Abstract
We identified a Cu accumulating structure with a dynamic role in intracellular Cu homeostasis. During Zn limitation, Chlamydomonas reinhardtii hyperaccumulated Cu, dependent on the nutritional Cu sensor CRR1, but was functionally Cu-deficient. Visualization of intracellular Cu revealed major Cu accumulation sites coincident with electron-dense structures that stained positive for low pH and polyphosphate, suggesting that they are lysosome-related organelles. NanoSIMS showed colocalization of Ca and Cu, and X-ray absorption spectroscopy (XAS) was consistent with Cu+ accumulation in an ordered structure. Zn resupply restored Cu homeostasis concomitant with reduced abundance of these structures. Cu isotope labeling demonstrated that sequestered Cu+ became bio-available for the synthesis of plastocyanin, and transcriptome profiling indicated that mobilized Cu became visible to CRR1. Cu trafficking to intracellular accumulation sites may be a strategy for preventing protein mis-metallation during Zn deficiency and enabling efficient cuproprotein (re)-metallation upon Zn resupply.
Introduction
Many proteins in cells are associated with metal ions, which provide structural stability and catalytic functionalities like electrophiles, reductants and oxidants that are not readily provided by functional groups of amino acids1. Nature has used the unique chemical properties of each metal ion -- such as ligand preferences, coordination geometries and redox potential -- to generate an amazing repertoire of catalytic abilities, such as the reduction of dinitrogen to ammonium and the oxidation of water, under gentle biological conditions. These catalytic activities are dependent on specific metal cofactors in unique active sites, and life is therefore dependent on the bioavailability of a combination of metal ions. It is critical that the right metal cofactor occupies specifically its dedicated active site. The divalent metal ions of Mn, Fe, Co, Ni, Cu and Zn bind to functional groups in proteins according to thermodynamic preferences described by the Irving-Williams series2, which means that without a mechanism for selectivity in vivo, metal binding would be driven by the concentration of the metal ion and these preferences toward the available binding sites3,4. The best understood mechanisms for ensuring selectivity of metal-protein association include the metallochaperones (prototyped by the Cu chaperones Atx1 and Ccs5,6), which effect transfer of the metal to its final target site via specific protein-protein interactions and ligand exchange, and compartmentalized assembly (prototyped by MncA)7, in which holoprotein formation is restricted to a compartment with the appropriate high concentration of the desired metal ion and low concentration of an undesired but thermodynamically preferred metal ion7–9. Kinetic barriers generally preclude metal exchange or dissociation once the metalloprotein is in its final sub-cellular location.
Since Cu2+ is at the top of the Irving-Williams series and Cu+ also has strong preferences for soft ligands like thiolates, most intracellular compartments maintain very low concentrations of “free” Cu ions10. For instance, Chlamydomonas reinhardtii (C. reinhardtii), a eukaryotic reference organism for studying metal homeostasis in the context of chloroplast biology, maintains a Cu quota that is determined strictly by the abundance of various cuproproteins required for its metabolism, especially plastocyanin for photosynthesis, cytochrome (Cyt) oxidase for respiration and a ferroxidase for iron assimilation, even when presented with 100-fold excess Cu in the growth milieu11,12. This is achieved by restricted Cu uptake. In Cu-deficient conditions, C. reinhardtii, like other algae and many cyanobacteria, can reduce the Cu quota substantially by substituting a heme protein, Cyt c6, for plastocyanin13,14. The reciprocal accumulation of Cyt c6 vs. plastocyanin as a function of medium Cu content serves as a readout of the Cu nutritional state, and is effected by a transcriptional program dependent on a Cu-sensing SBP-domain transcription factor called CRR115,16.
Cu homeostasis in C. reinhardtii is disrupted by nutritional Zn deficiency, which results in unprecedented Cu accumulation, up to 20 times the typical quota17. In this work, we used high resolution secondary ion mass spectrometry (SIMS) with a NanoSIMS 50 to localize Cu in intracellular compartments18,19, reminiscent of the acidocalcisome and the previously-described zincosomes20–22. The accumulated Cu+ was in a reproducibly organized chemical environment consisting of N, S and O ligands, but it became bio-available with priority over extracellular Cu for de-activating CRR1 and metalating apoplastocyanin. We hypothesize that compartmentalization would prevent mis-metallation of Zn enzymes but this would result in intracellular Cu deficiency, which would activate CRR1, resulting in feed-forward over-accumulation. Compartmentalized sequestration of accumulated Cu+ instead of cellular efflux, which dominates in bacterial systems as a means of detoxification23–25, allowed the storage of this metal ion for future use in a situation of challenging micronutrient availability.
Results
Zn-deficiency disrupts Cu homeostasis
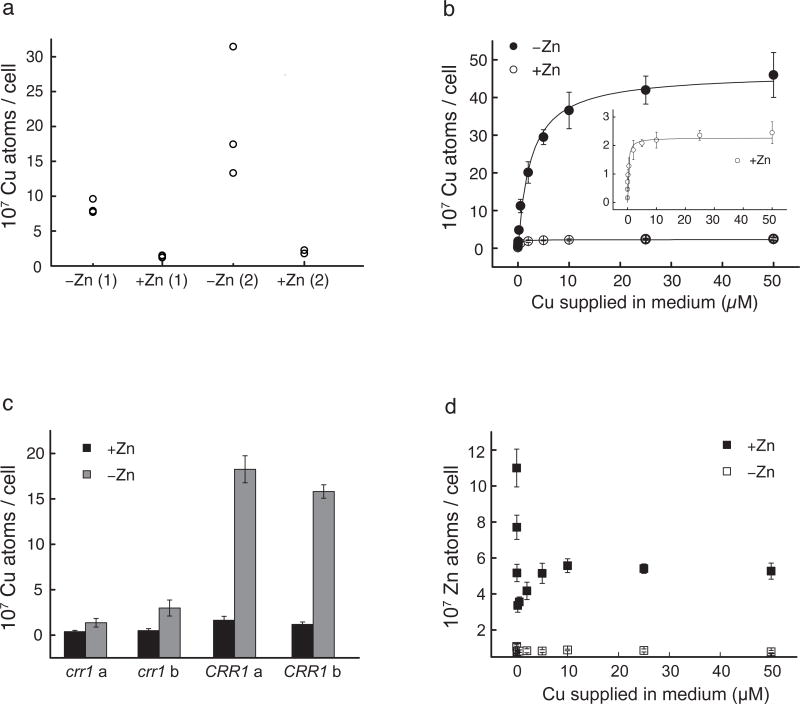
C. reinhardtii keeps intracellular Cu content relatively constant between ~ 1–2.5 × 107 atoms per cell when the external milieu contains chelated Cu ranging from 1 to 80 μM12, but this fine-tuned homeostatic mechanism is disrupted in Zn-limited cells17. Zn-limited cells showed a growth phenotype, especially in the second round of cultivation in limited medium (Supplementary Results, Supplementary Fig. 1). Inductively coupled plasma mass spectrometry (ICP-MS) analysis (Fig. 1) of these cells showed dramatic accumulation of Cu, up to ~ 30×107 atoms per cell in standard growth medium containing 2 μM Cu2+-EDTA (Fig. 1a). If the external supply was increased from 2 to 50 μM, intracellular Cu content was further enhanced to ~ 40 × 107 atoms per cell (Figure 1b). Hyper-accumulation occured only in CRR1 but not crr1 cells, indicating that the pathway is dependent on the nutritional Cu regulon (Fig. 1c). Indeed, the CTR transporters, which are the route for Cu+ assimilation were upregulated in Zn-limited cells despite adequate extracellular Cu2+ and excessive intracellular Cu+ (see below).
Figure 1. Zn deficiency induces CRR1-dependent Cu hyperaccumulation.
ICP-MS analysis of Cu and Zn contents in cells grown under Zn-limited (−Zn) or -replete (+Zn) conditions. (a) Cellular Cu contents in cells from three independent C. reinhardtii cultures (strain CC-4532) that were grown to mid-logarithmic phase in either −Zn or +Zn (containing 2.5 μM ZnCl2) TAP medium (in first (1) and second (2) round of transfer to indicated medium). Quantifications of Cu (b) and Zn (d) contents in cells grown under a concentration range of supplemented Cu (0, 0.02, 0.05, 0.2, 0.5, 2, 5, 10, 25, and 50 μM) in −Zn or +Zn TAP medium. The inset in b is the rescaled representation of Cu contents in the Zn-replete cells. The difference in total Cu contents between −Zn and +Zn cells is about 20-fold. (c) Cu contents in crr1-2 frameshift mutants16 (CC-3960) and CRR1 complemented strains (a and b representing two independent isolates). Quantification results from three different experiments per strain and condition are shown as separate data points (a) or as averages with corresponding standard deviations (b–d).
Biological but not chemical Cu deficiency
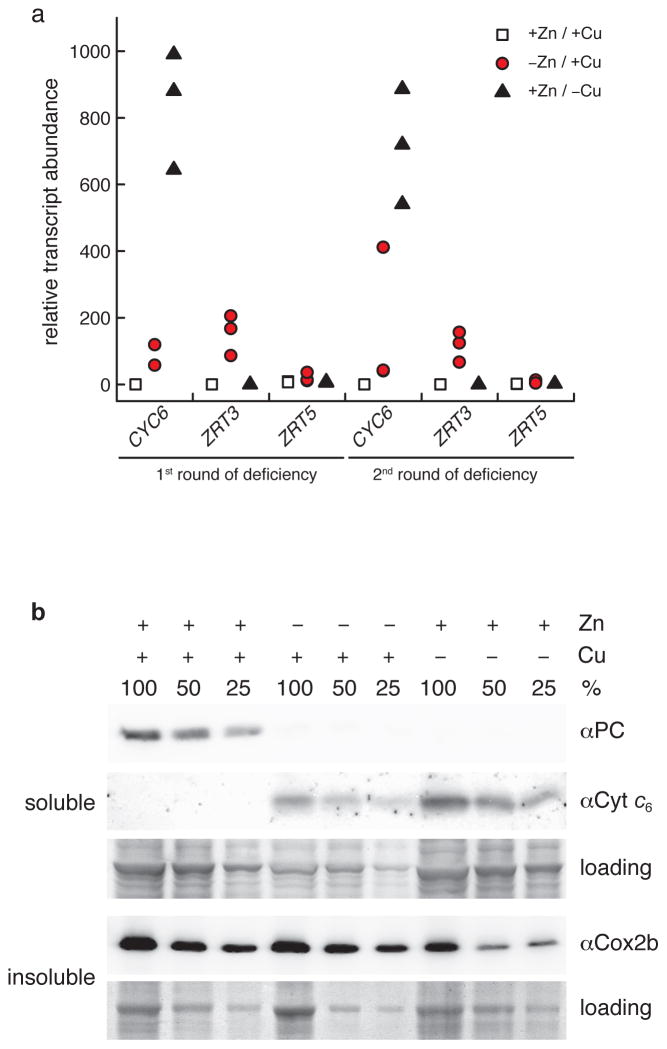
To distinguish the underlying mechanism, we monitored the expression of sentinel genes of the Cu regulon by quantitative reverse-transcriptase (RT)-PCR. Expression of CYC6 (encoding Cyt c6), whose transcripts are completely absent in Cu- and Zn-replete cells, was strikingly increased in Zn-limited cells (Figure 2a) as was the corresponding protein (Figure 2b). On the other hand, the abundance of plastocyanin was dramatically reduced in Zn-limited cells just as it was in Cu-deficient ones (Figure 2b), indicative of functional Cu-deficiency. This situation would activate the expression of the assimilatory CTR-family Cu+ transporters via CRR1 and in so doing would account for the concentration-dependent hyper-accumulation of Cu. We conclude that Zn-limited cells experience biological but not chemical Cu-deficiency.
Figure 2. Zn-limited cells also express Cu deficiency markers.
(a) Quantitative RT-PCR (qRT-PCR) to test for the expression of genes that react to altered Zn and Cu availabilities, respectively. The tested genes were CYC6 encoding cytochrome c6 (Cyt c6) as a marker for the Cu-deficiency regulon and ZRT3/ZRT5 encoding ZIP transporters as markers of the Zn-deficiency regulon. The relative transcript abundances as calculated with the LinRegPCR program are plotted as separate data points for triplicate cultures grown either under replete conditions (+Zn/+Cu), Zn deficiency (−Zn/+Cu), or under Cu deficiency (+Zn/−Cu) in first and second round of limitation, respectively. (b) Immunoblot analyses showed that Cyt c6 and plastocyanin (PC) were adjusted in Zn deficient cells to levels expected from Cu deficient cells. Shown are immunoblots with dilution series (100, 50, and 25 %) of soluble and insoluble protein extracts that were incubated with antiPC, antiCyt c6, and an antibody against a Cu-binding subunit of Cyt oxidase (antiCox2b), respectively. This experiment was carried out three times and a representative set of those is shown. Full Coomassie-stained gels of loading controls are shown in Supplementary Figure 18.
We observed that the Zn content was only slightly increased (about 2.5 fold) in Cu-depleted vs. -replete cells (Figure 1d), possibly because of increased expression of IRT2 (encoding iron responsive transporter 2 of the ZIP family26). This finding is consistent with the expression of sentinel genes of the Zn regulon, ZRT3 and ZRT517 (encoding Zn responsive transporters of the ZIP family) only in Zn-limited but not Cu-deficient cells (Figure 2a),
Cu accumulates in foci and colocalizes with Ca
In pilot survey experiments to screen whether Cu might be sequestered (i.e. biologically inaccessible) in Zn deficient C. reinhardtii cells, we used fluorescence imaging with a pair of structurally and functionally matched fluorophores (Fig. 3a), where one has the capability to respond to Cu and the other does not (Supplementary Figure 2a–c). Cu Sensor-3 (CS3)27, the Cu-responsive dye, showed a selective and high turn-on response to Cu (75-fold) and tighter Kd for Cu+ (Kd = 9×10−14) relative to glutathione, an abundant competing cellular ligand (apparent Kd = 9×10−12 28 assuming the major species is a 1:1 Cu:GSH complex)29. CS3 did not respond to pH alone and retained the ability to respond to Cu in the pH range of 4 to 7.5 (Supplementary Fig. 2b), which makes it suitable for staining intracellular compartments. Ctrl-CS3 (1) employed an identical BODIPY fluorophore and has the exact same number of receptor atoms (Supplementary Fig. 2a), except that the four sulfur atoms that are essential for metal interaction in CS3 are replaced by isosteric carbons that lack affinity for Cu binding (Fig. 3a). Indeed, Ctrl-CS3 did not respond to Cu or pH changes as demonstrated by Cu and pH titration assays (Supplementary Fig. 2c) and thus could be used in conjunction with CS3 to distinguish between Cu- vs. any potential dye-backbone-dependent responses.
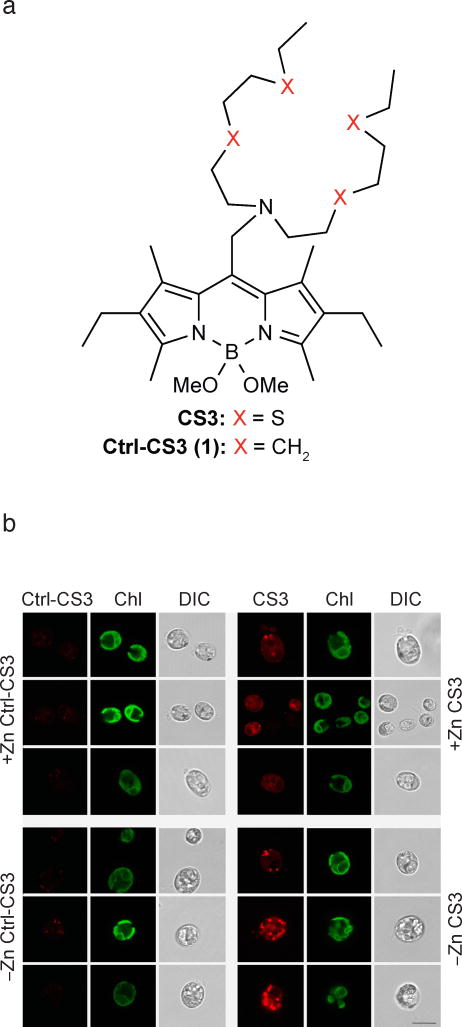
Figure 3. Cu(I)-sensitive CS3 staining suggests Cu accumulation in intracellular compartments.
(a) Chemical structure of the Cu+-binding fluorescent dye CS3 and the non-copper binding analog control-CS3 (Ctrl-CS3), where the four metal binding sulfur atoms (CS3) are replaced by isosteric carbons (Ctrl-CS3). For the detailed synthesis refer to the Supplementary note and Supplementary Figure 2a. (b) Zn-limited and Zn-replete wildtype C. reinhardtii (CC-4532) cells were stained with the cuprous dye CS325 to observe intracellular Cu distribution by confocal microscopy. For control, we stained cells from the same cultures with the control dye Ctrl-CS3. DIC = differential interference contrast; Chl = chlorophyll autofluorescence. The shown scale bar represents 10 μm for all images. We analyzed 55 Zn-deficient cells stained with CS3, 30 Zn-deficient cells treated with Ctrl-CS3, 54 Zn-replete cells stained with CS3, and 64 Zn-replete cells treated with Ctrl-CS3 in total.
Confocal fluorescence microscopy with CS3, but not its Ctrl-CS3 counterpart that does not respond to Cu, showed the presence of fluorescent foci that correlated with the Zn-deficiency status of the cells (Figure 3b); the foci were not as frequently observed in Zn-replete cells (Figure 3b). More importantly, fluorescent foci were seen only in situations where ICP-MS methods indicated high intracellular Cu content. Specifically, the fluorescent foci were not observed in cells of the crr1 mutant, which cannot hyperaccumulate Cu, but were readily visualized in Zn-limited cells of the complemented strain (Supplementary Fig. 3a). The CS3 signal was significantly reduced when cells were treated with a cell permeant Cu chelator, glutathione mono ethyl ester prior to staining with CS3 (Supplementary Fig. 3b). Multiple control experiments were performed to ensure the specificity of CS3 for Cu+ in this model system (Supplementary Note, Supplementary Fig. 20).
Because CS3 is based on a lipophilic BODIPY-platform27, there is concern that the probe might be staining lipid bodies, which do accumulate in Zn-deficient cells30, and we performed multiple lines of control experiments to address this potential issue. First, the Ctrl-CS3 dye, which lacks the Cu binding atoms but retains the same lipophilic dye platform, did not stain the putative Cu puncta or any other body in Zn-deficient cells (Figure 3b). We also tested whether CS3 would visualize lipid bodies in N-deficient Chlamydomonas cells, which hyper-accumulate lipids but do not hyper-accumulate Cu. We found that lipid bodies in N-deficient cells, which do stain well with Nile Red, did not stain with CS3 in parallel experiments with the same settings (Supplementary Fig. 4).
With these pilot data suggesting further investigation, we sought to use an unequivocal physical technique to identify the intracellular Cu foci. Direct metal detection techniques such as X-ray fluorescence microscopy have been valuable to support indirect metal imaging obtained using synthetic fluorophores27. In the present study, electron-dense structures were revealed in microtomed thin-sections of fixed cells by transmission electron microscopy (TEM) and these were candidate sites for Cu accumulation. Indeed, the abundance and size of the electron dense structures correlated with Zn nutrition (Figure 4a). Kruskal-Wallis One Way Analysis of Variance on ranks of the electron-dense areas in the electron micrographs in relation to total cell areas found a significant difference between the Zn-limited and Zn-replete sections (p-value P= <0.001) (Figure 4a, right panel). NanoSIMS imaging, which allows simultaneous spatial detection of elements based on their mass, indicated that regions containing calcium (Ca) and Cu correlated with electron-dense structures at the periphery of the cells visible in TEM images of the same sections (Figure 4b). Co-localization of Ca and Cu in Zn-limited cells was confirmed independently by confocal microscopy and dual staining of cells with CS3 and Fluo-4-AM, a Ca2+-specific fluorescent dye (Supplementary Fig. 5).
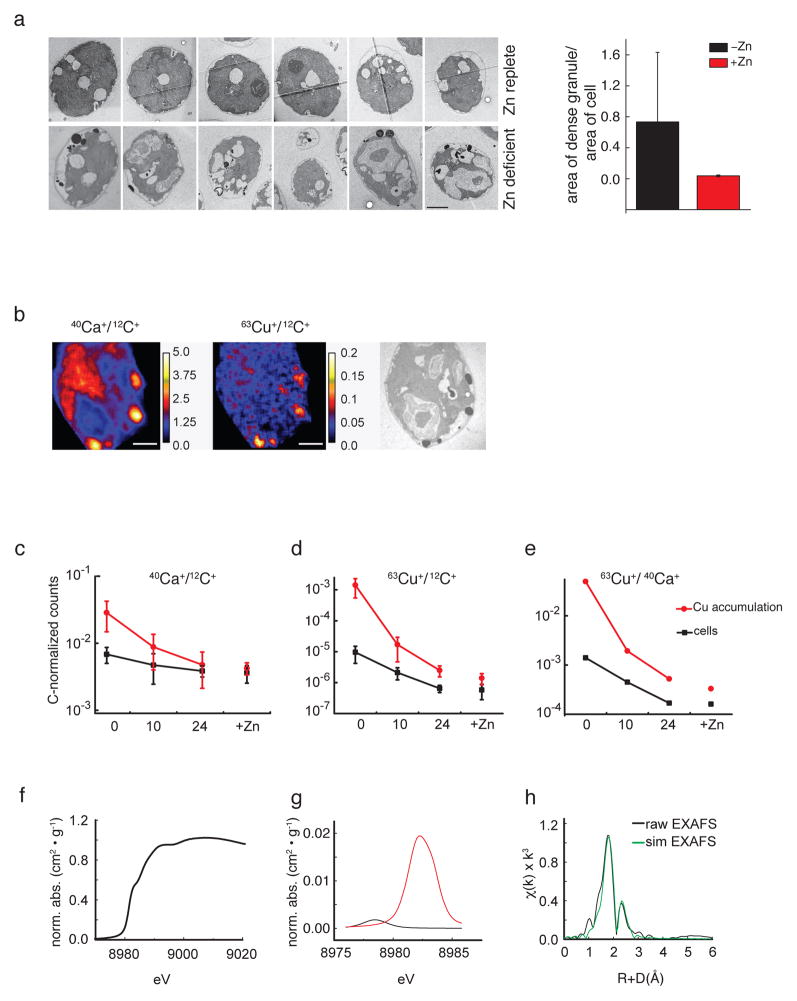
Figure 4. Intracellular Cu is traceable to Cu accumulating compartments.
(a) Transmission electron microscopy (TEM) revealed electron-dense structures in Zn-limited cells (scale bar 2 μm). Manually defined areas of cells and the contained electron-dense structures were measured with Image J. We analyzed the statistical significance of Zn-limited vs Zn-replete cells by Kruskal-Wallis One Way Analysis of Variance on ranks (P = <0.001). Error bar shows SD +/− from three independent experiments. (b) NanoSIMS shows 40Ca+ and 63Cu+ co-localize in Zn-limited cells, coinciding with electron-dense structures in TEM (scale bars 1 μm). (c–d) Relative intracellular Ca (c) and Cu (d) measured during Zn resupply (to −Zn compared to +Zn). Samples from three independent cultures were collected 0, 10, and 24 h after Zn addition, and ten cells per time point were examined by NanoSIMS. Average ion ratio values were plotted based on whole cell area (“cell”) and intracellular areas of Ca and Cu accumulations with their corresponding standard deviations. (e) Ratio of 40Ca+ over 63Cu+ at different time points. All NanoSIMS counts were normalized to 12C+. (f–h) XAS spectra for Cu in a representative Zn-limited C. reinhardtii sample. (f) Cu XANES spectrum with a predominant spectral feature at 8984 eV, which corresponds to a 1s → 4p electronic transition typically seen in centrosymmetric Cu+ samples. (g) Expansion of the Cu pre-edge spectral features (red) offset and compared to the Cu2+SO4 (black) model. (h) Fourier transforms of the raw Cu EXAFS (black) with best fit simulation (green).
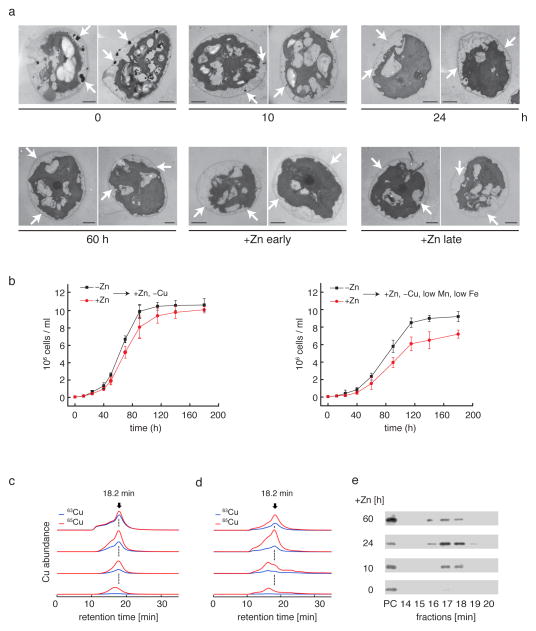
To test the relationship between sites of Cu concentration and Zn-deficiency, we re-supplied Zn2+ to cultures of Zn-limited cells, and sampled over a 24 h time course for NanoSIMS analysis of thin sections. We noted a decrease in Cu abundance over time at the foci of Cu concentration as well as over the total cell area (Figure 4c, d, e), which coincided with the ICP-MS measurements showing Cu accumulation only under strict Zn limitation. This observation suggests further that Cu can be mobilized from the sites of concentration (see below).
Compartmentalized Cu+ is stabilized by O/N and S ligands
We recorded Cu K-edge XANES spectra, which provide information about the average electronic state and ligand symmetry of the metal, on Zn-limited C. reinhardtii cells (Figure 4f). The spectra showed a highly reproducible pre-edge feature centered at 8984 eV, which is typically attributed to 1s-4p electronic transitions in centrosymmetric Cu+ environments: the intensity of this feature also provides insight into the cuprous coordination geometry, which was indicative of 2 to 3 coordinate Cu+. The absence of any appreciable feature at 8980 eV in the Cu pre-edge XANES, a feature attributed to 1s-3d transition in Cu2+, suggests Cu2+ in these samples was limited (Figure 4g). Based on model pre-edge XANES analysis, our expectation is that Cu2+ contamination in the samples would be noticeable, had it been present at 15% or greater proportion. We therefore estimate that these structures are at least 85% Cu+31.
EXAFS on the same sample allowed us to determine the average metrical parameters for the ligand environment of the cuprous metal. EXAFS simulations showed that Cu is coordinated in its nearest neighbor environment by a mixture of oxygen/nitrogen and sulfur ligands, with long-range scattering attributed to carbon only. Parameters from the best-fit simulation of the Cu EXAFS (Supplementary Fig. 6) indicate the average metal-ligand coordination environment is constructed by ca. 2 O/N ligands with an average bond length of 2.11 Å and a single resolvable S ligand at 2.3 Å (Supplementary Table 1). Long-range scattering (R > 3Å) could be best fit with carbon scattering at 2.75 Å. Attempts to include Cu-Cu scattering were unsupported in these data. These simulation parameters closely match the projected ligand environments observed in the Fourier transform of the sample Cu EXAFS (Figure 4h) and the coordination geometry similar to that seen in the M-site in peptidylglycine monooxygenase at low pH32.
Biochemical enrichment of the Cu containing compartment
To determine if Cu+ is concentrated in a compartment, we used biochemical fractionation methods to enrich cell fractions based on Cu and Ca content. Zn-replete and Zn-limited cells were disrupted by grinding in the presence of silicon carbide particles, followed by density gradient centrifugation. The Cu and Ca containing fractions contained small “bodies” with an average diameter of 0.2 μm after they were dispersed in detergent-containing buffered solutions. Without detergent, the bodies, which were much more abundant in extracts of Zn-limited cells, had a tendency to form large aggregates, even upon vigorous vortexing. Elemental analysis of EDTA-washed bodies indicated that they contained also P, independent of cellular Zn content, and were indeed enriched in Cu and Ca when isolated from Zn-limited cells (Supplementary Fig. 7). Staining with CS3 also indicated that the bodies isolated from Zn-limited cells were highly enriched in Cu+ compared to Zn-replete cells (Supplementary Fig. 8).
Staining with 4′,6-diamidino-2-phenylindole (DAPI) and monitoring polyphosphate-specific fluorescence emission between 500 and 550 nm33 suggested the presence of this polymer in the isolated fraction (Supplementary Fig. 9), which may represent the electron dense material visible by TEM. Fixed cells from Zn-limited cultures showed bright and larger areas of DAPI-staining in the polyphosphate-specific emission range, while smaller foci were present at lower abundance in Zn-replete cells (Supplementary Fig. 9a). Hence, size and abundance of intracellular polyphosphate containing bodies, which were purified on the basis of Cu and Ca content, correlated with the electron-dense TEM structures observed in C. reinhardtii cells (Figure 4a).
When we stained the fraction enriched for the Cu- and Ca-containing bodies with LysoSensor Green DND-189 (pKa ~ 5.2), which has increased fluorescence intensity at low pH, we noted staining in the isolated bodies, with higher intensity in Cu-containing compartments isolated from Zn-limited cells, suggestive of lower pH in the bodies during low Zn growth (Supplementary Fig. 8). Indeed, the specific activity of a characteristic marker of the acidocalcisomes, H+-PPase20,34, was slightly higher in the fraction from Zn-limited cells (Supplementary Fig. 10a) and had a dose-dependent response to specific inhibitors34,35 (Supplementary Fig. 10b). Further, co-staining of whole cells with CS3 and the membrane permeable pH sensor ageladine A36 confirmed a general co-localization of Cu+ accumulation and acidic intracellular compartments, which were more abundant in Zn-limited cells (Supplementary Fig. 9b). Taken together, we conclude that the Cu-containing compartment is analogous to the previously-described acidocalcisome, which is a lysosome-related organelle20,37. The metal content of this compartment was dynamic, as evidenced by increased Mg and Zn in the fraction purified from Zn-replete cultures relative to the Zn-limited culture (Supplementary Fig. 7c).
Accumulated Cu was bio-available
To test whether Cu in the compartment could be released, we re-supplied Zn-limited cells with ZnCl2 and used CS3 in a pilot experiment to monitor intracellular Cu-containing compartments. Fluorescent foci decreased starting about 3 to 4.5 h after Zn2+ resupply and fluorescence became more homogenously distributed throughout the cell by about 24 h (Supplementary Fig. 11). This correlated with a decrease in the abundance and size characteristic of electron-dense structures in Zn-limited cells (Figure 5a), consistent with these structures being related to sites of Cu accumulation. To test whether the released Cu+ was bio-available, we moved Zn-limited (Cu+-accumulating) cells to Zn-replete but Cu-deficient growth medium and found that these cells had higher growth rates compared to cells shifted from Zn-replete (not Cu+-accumulating) conditions to Cu deficiency, presumably because the former had access to an internal Cu+ reservoir (Figure 5b, left panel). This effect was even more significant when the new medium was depleted also for manganese and iron, which also accumulated slightly (2–3-fold) in Zn-limited cells (Figure 5b, right panel, Supplementary Figure 12).
Figure 5. Changes in abundance of electron dense bodies and Cu redistribution upon Zn resupply.
(a) Electron microscopy of Zn-limited cells during Zn resupply (0, 10, 24, 60 h after addition of ZnCl2) and Zn-replete cells in early and late logarithmic growth phases (+Zn early and +Zn late). Two representative specimens from three independent cultures are shown for each condition. Arrows point to electron-dense bodies, mainly in the periphery of the cell (scale bars 2 μm). (b) Growth curves of Zn-limited or replete cells, which were inoculated into fresh TAP medium containing ZnCl2 but no Cu (left panel) or no Cu and 0.1 μM Mn and 0.5 μM Fe (right panel). We show mean values of three independent experiments with corresponding standard deviations. (c–e) 65Cu-labelled cells grown under Zn limitation were supplied with 63Cu (10 μM 63CuCl2) either together with Zn resupply (c) or 5h after Zn resupply (d). Soluble proteins were extracted anaerobically at 0, 10, 24 and 60 h after Zn addition, and analyzed by LC-ICP-MS to monitor the Cu isotope content of the protein species. This experiment was performed twice with 65Cu as label and chased with 63Cu and twice with 63Cu as label and chased with 65Cu. Shown are the ion chromatograms for Cu isotopes 63 and 65. The emerging Cu isotope peak at 18.2 min corresponds to plastocyanin-associated Cu. The corresponding LC fractions were analyzed by immunoblotting regarding their plastocyanin contents (e). Immunoblot analysis of respective fractions with purified plastocyanin (PC) from C. reinhardtii as standard.
To document the utilization of accumulated Cu+ for de novo synthesis of a cuproprotein, we labelled Zn-limited cells with isotopically enriched Cu (63Cu or 65Cu) and then we added five-fold excess of the other Cu isotope, either simultaneously with Zn-resupply (Figure 5c), or 5 h after Zn resupply (Figure 5d). We separated plastocyanin and monitored the polypeptide abundance by immunoblotting (Figure 5e) and the Cu isotope abundances by ICP-MS. The peak of eluted plastocyanin with a retention time of 16.6 min (17 min fraction) compared to the plastocyanin associated peak with a retention time of 18.2 min was due to the time lag between protein detection by the LC multiwavelength detector and Cu detection by the ICP-MS mass analyzer (Supplementary Fig. 13). MSE based quantifications of plastocyanin (PC) and Cyt c6 abundances in LC fractions obtained from reciprocal Cu isotope labeling experiments are shown in Supplementary Figure 14. Newly synthesized proteins potentially have two different pools of Cu they can access: one derived from intracellular Cu accumulating sites and the other from the growth medium. Resupply of Zn resulted in strong stimulation of growth (Supplementary Fig. 15a) and a 200-fold increase in plastocyanin accumulation (Supplementary Fig. 14). The Cu associated with newly synthesized plastocyanin showed a 2:1 preference for the isotope species from the intracellular store (Figure 5c, Supplementary Figure 15b). However, at 24 h post Zn resupply, the Cu content of the cell included both the intracellular prior accumulation and extracellular over-supplied species in about equal proportion (Supplementary Fig. 16). At 60 h post Zn resupply, the extracellularly over-supplied species represented about 2/3 of the intracellular Cu pool, but plastocyanin remained relatively enriched (50/50 mix of both isotopes) for Cu from the intracellular accumulation site (Figure 5c, Supplementary Figure 15b, 16). We conclude that upon restoration of Cu homeostasis, Cu+ from the internal Cu accumulating compartment is more readily accessible for metallation of de novo synthesized cuproproteins.
When we supplied the other Cu isotope to cells 5 h after Zn-resupply rather than simultaneously, we found that the Cu isotope in plastocyanin is 2:1 enriched for the prior compartment-accumulated form even 60 h later (Figure 5d, Supplementary Figure 15c). We attribute this result to the reduced expression of CTR transporters (see below) upon restoration of Cu homeostasis, resulting from Cu visibility to CRR1, and hence reduced uptake of externally supplied Cu.
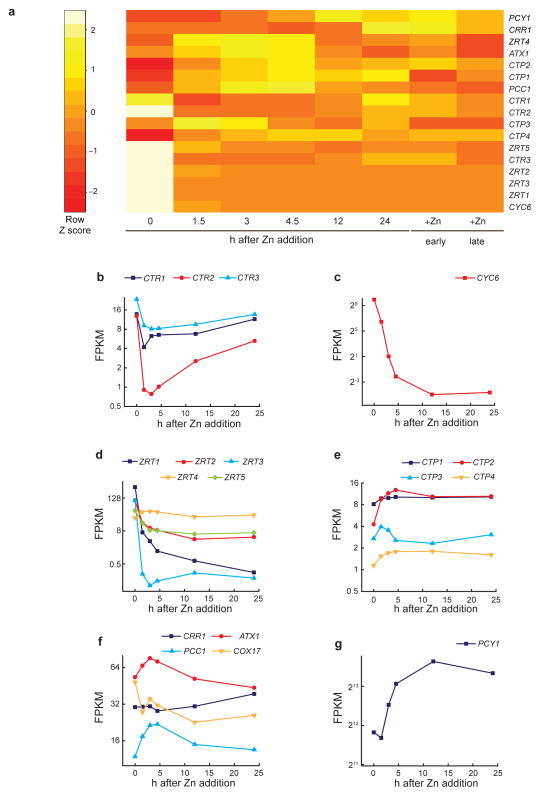
To assess the operation of CRR1, we monitored the transcriptome as a function of Zn re-supply (between 0 and 24 h) by RNA-Seq. A heat map showing individual changes in transcript abundance for 17 genes involved in Cu and Zn homeostasis is presented (Figure 6a and with absolute scaling in Supplementary Figure 17a). The P-values indicate the significance of FPKM changes between time points (Supplementary Table 2). CTR transcripts were abundant in Zn-limited cells, supporting the contention that Cu+-hyper-accumulation was driven by high CTR expression (Figure 6b). Transcript abundance for all three CTR genes decreased rapidly (comparable to the behavior of CYC6, a sentinel gene of nutritional Cu signaling) during the first 3–5 h, which is consistent with Cu+ mobilization from the internal compartment (Supplementary Fig. 11) and hence de-activation of CRR116(Figure 6c). Interestingly, as intracellular Cu was depleted for synthesis of cuproproteins during growth and division, CRR1 and hence expression of its target genes were re-activated. RNAs encoding the candidate Zn2+ transporters, members of the ZIP family, were also rapidly reduced upon Zn2+ addition and those genes remained repressed throughout the time course (Figure 6d).
Figure 6. Transcriptome response to Zn resupply monitored by RNAseq analysis.
(a) Heatmap showing Z scores (interpreted as a measure of standard deviation away from the mean) for the changes of FPKM (fragments per kilobase of exon per million fragments mapped) of select genes indicated at the right margin. The time points 0–24 h after Zn addition indicate the sampling during Zn resupply to a Zn-limited culture. Columns “+Zn early” and “+Zn late” indicate samples from Zn-replete cultures taken in the stages of early logarithmic and beginning stationary growth phase, respectively. (c–g) The mRNA abundances in FPKM are given for (b) members of the CTR family, (c) CYC6, (d) genes encoding members of the ZIP family, (e) genes encoding members of the P1B-type ATPase family, (f) CRR1 and Cu chaperones ATX1, PCC1 and COX17, and (g) PCY1 (plastocyanin). The y-axes of the diagrams are log2 scaled.
Cu homeostasis
The time course experiment also revealed a previously unappreciated effect on the expression of genes for intracellular Cu movement. Transcripts encoding the intracellular “distributive” CTP transporters, which use ATP hydrolysis to transport copper ions against a concentration gradient, as well as those encoding Cu+ chaperones ATX1 and PCC138 increased in abundance soon after Zn re-supply (Figure 6e, f), presumably to facilitate Cu distribution to the chloroplast for plastocyanin biosynthesis (see above) and to the secretory pathway for biosynthesis of a ferroxidase-ferric transporter complex required for iron uptake39,40 (Supplementary Fig. 17b). We note that transcripts PCY1, FOX1, FTR1, encoding plastocyanin and the iron uptake pathway also increased in parallel with increased Cu+ bio-availability (Figure 5c, 6g, Supplementary Figure 17b). In contrast, the abundance of COX17 transcripts encoding functions required for Cyt oxidase assembly41,42 decreased, which is consistent with the lack of change in COX2A and COX3 transcript abundance upon Zn resupply (Supplementary Fig. 17b) and the relative stability of Cyt oxidase even in a Cu+-challenged growth condition (Figure 2b).
Discussion
Typically, the Cu quota of a cell is tightly regulated and determined by the abundance of various cuproproteins, but poor nutrition or genetic mutation can result in mis-regulation and hence overload of metals. Here, we took advantage of Cu mis-regulation in Zn-limited cells to analyze sites of Cu sequestration in C. reinhardtii. Pilot survey experiments with a pair of matched fluorescent dyes that can (CS3) or cannot (Ctrl-CS3) respond to Cu+ in live-cell imaging experiments led to direct detection of sites of Cu accumulation by imaging mass spectrometry of fixed, sectioned cells, which localized bound Cu only44. In this context, we note that fluorescent probes are a developing technology that should be employed with caution and appropriate controls, both genetic and chemical. These reagents are designed and physically characterized by chemists in tightly controlled in vitro environments but biologists may use them in vastly divergent cells and organisms where solubility, intracellular distribution and reactivities can vary.
To help address some of these potential issues, we designed an iso-structural control compound that can be employed to distinguish between dye-dependent and receptor- dependent fluorescence responses in an intracellular environment. Replacement of metal- interacting sulfurs in CS3 with isosteric carbons yields a matched Ctrl-CS3 dye that lacks responsiveness to Cu and pH and represents a new chemical approach for the use of small molecule sensors in vivo. Yet, the presence of more reactive functionalities in CS3 vs. Ctrl-CS3 may create unanticipated chemical reactions in vivo. The use of appropriate controls cannot be over-emphasized. For instance, while CS3 does not by itself stain lipid bodies in Chlamydomonas, CS3 (but not Ctrl-CS3) will fluoresce in co-staining experiments with some dyes (e.g. DPH), possibly because of an interaction between the two dyes dependent on the 4 S atoms. The precise mechanism by which CS3 responds in Chlamydomonas compartments is insufficiently understood, whether by direct binding or by an unknown Cu+-mediated process, and the fluorescence output is not quantitative. Nevertheless, the dependence of the signal on intracellular Cu content, as directly measured by ICP-MS and Nano-SIMS, and a functional CRR1 locus, along with the fact that the isosteric Ctrl-CS3 does not respond to changes in intracellular Cu in these same strains, strengthens the interpretation of the CS3 signal in the models used in this study (Figure 3b). The use of fluorescent probes as a tool for pilot screening in live samples is potentially useful in downstream high throughput genetic screens now that their validity has been established in this organism with more resource–intensive physical techniques. We note that in vitro experiments with small molecule sensors (see Supplementary Note) clearly do not faithfully mimic all aspects of their chemistry in a biological setting. Therefore, each probe must be tested in each experimental model accompanied by other chemical and/or genetic controls in that model.
Imaging mass spectrometry has the caveat that it requires chemical fixation and sectioning of cells and is therefore restricted to analysis of bound elements in that section. We cannot rule out the possibility that there is another pool of Cu+ that neither technique reached. Taken together, both methods support the conclusion that C. reinhardtii sequestered Cu under Zn limitation, causing Cu to be unavailable for cuproprotein biosynthesis and hence C. reinhardtii plastocyanin levels (a biomarker for intracellular Cu) were low (Figure 2b). Sequestered Cu+ was also invisible to a Cu-sensing transcription factor, CRR1, responsible for maintaining cellular Cu quota16. Accordingly, C. reinhardtii expressed the nutritional Cu regulon, including the CTR genes encoding plasma membrane-localized Cu+ assimilation components12. The transporters drew more Cu+ into the cell and hence Cu+ hyper-accumulated dependent on the external supply and CRR1 (Figure 1b–c).
We probed the identity of the Cu accumulation sites based on prior description of acidocalcisomes20,34. The acidocalcisome is a low pH, lysosome-related organelle that contains polyphosphate and metal ions, especially Ca and Fe37. Its Cu content had not previously been described, potentially because of the common use of Cu grids for microscopic techniques for metal visualization34. Staining of intact cells or enriched compartments with fluorescent probes confirmed co-localization of Cu, Ca and P, and we conclude that this compartment is indeed related to the previously-described acidocalcisome, although we wonder whether in any one cell, there may be different types of acidocalcisomes housing different concentrations of individual metals. By analogy to zincosomes described in worms and animal cells and Zn-accumulating lysosome-related organelles, it may be useful to refer to the Cu+ containing compartments as cuprosomes22,43–46.
Researchers have previously found foci of Cu accumulation, including in cells of Wilson Disease patients, in the Ctrint/int mouse where intestinal epithelial cells contain sites of Cu concentration and in mouse fibroblasts lacking metallothionein and Atox147,48. In these situations as well, there were high concentrations of biologically unavailable Cu. The recapitulation of this phenotype in C. reinhardtii provided an excellent opportunity to use a genetic approach to dissect the mechanism of formation of the Cu-loaded compartment and the pathways of Cu mobilization from these compartments. At present, the identities of the transporters that load and unload Cu+ into and from this lysosome-related organelle are unknown. A proteomic analysis of a purified acidocalcisome from a red alga did identify metal transporters in the boundary membrane and these are certainly candidates that can be tested49.
Another important question is the chemical speciation of the stored Cu. The microbial experimental system used in this work allowed the generation of a homogenous population of cells with very high Cu content, suitable for application of X-ray spectroscopic approaches. Independent samples generated months apart yielded superimposable XANES spectra, indicating that sequestered Cu was reproducibly organized. The EXAFS analysis suggests mononuclear Cu associated with organic ligands (O/N and S) rather than with polynuclear Cu species. We compared the spectra to those of known compounds (including glutathione, polyphosphate, phytochelatins) but the Cu species in this work was distinct from previously characterized species. While transcriptome profiling indicated up-regulation of a phytochelatin synthase, this may be a pre-emptive mechanism against Zn shock43 rather than a means for chelating the sequestered Cu+.
Aside from the known function of acidocalcisomes to house elements, such as Ca, P, Fe, and Zn, we showed here that this compartment can also house Cu+. More importantly, the compartment served as a reservoir of Cu+ that could become bio-available when the cell required Cu+. The vacuole in yeast is perhaps the most analogous compartment: it stores Zn2+ in a situation of excess but in deficiency it can mobilize the Zn2+ and support several rounds of replication in the absence of external Zn43. We showed that the same is true for sequestered Cu+. Indeed, even in the presence of extracellular Cu, compartmentalized Cu+ was used with priority over assimilated Cu+ for the biosynthesis of holoplastocyanin. Furthermore, accumulated Cu+ offered a selective advantage to cells when they were faced with nutritional deficiency, consistent with bio-availability of compartmentalized Cu+. We conclude that the metal content of the Cu accumulating compartment is dynamic and can be used for productive metallation of apoproteins.
Sequestered Cu+ was released when we removed the condition prompting sequestration, namely Zn2+ limitation (Figure 5a, Supplementary Figure 11). The released Cu+ was not only available for holoplastocyanin formation (Figure 5d) but it also became visible to CRR1. Transcriptome analysis indicated rapid de-activation of the Cu regulon coincident with accumulation of holoplastocyanin (Figure 6). This accounted for decreased Cu+ uptake capacity as a function of time after Zn addition (Figure 5d). On the other hand, Cu+ handling components (PCC1, ATX1 for delivery of the metal to the chloroplast or secretory pathway) were instead up-regulated, consistent with redistribution of intracellular Cu+.
Why was Cu+ sequestered in Zn-limited cells? One possibility is that this occured to prevent mis-metallation of Zn2+ sites in enzymes. In the absence of a specific mechanism for Zn2+ delivery to these enzymes, low intracellular concentrations of Zn2+ might allow binding of Cu+, which would be more competitive for binding to these sites. This is true for the SBP domain of CRR1, which binds Cu+ with 105 higher affinity relative to Zn2+16. Indeed, in Euglena gracilis, Zn-deficiency results in mis-incorporation of Cu instead of Zn into an endonuclease, rendering it inactive50. Sequestration of Cu+ would remove the more competitive metal species and allow trace levels of Zn2+ to metalate critical proteins, such as those involved in nucleic acid transactions. Another possibility is that Cu is transiently delivered to this compartment en route to its final destination, but in Zn-deficiency egress is blocked or inhibited. This may occur if the transport from the compartment requires Zn2+ or if the cytoplasmic Zn/Cu ratio regulates efflux. Regardless, this work underscores the interplay between multiple metal-homeostasis pathways.
Online methods
Strains and cultures
The Chlamydomonas reinhardtii strains CC-4532 (wild-type - wt) and CC-3960 (crr1-2) are available at the C. reinhardtii culture collection (http://www.chlamy.org). We also used two independent transformants of crr1-2 with pARG7.8 (crr1a and crr1b), and two independent transformants of the complementation strain containing both pARG7.8 and pCRR1 (CRR1a and CRR1b) were used as a control16.
Unless noted otherwise, algae were grown from an inoculum of 1×105 cells/mL in Tris-acetate-phosphate (TAP) medium containing our revised micronutrient composition30 at 24°C under continuous light (~90 μmol m−2 s−1) with shaking (180 rpm)51. To establish the light spectrum as shown in Supplementary Figure 19, we used 2 cool white fluorescent bulbs at 4100K for each 1 warm white fluorescent bulbs at 3000K.
For studying metal-defined conditions, all glassware was triple washed in 6 N hydrochloric acid followed by at least six rinses in Milli-Q-purified (Millipore) water. Zn limitation was induced by inoculating cells from a nutrient-replete culture to a number of 105 cells/mL into 100 mL culture medium without supplemented Zn (“first round culture”), and then into medium without supplemented Zn (“second round culture”). ZnCl2 was resupplied at 2.5 μM to these limited cultures where described when they reached a density of 1×106 cells/mL (“t0h” of Zn resupply). For control samples, Cu deficiency was induced similarly by omitting Cu during media preparation.
Growth rate determination
Triplicate first and second round cultures of the wt strain were grown as described above, and samples were counted every 24 h by light microscopy using a hemocytometer (Hausser Scientific).
Quantitative metal and phosphorus content analysis of cells and Cu accumulating compartments
A total number of 1×108 cells was collected by centrifugation at 1,700 xg for 10 min and washed once in 1 mM Na2-EDTA to remove cell surface-associated metals and once in Milli-Q water. The washed cell paste was overlaid with nitric acid of 24 % (v/v) in 1 ml and digested at 65 °C for about 12 h. The cell hydrolysate was diluted in corresponding to a final concentration MilliQ-water to a final nitric acid concentration of 2.4 % (v/v). These measurements were repeated routinely to determine the cellular Cu content of all cells that were used for other experiments, such as NanoSIMS analysis, XAS, and confocal microscopy.
Isolated compartments were collected at 5,000 xg for 20 min to a total number of 109 per analyzed condition and were treated as described above. Aliquots of fresh or spent culture medium were treated with nitric acid to a final concentration of 2.4 % (v/v). To obtain corresponding blank samples, the volume of cell hydrolysate, Cu enriched hydrolysate or culture medium was replaced by MilliQ-water and treated as described above. Total metal and phosphorous contents were measured by inductively-coupled plasma mass spectrometry (ICP-MS) on an Agilent 7500ce instrument, and quantification was done by using appropriate calibration standards (Icpsupplies).
NanoSIMS analysis
We used the LLNL Cameca NanoSIMS 50 (Gennevilliers, France) to image the intracellular distribution of bound Ca, Cu, and Zn18,19,52. Cells three independent cultures that were grown for three days in Zn-limited medium or cultured during Zn resupply were collected by centrifugation and immersed in a solution containing 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M PBS, pH 7.4, for 2 h at room temperature and then incubated at 4°C. Subsequently, 0.5% of tannic acid was added to the cells and incubated for an hour at room temperature. The cells were washed five times in 0.1 M PBS buffer and postfixed in a solution of 1% OsO4 in PBS, pH 7.2–7.4. The samples were washed four times in Na acetate buffer, pH 5.5. The samples were dehydrated in an ethanol gradient (50, 75, 95, 100, 100, and 100%) for 10 min each, passed through propylene oxide, and infiltrated for 2 h first in a 1:1 mixture of Epon 812 and propylene oxide, and then 2:1 for 2 h. The cells were then infiltrated in pure Epon 812 overnight and cured in an oven at 60 °C for 48 hr. Sections of 200 nm thickness (gray interference color) were cut on an ultramicrotome (RMC MTX) using a diamond knife and deposited on 200 mesh carbon-coated molybdenum grids. Sectioned cells were first located and imaged by transmission electron microscopy (TEM). Then they were relocated and analyzed in the NanoSIMS 50. A focused negative oxygen ion primary beam was scanned over the sample (256×256 pixels, 25–100 μm2 raster) to generate secondary ions. The secondary ion mass spectrometer was tuned for ~5000 and 12C+, 40Ca+, 63Cu+ and 66Zn+ were detected simultaneously by electron multipliers in pulse counting mode. The correct metal ion peaks were identified using NBS610 glass (National Institute of Standards and Technology, USA). Each analysis area was scanned 10 to 20 times to collect serial secondary ion images for quantification.
The NanoSIMS ion image data were processed using custom software (LIMAGE, L.R. Nittler, Carnegie Institute of Washington, USA). The ion images were corrected for detector dead time and image shifts between scans, and then used to produce ion ratio images. For defined regions of interest (either whole cell areas or areas of Cu accumulation) ion ratios were calculated for 40Ca+/12C+, 63Cu+/12C+ and 66Zn+/12C+ by averaging the replicate scans18,19,52. These data provide relative quantitative composition, but they were not standardized to provide concentration data.
Cu isotope labeling
For labeling with >98.5 % enriched 63Cu or 65Cu isotopes (Cambridge Isotope Laboratories, Inc.), all media were prepared by using either 63CuCl2 or 65CuCl2 (at 2 μM initial concentration) as the sole Cu source. Cells were grown for at least 12 generations in the presence of the enriched Cu source. Upon resupply of Zn in the second round cultures, the other enriched Cu isotope was added at 5-fold molar excess (10 μM) to these cultures either directly or 5 h after the addition of Zn. Cells were collected at time points 0, 10, 24 and 60 h for LC-ICP-MS analysis of soluble proteins (see growth curves and sampling scheme in Supplementary Fig. 15a) by centrifugation (4,000 xg at 4°C for 10 min) and washed twice with Millipore water. Further samples from the same cultures were taken for quantitative ICP-MS to analyze the Cu isotope content of both cell pellets (aliquots of 108 cells) and medium supernatant. The cell pellets were washed twice with 2.5 mM Na2-EDTA and once with Millipore water to remove remaining metal salts from the cell surface.
Work under anaerobic conditions
All anaerobic work was carried out in an airtight glove box (Coy Laboratories) containing a controlled H2 (3%)/N2 (97%) atmosphere. Solutions and plastic ware were allowed to equilibrate for at least 6 h inside the anaerobic chamber before usage. Buffer solutions for HPLC were made anoxic by purging them with forming gas overnight.
Protein preparation for metalloprotein analysis
Collected cells were resuspended under anaerobic conditions in anoxic chromatographic analysis buffer (20 mM Tris, pH 7.5, 20 mM KCl) to equal densities of 108 cells/mL. Work under anaerobic atmosphere was continued during the whole experimental process to preserve the native metal oxidation states. For the quantitative release of soluble proteins including plastocyanin53,54, two slow freeze (−80 °C) and thaw (20 °C) cycles for cell lysis were carried out in closed and sealed tubes. Supernatant and cell pellets were separated by centrifugation at 20,000 xg at 4°C for 15 min. To remove insoluble membrane fractions remaining in the supernatant, ultracentrifugation (250,000 xg at 4°C for 1 h) was done in tubes closed by heat-melting. The clear supernatant (with a total protein content of ~1 mg/mL) was then subjected to subsequent protein fractionation and metal content analysis by HPLC-ICP-MS (see below).
Metalloprotein analysis by HPLC-ICP-MS
Isotope-specific metalloprotein analysis was performed by high performance liquid chromatography (HPLC) linked with inductively-coupled plasma mass spectrometry (ICP-MS). Aliquots of total soluble protein (~0.5 mg) prepared from different cell samples were injected manually into a bioinert Agilent 1260 HPLC system (Agilent Technologies) that was equilibrated inside an anaerobic chamber (Coy Laboratories). The protein crude extracts were separated anaerobically in 20 mM Tris-Cl, pH 7.5, 20 mM KCl at 25 °C and at a flow rate of 0.5 mL/min via size exclusion chromatography on a 7.5 mm×30 cm TSKgel G3000SW column (Tosoh). The eluent stream was subjected to a split flow directing it to the source of the ICP-MS for online metal analysis using the high matrix interface as well as to the HPLC fraction collector for subsequent protein species analysis by immunoblotting and mass spectrometry. ICP-MS analysis was run in the helium gas mode with time-resolved analysis to minimize signal suppression in presence of higher salt contents. Isotope-specific ion chromatograms were obtained for 65Cu and 63Cu upon extraction from the total ion signals. The protein elution profiles were recorded by a multi wavelength detector at 280 nm using a 500 nm reference signal. The delay time between UV/vis detection of the protein species and the detection of associated metal cofactor(s) by ICP-MS was ~1.6 min, which was also confirmed by using Cu/Zn superoxide dismutase (Sigma Aldrich) as well as purified C. reinhardtii plastocyanin as metalloprotein standards (see Supplementary Figure 13).
Electron microscopy
Cells and Cu accumulating compartments were prepared for electron microscopy as described for NanoSIMS analysis above. Sections of 60 nm thickness (gray interference color) were cut on an ultramicrotome (RMC MTX) using a diamond knife. The sections were deposited on single-hole grids coated with Formvar and carbon and double-stained in aqueous solutions of 8% uranyl acetate for 25 min at 60°C and lead citrate for 3 min at room temperature. Thin sections were examined with a 100CX JEOL (80 kV) electron microscope. For NanoSIMS studies, samples were prepared without uranyl acetate and lead citrate stainings, and 200 nm sections were cut and deposited on molybdenum grids, which were carbon-coated prior to analysis. Image J software was used to manually define and measure whole cell areas and electron dense structures. The ratios of electron dense structure to whole cell area are given in percentage. Cells from three independent Zn-replete or–limited cultures were analyzed. 55 Zn-limited cells and 79 Zn-replete cells were analyzed. Kruskal-Wallis One Way Analysis of Variance on ranks was used to determine the statistical significance of the data (P = <0.001). For TEM analysis of Zn resupply time course, we analyzed 45 cells at t = 0 h, 32 cells at t = 10 h, 40 cells at t = 24 h, 42 cells at t = 60 h, 35 cells from the Zn-replete culture (early), and 28 cells from the Zn-replete culture (late).
RNA-Seq
RNA was extracted from Zn-limited cells and at 1.5, 3, 4.5, 12, and 24 h after Zn resupply as described above. Two Zn-replete samples were collected as controls: in mid-logarithmic and in stationary phase.
Quadruplicate cDNA libraries from each time point were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina) following the manufacturer’s instructions (Revision D). Sequencing was performed on a HiSeq 2000 high throughput sequencer (Illumina) according to the manufacturer’s instructions. The libraries were loaded 8 per lane and sequenced for 50 nt. After demultiplexing, this yielded 2.5×107 reads on average per library. Quality scores were high, averaging between 33 and 40 over the length of the reads. The resulting reads were aligned to v5.3.1 of the C. reinhardtii genome assembly1 available from the Joint Genome Institute at www.phytozome.net. Alignments were performed using TopHat255 with guidance from the Augustus 11.6 gene models (www.phytozome.net). Expression was quantified and standardized in terms of fragments per kb of exon per million fragments (FPKMs) by Cufflinks, and differential expression testing was performed by Cuffdiff56.
We deposited the raw reads and expression estimates from the RNAseq experiment at the NCBI GEO database with the accession number GSE58786.
Supplementary Material
Acknowledgments
This work is supported, in part, by grants from the NIH (GM42143 and GM092473 to S.S.M., DK068139 to T.L.S. and GM079465 to C.J.C.), the United States Department of Energy Cooperative Agreement (DE-FC02-02ER63421 to David Eisenberg for support of J.A.L.), and the German Academic Exchange Service DAAD (A.H.H. D0847579 and M.M. D1242134). Work at LLNL was performed under the auspices of the U.S. Department of Energy at Lawrence Livermore National Laboratory under Contract DE-AC52- 07NA27344, with funding provided by the US Department of Energy (DOE) Genomic Science Program under contract SCW1039. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL). SSRL is a national user facility operated by Stanford University and the SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program. D.B. is supported by NIH (T32HL120822) and C.J.C. is an Investigator with the Howard Hughes Medical Institute. Electron microscopy was performed at the Electron Microscopy Services Center of the University of California, Los Angeles, Brain Research Institute. We thank Allegra Aron and Karla M. Ramos-Torres for their help with re-synthesis and optical spectroscopy of fresh CS3 and Ctrl-CS3 for control experiments suggested by a reviewer.
Footnotes
Authors’ contributions
S.S.M., A.H.H., and M.M. designed experiments. A.H.H., M.M. and J.K. cultured cells and supplied samples for NanoSIMS, XAS, RNAseq. M.M. and J.K. measured cellular metal contents by ICP-MS. A.H.H. performed immunoblotting and qRT-PCR for expression analysis. A.H.H. and M.M. imaged cells by confocal and electron microscopy and analyzed the resulting data. J.P.R., P.K.W. and M.M. analyzed intracellular metal distribution by NanoSIMS; D.B. and T.L.S. collected and analyzed XAS data. M.M. isolated Cu containing compartments and did the Cu isotope labeling experiments in conjunction with LC-ICP-MS analysis. D.I.S. and M.M. performed quantitative mass spectrometry of protein fractions under the supervision of J.A.L. S.D.G. prepared RNAseq libraries and analyzed resulting data. S.C.D., D.W.D., J.C. synthesized the Cu+-sensitive CS3 dye (and control) under the supervision of C.J.C. M.M., S.S.M., and A.H.H wrote the manuscript with input from J.C. and P.K.W.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Anne Hong-Hermesdorf, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.
Marcus Miethke, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.
Sean D Gallaher, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.
Janette Kropat, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.
Sheel C Dodani, Department of Chemistry and Howard Hughes Medical Institute, University of California, Berkeley, USA.
Jefferson Chan, Department of Chemistry and Howard Hughes Medical Institute, University of California, Berkeley, USA.
Dulmini Barupala, Department of Pharmaceutical Sciences, Wayne State University, Detroit, USA.
Dylan W Domaille, Department of Chemistry and Howard Hughes Medical Institute, University of California, Berkeley, USA.
Dyna I Shirasaki, Department of Biological Chemistry, University of California, Los Angeles, USA.
Joseph A Loo, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.Institute for Genomics and Proteomics, University of California, Los Angeles, USA.Department of Biological Chemistry, University of California, Los Angeles, USA.
Peter K Weber, Chemical Sciences Division, Lawrence Livermore National Laboratory, Livermore, USA.
Jennifer Pett-Ridge, Chemical Sciences Division, Lawrence Livermore National Laboratory, Livermore, USA.
Timothy L Stemmler, Department of Pharmaceutical Sciences, Wayne State University, Detroit, USA.
Christopher J Chang, Department of Chemistry and Howard Hughes Medical Institute, University of California, Berkeley, USA.
Sabeeha S Merchant, Department of Chemistry and Biochemistry, University of California, Los Angeles, USA.Institute for Genomics and Proteomics, University of California, Los Angeles, USA.
References
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–18. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Irving H, Williams RJP. The stability of transition-metal complexes. J Chem Soc. 1953;637:3192–3210. [Google Scholar]
- 3.Dudev T, Lim C. Metal binding affinity and selectivity in metalloproteins: insights from computational studies. Annu Rev Biophys. 2008;37:97–116. doi: 10.1146/annurev.biophys.37.032807.125811. [DOI] [PubMed] [Google Scholar]
- 4.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–30. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 5.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–8. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 6.Valentine JS, Gralla EB. Delivering Copper Inside Yeast and Human Cells. Science (80-) 1997;278:817–818. doi: 10.1126/science.278.5339.817. [DOI] [PubMed] [Google Scholar]
- 7.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–42. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 8.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 9.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–79. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster AW, Robinson NJ. Promiscuity and preferences of metallothioneins: the cell rules. BMC Biol. 2011;9:25. doi: 10.1186/1741-7007-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merchant SS, et al. Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim Biophys Acta. 2006;1763:578–94. doi: 10.1016/j.bbamcr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Page MD, Kropat J, Hamel PP, Merchant SS. Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell. 2009;21:928–43. doi: 10.1105/tpc.108.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant S, Bogorad L. Metal ion regulated gene expression: use of a plastocyanin-less mutant of Chlamydomonas reinhardtii to study the Cu(II)-dependent expression of cytochrome c-552. EMBO J. 1987;6:2531–5. doi: 10.1002/j.1460-2075.1987.tb02540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merchant S, Hill K, Howe G. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 1991;10:1383–9. doi: 10.1002/j.1460-2075.1991.tb07658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kropat J, et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer F, et al. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell. 2010;22:4098–113. doi: 10.1105/tpc.110.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malasarn D, et al. Zinc deficiency impacts CO2 assimilation and disrupts copper homeostasis in Chlamydomonas reinhardtii. J Biol Chem. 2013;288:10672–83. doi: 10.1074/jbc.M113.455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosal S, et al. Imaging and 3D elemental characterization of intact bacterial spores by high-resolution secondary ion mass spectrometry. Anal Chem. 2008;80:5986–92. doi: 10.1021/ac8006279. [DOI] [PubMed] [Google Scholar]
- 19.Slaveykova VI, Guignard C, Eybe T, Migeon HN, Hoffmann L. Dynamic NanoSIMS ion imaging of unicellular freshwater algae exposed to copper. Anal Bioanal Chem. 2009;393:583–9. doi: 10.1007/s00216-008-2486-x. [DOI] [PubMed] [Google Scholar]
- 20.Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SNJ. Acidocalcisomes - conserved from bacteria to man. Nat Rev Microbiol. 2005;3:251–61. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 21.Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochim Biophys Acta. 2006;1763:711–22. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Roh HC, Collier S, Guthrie J, Robertson JD, Kornfeld K. Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab. 2012;15:88–99. doi: 10.1016/j.cmet.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett. 2012;330:30–7. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat Chem Biol. 2013;9:177–83. doi: 10.1038/nchembio.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen BP. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:689–93. doi: 10.1016/s1095-6433(02)00201-5. [DOI] [PubMed] [Google Scholar]
- 26.Castruita M, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–92. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodani SC, et al. Calcium-dependent copper redistributions in neuronal cells revealed by a fluorescent copper sensor and X-ray fluorescence microscopy. Proc Natl Acad Sci U S A. 2011;108:5980–5. doi: 10.1073/pnas.1009932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: a mechanistic systems biology approach. Cell Mol Life Sci. 2010;67:2563–89. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corazza A, Harvey I, Sadler PJ. 1H, 13C-NMR and X-ray Absorption Studies of Copper(I) Glutathione Complexes. Eur J Biochem. 1996;236:697–705. doi: 10.1111/j.1432-1033.1996.0697d.x. [DOI] [PubMed] [Google Scholar]
- 30.Kropat J, et al. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–80. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman RL, et al. Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg Chem. 2006;45:8372–81. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauhan S, Kline CD, Mayfield M, Blackburn NJ. Binding of copper and silver to single-site variants of peptidylglycine monooxygenase reveals the structure and chemistry of the individual metal centers. Biochemistry. 2014;53:1069–80. doi: 10.1021/bi4015264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aschar-Sobbi R, et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J Fluoresc. 2008;18:859–66. doi: 10.1007/s10895-008-0315-4. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz F, Marchesini N, Seufferheld M, Govindjee &, Docampo R. The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem. 2001;276:46196–203. doi: 10.1074/jbc.M105268200. [DOI] [PubMed] [Google Scholar]
- 35.Rea PA, Poole RJ. Vacuolar H+-translocating pyrophosphatase. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:157–180. [Google Scholar]
- 36.Bickmeyer U, Grube A, Klings KW, Köck M. Ageladine A, a pyrrole-imidazole alkaloid from marine sponges, is a pH sensitive membrane permeable dye. Biochem Biophys Res Commun. 2008;373:419–22. doi: 10.1016/j.bbrc.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 37.Huang G, et al. Adaptor protein-3 (AP-3) complex mediates the biogenesis of acidocalcisomes and is essential for growth and virulence of Trypanosoma brucei. J Biol Chem. 2011;286:36619–30. doi: 10.1074/jbc.M111.284661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaby-Haas CE, Merchant SS. The ins and outs of algal metal transport. Biochim Biophys Acta. 2012;1823:1531–52. doi: 10.1016/j.bbamcr.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontaine, La S, et al. Copper-Dependent Iron Assimilation Pathway in the Model Photosynthetic Eukaryote Chlamydomonas reinhardtii. Eukaryot Cell. 2002;1:736–757. doi: 10.1128/EC.1.5.736-757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JC, Hsieh SI, Kropat J, Merchant SS. A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot Cell. 2008;7:541–545. doi: 10.1128/EC.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J Biol Chem. 2004;279:35334–40. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 42.Remacle C, et al. Knock-down of the COX3 and COX17 gene expression of cytochrome c oxidase in the unicellular green alga Chlamydomonas reinhardtii. Plant Mol Biol. 2010;74:223–33. doi: 10.1007/s11103-010-9668-6. [DOI] [PubMed] [Google Scholar]
- 43.Simm C, et al. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot Cell. 2007;6:1166–77. doi: 10.1128/EC.00077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmiter RD, Huang L. Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Pflugers Arch. 2004;447:744–51. doi: 10.1007/s00424-003-1070-7. [DOI] [PubMed] [Google Scholar]
- 45.MacDiarmid CW, Gaither L, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–55. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyabe S, Izawa S, Inoue Y. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2001;282:79–83. doi: 10.1006/bbrc.2001.4522. [DOI] [PubMed] [Google Scholar]
- 47.Miyayama T, Suzuki KT, Ogra Y. Copper accumulation and compartmentalization in mouse fibroblast lacking metallothionein and copper chaperone, Atox1. Toxicol Appl Pharmacol. 2009;237:205–13. doi: 10.1016/j.taap.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 48.Ralle M, et al. Wilson disease at a single cell level: intracellular copper trafficking activates compartment-specific responses in hepatocytes. J Biol Chem. 2010;285:30875–83. doi: 10.1074/jbc.M110.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yagisawa F, et al. Identification of novel proteins in isolated polyphosphate vacuoles in the primitive red alga Cyanidioschyzon merolae. Plant J. 2009;60:882–93. doi: 10.1111/j.1365-313X.2009.04008.x. [DOI] [PubMed] [Google Scholar]
- 50.Czupryn M, Falchuk KH, Stankiewicz A, Vallee BL. A Euglena gracilis endonuclease Zinc Endonuclease. Biochemistry. 1993;32:1204–1211. doi: 10.1021/bi00056a002. [DOI] [PubMed] [Google Scholar]
- 51.Harris EH, Stern DB, Witman GB. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and its laboratory use. Academic press; 2009. [Google Scholar]
- 52.Chandra S. In: Encycl Mass Spectrom. Gross M, Caprioli R, editors. Elsevier; 2010. pp. 469–480. [Google Scholar]
- 53.Merchant S, Bogorad L. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol Cell Biol. 1986;6:462–9. doi: 10.1128/mcb.6.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li HH, Merchant S. Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii Evidence for a protease-susceptible conformation of the apoprotein and regulated proteolysis. J Biol Chem. 1995;270:23504–23510. doi: 10.1074/jbc.270.40.23504. [DOI] [PubMed] [Google Scholar]
- 55.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.