Abstract
The functions of the proteins encoded by the Bardet–Biedl syndrome (BBS) genes are unknown. Mutations in these genes lead to the pleiotropic human disorder BBS, which is characterized by obesity, retinopathy, polydactyly, renal and cardiac malformations, learning disabilities, and hypogenitalism. Secondary features include diabetes mellitus and hypertension. Recently, it has been suggested that the BBS phenotypes are the result of a lack of cilia formation or function. In this study, we show that mice lacking the Bbs4 protein have major components of the human phenotype, including obesity and retinal degeneration. We show that Bbs4-null mice develop both motile and primary cilia, demonstrating that Bbs4 is not required for global cilia formation. Interestingly, male Bbs4-null mice do not form spermatozoa flagella, and BBS4 retinopathy involves apoptotic death of photoreceptors, the primary ciliated cells of the retina. These mutation data demonstrate a connection between the function of a BBS protein and cilia. To further evaluate an association between cilia and BBS, we performed homology comparisons of BBS proteins in model organisms and find that BBS proteins are specifically conserved in ciliated organisms.
Bardet–Biedl syndrome (BBS) is a genetically heterogeneous disorder with linkage to eight loci. Six BBS genes (BBS1, BBS2, BBS4, BBS6, BBS7, and BBS8) have been identified (1–7). With the exception of BBS6, which has similarity to type II chaperonins (8), BBS proteins do not show extensive homology with proteins of known function. BBS4 and BBS8 contain tetratricopeptide repeat domains, and a region of BBS8 shows similarity to the prokaryotic pilF domain. Based on the observations that BBS8 localizes to the basal body of ciliated cells and expression of the Caenorhabditis elegans orthologues of several BBS proteins are limited to ciliated cells, it has been hypothesized that BBS is the result of a defect in cilia assembly or function (7). We previously used positional cloning to identify the human BBS4 gene (3). To help elucidate the function of the BBS proteins, we have now targeted the Bbs4 gene in mice. Bbs4–/– mice recapitulate aspects of the human phenotype: they become obese, fail to reproduce, and display retinal degeneration. We show the presence of both motile and primary cilia in these mice, demonstrating that Bbs4 deficiency does not prevent global ciliogenesis. Interestingly, male knockout mice have a complete lack of flagella, demonstrating that the Bbs4 protein is necessary for flagella formation during spermatogenesis. Furthermore, Bbs4-null mice undergo retinal degeneration due principally to photoreceptor cell loss associated with outer segment attenuation, suggesting a role for BBS proteins in maintenance of sensory cilia. These data demonstrate a role for BBS proteins in facets of cilia function.
Materials and Methods
Construction of the Bbs4 Gene Targeting Vector. blast analysis of the Celera mouse fragment database with the human BBS4 cDNA sequence identified sequences corresponding to the mouse Bbs4 exons. A Bbs4 replacement vector was constructed in two cloning steps. For the 5′ homology, a 3-kb PCR fragment containing the genomic DNA sequence corresponding to exons 3–5 was subcloned into the NheI and XhoI sites of pOSdupdel (a gift of O. Smithies, University of North Carolina, Chapel Hill). For the 3′ homology, a 3.8-kb PCR fragment containing exons 12–15 was subcloned into the SalI and NotI sites.
Generation of Bbs4–/– Mice. The targeting construct was linearized at a unique NotI site and electroporated into R1 embryonic stem (ES) cells (129 × 1/SvJ3129S1/Sv). The ES cells were selected in G418 and Ganciclovir, and surviving colonies were expanded into 96-well plates for screening. Genomic DNA was isolated from ES cells by incubating in 50 μl of lysis buffer (50 mM Tris·HCl, pH 8.5/2.5 mM EDTA/100 mM NaCl/0.1% SDS/500 μg/ml proteinase K) for 12–24 h at 55°C. After incubation, cell lysates were diluted into 500 μlofddH2O, and 2-μl aliquots were used as templates for PCR amplification. Primers specific for the 5′ knockout allele (forward, 5′-CGTGTACCCACACATGGGGAGCCA-3′; reverse, 5′-CCGGAGAACCTGCGTGCAATCCAT-3′) and 3′ knockout allele (forward, 5′-GGGCTGACCGCTTCCTCGTGCTTT-3′; reverse, 5′-TGCACCCAGGAGGGATGCCCATAA-3′) were used in long-range PCRs to identify correct targeting events. PCR was performed in 25-μl reactions containing 5 μl of 5× PCR buffer [300 mM Tris·SO4, pH 9.1/90 mM (NH4)2SO4/10 mM MgSO4], 250 μM each of dATP, dCTP, dGTP, and dTTP, 2.5 pmol of each primer, and 1 unit of Elongase (Invitrogen). Samples were subjected to 35 cycles of 94°C for 30 s and 68°C for 6 min. Amplification products were visualized on a 0.8% agarose gel. ES cells heterozygous for the mutant Bbs4 allele were injected into C57BL/6J blastocysts. Two targeted clones were injected into blastocysts, and both produced chimeras. Chimeric animals were used to generate Bbs4 heterozygous (Bbs4+/–) mice on a mixed genetic background by mating with C57BL/6J mice. Mice on the mixed genetic background were backcrossed for at least two generations to enrich for the C57BL/6J background. Heterozygous mice were intercrossed, and the progeny were genotyped by PCR using the primers indicated above to identify the presence of the targeted allele. Presence of the wild-type allele was determined by using primers specific for the 5′ wild-type allele (forward, 5′-CGTGTACCCACACATGGGGAGCCA-3′; reverse, 5′-GGATCCGCAGGCCATGTGCTCAAA-3′) and 3′ wild-type allele (forward, 5′-CACCGGACCTTCCAGACCGACCTC-3′; reverse, 5′-TGCACCCAGGAGGGATGCCCATAA-3′). PCR conditions were as described above.
RNA Isolation and Northern Blot Analysis. Tissues from 8-month-old female mice were rapidly frozen in liquid nitrogen and stored at –80°C until use. Total cellular RNA extraction, gel electrophoresis, blotting, hybridization, and autoradiography were carried out as described (9). The mouse Bbs4 cDNA probe was generated by using primers specific for the 3′ UTR (forward, 5′-CCATAACCTGGGAGTGTGCT-3′; reverse, 5′-AGCAGATTTGCTGCCTGAAT-3′). The blot was stripped of radioactivity and rehybridized with a cDNA probe for β-actin to verify equal loading of RNA.
Histological, Immunohistological, and Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL) Analysis. After death, eyes were enucleated and fixed by immersion in a solution of 4% paraformaldehyde in 100 mM cacodylate buffer, pH 7.4. After 2–4 h of fixation, eyes were washed three times in 10 mM PBS followed by infiltration and embedment in acrylamide, as described (10). When possible, the blocks were oriented such that sagittal sections included the superior and inferior axes. Cryostat sections were collected at a thickness of 5–7 μm, and stored at 4°C until use. Testes were isolated and fixed in 4% paraformaldehyde in PBS overnight. The tissues were then blocked in paraffin and sectioned for hematoxylin/eosin staining. Labeling of rod photoreceptor cells was performed with the RET-P1 monoclonal antibody (Santa Cruz Biotechnology) at a 1:1,000 dilution, followed by visualization with Alexa-488-conjugated antibodies directed against mouse IgG (Molecular Probes). For TUNEL staining, the In Situ Cell Death Detection kit, TMR red (Roche Applied Science), was used according to the manufacturer's instructions. Nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; Molecular Probes). Sections were viewed on an Olympus BX-51 microscope and images were collected with a SPOT RT digital camera (Diagnostic Instruments).
Electron Microscopy Analysis. Trachea from Bbs4–/– animals was fixed in half-strength Karnovsky's fixative (11) for several hours before osmium postfixation, dehydration, and embedding in Epon 812. Ultrathin sections were observed on a transmission electron microscope, and photographs were taken at ×35,000 and ×2,500 through the cilia. Testes from Bbs4+/+ and Bbs4–/– animals were embedded in Spurr resin and stained with 1% toluidine blue. Kidneys from Bbs4+/+ and Bbs4–/– animals were dehydrated in ethanol and dried by using a critical point drier. The samples were then mounted on an aluminum stub by using double-stick carbon tape, coated with gold/palladium by using a sputter coater, and then examined on a Philips XL 30 scanning electron microscope.
Results
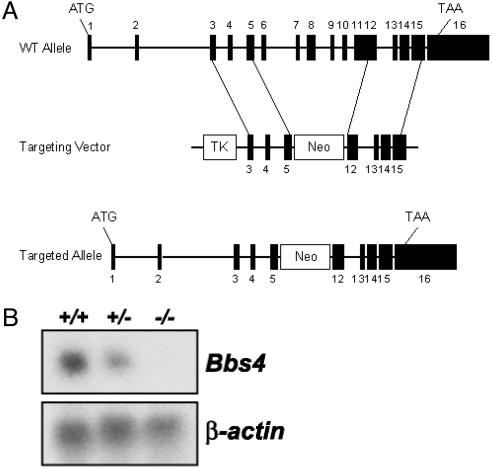
Bbs4 Targeting. We targeted the Bbs4 gene in mice by constructing a vector containing regions of the mouse Bbs4 gene that, upon homologous recombination, replaced exons 6–11 with a neomycin cassette (Fig. 1A). This event removes approximately one-third of the Bbs4 coding sequence, resulting in a frameshift expected to create a null allele. Northern blot analysis was performed by using RNA isolated from Bbs4+/+, Bbs4+/–, and Bbs4–/– mice to confirm that the gene targeting resulted in a null allele (Fig. 1B). Genotyping of litters from Bbs4+/– intercrosses resulted in F2 ratios for Bbs4+/+/Bbs4+/–/Bbs4–/– animals of 0.36:0.51:0.13, significantly different from the predicted 0.25:0.5:0.25 Mendelian ratios (χ2 = 7.21, P < 0.05).
Fig. 1.
Targeting of the Bbs4 gene. (A) Diagram of the Bbs4 targeting vector and the resulting recombination product. Upon homologous recombination, exons 6–11 are replaced with a neomycin cassette. (B)(Upper) Northern blot analysis of Bbs4 expression in kidney total cellular RNA from wild-type (+/+), heterozygous (+/–), and homozygous (–/–) animals. The probe is a 960-bp Bbs4 partial 3′ cDNA. (Lower) The same blot probed with β-actin as a loading control.
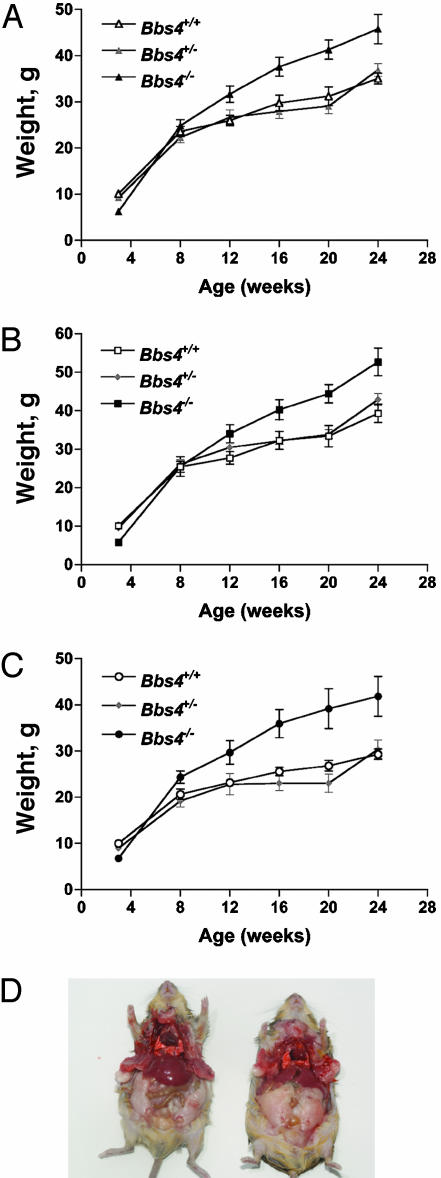
Bbs4-Null Mice Become Obese. Bbs4–/– mice appear to be small at birth, weigh less than their littermates at 3 weeks of age, and become obese over time (Fig. 2 A–C). After weaning, Bbs4–/– mice begin to gain more weight than wild-type mice, concurrent with greater food intake (data not shown). By 12 weeks of age, Bbs4-null mice weigh significantly more than their control littermates. The weight difference between Bbs4-null animals and controls becomes greater in older animals and a similar weight gain is seen for both male and female animals. Examination of the fat distribution in older animals demonstrates that the increased weight corresponds to an increase in centrally deposited adipose tissue (Fig. 2D).
Fig. 2.
Comparison of weight and adipose tissue deposition in Bbs4 mice. (A) The weights of all wild-type (Bbs4+/+; n = 40), heterozygous (Bbs4+/–; n = 40), and homozygous (Bbs4–/–; n = 36) mice at 3, 8, 12, 16, 20, and 24 weeks of age. Values are expressed as mean ± SEM. (B) The weights of male wild-type (Bbs4+/+; n = 20), heterozygous (Bbs4+/–; n = 20), and homozygous (Bbs4–/–; n = 20) mice. Values are expressed as mean ± SEM. (C) The weights of female wild-type (Bbs4+/+; n = 20), heterozygous (Bbs4+/–; n = 20), and homozygous (Bbs4–/–; n = 16) mice. Values are expressed as mean ± SEM. (D) Examination of adipose tissue deposition demonstrates an increase in the amount of central adipose tissue in a Bbs4–/– mouse (Right), as compared to a Bbs4+/+ mouse (Left). The mice are ≈10-month-old males.
Bbs4-Null Mice Undergo Apoptotic Retinal Degeneration. As retinopathy is a primary feature of BBS, we examined the retinas from Bbs4–/– mice. Bbs4–/– mice of ages 7 months or older exhibit a complete absence of photoreceptor cells as assessed by a lack of the outer nuclear layer and the absence of photoreceptor inner and outer segments (Fig. 3 A–D). The inner retina of these animals appears to be intact with normal inner nuclear and ganglion cell layers.
Fig. 3.
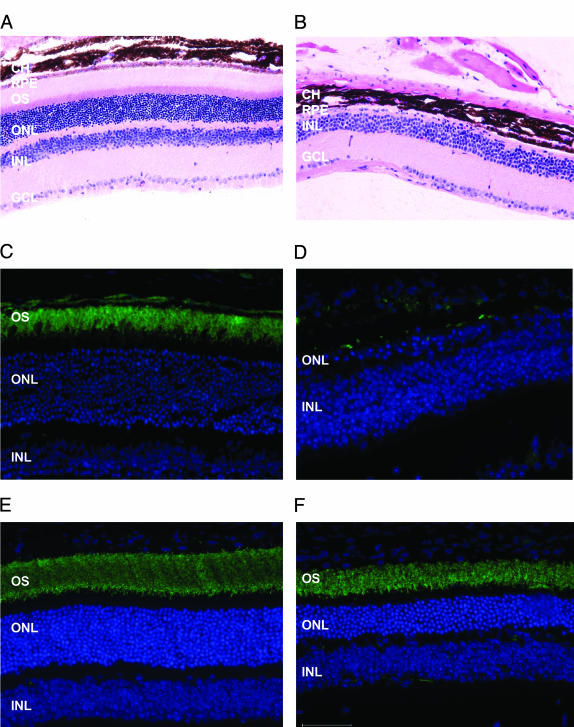
Examination of mouse retina. (A and B) Hematoxylin and eosin staining of retinal sections from a Bbs4+/+ (A) and Bbs4–/– (B) mouse. The entire photoreceptor layer is absent in the Bbs4–/– section. CH, choroids; RPE, retinal pigment epithelium; OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (C and D) Labeling of rods with anti-Opsin (green) from a Bbs4+/+ (C) and Bbs4–/– (D) mouse. The lack of labeling in the Bbs4–/– retinal section confirms the absence of the photoreceptors. (E and F) Labeling of rods with anti-Opsin (green) from a 6-week-old Bbs4+/+ (E) and Bbs4–/– (F) mouse, demonstrating the presence of photoreceptors. The outer nuclear and photoreceptor layers in the Bbs4–/– section are approximately one-half the thickness of the Bbs4+/+ section, indicating significant photoreceptor cell degeneration.
To determine whether the absence of photoreceptors in the older animals is the result of retinal degeneration or represents a developmental defect, we evaluated eyes from young animals. Two-week-old Bbs4–/– mice appear to have normal retinas. However, at 6 weeks of age, Bbs4–/– mice show attenuation of the photoreceptor outer segment and the outer nuclear layer is one-half the thickness of that of control animals (Fig. 3 E and F). By six weeks of age, an abnormal electroretinogram is observed, demonstrating abnormal retinal function. In human BBS patients, the retinal degeneration is similarly progressive with loss of visual acuity usually beginning before 10 years of age and resulting in legal blindness by 20 years of age.
We examined the mechanism of photoreceptor cell death in 6-week-old Bbs4–/– mice by using TUNEL analysis. Control animals possess extremely rare TUNEL-positive nuclei in the retina, in contrast to Bbs4–/– mice, which exhibit multiple labeled nuclei per section (data not shown). TUNEL-positive nuclei were not observed in the inner nuclear and ganglion cell layers, indicating that only the outer nuclear layer is affected. These results demonstrate that Bbs4–/– mice undergo retinal degeneration through an apoptotic mechanism that specifically targets the photoreceptor cells and involves attenuation of the photoreceptor outer segments.
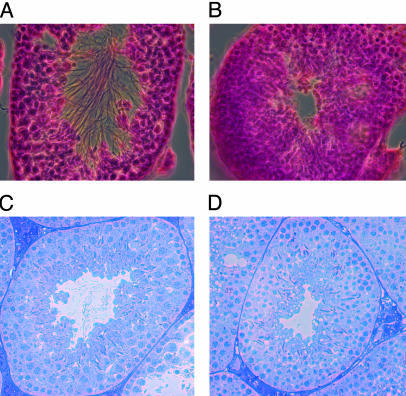
Male Bbs4-Null Mice Do Not Develop Flagella. We observed that Bbs4–/– males failed to sire offspring. To investigate the male infertility, we examined the seminiferous tubules from Bbs4–/– mice including young sexually mature animals as well as older animals. In Bbs4+/+ animals, spermatozoa with normal morphology were observed in all animals examined as evidenced by the presence of flagella and condensed sperm nuclei (Fig. 4 A and C). However, in the Bbs4–/– males, there is a complete lack of flagella (Fig. 4B), even on cells with a condensed sperm head and acrosome (Fig. 4D). The inability to detect any flagella in the seminiferous tubules of male Bbs4–/– mice of different ages, despite evidence of developing spermatozoa, indicates that flagella are not formed.
Fig. 4.
Histological examination of mouse seminiferous tubules. (A) Hematoxylin and eosin staining of a testes section from a Bbs4+/+ mouse. Flagella are apparent in the lumen of the tubule. (B) Hematoxylin and eosin staining of a testes section from a Bbs4–/– mouse showing the absence of flagella in the lumen. (C) Toluidine blue staining of a 1-μm testes section from a Bbs4+/+ mouse showing spermatozoa with normal morphology as evidenced by the presence of flagella and condensed sperm nuclei. (D) Toluidine blue staining of a 1-μm testes section from a Bbs4–/– mouse showing the presence of condensed sperm nuclei in the absence flagella.
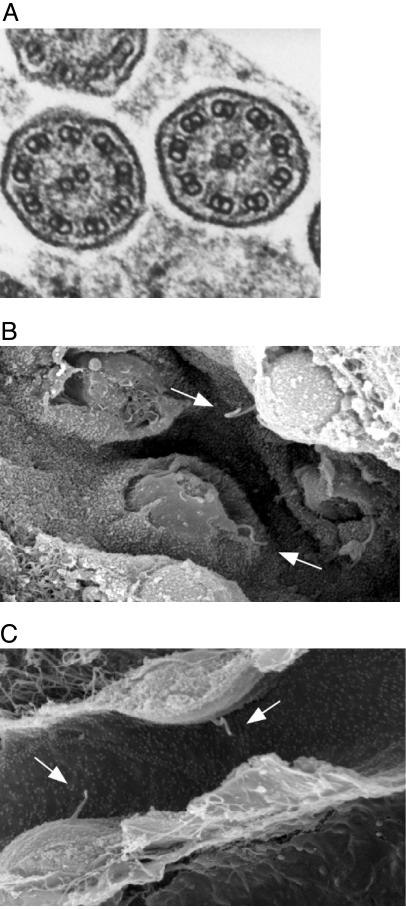
Lack of Bbs4 Does Not Prevent Motile or Primary Cilia Formation. Because of the observed attenuation of photoreceptor outer segments, absence of spermatozoa flagella, and the recently reported evidence that some BBS proteins localize to basal bodies, we used electron microscopy to examine the structure of both motile and primary cilia in Bbs4–/– mice. Transmission electron microscopy was used to examine the structure of motile cilia from the trachea of Bbs4–/– mice. The normal axoneme structure with the corresponding 9 + 2 microtubule arrangement was observed in Bbs4–/– mice (Fig. 5A). To assess the structure of primary cilia, we performed scanning electron microscopy on kidney epithelial cells. The presence of primary cilia extending from the surface of renal tubular cells was observed in Bbs4+/+ (Fig. 5B, arrows) and Bbs4–/– (Fig. 5C, arrows) animals. Therefore, Bbs4–/– mice possess both motile and primary cilia that appear grossly normal in length, number, and structure.
Fig. 5.
Electron microscopy analysis of motile and primary cilia. (A) Transmission electron microscopic image of a cross-section of cilia in the trachea of a Bbs4–/– mouse. The nine outer and two inner microtubules are clearly defined, indicating that the internal structure of motile cilia appears normal. (B and C) Scanning electron microscopic image of renal tubule cells from a Bbs4+/+ (B) and a Bbs4–/– (C) mouse. Normal appearing primary cilia (arrows) are evident in both.
Phylogenetic Conservation of BBS Proteins. To determine whether phylogenetic data support a role for BBS proteins in cilia function, we performed blast analysis of BBS4 and the other known BBS proteins against available genomic sequence databases from EnsEMBL (Mus musculus, Danio rerio, Drosophila melanogaster, and Caenorhabditis elegans), JGI (Ciona intestinalis and Chlamydomonas reinhardtii), TIGR (Trypanosoma brucei and Trypanosoma cruzi), SGD (Saccharomyces cerevisiae), Sanger (Schizosaccharomyces pombe), Broad (Aspergillus nidulans), and TAIR (Arabidopsis thaliana). Organisms for which putative BBS homologues were identified were further evaluated by blast analysis against the predicted proteins of those organisms. A striking finding is that, in general, ciliated organisms have highly homologous sequences for BBS proteins, whereas nonciliated organisms do not (Table 1). For example, even lower single-cell ciliated organisms such as T. brucei, T. cruzi, and C. reinhardtii have highly homologous sequences to the known BBS proteins. In addition, the ciliated invertebrate, Ciona intestinalis, has highly homologous sequences to BBS proteins. In contrast, organisms without cilia (i.e., S. cerevisiae, Schizosaccharomyces pombe, Aspergillus nidulans, and A. thaliana) do not have sequences that are highly homologous to BBS protein sequences. Interestingly, D. melanogaster has homologues only to BBS1, BBS4, and BBS8. A notable exception to the general conservation of BBS proteins in ciliated organisms is BBS6, which has only low-level homology across most nonvertebrate organisms evaluated.
Table 1. Phylogenetic profile of BBS protein homologies across various genomes showing expected values obtained from blast analysis of human BBS proteins against predicted protein databases of a set of ciliated (MM, DR, CI, CE, CR, TC, TB, DM) and nonciliated (SC, SP, AN, AT) organisms.
| MM | DR | CI | CE | CR | TC | TB | DM | SC | SP | AN | AT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBS1 | 0.0 | 2e-82 | 1e-138 | 7e-64 | 3e-75 | 2e-73 | 9e-64 | 5e-64 | 3.1 | 2.5 | 1.1 | 3.2 |
| BBS2 | 0.0 | 0.0 | 0.0 | 1e-85 | 6e-55 | 4e-98 | 1e-87 | 0.24 | 0.024 | 0.13 | 3.0 | 1.1 |
| BBS4 | 0.0 | 0.0 | 1e-134 | 8e-11 | 1e-14 | 2e-80 | 3e-72 | 3e-55 | 5e-9 | 2e-7 | 1e-6 | 6e-13 |
| BBS6 | 0.0 | 1e-130 | 6e-26 | 4e-8 | 2e-7 | 5e-18 | 2e-11 | 2e-11 | 5e-13 | 3e-15 | 9e-13 | 2e-8 |
| BBS7 | 0.0 | 0.0 | 0.0 | 1e-110 | 1e-115 | 3e-50 | 8e-41 | 2.0 | 0.16 | 0.62 | 1.3 | 1.8 |
| BBS8 | 0.0 | 0.0 | 0.0 | 1e-109 | 1e-135 | 4e-92 | 1e-79 | 4e-53 | 2e-4 | 2e-5 | 4e-5 | 2e-7 |
| Estimated genome size, GB | 2.5 | 1.6 | 0.16 | 0.1 | 0.1 | 0.035 | 0.035 | 0.137 | 0.012 | 0.014 | 0.031 | 0.115 |
MM, Mus musculus; DR, Danio rerio; CI, Ciona intestinalis; CE, C. elegans; CR, Chlamydomonas reinhardtii; TC, T. cruzi; TB, T. brucei; DM, D. melanogaster; SC, S. cerevisiae; SP, Schizoaccharomyces pombe; AN, Aspergillus nidulans; AT, A. thaliana.
Discussion
The findings described here contribute to our understanding of the function of the BBS4 protein. We show that mice lacking Bbs4 develop obesity, display retinal degeneration, and fail to synthesize flagella during spermatogenesis. Based on localization of some BBS proteins to the basal body of ciliated cells (7), BBS proteins could in principle be involved in ciliogenesis, cilia maintenance, intraflagellar transport (IFT), and/or microtubule dependent intracellular transport. Our results show that absence of the Bbs4 protein does not prevent initial formation of either motile or primary cilia, although absence of this protein does result in failure of flagella formation in mature spermatozoa. These findings suggest that there are differences in the requirement for Bbs4 between the assembly of flagella and cilia. It has been shown that some components of IFT are required for cilia and flagella assembly (12). However, some differences in these processes have also been reported. For example, it has been demonstrated in Drosophila that IFT is required to differentiate sensory cilia, but not spermatozoa (13). Further study will be necessary to determine whether the functions of the BBS proteins are limited to a subset of cilia.
It has been suggested that BBS proteins are unlikely to play a role in IFT because they do not localize to the axoneme (7). However, these localization data are consistent with the possibility that the BBS proteins serve as mediators for communication and/or intracellular transport between the cilium and the interior of the cell. Rather than playing a direct role in IFT, BBS proteins may play an auxiliary role by preparing molecules for transport, or by playing a role in microtubule-associated intracellular transport.
A defect in IFT is consistent with the retinal degeneration that is seen in Bbs4–/– mice. The connecting cilium between the inner and outer segment of the retina is thought to be the exclusive route of transport of newly synthesized proteins from the inner to outer segment. Disruption of this process in a conditional kinesin-II knockout mouse model has been shown to cause retinal degeneration, demonstrating that IFT is required for maintaining the outer segment of photoreceptor cells (14). We show that the loss of Bbs4 does not prevent initial formation of photoreceptor outer segments, and we propose that its absence compromises the intracellular transport that is required for maintaining the rapidly regenerating outer segments of these cells. The failure of flagella formation and photoreceptor outer segment maintenance in Bbs4–/– animals provides the first direct mutational data indicating a ciliary role for BBS proteins.
In addition to the mouse mutation data, we provide phylogenetic data comparing human BBS proteins and the genomes of model organisms. These data demonstrate statistically significant conservation of BBS proteins in lower ciliated organisms, supporting a role for BBS proteins in cilia function. The one exception to this conservation is BBS6. This may indicate that this protein is not directly necessary for cilia function in these organisms. Of interest is the observation that Drosophila have homologues to BBS1, BBS4, and BBS8, but not to BBS2, and BBS7, suggesting that BBS1, BBS4, and BBS8 are required for function. Of note, the phylogenetic data provide a strategy for identifying other human genes involved in cilia function, and hence additional candidate BBS genes. By identifying human genes that are specifically conserved in ciliated organisms (particularly organisms with relatively small genome sizes) but not conserved in nonciliated organisms, one can determine a subset of genes that are enriched for genes serving cilia related functions. The power of this approach is evident when one considers that, although the genome sizes of two trypanosome species are ≈1% of the human genome, they both have homologues for the BBS genes.
It is unclear why a lack of Bbs4 leads to obesity. This may represent an effect that is independent of the role of Bbs4 in ciliary function. Alternatively, the presence of obesity as a major component of the BBS phenotype may indicate a previously unknown connection between ciliary function and the maintenance of appropriate body weight. Bbs4–/– mice demonstrate increased food intake compared to control animals; thus, whatever role Bbs4 plays in regulation of body mass appears to involve satiety regulation. Bbs4 deficiency may result in loss of specific hypothalamic neurons involved in satiety, or may result in failure of intracellular transport of key components of hypothalamic neurons.
It will be of interest to determine whether genetic interaction of BBS proteins occur, whether BBS genes form complementation groups, and whether combinations of partial deficits (i.e., heterozygous mutations) at two or more loci lead to BBS phenotypes. In humans, there is wide inter- and intrafamilial phenotypic variability, suggesting the presence of modifier loci (15, 16). The generation of Bbs4 knockout mice is a step toward investigating the role of genetic modifiers in BBS. It should be noted that components of the phenotype (obesity, retinal degeneration, and infertility) appear to be highly penetrant in the animals studied to date. This result supports the hypothesis that, in humans, BBS is inherited as an autosomal recessive disease with variable expressivity (17). It is also of interest that Bbs4–/– mice do not have some components of the human phenotype, most notably the polydactyly. It will be of interest to determine whether the lack of polydactyly is specific for Bbs4-null mice and whether there are gene-specific components of the BBS phenotype.
Acknowledgments
We thank K. Bugge, M. Olvera, C. Searby, C. Eastman, G. Beck, M. Shastri, R. Burry, A. Fischer, and K. Wolken for technical assistance and D. Aguiar Crouch for administrative assistance. This work was supported by National Institutes of Health Grants P50-HL-55006 (to V.C.S.) and R01-EY-11298 (to V.C.S. and E.M.S.), the Carver Endowment for Molecular Ophthalmology (E.M.S. and V.C.S.), Research to Prevent Blindness, New York (Department of Ophthalmology, University of Iowa). V.C.S. and E.M.S. are Investigators of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BBS, Bardet–Biedl syndrome; ES, embryonic stem; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; IFT, intraflagellar transport.
References
- 1.Mykytyn, K., Nishimura, D. Y., Searby, C. C., Shastri, M., Yen, H. J., Beck, J. S., Braun, T., Streb, L. M., Cornier, A. S., Cox, G. F., et al. (2002) Nat. Genet. 31, 435–438. [DOI] [PubMed] [Google Scholar]
- 2.Nishimura, D. Y., Searby, C. C., Carmi, R., Elbedour, K., Van Maldergem, L., Fulton, A. B., Lam, B. L., Powell, B. R., Swiderski, R. E., Bugge, K. E., et al. (2001) Hum. Mol. Genet. 10, 865–874. [DOI] [PubMed] [Google Scholar]
- 3.Mykytyn, K., Braun, T., Carmi, R., Haider, N. B., Searby, C. C., Shastri, M., Beck, G., Wright, A. F., Iannaccone, A., Elbedour, K., et al. (2001) Nat. Genet. 28, 188–191. [DOI] [PubMed] [Google Scholar]
- 4.Slavotinek, A. M., Stone, E. M., Mykytyn, K., Heckenlively, J. R., Green, J. S., Heon, E., Musarella, M. A., Parfrey, P. S., Sheffield, V. C. & Biesecker, L. G. (2000) Nat. Genet. 26, 15–16. [DOI] [PubMed] [Google Scholar]
- 5.Katsanis, N., Beales, P. L., Woods, M. O., Lewis, R. A., Green, J. S., Parfrey, P. S., Ansley, S. J., Davidson, W. S. & Lupski, J. R. (2000) Nat. Genet. 26, 67–70. [DOI] [PubMed] [Google Scholar]
- 6.Badano, J. L., Ansley, S. J., Leitch, C. C., Lewis, R. A., Lupski, J. R. & Katsanis, N. (2003) Am. J. Hum. Genet. 72, 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansley, S. J., Badano, J. L., Blacque, O. E., Hill, J., Hoskins, B. E., Leitch, C. C., Kim, J. C., Ross, A. J., Eichers, E. R., Teslovich, T. M., et al. (2003) Nature 425, 628–633. [DOI] [PubMed] [Google Scholar]
- 8.Stone, D. L., Slavotinek, A., Bouffard, G. G., Banerjee-Basu, S., Baxevanis, A. D., Barr, M. & Biesecker, L. G. (2000) Nat. Genet. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- 9.Swiderski, R. E., Ying, L., Cassell, M. D., Alward, W. L., Stone, E. M. & Sheffield, V. C. (1999) Brain Res. Mol. Brain Res. 68, 64–72. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, L. V. & Blanks, J. C. (1984) Curr. Eye Res. 3, 969–974. [DOI] [PubMed] [Google Scholar]
- 11.Russell, S. R., Mullins, R. F., Schneider, B. L. & Hageman, G. S. (2000) Am. J. Ophthalmol. 129, 205–214. [DOI] [PubMed] [Google Scholar]
- 12.Pazour, G. J., Dickert, B. L., Vucica, Y., Seeley, E. S., Rosenbaum, J. L., Witman, G. B. & Cole, D. G. (2000) J. Cell Biol. 151, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, Y. G., Kwok, B. H. & Kernan, M. J. (2003) Curr. Biol. 13, 1679–1686. [DOI] [PubMed] [Google Scholar]
- 14.Marszalek, J. R., Liu, X., Roberts, E. A., Chui, D., Marth, J. D., Williams, D. S. & Goldstein, L. S. (2000) Cell 102, 175–187. [DOI] [PubMed] [Google Scholar]
- 15.Riise, R., Andreasson, S., Borgastrom, M. K., Wright, A. F., Tommerup, N., Rosenberg, T. & Tornqvist, K. (1997) Br. J. Ophthalmol. 81, 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riise, R., Tornqvist, K., Wright, A. F., Mykytyn, K. & Sheffield, V. C. (2002) Arch. Ophthalmol. 120, 1364–1367. [DOI] [PubMed] [Google Scholar]
- 17.Mykytyn, K., Nishimura, D. Y., Searby, C. C., Beck, G., Bugge, K., Haines, H. L., Cornier, A. S., Cox, G. F., Fulton, A. B., Carmi, R., et al. (2003) Am. J. Hum. Genet. 72, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]