, E) Scale bar, 250 nm. (F) Scale bar, 100 nm. Arrows indicate postsynaptic densities and arrowhead points to membrane-bound vesicles in presynaptic terminal.
We found that Cdh8 co-localized with ~35% of excitatory presynaptic vGlut+ terminals and ~40% of synGAP+ postsynaptic sites (n = 10 neurons). Triple-labeled preparations showed that many of the Cdh8 immunopositive puncta corresponded to vGlut/synGAP appositions, but non-synaptic puncta were also observed (Fig 7B). These data indicate that Cdh8 is likely present on both sides of many developing excitatory synapses.
To determine if Cdh8 localized to excitatory synapses in the striatum, we examined the ultrastructural localization of Cdh8 by postembedding immunogold labeling of P30 striatal slices following aldehyde-perfusion. We found that clusters of Cdh8 immunogold particles were concentrated within active zones on pre- and postsynaptic membranes at asymmetric synaptic junctions, consistent with homophilic binding across synaptic clefts (Fig 7C). This synaptic localization pattern of Cdh8 immunogold labeling was duplicated using a different Cdh8 antibody (Fig 7D-F). We did not attempt to quantify the percentage of Cdh8 labeling in striatum that was found at synapses, nor did we attempt to determine how much of the total Cdh8 label was found pre- or postsynaptically. However, while both antibodies showed that Cdh8 gold particles were primarily found clustered on both sides of asymmetric synaptic junctions, we also observed Cdh8 gold particles within presynaptic terminals along membrane-bound vesicles (Fig 7D) and perisynaptically (Fig 7E). Additionally, qualitative observations suggested that Cdh8 was not localized to symmetric (inhibitory) synapses. Together, these data suggest that Cdh8 participates in synapse adhesion in dorsal striatum and may play a role in regulating the function, plasticity and/or targeting of excitatory synapses on MSNs.
Cdh8 knockdown impairs dendritic arborization
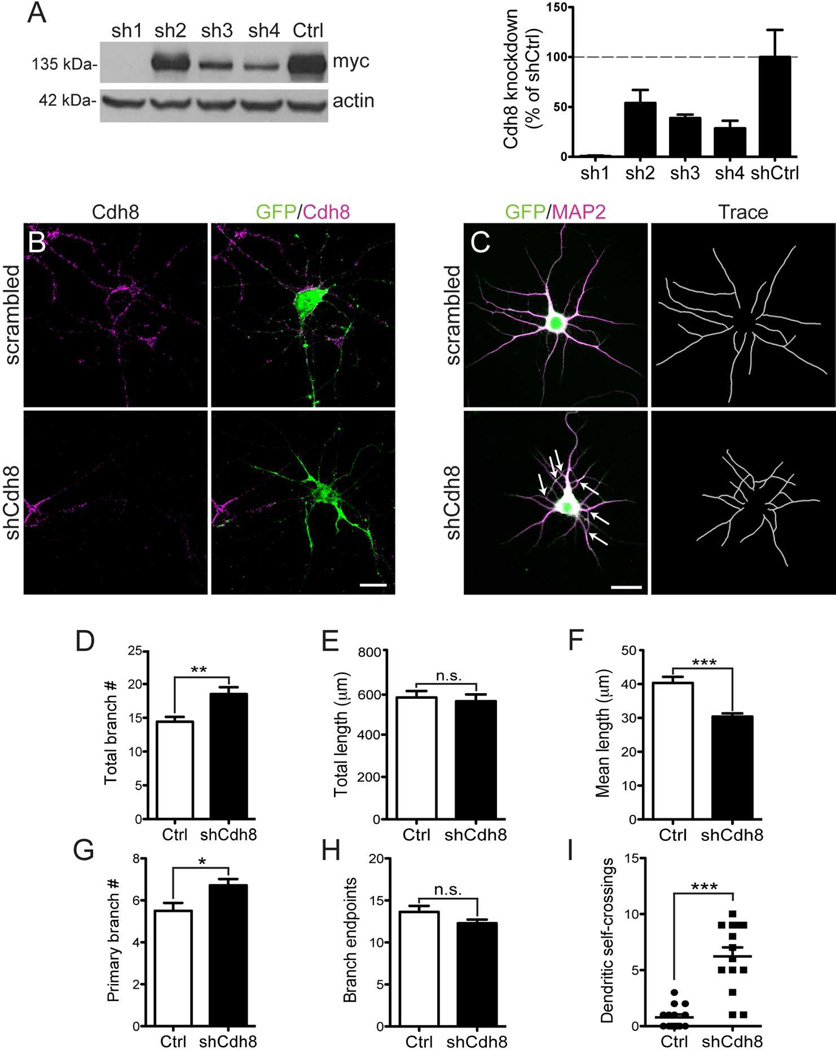
Classic cadherins can regulate dendritic growth during development (Riehl et al., 1996, Esch et al., 2000, Zhu and Luo, 2004, Bekirov et al., 2008). To test whether Cdh8 functions in this way, we examined dendritic growth and arborization in primary cortical neurons following Cdh8 knockdown using a shRNA directed against Cdh8. To test knockdown efficiency, one of four shRNAs directed against Cdh8 or a scrambled negative control plasmid was expressed in HEK293 cells along with myc-tagged-Cdh8 (see methods for details). Western blot with anti-c-myc antibody revealed that two shRNAs – shRNA1 and shRNA4 – produced nearly 100% and 70% knockdown of Cdh8, respectively (Fig 8A).
Figure 8. Cdh8 knockdown impairs dendritic outgrowth and self-avoidance in cortical neurons.
(A) Western analysis of HEK293 cells co-expressing myc-tagged-Cdh8, tTa transactivator, and either shRNA plasmids (sh1-4) or a scrambled negative control (shCtrl). Representative immunoblots using anti-c-myc antibody detect Cdh8 at the predicted size of ~135 kDa. Actin was used as a loading control. Densitometric quantification of anti-c-myc antibody shown to the right (n = 3). (B) Knockdown of endogenous Cdh8 in cultured cortical neurons at 14 DIV. Neurons co-transfected with pCAGGs-GFP and scrambled shRNA (scrambled) or Cdh8-shRNA (shCdh8) plasmids were immunolabeled for GFP (green) and Cdh8 (magenta). Cdh8 was visualized using a live staining protocol. (C) GFP-transfected neurons (green) labeled with somatodendritic marker, MAP2 (magenta) in cortical neurons at 5 DIV. Neurolucida reconstructions of dendrites shown in the right panels. Arrows indicate dendrite self-crossings. Scale bar, 20 µm. (D-I) Quantitative comparisons of dendrites in Ctrl and shCdh8 cultured cortical neurons. (D) The total number of dendritic branches, (E) total dendritic length, and (F) mean dendritic length (n = 14). (G) The number of primary branches and (H) branch endpoints (n = 14). (I) Scatter plot of dendritic self-crossings (n = 14). Error bars represent S.E.M. * p < 0.05, ** p < 0.001, *** p < 0.0001
Rat primary neurons consistently produce higher transfection efficiencies than mouse neurons, so we used shRNA4, which shows 100% sequence homology with rat mRNA to knockdown Cdh8 in cultured rat cortical neurons. Transfected neurons were identified by GFP fluorescence and levels of endogenous Cdh8 were assessed by immunolabeling intensity using the live-cell staining protocol (see Materials and Methods). Cdh8 immunofluorescence was reduced by ~50% in shRNA - transfected neurons at 3 DIV, and knockdown was sustained as neurons developed through 7 and 14 DIV, compared with those transfected with scrambled plasmid at the same time points (Mann-Whitney, % of control - 3 DIV: 48.8±9.43, n = 39-40, p = 0.019; 7 DIV: 19.1±5.23, n = 18, p < 0.0001; 14 DIV: 14.3±4.35, n = 15-20, p < 0.0001; Fig 8B).
To evaluate how the loss of Cdh8 affects dendritic morphology in cultured neurons, we screened Cdh8-deficient neurons at 5 DIV for abnormal outgrowth phenotypes. GFP-expressing neurons were immunolabeled with the somatodendritic marker MAP2 (Fig 8C), and confocal images of MAP2 labeling were traced using Neurolucida. Cdh8 knockdown significantly increased the total number of dendritic branches per neuron (n = 14, p = 0.003; Fig 8D). This was driven, in part, by an increase in the number of primary dendrites (defined as those emanating directly from the soma) (n = 14, p = 0.018; Fig 8G), as well as by a significant reduction in the mean length of dendritic segments (Welch’s corrected t-test, n =14, p = 0.0001; Fig 8F). There was no change in overall dendritic length (n = 14, p = 0.697; Fig 8E) or in the number of dendritic branch endpoints compared with control neurons (n= 14, p = 0.114; Fig 8H). While an increase in branch number with no change in endpoints seems puzzling, Cdh8-deficient neurons consistently showed a population of dendrites that looped back on their sister dendrites with no obvious branch endpoint (Fig. 8C, arrows). This is abnormal, as sister neurites from the same neuron typically avoid each other (Kramer et al., 1985). To quantify the degree of dendritic self-crossings, we subtracted the total number of branches by the number of branch endpoints. This analysis showed a significantly greater number of dendrite self-crossing in Cdh8 shRNA knockdown neurons in comparison with controls (Mann Whitney, n = 14, p < 0.0001; Fig 8I), indicating a defect in dendritic self-avoidance. Taken together, our results show that the loss of Cdh8 impairs normal dendritic outgrowth and self-avoidance, and suggests that such early loss of Cdh8 could lead to abnormal synaptic connections.
Discussion
We show here that Cdh8 is highly expressed by corticostriatal projection neurons in mPFC and by MSNs in dorsal striatum, with levels of Cdh8 mRNA and protein peaking in both structures at the beginning (~P10) of an early postnatal period of rapid synaptogenesis in striatum (Sharpe and Tepper, 1998, Tepper et al., 1998). Further, we find that Cdh8 is highly concentrated at asymmetric (excitatory) synaptic junctions within striatum, and is expressed equally by MSNs that form the direct- or the indirect-pathways. Functional analyses demonstrated that Cdh8 contributes to the generation of cortical neuron dendritic arbors and dendrite self-recognition. Together, our findings suggest multifaceted roles for Cdh8 in the molecular control of developing neurons and circuits that comprise corticostriatal pathways.
Our data showing that Cdh8 is synaptically localized in striatum is consistent with previous studies showing synaptic localization of Cdh8 in S1 (Gil et al., 2002) and hippocampus (Huntley et al., 2012), and suggest that Cdh8 participates in the assembly and maturation of corticostriatal synapses. It is well known that N-cadherin, a type I cadherin, plays an important role in the initial assembly of synapses (Togashi et al., 2002, Bozdagi et al., 2004), and can confer laminar identity (Yamagata et al., 1995, Poskanzer et al., 2003). However, its very broad distribution at nearly all excitatory synapses makes it unlikely to contribute extensively to synapse specificity. The type II subfamily of cadherins, of which Cdh8 is a member, shows more restricted distribution patterns than type I cadherins (Suzuki et al., 1997), making them more likely candidates to confer elements of a multi-component recognition/adhesive code. Consistent with this role, another type II cadherin, Cdh9, is expressed in presynaptic dentate gyrus granule cells and postsynaptic CA3 hippocampal neurons and is required for the normal development of mossy fiber synapses (Williams et al., 2011), while Cdh8 expression in presynaptic dorsal root ganglia and postsynaptic spinal cord is required for normal transmission of cold-sensation sensory input (Suzuki et al., 2007). Alternatively, Cdh8 may be responsive to, and/or enable, forms of activity-dependent synaptic plasticity such as long-term potentiation (LTP) displayed by corticostriatal synapses (Di Filippo et al., 2009). In rat hippocampus, for example, LTP of synapses between perforant path axons and dentate gyrus granule neurons results in a rapid redistribution of synaptic Cdh8 which may facilitate structural spine remodeling at potentiated synapses (Huntley et al., 2012). In any event, Cdh8 adhesion would be anticipated to recruit F-actin to sites of adhesion and promote differentiation, stabilization, and normal synapse function (Zhang and Benson, 2001, Togashi et al., 2002, Jungling et al., 2006). In the future, functional studies will be required to determine definitively if Cdh8 contributes to synaptogenesis and striatal synaptic plasticity.
Recent work suggests a modest bias in the extent to which particular cortical areas target direct- or indirect-pathway MSNs (Wall et al., 2013). While Cdh8 may help to specify interactions between cortex and striatum, it appears unlikely that it encodes selective targeting of cortical input to direct- or indirect-MSN populations, as we found that Cdh8 is expressed equally by both direct- and indirect-MSNs. On the other hand, MSNs receive afferent input from two distinct classes of corticostriatal projection neurons – intratelencephalic (IT-type) and pyramidal tract (PT-type) neurons (Reiner et al., 2010) – which are distinguished by the projections of their axon collaterals and are differentially vulnerable to Parkinson’s disease and autism spectrum disorder (ASD)-related pathogenesis (Ballion et al., 2008, Shepherd, 2013). It will be important in future studies to determine whether Cdh8 is differentially expressed by either of these cortical types.
Our loss-of-function studies in primary cortical neurons show that Cdh8 knockdown leads to significant dendritic abnormalities. Strikingly, Cdh8-deficient neurons display self-crossing dendrites, suggesting a defect in dendrite self-avoidance, a property that maximizes coverage of the neuropil and interaction with incoming axons (Kramer and Kuwada, 1983, Montague and Friedlander, 1989, Grueber and Sagasti, 2010). While the mechanisms by which homophilic cadherin-based adhesive interactions can mediate repulsion are not known, several lines of research show that the protein families mediating self-recognition in dendrites all participate in homophilic adhesion. Deletion or loss-of-function mutations of Down’s syndrome cell adhesion molecule (DsCAM) genes in a variety of neurons in Drosophila or in mouse retina (Hughes et al., 2007, Matthews et al., 2007, Soba et al., 2007) or the entire family of γ-protocadherins in mouse amacrine or Purkinje cells (Lefebvre et al., 2012) promotes excessive dendritic bundling or overlap. These effects are cell-autonomous and can be rescued by the introduction of any single family member (Matthews et al., 2007, Lefebvre et al., 2012). However, different cell adhesion families appear to confer this property in different cell types, as the loss of either DSCAM or γ-protocadherins in mouse cortical neurons does not enhance interactions between sister dendrites (Garrett et al., 2012, Maynard and Stein, 2012). It is possible that Cdh8 acts collaboratively to generate dendrite repulsion. Classic cadherins and protocadherins can interact in cis and can both positively (Emond et al., 2011) or negatively (Yasuda et al., 2007) regulate adhesive interactions generated by either family. Moreover, overexpression and mis-localization of the normally axonally restricted Nectin 1, an Ig superfamily member that partners with cadherins via shared intracellular binding partners, promotes interactions between sister dendrites as well as excessive interactions between axons and dendrites (Togashi et al., 2006). It will be important for future studies to identify the molecular details that enable Cdh8-mediated dendrite self-avoidance, as well as to pursue effects of Cdh8 on dendrite self-avoidance in vivo in cortical and striatal neurons. Finally, while there are multiple adhesion systems operative at synapses, the present study adds to the growing recognition that each has unique roles in development and/or maintenance of synaptic structure and function, which bears importantly on their contribution to disorders of brain circuitry (Dalva et al., 2007, Friedman et al., 2014).
Recently, CDH8 has been identified as an ASD and learning disability susceptibility gene. Data from the Autism Genome Project Consortium shows a rare familial microdeletion on chromosome 16q21, containing only the gene that encodes Cdh8. In one family, individuals harboring the 16q21 deletion had autism and learning disabilities, while their siblings were unaffected. In a second family, an overlapping deletion transmitted from a parent exhibiting moderate learning disability was observed in an individual with severe learning disability (Pagnamenta et al., 2011). Another study found that two patients harboring deletions immediately upstream of CDH8 have autism (Walker and Scherer, 2013). Our data showing that Cdh8 controls cortical dendritic organization and is enriched in developing corticostriatal neurons and MSN synapses suggest that such cognitive abnormalities in humans reflect defects in Cdh8-mediated molecular control over developing corticostriatal neurons and circuits. This is further supported by numerous studies implicating abnormal development of PFC and PFCstriatal projections in the underlying neurobiology of ASDs (Rinaldi et al., 2008, Courchesne et al., 2011, Peca et al., 2011).
Taken together our studies have provided novel insight into the molecular constituents of corticostriatal circuits – pathways that are essential for high-order executive function and implicated in cognitive deficits associated with ASDs, as well as in Parkinson’s and Huntington’s disease. These studies provide the framework for future loss-of-function studies associated with disrupted corticostriatal circuit development in ASD-related pathologies and provide a means by which to target approaches to particular circuits disrupted in neurodegenerative disease.
Acknowledgments
We thank Bill Janssen, Rishi Puri and Frank Yuk for assistance with electron microscopy, Esther Kim and Fumiko Isoda for assistance with qRT-PCR, and Leigh Needleman for assistance with isotopic in situ hybridization and analysis. Some of the microscopy was performed at the Microscopy Shared Resource Facility at the Icahn School of Medicine at Mount Sinai.
Grant Sponsor: National Institute of Mental Health grant MH100600; Simons Foundation; Michael J Fox Foundation; NIMH T32 Training Program in Mental Health grant (T32 MH096678).
Footnotes
The authors declare no competing financial interests.
Author contributions
LGF, GWH, and DLB designed research; LGF, FWR, JMS, RM, and FMW performed research; LGF, FWR, and JMS analyzed data; and LGF, GWH, and DLB wrote the paper.
References
- Alexander GE, Crutcher MD. Functional architecture of basal ganglia circuits: neural substrates of parallel processing. Trends in Neurosci. 1990;13:266–271. doi: 10.1016/0166-2236(90)90107-l. [DOI] [PubMed] [Google Scholar]
- Anderson TR, Shah PA, Benson DL. Maturation of glutamatergic and GABAergic synapse composition in hippocampal neurons. Neuropharm. 2004;47:694–705. doi: 10.1016/j.neuropharm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Andre VM, Fisher YE, Levine MS. Altered balance of activity in the striatal direct and indirect pathways in mouse models of Huntington's disease. Front Systems Neurosci. 2011;5:46. doi: 10.3389/fnsys.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla PK, Huntley GW. Early postnatal expression and localization of matrix metalloproteinases-2 and -9 during establishment of rat hippocampal synaptic circuitry. J Comp Neurol. 2014;522:1249–1263. doi: 10.1002/cne.23468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballion B, Mallet N, Bezard E, Lanciego JL, Gonon F. Intratelencephalic corticostriatal neurons equally excite striatonigral and striatopallidal neurons and their discharge activity is selectively reduced in experimental parkinsonism. Eur J Neurosci. 2008;27:2313–2321. doi: 10.1111/j.1460-9568.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- Bekirov IH, Nagy V, Svoronos A, Huntley GW, Benson DL. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus. 2008;18:349–363. doi: 10.1002/hipo.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekirov IH, Needleman LA, Zhang W, Benson DL. Identification and localization of multiple classic cadherins in developing rat limbic system. Neuroscience. 2002;115:213–227. doi: 10.1016/s0306-4522(02)00375-5. [DOI] [PubMed] [Google Scholar]
- Benson DL, Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J Neurosci. 1998;18:6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendse HW, Galis-de Graaf Y, Groenewegen HJ. Topographical organization and relationship with ventral striatal compartments of prefrontal corticostriatal projections in the rat. J Comp Neurol. 1992;316:314–347. doi: 10.1002/cne.903160305. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosi. 2004;27:509–521. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock JH, Elste A, Huntley GW. Distribution and injury-induced plasticity of cadherins in relationship to identified synaptic circuitry in adult rat spinal cord. J Neurosci. 2004;24:8806–8817. doi: 10.1523/JNEUROSCI.2726-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Sorensen JC, Ostergaard K, Zimmer J. Early postnatal development of the rat corticostriatal pathway: an anterograde axonal tracing study using biocytin pellets. Anat and Embryol. 1999;200:73–80. doi: 10.1007/s004290050261. [DOI] [PubMed] [Google Scholar]
- Corbit LH, Balleine BW. The role of prelimbic cortex in instrumental conditioning. Behav Brain Res. 2003;146:145–157. doi: 10.1016/j.bbr.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, Barnes CC, Pierce K. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima AD, Merten MD, Voigt T. Neuritic differentiation and synaptogenesis in serumfree neuronal cultures of the rat cerebral cortex. J Comp Neurol. 1997;382:230–246. doi: 10.1002/(sici)1096-9861(19970602)382:2<230::aid-cne7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Delint-Ramirez I, Fernandez E, Bayes A, Kicsi E, Komiyama NH, Grant SG. In vivo composition of NMDA receptor signaling complexes differs between membrane subdomains and is modulated by PSD-95 and PSD-93. J Neurosci: the. 2010;30:8162–8170. doi: 10.1523/JNEUROSCI.1792-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Filippo M, Picconi B, Tantucci M, Ghiglieri V, Bagetta V, Sgobio C, Tozzi A, Parnetti L, Calabresi P. Short-term and long-term plasticity at corticostriatal synapses: implications for learning and memory. Behavi Brain Res. 2009;199:108–118. doi: 10.1016/j.bbr.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Ding JB, Oh WJ, Sabatini BL, Gu C. Semaphorin 3E-Plexin-D1 signaling controls pathway-specific synapse formation in the striatum. Nat Neurosci. 2012;15:215–223. doi: 10.1038/nn.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Blevins CJ, Jontes JD. A complex of Protocadherin-19 and Ncadherin mediates a novel mechanism of cell adhesion. J Cell Biol. 2011;195:1115–1121. doi: 10.1083/jcb.201108115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch T, Lemmon V, Banker G. Differential effects of NgCAM and N-cadherin on the development of axons and dendrites by cultured hippocampal neurons. J Neurocytol. 2000;29:215–223. doi: 10.1023/a:1026515426303. [DOI] [PubMed] [Google Scholar]
- Friedman LG, Benson DL, Huntley GW. Cadherin-based trans-synaptic networks in establishing and modifying neural connectivity. Curr Topics in Dev Biol. 2014 doi: 10.1016/bs.ctdb.2014.11.025. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbott PL, Warner TA, Jays PR, Salway P, Busby SJ. Prefrontal cortex in the rat: projections to subcortical autonomic, motor, and limbic centers. J Comp Neurol. 2005;492:145–177. doi: 10.1002/cne.20738. [DOI] [PubMed] [Google Scholar]
- Garrett AM, Schreiner D, Lobas MA, Weiner JA. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74:269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Ann Rev Neurosci. 1992;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil OD, Needleman L, Huntley GW. Developmental patterns of cadherin expression and localization in relation to compartmentalized thalamocortical terminations in rat barrel cortex. J Comp Neurol. 2002;453:372–388. doi: 10.1002/cne.10424. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Sagasti A. Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb Perspect Biol. 2010;2:a001750. doi: 10.1101/cshperspect.a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KJ, Craig AM. Synapse composition and organization following chronic activity blockade in cultured hippocampal neurons. J Comp Neurol. 2005;490:72–84. doi: 10.1002/cne.20635. [DOI] [PubMed] [Google Scholar]
- Hertel N, Krishna K, Nuernberger M, Redies C. A cadherin-based code for the divisions of the mouse basal ganglia. J Comp Neurol. 2008;508:511–528. doi: 10.1002/cne.21696. [DOI] [PubMed] [Google Scholar]
- Heyers D, Kovjanic D, Redies C. Cadherin expression coincides with birth dating patterns in patchy compartments of the developing chicken telencephalon. J Comp Neurol. 2003;460:155–166. doi: 10.1002/cne.10631. [DOI] [PubMed] [Google Scholar]
- Hsiao K, Bozdagi O, Benson DL. Axonal cap-dependent translation regulates presynaptic p35. Dev Neurobiol. 2014;74:351–364. doi: 10.1002/dneu.22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley GW, Benson DL. Neural (N)-cadherin at developing thalamocortical synapses provides an adhesion mechanism for the formation of somatopically organized connections. J Comp Neurol. 1999;407:453–471. [PubMed] [Google Scholar]
- Huntley GW, Elste AM, Patil SB, Bozdagi O, Benson DL, Steward O. Synaptic loss and retention of different classic cadherins with LTP-associated synaptic structural remodeling in vivo. Hippocampus. 2012;22:17–28. doi: 10.1002/hipo.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Sanes JR. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science. 1997;276:1428–1431. doi: 10.1126/science.276.5317.1428. [DOI] [PubMed] [Google Scholar]
- Jungling K, Eulenburg V, Moore R, Kemler R, Lessmann V, Gottmann K. N-cadherin transsynaptically regulates short-term plasticity at glutamatergic synapses in embryonic stem cell-derived neurons. J Neurosci. 2006;26:6968–6978. doi: 10.1523/JNEUROSCI.1013-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Korematsu K, Nishi T, Okamura A, Goto S, Morioka M, Hamada J, Ushio Y. Cadherin-8 protein expression in gray matter structures and nerve fibers of the neonatal and adult mouse brain. Neuroscience. 1998;87:303–315. doi: 10.1016/s0306-4522(98)00077-3. [DOI] [PubMed] [Google Scholar]
- Korematsu K, Redies C. Expression of cadherin-8 mRNA in the developing mouse central nervous system. J Comp Neurol. 1997;387:291–306. [PubMed] [Google Scholar]
- Kramer AP, Goldman JR, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. I. Origin of natural variations in the branching pattern. J Neurosci. 1985;5:759–767. doi: 10.1523/JNEUROSCI.05-03-00759.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer AP, Kuwada JY. Formation of the receptive fields of leech mechanosensory neurons during embryonic development. J Neurosci. 1983;3:2474–2486. doi: 10.1523/JNEUROSCI.03-12-02474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Tye LD, Kreitzer AC. Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat Neurosci. 2012;15:816–818. doi: 10.1038/nn.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krettek JE, Price JL. Projections from the amygdaloid complex to the cerebral cortex and thalamus in the rat and cat. J Comp Neurol. 1977;172:687–722. doi: 10.1002/cne.901720408. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Maynard KR, Stein E. DSCAM contributes to dendrite arborization and spine formation in the developing cerebral cortex. J Neurosci. 2012;32:16637–16650. doi: 10.1523/JNEUROSCI.2811-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz CD, Carcea I, McNickle DG, Dickson TC, Ge Y, Salton SR, Benson DL. ERM proteins regulate growth cone responses to Sema3A. J Comp Neurol. 2008;510:351–366. doi: 10.1002/cne.21799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskevich F, Zhu Y, Ranscht B, Sanes JR. Expression of multiple cadherins and catenins in the chick optic tectum. Mol Cell Neurosci. 1998;12:240–255. doi: 10.1006/mcne.1998.0718. [DOI] [PubMed] [Google Scholar]
- Montague PR, Friedlander MJ. Expression of an intrinsic growth strategy by mammalian retinal neurons. Proc Natl Acad Sci U S A. 1989;86:7223–7227. doi: 10.1073/pnas.86.18.7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortillo S, Elste A, Ge Y, Patil SB, Hsiao K, Huntley GW, Davis RL, Benson DL. Compensatory redistribution of neuroligins and N-cadherin following deletion of synaptic beta1-integrin. J Comp Neurol. 2012;520:2041–2052. doi: 10.1002/cne.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitczuk JS, Patil SB, Matikainen-Ankney BA, Scarpa J, Shapiro ML, Benson DL, Huntley GW. N-cadherin regulates molecular organization of excitatory and inhibitory synaptic circuits in adult hippocampus in vivo. Hippocampus. 2014;24:943–962. doi: 10.1002/hipo.22282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Oh JS, Manzerra P, Kennedy MB. Regulation of the neuron-specific Ras GTPase-activating protein, synGAP, by Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 2004;279:17980–17988. doi: 10.1074/jbc.M314109200. [DOI] [PubMed] [Google Scholar]
- Pagnamenta AT, Khan H, Walker S, Gerrelli D, Wing K, Bonaglia MC, Giorda R, Berney T, Mani E, Molteni M, Pinto D, Le Couteur A, Hallmayer J, Sutcliffe JS, Szatmari P, Paterson AD, Scherer SW, Vieland VJ, Monaco AP. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J Med Genet. 2011;48:48–54. doi: 10.1136/jmg.2010.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskanzer K, Needleman LA, Bozdagi O, Huntley GW. N-cadherin regulates ingrowth and laminar targeting of thalamocortical axons. J Neurosci. 2003;23:2294–2305. doi: 10.1523/JNEUROSCI.23-06-02294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reep RL, Cheatwood JL, Corwin JV. The associative striatum: organization of cortical projections to the dorsocentral striatum in rats. J Comp Neurol. 2003;467:271–292. doi: 10.1002/cne.10868. [DOI] [PubMed] [Google Scholar]
- Reiner A, Hart NM, Lei W, Deng Y. Corticostriatal projection neurons - dichotomous types and dichotomous functions. Front Neuroanat. 2010;4:142. doi: 10.3389/fnana.2010.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehl R, Johnson K, Bradley R, Grunwald GB, Cornel E, Lilienbaum A, Holt CE. Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron. 1996;17:837–848. doi: 10.1016/s0896-6273(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Perrodin C, Markram H. Hyper-connectivity and hyper-plasticity in the medial prefrontal cortex in the valproic Acid animal model of autism. Front Neural Circuits. 2008;2:4. doi: 10.3389/neuro.04.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda B, Mesias R, Li X, Yue Z, Benson DL. Short- and long-term effects of LRRK2 on axon and dendrite growth. PloS one. 2013;8:e61986. doi: 10.1371/journal.pone.0061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148:579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe NA, Tepper JM. Postnatal development of excitatory synaptic input to the rat neostriatum: an electron microscopic study. Neurosci. 1998;84:1163–1175. doi: 10.1016/s0306-4522(97)00583-6. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JR, Macklis JD. Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex. 2012;24:293–303. doi: 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JR, Macklis JD. Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex. 2014;24:293–303. doi: 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SC, Furue H, Koga K, Jiang N, Nohmi M, Shimazaki Y, Katoh-Fukui Y, Yokoyama M, Yoshimura M, Takeichi M. Cadherin-8 is required for the first relay synapses to receive functional inputs from primary sensory afferents for cold sensation. J Neurosci. 2007;27:3466–3476. doi: 10.1523/JNEUROSCI.0243-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol Cell Neurosci. 1997;9:433–447. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Sharpe NA, Koos TZ, Trent F. Postnatal development of the rat neostriatum: electrophysiological, light- and electron-microscopic studies. Dev Neurosci. 1998;20:125–145. doi: 10.1159/000017308. [DOI] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Togashi H, Miyoshi J, Honda T, Sakisaka T, Takai Y, Takeichi M. Interneurite affinity is regulated by heterophilic nectin interactions in concert with the cadherin machinery. J Cell Biol. 2006;174:141–151. doi: 10.1083/jcb.200601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Scherer SW. Identification of candidate intergenic risk loci in autism spectrum disorder. BMC Genom. 2013;14:499. doi: 10.1186/1471-2164-14-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Callaway EM, Kreitzer AC. Differential innervation of directand indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, Ghosh A. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron. 2011;71:640–655. doi: 10.1016/j.neuron.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SP, Jones EG. Cells of origin and terminal distribution of descending projections of the rat somatic sensory cortex. J Comp Neurol. 1977;175:129–157. doi: 10.1002/cne.901750202. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Herman JP, Sanes JR. Lamina-specific expression of adhesion molecules in developing chick optic tectum. J Neurosci. 1995;15:4556–4571. doi: 10.1523/JNEUROSCI.15-06-04556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, De Robertis EM, Yamagata K. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ, Balleine BW. Lesions of dorsolateral striatum preserve outcome expectancy but disrupt habit formation in instrumental learning. Eur J Neurosci. 2004;19:181–189. doi: 10.1111/j.1460-9568.2004.03095.x. [DOI] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ, Balleine BW. Blockade of NMDA receptors in the dorsomedial striatum prevents action-outcome learning in instrumental conditioning. Eur J Neurosci. 2005;22:505–512. doi: 10.1111/j.1460-9568.2005.04219.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Benson DL. Stages of synapse development defined by dependence on F-actin. J Neurosci. 2001;21:5169–5181. doi: 10.1523/JNEUROSCI.21-14-05169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Luo L. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron. 2004;42:63–75. doi: 10.1016/s0896-6273(04)00142-4. [DOI] [PubMed] [Google Scholar]