Abstract
During lung infection with virus, airway-derived dendritic cells (DC) have been thought to be the dominant cell type involved in acquisition, transport, and direct antigen presentation for cytotoxic T lymphocyte priming. Contrary to this view, we have found that both an airway-derived CD8α–CD11b– DC subset and distinct CD8α+ lymph node resident DC can present class I-restricted antigens after lung infection with influenza virus or herpes simplex virus 1. Presentation by a nonairway-derived DC population argues that cytotoxic T lymphocyte priming may involve interplay between different DC subsets, not all of which originate within the site of infection.
Keywords: T lymphocyte, influenza A virus
The classic paradigm of dendritic cell (DC) involvement in T cell responses revolves around a linear progression of events, starting with their capture of antigen in peripheral tissues and followed by migration to draining lymphoid organs and presentation for the purpose of T cell priming (1). There is ample evidence that DC can acquire antigen in peripheral tissues and transport this to draining lymph nodes (LNs). Separate from this, DC are particularly adept at T cell priming, with their expression of an array of molecules required for this event. However, it is becoming increasingly clear that DC represent a heterogeneous population of cells whose diversity lends itself to subset specialization. There is growing evidence that only certain subsets are involved in T cell priming, possibly even in the distinct type of T cell subset that they engage (2–5). Thus, it can no longer be assumed that DC found to have originated from any particular site of infection are automatically the subset directly implicated in T cell stimulation.
We recently have used a model of cutaneous infection with herpes simplex virus (HSV) to show that the dominant skin migrating DC population, the Langerhans cells, were not involved directly in cytotoxic T lymphocyte (CTL) priming to this virus (3). Indeed, it seems likely that the actual priming DC, which belonged to the CD8α+ DC subset, did not originate within the epidermal layer of skin that harbored the infection, although this could not be proven definitively in this case. Given this finding, we sought to examine other routes of infection to determine whether we could find definitive examples where nonmigrating DC were involved in CTL priming. To this end, we have used a combination of lung infection with virus and direct labeling of tissue-resident DC with a fluorescent dye to show that although there is a population of migrating DC capable of class I-restricted presentation of viral antigen, a dominant nonairway-derived CD8α+ DC subset previously found to be involved in a number of other CTL priming events also was called into play (2–4, 6, 7).
Materials and Methods
Mice, Virus, and Infections. C57BL/6 and TAP-10/0 mice (8) were bred and maintained at The Walter and Eliza Hall Institute of Medical Research. Experiments with all mice began when they were between 6 and 10 weeks of age. gBT-I.1 (H-2b) transgenic mice (gBT-I) express a T cell antigen receptor (TCR) (Vα 2Vβ 8.1) specific for the immunodominant MHC class I-restricted epitope of HSV glycoprotein B (gB498–505) (9).
Mice were anaesthetized with methoxyfluorane and then infected with virus diluted in 25 μl of PBS for intranasal (i.n.) infection. Infections were undertaken with HSV, HKx31 influenza A (H3N2), or WSN-gB (H1N1) influenza virus (Flu.gB), the latter of which contains the gB498–505 Kb-restricted epitope of HSV inserted into the neurominidase stalk (10). For i.n. infection, 102 plaque-forming units (pfu) of HKx31, 102.6 pfu of Flu.gB influenza A virus, or 4 × 104 pfu of HSV were used.
Detection of Antigen Presentation by LN DC. The lacZ-inducible hybridoma specific for DbNP366 (BWZ-IFA.NP4) was used to analyze antigen presentation by mediastinal LN cells that were released by collagenase/DNase digestion for 20 min followed by a 5-min incubation with 0.099 M EDTA. Some preparations were depleted of cells expressing CD11c, CD11b, CD8α, CD4, CD3, B220, or CD205 by using antibodies and magnetic beads as described (2, 4, 11). LN cells then were cultured with 105 BWZ-hybridoma cells for 17 h to detect antigen presentation as measured by the number of lacZ+ cells, as described (4, 11, 12). The number of lacZ+ cells in the presence of naive stimulators has been subtracted from the number of lacZ+ cells cultured with stimulators from virally infected mice to derive the specific number of lacZ+ cells.
DC Isolation from LNs. DC were isolated essentially as described (13, 14). Briefly, mediastinal LN were digested for 20 min at room temperature with collagenase/DNase and then treated for 5 min with EDTA to disrupt T cell–DC complexes. Cells not of the DC lineage were depleted by incubating in predetermined optimal concentrations of purified antibodies [anti-CD3 (KT3), anti-Thy1 (T24/31.7), anti-CD19 (ID3), anti-GR-1 (RB6–8C5), and anti-erythrocyte (TER-119)] and then by removing the antibody-binding cells with anti-rat Ig-coupled magnetic beads (Dynabeads, Dynal, Oslo). Note that in our hands plasmacytoid DC are not depleted by using anti-GR-1 mAb (15). For some preparations of DC, B220+ and CD8α+ populations also were removed by substituting anti-B220 (RA3–6B2) for anti-CD19 and including anti-CD8α (53–6.7) mAb in the depletion mixture. The DC in the enriched populations were gated on CD11c+ cells before sorting into specific subsets.
5,6-Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) Labeling of Transgenic T Cells. LNs (inguinal, brachial, axillary, sacral, superficial cervical, iliac, and mesenteric) were obtained from gBT-I TCR transgenic mice and CD8+ T cells purified by using a mixture of optimally titered antibodies to deplete cells expressing Mac-1 (M1/70), F4/80, Ter 119, GR-1, MHC class II (M5/114), and CD4 (GK1.5) followed by sheep anti-mouse and anti-rat Dynabeads (Dynal). Enriched cells contained 87–96% specific CD8+ TCR transgenic T cells. These were labeled with CFSE (Molecular Probes) (4).
Analysis of in Vitro Proliferation of Naïve T Cells by DC. CFSE-labeled gBT-I CD8+ T cells (5 × 104) were added to 1.25 × 104 fluorescence-activated cell sorter (FACS)-sorted DC in 200 μl of mouse tonicity RPMI medium 1640 containing 10% FCS, 50 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (complete medium) in 96-well V-bottom plates (Costar, Corning). Each culture was performed in duplicate. Cultures were analyzed for proliferation after 60 h. Cells were stained with anti-CD8α-APC (53–6.7; BD Pharmingen) and anti-Vα2-PE (B20.1; BD Pharmingen). CD8α+Vα 2+PI– cells from the entire well were analyzed for proliferation by flow cytometry. In antigen-transfer assays, CFSE-labeled gBT-I cells were cocultured with 3 × 104 CD8α+ DC from the mediastinal LN of naïve C57BL/6 mice together with 3 × 105 CD8α–CD11b– (CD45RA–) DC from TAP-10/0 mice infected i.n. 3 days previously with Flu.gB. Because of the difficulty in obtaining the number of DC required for antigen-transfer experiments, these experiments were performed as single samples, with three separate experiments showing similar results.
In Vivo Staining of DC with CFSE. CFSE was dissolved at 25 mM in DMSO and subsequently diluted to 8 mM in PBS. CFSE (50 μl) was administered i.n. to each mouse after anesthesia.
Results
More Than One DC Subset Is Involved in Class I-Restricted Presentation After Lung Infection with Virus. We previously had shown that CD8α+ DC were solely involved in antigen presentation after skin or i.v. infection with HSV (2–4). Before determining whether this was also the case after pulmonary infection with influenza virus, the peak of presentation was identified by examining the kinetics of class I-restricted antigen presentation. This was quantitated by using an in vitro assay employing an inducible β-galactosidase expressing T cell hybridoma specific for the immunodominant determinant from influenza nucleoprotein (NP) (12). Antigen presentation by cells released from lung-draining mediastinal LNs was first detected 1 day after infection (Fig. 1A), peaked at day 3, and was largely extinguished by day 10. To determine whether CD8α+ DC were the only subset contributing to this activity, single-cell suspensions prepared from mediastinal LNs at the day 3 peak of antigen presentation were depleted of various subsets of cells after incubation with individual antibodies specific for leukocyte markers. Fig. 1B shows that depletion of CD11c+ cells abrogated all presentation, suggesting that non-DC did not contribute significantly to the observed stimulation of the NP-specific T cell hybridoma. Depletion with anti-CD8α antibody resulted in an ≈60% reduction in presentation, showing that although CD8α+ DC contributed significantly to this response, at least one other DC subset also was involved in class I-restricted presentation after lung infection with influenza. Similar findings were made on days 1, 2, 3, and 4 after infection (data not shown).
Fig. 1.
Kinetic analysis of antigen presentation in mediastinal LNs during primary HKx31 influenza infection. (A) Mediastinal LNs from influenza-infected C57BL/6 mice were treated with collagenase/DNase to form single-cell suspensions. These cells were cultured with a lacZ-inducible hybridoma specific for influenza NP (BWZ-IFA.NP4) and enumerated in duplicate for β-galactosidase-producing cells. Each time point represents the mean of three to five mice. (B) A CD11c+ cell is responsible for presentation of DbNP366 after influenza infection. Three days after i.n. infection with HKx31 influenza, mediastinal LNs were pooled (15 mice), and collagenase/DNase was digested to form single cells. These cells either were left undepleted or were depleted of specific cellular subsets by using mAb staining followed by anti-rat Dynabeads. They were then cultured with a lacZ-inducible hybridoma specific for a class I-restricted epitope of influenza NP (BWZ-IFA.NP4). Data show the mean and SD of three separate experiments. Data for CD4 depletion show the mean (bar) and independent values (open circles) of two experiments. *, P < 0.01 or less.
Conventional CD8α+ and a Previously Uncharacterized CD8α–CD11b– DC Subset Are Involved in Class I-Restricted Presentation After Lung Infection with HSV and Influenza Virus. At the beginning of our investigation, it had been reported that mouse DC could be divided into at least six different subsets based on expression of a variety of markers such as CD11b, CD205, and CD8α (13–18). For the purposes of initial determination of which subsets were involved in presentation, we divided DC into three broad groups: the CD8α+CD45RA– DC (CD8α DC) found to present antigen as described above, the CD45RA+ plasmacytoid DC previously shown to respond to a variety of different viruses including influenza (15, 19–21), and a mixture of the remaining DC that expressed neither the CD8α nor CD45RA markers. The latter were simply termed double-negative DC (DN DC). In these experiments, we used a pulmonary infection with a recombinant influenza virus (Flu.gB) expressing the immunodominant determinant from the HSV glycoprotein B (gB), which allowed presentation to be identified as the ability of purified DC to stimulate CSFE-labeled resting T cells from the HSV gB-specific gBT-I TCR transgenic animal (9). Fig. 2A shows that on day 3 after infection gB-specific T cell stimulatory activity was found in both the CD8α DC subset and in the DN DC mixture with little or no activity residing with plasmacytoid DC. Similar patterns of DC subset presentation of Flu.gB were seen on days 2, 3, 5, 7, and 9 (data not shown). Activation of naïve T cells was not detected on day 1 after infection. Because previous experiments involving cutaneous infection with HSV showed all presentation resided solely with the CD8α DC, we wanted to determine whether the recruitment of other DC seen here reflected the use of a different virus (influenza versus HSV) or use of a different route of infection (lung versus skin). To this end, we infected C57BL/6 mice i.n. with HSV and determined the pattern of subset presentation of the gB determinant (Fig. 2B). As with Flu.gB, presentation was found in both the CD8α DC and DN DC, arguing that this pattern of DC involvement was attributable to the route of infection and not the particular virus under investigation.
Fig. 2.
Two subsets of DC prime naïve CD8+ T cells after i.n. virus infection. Three days after i.n. infection with either recombinant influenza A virus Flu.gB (A) or HSV (B), DC were enriched from the mediastinal LNs, stained for CD8α and CD45RA expression, and sorted by fluorescence-activated cell sorter (FACS) into CD8α DC, DN DC, or plasmacytoid DC before culturing with CFSE-labeled gBT-I CD8+ T cells. Proliferation was analyzed at 60 h of culture. The percentage of proliferating cells for each culture is indicated in the upper left corner of each histogram. Shown are representative experiments from at least three separate experiments.
Because the DN DC represented a mixture of cells, they were further subfractionated on the basis of expression of CD11b, F4/80, CD24, or CD205 (Fig. 3A). This division revealed that their presenting capacity resided with cells that were CD11b–, F4/80+, CD24hi, and CD205+. Expression of F4/80 on DC was significantly lower than for macrophages (data not shown). To determine whether two of these markers colocalize to the same subset of DC, the DN DC were costained for CD11b and F4/80 and then sorted into three groups: the CD11b–F4/80+, CD11b+F4/80+, and the F4/80–. This sorting showed that virtually all presentation by DN DC colocalized with a subset that was CD11b–F4/80+ (Fig. 3B). This phenotype is distinct from that of the six subsets previously identified in the spleen and LNs (13–18) and for simplicity will be referred to here as CD8α–CD11b– DC.
Fig. 3.
Phenotypical analysis of the DN DC presenting viral antigen after i.n. infection. (A) DC from C57BL/6 mice infected 3 d earlier with Flu.gB (10 mice per group) were enriched as described in Materials and Methods. Enriched DC were stained for CD11c, CD8α, and CD45RA in addition to one of the following: CD24, F4/80, CD11b, or CD205. Cells staining for CD8α and CD45RA were excluded, and remaining CD11c+ cells were sorted according to their level of expression of each additional marker (expression levels are indicated on flow cytometry histograms). Sorted DC were cultured with 5 × 104 CFSE-labeled gBT-I cells, and proliferation was assessed by loss of CFSE staining. (B) Mice were infected with either Flu.gB (Upper) or HSV (Lower), and DC populations were enriched as above but were additionally depleted of CD8+ and CD45RA+ cells and then stained for CD11c, CD11b, and F4/80 expression before sorting for CD11b–F4/80+, CD11b+F4/80+, and F4/80– cells. CFSE-labeled gBT-I CD8+ T cells (5 × 104) were cultured with these DC subsets, and proliferation was analyzed at 60 h of culture. The percentage of proliferating cells for each culture is indicated in each histogram. Data are representative of three separate experiments. (C) Analysis of expression of F4/80 and CD11b on purified DN DC subsets from various LNs after depletion of CD8α+ and CD45RA+ cells. Profiles are gated on CD11c+PI– cells.
Given that DN DC were not found to present antigen after skin infection (3), we sought to determine whether the CD11b–F4/80+ subset was missing from skin-draining LNs. Fig. 3C shows that this subset of DC indeed was absent from these LNs. When we examined LNs draining other internal organs, such as the liver and kidney, it became clear that the CD11b–F4/80+ population, missing in skin-draining LNs, was present in a range of different LNs (Fig. 3B). Whether this population is identical to that within mediastinal LNs presenting the class I-restricted virus antigen remains to be shown.
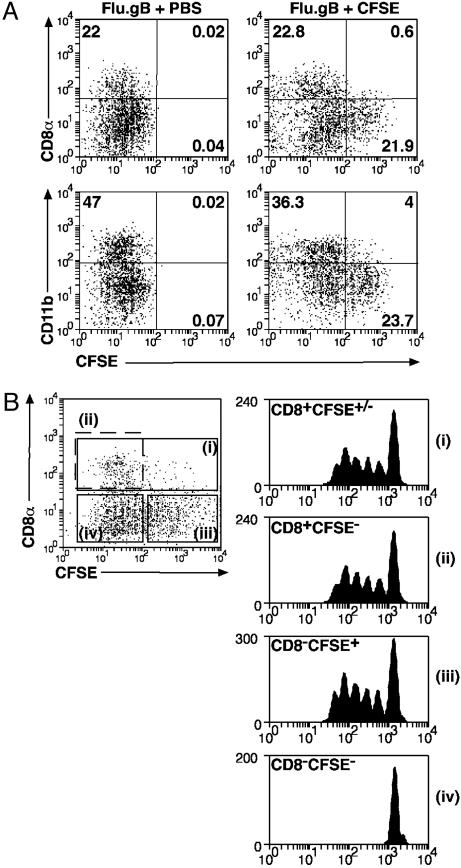
Of Those DC Presenting Viral Antigens, Only CD8α–CD11b– DC Appear to Originate Within the Airways. We have shown previously that CD8α DC are involved in the response to cutaneous infection with HSV but were unable to ascertain whether they trafficked from the site of infection or were LN-resident. To assign the origin of the different subsets, animals were instilled i.n. with a CFSE solution 6 h before infection with Flu.gB, and then 3 d later the DC in draining mediastinal LNs were examined for the presence of label. DC labeled with CFSE were deemed to have originated within the airways (22, 23). Subset analysis of the migrating population showed that very few of the CD8α DC were airway-derived because almost all were CSFE– (Fig. 4A). In contrast, approximately half of the CD8α–CD11b– DC stained with this dye. Therefore, of the two DC subsets shown to present class I-restricted virus antigen, only cells within the CD8α–CD11b– subset appeared to originate within the lung. This finding also excludes the possibility that migratory CD8α– DC transformed into the CD8α+ DC. As reported by Legge and Braciale (23), we found that administration of CpG inhibited the appearance of CFSE+CD8α–CD11b– DC in the mediastinal LN (data not shown). This finding supports the view that the vast majority of these DC originate in the lung and do not acquire staining by means of free CFSE or CFSE-labeled molecules in lymph.
Fig. 4.
Trafficking and antigen presentation by DC subsets after influenza infection. (A) Mice were inoculated i.n. with CFSE or carrier 6 h before i.n. infection with Flu.gB. Three days later, mediastinal LNs were pooled from four mice per group and digested with collagenase/DNase. Cell suspensions were enriched for conventional DC by labeling cells with antibodies anti-CD3 (KT3), anti-Thy1 (T24/31.7), anti-B220 (RA3–6B2), anti-GR-1 (RB6–8C5), and anti-erythrocyte (TER-119) followed by depletion with anti-rat Ig-coupled magnetic beads. DC preparations in Lower also were depleted of CD8α+ cells. Cells were stained with anti-CD11c-PE and anti-CD8α-APC (Upper) or anti-CD11c-PE and anti-CD11b-bio/SA-APC (Lower), and CD11c+ cells were examined for CFSE expression. CFSE+ DC represented 2–4% of CD11c+ cells in naïve mice. (B) DC were prepared from CFSE-treated mice as in A and stained with anti-CD11c-PE and CD8α-APC (Left). These cells then were sorted into four groups: (i) CD8α+ DC, (ii) CD8α+CFSE– DC, (iii) CD8α–CFSE+ DC, and (iv) CD8α–CFSE– DC; and 12,500 DC of each population were cultured with 5 × 104 CFSE-labeled gBT-I CD8+ T cells. Proliferation was analyzed at 60 h of culture. Data are representative of two experiments.
Because not all CD8α–CD11b– DC were labeled with dye, we wanted to determine whether presentation was confined to the migrating DC pool or whether nonmigrating DC apart from the CD8α DC subset also were involved in class I-restricted presentation. To this end, we isolated DC from draining mediastinal LNs after CFSE instillation and influenza infection and determined the pattern of presentation based on CD8α expression and labeling with dye. The results in Fig. 4B argue that, apart from the CD8α DC, all other presentation is confined to the CFSE+ DC population. This finding suggests that although the class I-presenting CD8α DC do not originate within the lung, all other presentation is confined to DC directly emigrating from infected tissues.
The observation that CD8α DC do not traffic from the lung but are able to present viral antigens after lung infections raises the question of where these DC obtain their antigen. It is extremely unlikely that viral antigens were acquired during cell purification as (i) no infectious virus could be detected at this day 3 time point (Table 1) and (ii) exogenous addition of virus to DC led to presentation by all subsets including the plasmacytoid DC (ref. 2 and data not shown). Another possibility is that virus drains directly from the lung to the LN and infects CD8α DC. This possibility again is highly unlikely because for Flu.gB infection, no infectious virus could be detected on days 1, 2, or 3 after infection, and only sporadic virus could be detected for HSV infection (Table 1). Furthermore, this would require selective infection of the CD8α DC within the LN, because several other DC subsets resident within this site failed to present viral antigens. This observation leaves open the most likely explanation that CD8α DC obtain their antigen from the trafficking CD8α–CD11b– DC, although we have not strictly excluded the possibility that viral debris flowing to the LN through lymphatic vessels could act as a source of antigen.
Table 1. Virus isolated from lung and mediastinal LN after i.n. infection with Flu.gB or HSV-1.
| Viral titer (pfu per organ)
|
|||||||
|---|---|---|---|---|---|---|---|
| Flu.gB days after infection
|
HSV-1 days after infection
|
||||||
| Tissue | Mouse | 1 | 2 | 3 | 1 | 2 | 3 |
| Lung | 1 | 7.6 × 103 | 15.6 × 104 | 14.2 × 103 | 12.2 × 103 | 40.6 × 104 | 7.6 × 103 |
| 2 | 4.8 × 103 | 9.4 × 103 | 28 × 104 | 3.3 × 103 | 38 × 104 | 2.7 × 103 | |
| 3 | 8.2 × 103 | 22.6 × 103 | 23.6 × 104 | 40 × 105 | 37.6 × 104 | 23 × 104 | |
| 4 | 4.3 × 105 | nt | nt | nt | |||
| 5 | 2.35 × 105 | nt | nt | nt | |||
| LN | 1 | 0 | 0 | 0 | 17.8 | 50 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 4 | nt | 16 | 0 | 0 | |||
| 5 | nt | 0 | 0 | 0 | |||
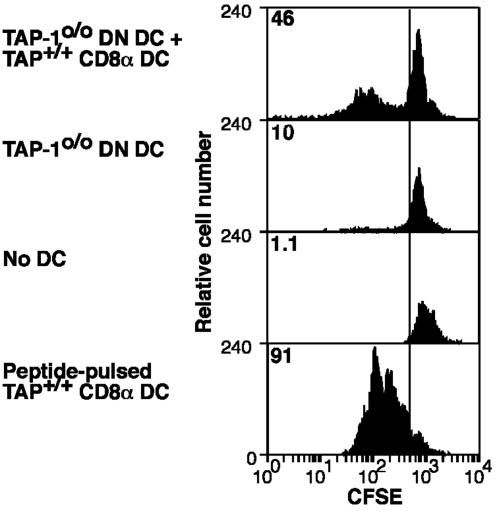
To determine whether CD8α–CD11b– DC could act as a source of antigen for CD8α DC, TAP-10/0 mice were infected i.n. with Flu.gB, and then their CD8α–CD11b– DC were isolated from the mediastinal LN on day 3. These DC alone, or mixed with CD8α DC from the mediastinal LNs of normal uninfected C57BL/6 mice, were examined for the ability to stimulate gBT-I cells (Fig. 5). As expected, TAP-10/0 CD8α–CD11b– DC from infected mice, on their own, could not present viral antigens (because of their TAP-1 deficiency). However, addition of normal CD8α DC allowed stimulation of naïve gB-specific T cells, implying that CD8α–CD11b– DC could act as an antigen source for the CD8α DC.
Fig. 5.
CD8α–CD11b– DC can act as a source of antigen for CD8α DC. Three days after i.n. infection of TAP-10/0 mice with the recombinant influenza A virus Flu.gB, DC were enriched from the mediastinal LNs (CD45RA+ cells were depleted), stained for CD11c, CD11b, and CD8α expression, and sorted by fluorescence-activated cell sorter (FACS) for CD8α–CD11b– (DN) DC. These antigen-bearing DN DC then were cocultured with CD8α+ DC purified from mediastinal LN of naïve C57BL/6 mice at a 10:1 ratio together with CFSE-labeled gBT-I CD8+ T cells (first histogram). Control cultures included TAP-10/0 DN DC from infected mice together with gBT-I CD8+ T cells (second histogram), gBT-I CD8+ T cells alone (third histogram), and gBT-I cells with splenic CD8α DC pulsed with 0.001 μM gB peptide (fourth histogram). Proliferation of gBT-I T cells was analyzed at 60 h of culture. The percentage of proliferating cells for each culture is indicated in the upper left corner of each histogram. Shown is a representative experiment from three separate experiments.
Discussion
By ex vivo isolation of DC during viral infection, we have found that of the many DC subsets so far identified, only two appear to be involved in class I-restricted antigen presentation after lung infection. These are the CD8α DC and a previously uncharacterized CD8α–CD11b– subset that appears unique to LNs draining certain internal organs such as the lung. Interestingly, only the CD8α–CD11b– DC originates within the infected airways. This DC subset is unlikely to express CD8α in the lung, as there do not appear to be any lung-resident DC subsets that express this molecule (22, 24–26). The finding that migrating lung-derived DC are involved in presentation within the LNs is perhaps not surprising, because in nonviral systems the direct introduction of DC into the airways, or instillation with labeled protein, results in migration of antigen-bearing DC to the lung-draining LNs (22, 24–26). What is surprising is the finding that presentation can extend beyond the airway-derived DC population. Vermaelen et al. (22) used a system of intratracheal instillation of fluorescein-derivatized ovalbumin to distinguish labeled lung-origin DC from unlabeled nonairway-derived populations. Although these investigators also found some level of class II-restricted T cell stimulation within the LN-resident DC population, this activity was orders of magnitude less effective than that attributed to the migrating DC pool. In contrast, our antibody depletion study (Fig. 1) shows that the LN-resident CD8α DC appear to contribute substantially to class I-restricted presentation at its peak, 3 d after infection. Our observations on class I-restricted presentation complement recent findings that show migrating and LN-resident DC can play a role in class II-restricted presentation (27–29).
In terms of lung infection, although various investigations have attributed T cell priming solely to the migratory DC population (22–26, 30, 31), our study suggests that these results need to be interpreted with caution. Direct introduction of antigen-pulsed DC into the airways has been shown to effectively prime helper T cell responses to soluble protein (24–26), although as discussed above, even when free protein is introduced into the lung, presentation by endogenous DC is likely dominated by migratory cells. In contrast, class I-restricted CTL responses to virus appear more complex in origin as shown here, with two DC subsets potentially playing an active role in this event.
Finally, it is interesting to compare this study with our previous investigations on skin infection with HSV (3, 4). In that case, we found that only the CD8α DC were capable of presenting class I-restricted antigen for CTL priming. Given that we show here the antigen-bearing migratory CD8α–CD11b– DC appear absent from skin-draining LNs, it is no surprise that they were not seen to contribute to CTL priming after skin infection. However, it may be that other cells unique to that tissue substitute for the CD8α–CD11b– subset found in the lung. The obvious candidates are the migrating Langerhans cells within the skin, known to acquire and transport antigen from this site (32). It remains unclear why these skin-derived DC were never found capable of presenting HSV in the cutaneous form of infection (3). Conversely, it may be that direct presentation by some but not other migrating DC populations is subordinate to the apparently more ubiquitous class I presentation involving the CD8α DC population.
In conclusion, our study highlights the involvement of both migrating and nonmigrating DC populations in class I-restricted antigen presentation after lung infection with virus. It also raises the possibility that CTL priming represents a complex interplay between distinct DC subsets involved in the processes of antigen transport and presentation.
Acknowledgments
We thank David Vremec, the staff of The Walter and Eliza Hall Institute Flow Cytometry Facility, Katherine Jordan, and Jiang-Li Tan for technical assistance. This work was supported by grants from the National Health and Medical Research Council of Australia. G.T.B. is a Wellcome Trust Senior Overseas Fellow, and W.R.H. is a Howard Hughes Medical Institute International Fellow.
Abbreviations: DC, dendritic cells; LN, lymph node; HSV, herpes simplex virus; TCR, T cell antigen receptor; i.n., intranasal; pfu, plaque-forming units; CFSE, 5,6-carboxyfluorescein diacetate succinimidyl ester; NP, nucleoprotein; DN, double-negative; CTL, cytotoxic T lymphocyte.
References
- 1.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245–252. [DOI] [PubMed] [Google Scholar]
- 2.Belz, G. T., Smith, C. M., Eichner, D., Shortman, K., Karupiah, G. & Heath, W. R. (2004) J. Immunol. 172, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 3.Allan, R. S., Smith, C. M., Belz, G., van Lint, A. L., Wakim, L. M., Heath, W. R. & Carbone, F. R. (2003) Science 301, 1925–1928. [DOI] [PubMed] [Google Scholar]
- 4.Smith, C. M., Belz, G. T., Wilson, N. S., Villadangos, J. A., Shortman, K., Carbone, F. R. & Heath, W. R. (2003) J. Immunol. 170, 4437–4440. [DOI] [PubMed] [Google Scholar]
- 5.Zhao, X., Deak, E., Soderberg, K., Linehan, M., Spezzano, D., Zhu, J., Knipe, D. M. & Iwasaki, A. (2003) J. Exp. Med. 197, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Haan, J. M., Lehar, S. M. & Bevan, M. J. (2000) J. Exp. Med. 192, 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moron, G., Rueda, P., Casal, I. & Leclerc, C. (2002) J. Exp. Med. 195, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kaer, L., Ashton-Rickardt, P. G., Ploegh, H. L. & Tonegawa, S. (1992) Cell 71, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 9.Mueller, S. N., Heath, W. R., Carbone, F. R. & Jones, C. M. (2002) Immunol. Cell Biol. 80, 156–163. [DOI] [PubMed] [Google Scholar]
- 10.Blaney, J. E., Jr., Nobusawa, E., Brehm, M. A., Bonneau, R. H., Mylin, L. M., Fu, T. M., Kawaoka, Y. & Tevethia, S. S. (1998) J. Virol. 72, 9567–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belz, G. T., Behrens, G. M., Smith, C. M., Miller, J. F., Jones, C., Lejon, K., Fathman, C. G., Mueller, S. N., Shortman, K., Carbone, F. R. & Heath, W. R. (2002) J. Exp. Med. 196, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karttunen, J., Sanderson, S. & Shastri, N. (1992) Proc. Natl. Acad. Sci. USA 89, 6020–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vremec, D., Zorbas, M., Scollay, R., Saunders, D. J., Ardavin, C. F., Wu, L. & Shortman, K. (1992) J. Exp. Med. 176, 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henri, S., Vremec, D., Kamath, A., Waithman, J., Williams, S., Benoist, C., Burnham, K., Saeland, S., Handman, E. & Shortman, K. (2001) J. Immunol. 167, 741–748. [DOI] [PubMed] [Google Scholar]
- 15.O'Keeffe, M., Hochrein, H., Vremec, D., Caminschi, I., Miller, J. L., Anders, E. M., Wu, L., Lahoud, M. H., Henri, S., Scott, B., et al. (2002) J. Exp. Med. 196, 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shortman, K. & Liu, Y. J. (2002) Nat. Rev. Immunol. 2, 151–161. [DOI] [PubMed] [Google Scholar]
- 17.O'Keeffe, M., Hochrein, H., Vremec, D., Scott, B., Hertzog, P., Tatarczuch, L. & Shortman, K. (2003) Blood 101, 1453–1459. [DOI] [PubMed] [Google Scholar]
- 18.Vremec, D., Pooley, J., Hochrein, H., Wu, L. & Shortman, K. (2000) J. Immunol. 164, 2978–2986. [DOI] [PubMed] [Google Scholar]
- 19.Lund, J., Sato, A., Akira, S., Medzhitov, R. & Iwasaki, A. (2003) J. Exp. Med. 198, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asselin-Paturel, C., Boonstra, A., Dalod, M., Durand, I., Yessaad, N., Dezutter-Dambuyant, C., Vicari, A., O'Garra, A., Biron, C., Briere, F. & Trinchieri, G. (2001) Nat. Immunol. 2, 1144–1150. [DOI] [PubMed] [Google Scholar]
- 21.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P. & Reis e Sousa, C. (2003) Nature 424, 324–328. [DOI] [PubMed] [Google Scholar]
- 22.Vermaelen, K. Y., Carro-Muino, I., Lambrecht, B. N. & Pauwels, R. A. (2001) J. Exp. Med. 193, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legge, K. L. & Braciale, T. J. (2003) Immunity 18, 265–277. [DOI] [PubMed] [Google Scholar]
- 24.Havenith, C. E., Breedijk, A. J., Betjes, M. G., Calame, W., Beelen, R. H. & Hoefsmit, E. C. (1993) Am. J. Respir. Cell Mol. Biol. 8, 319–324. [DOI] [PubMed] [Google Scholar]
- 25.Havenith, C. E., Breedijk, A. J., Calame, W., Beelen, R. H. & Hoefsmit, E. C. (1993) Adv. Exp. Med. Biol. 329, 571–575. [DOI] [PubMed] [Google Scholar]
- 26.Lambrecht, B. N., Pauwels, R. A. & Fazekas De St Groth, B. (2000) J. Immunol. 164, 2937–2946. [DOI] [PubMed] [Google Scholar]
- 27.Manickasingham, S. & Reis e Sousa, C. (2000) J. Immunol. 165, 5027–5034. [DOI] [PubMed] [Google Scholar]
- 28.Inaba, K., Turley, S., Yamaide, F., Iyoda, T., Mahnke, K., Inaba, M., Pack, M., Subklewe, M., Sauter, B., Sheff, D., et al. (1998) J. Exp. Med. 188, 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itano, A., McSorley, S., Reinhardt, R., Ehst, B., Ingulli, E., Rudensky, A. & Jenkins, M. (2003) Immunity 19, 47–57. [DOI] [PubMed] [Google Scholar]
- 30.McWilliam, A. S., Nelson, D., Thomas, J. A. & Holt, P. G. (1994) J. Exp. Med. 179, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia, W., Pinto, C. E. & Kradin, R. L. (1995) J. Exp. Med. 181, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romani, N., Ratzinger, G., Pfaller, K., Salvenmoser, W., Stossel, H., Koch, F. & Stoitzner, P. (2001) Int. Rev. Cytol. 207, 237–270. [DOI] [PubMed] [Google Scholar]
- 33.Anders, E. M., Hartley, C. A., Reading, P. C. & Ezekowitz, R. A. (1994) J. Gen. Virol. 75, 615–622. [DOI] [PubMed] [Google Scholar]
- 34.Jones, C. M., Cose, S. C., Coles, R. M., Winterhalter, A. C., Brooks, A. G., Heath, W. R. & Carbone, F. R. (2000) J. Virol. 74, 2414–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]