Abstract
The green anole, Anolis carolinensis (ACA), is the model reptile for a vast array of biological disciplines. It was the first nonavian reptile to have its genome fully sequenced. During the genome project, the XX/XY system of sex chromosomes homologous to chicken chromosome 15 (GGA15) was revealed, and 106 X-linked genes were identified. We selected 38 genes located on eight scaffolds in ACA and having orthologs located on GGA15, then tested their linkage to ACA X chromosome by using comparative quantitative fluorescent real-time polymerase chain reaction applied to male and female genomic DNA. All tested genes appeared to be X-specific and not present on the Y chromosome. Assuming that all genes located on these scaffolds should be localized to the ACA X chromosome, we more than doubled the number of known X-linked genes in ACA, from 106 to 250. While demonstrating that the gene content of chromosome X in ACA and GGA15 is largely conserved, we nevertheless showed that numerous interchromosomal rearrangements had occurred since the splitting of the chicken and anole evolutionary lineages. The presence of many ACA X-specific genes localized to distinct contigs indicates that the ACA Y chromosome should be highly degenerated, having lost a large amount of its original gene content during evolution. The identification of novel genes linked to the X chromosome and absent on the Y chromosome in the model lizard species contributes to ongoing research as to the evolution of sex determination in reptiles and provides important information for future comparative and functional genomics.
Keywords: gene dosage, lizard, qPCR, sex chromosomes, sex determination, genetics of sex
The green anole, Anolis carolinensis (ACA), is a member of the highly diversified genus Anolis (family Dactyloidae), which traditionally serves as a model group for a vast spectrum of research, recently including, for example, genetics (Novick et al. 2009; Piskurek et al. 2009; Alföldi et al. 2011; Rovatsos et al. 2014a), cytogenetics (Castiglia et al. 2013; Gamble et al. 2014), physiology (Dalla Valle et al. 2010; Wu et al. 2010; Johnson et al. 2011; Murphy and Thompson 2011), behavior (Johnson and Wade 2010), developmental biology (Koshiba-Takeuchi et al. 2009; Eckalbar et al. 2012), and evolutionary ecology (Losos et al. 1998; Losos 2009; Kolbe et al. 2011). The green anole was the first nonavian reptile selected for whole-genome sequencing (Alföldi et al. 2011), and draft genomes of other reptiles followed shortly thereafter (Castoe et al. 2011; St John et al. 2012; Vonk et al. 2013; Wang et al. 2013). Public access to the annotated genome through the Ensembl (http://www.ensembl.org; Flicek et al. 2014) and National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) databases have promoted ACA to the status of a key model reptile species, attracted the attention of the global scientific community, and contributed to a recent increase in publications addressing comparative and functional genomics in squamate reptiles (Fujita et al. 2011; Miller et al. 2012; Bar-Yaacov et al. 2013; Eckalbar et al. 2013; Ezaz et al. 2013; Gautam et al. 2013).
The ACA karyotype consists of 2n = 36 chromosomes, with six pairs of macrochromosomes and 12 pairs of microchromosomes (Matthey 1931). Although pioneering studies had indicated that ACA possesses a genotypic sex determination (Viets et al. 1994), and because the sex chromosomes are homomorphic, these chromosomes had long remained unidentified. During the whole-genome sequencing, it was proven that ACA possesses the XX/XY system of sex chromosomes. The X chromosome (ACAX) was identified at that time using fluorescent in situ hybridization of 11 bacterial artificial chromosomes (BACs) containing loci from two contigs. These BACs hybridized to the p arms of two microchromosomes in females but only to the p arm of a single microchromosome in males (Alföldi et al. 2011), thus suggesting that these sequences are specific to the X and absent on the Y chromosome. The Y chromosome has not yet been identified, but it is assumed to be another microchromosome (Alföldi et al. 2011). The known X-linked region of ACA includes approximately 106 genes (National Center for Biotechnology Information) with orthologs linked to chromosome 15 of the chicken (Gallus gallus, GGA). Alföldi et al. (2011) speculated that additional X-linked genes might be present on unanchored scaffolds in the AnoCar 2.0 assembly.
Recent studies have demonstrated that the ACA X-linked region is X-linked not only among anoles (Gamble et al. 2014; Rovatsos et al. 2014a) but also across most phylogenetic lineages of iguanas (Pleurodonta; Rovatsos et al. 2014b). The origin of sex chromosomes in iguanas can in fact be traced back to the basal splitting of this group that occurred during the Cretaceous period, and their age can therefore be comparable with the age of sex chromosomes in birds or viviparous mammals. Taking into account that female heterogamety and environmental sex determination have been reported from dragon lizards and chameleons (Acrodonta), the closest outgroup of iguanas (Ezaz et al. 2005; Pokorná and Kratochvíl 2009; Young et al. 2013), it seems that these XX/XY sex chromosomes constitute a synapomorphy of iguanas.
In contrast to mammals and birds (Ferguson-Smith and Trifonov 2007; Griffin et al. 2007), our knowledge about ancestral karyotypes and rates of chromosomal evolution in the majority of the other lineages of amniotes is still limited. Only recent analyses based on gene mapping, chromosomal painting, and whole-genome sequencing have shown that the slow rate of interchromosomal rearrangements is likely to be characteristic for all sauropsids and not just for birds, which constitute their inner group (Matsuda et al. 2005; Srikulnath et al. 2009; Alföldi et al. 2011; Pokorná et al. 2011, 2012; Uno et al. 2012; Srikulnath et al. 2013; Young et al. 2013). All X-linked genes in ACA known before the current study have orthologs on chicken chromosome 15 (GGA15). We therefore can assume that the entire GGA15 could be highly homologous to ACAX. Nevertheless, although sauropsids possess a relatively low rate of interchromosomal rearrangement, it has been shown in birds that their intrachromosomal rearrangements occur rather frequently (Völker et al. 2010; Skinner and Griffin 2012; Lithgow et al. 2014).
In this study, we identified numerous novel X-linked genes in ACA using quantitative fluorescent real-time polymerase chain reaction (qPCR) to compare relative gene doses between male and female genomic DNA (Nguyen et al. 2013; Gamble et al. 2014; Gamble and Zarkower 2014; Rovatsos et al. 2014a,b). The putative X-linked genes were selected from eight unanchored scaffolds sharing homology to GGA15, as predicted by the Genomicus database (http://www.genomicus.biologie.ens.fr/genomicus-75.02/cgi-bin/search.pl; Louis et al. 2013). We tested whether all these scaffolds are X-linked in ACA and how frequent were intrachromosomal rearrangements of this chromosome during the independent evolutionary histories of chicken and green anole.
Materials and Methods
Total genomic DNA was isolated using the DNeasy Blood & Tissue kit (QIAGEN) from the blood of a male and a female of ACA. Primer pairs were designed on Primer3 software (Ye et al. 2012) according to ACA sequences from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank; Benson et al. 2013) for amplifying putative X-linked loci on scaffolds with homology to GGA15 and the known X-linked regions of ACA (Alföldi et al. 2011). Additional primer pairs were designed for autosomal genes localized to ACA chromosome 6, chromosome 5, and for the single-copy gene EF1a, which was used for normalization of the gene dosages in the qPCR analyses (Table 1 and Supporting Information, Table S1).
Table 1. Relative gene dosage ratios between sexes of Anolis carolinensis revealed by quantitative polymerase chain reaction.
| Gene Symbol | GenBank Gene ID | Topology in Anolis carolinensis | Relative gene dosage Between Sexes |
|---|---|---|---|
| EF1a | 100566578 | NW_003338888.1 | − |
| FBXW7 | 100554110 | Chromosome 5; NC_014780.1 | 1.08 |
| NHP2L1 | 100552266 | Chromosome 5; NC_014780.1 | 1.08 |
| ADARB2 | 100560912 | Chromosome 6; NC_014781.1 | 1.03 |
| ACAD10 | 100557969 | 0.53 | |
| CMKLR1 | 100559738 | 0.48 | |
| SNAP29 | 100554635 | NW_003338829.1 | 0.38 |
| SDF2L1 | 100562697 | 0.47 | |
| HIRA | 100556463 | 0.62 | |
| SEPT5 | 100563096 | 0.48 | |
| MLEC | 100557313 | 0.31 | |
| TBC1D10A | 100558296 | 0.38 | |
| CIT | 100553665 | 0.44 | |
| DTX1 | 100559683 | NW_003338885.1 | 0.53 |
| DDX54 | 100560281 | 0.49 | |
| IQCD | 100560479 | 0.51 | |
| PLBD2 | 100561071 | 0.51 | |
| LHX5 | 100555029 | 0.64 | |
| PUS1 | 100562702 | 0.48 | |
| EP400 | 100562901 | 0.52 | |
| FBRSL1 | 100563489 | 0.52 | |
| GOLGA3 | 100563881 | NW_003338911.1 | 0.46 |
| ZDHHC8 | 100561780 | 0.48 | |
| TRMT2A | 100564671 | 0.53 | |
| DGCR8 | 100561976 | 0.48 | |
| GAS2L1 | 100567612 | 0.32 | |
| SMTN | 100553012 | NW_003338964.1 | 0.33 |
| SLC7A4 | 100554971 | 0.47 | |
| B3GNT4 | 100566391 | 0.45 | |
| CLIP1 | 100566771 | NW_003338970.1 | 0.63 |
| KNTC1 | 100567355 | 0.52 | |
| KDM2B | 100557128 | 0.61 | |
| ORAI1 | 100556544 | 0.53 | |
| WDR66 | 100557915 | NW_003339097.1 | 0.50 |
| LRRC43 | 100558501 | 0.54 | |
| MLXIP | 100558305 | 0.54 | |
| FICD | 100557652 | 0.38 | |
| SART3 | 100557456 | NW_003339461.1 | 0.53 |
| TMEM119 | 100557060 | 0.54 | |
| BCR | 100554393 | 0.44 | |
| SPECC1L | 100554592 | NW_003339495.1 | 0.32 |
| ADORA2A | 100554785 | 0.37 |
Gene names and chromosomal position data follow GenBank database (http://www.ncbi.nlm.nih.gov/genbank).
The qPCR was carried out in a LightCycler II 480 (Roche Diagnostics). All samples were run in triplicates. A 15-μL reaction was performed, containing 2 ng of genomic DNA, 7.5 μL of SYBR Premix Ex Taq II (Takara), and 0.3 mM of each primer. The cycling conditions were 95° for 3 min, followed by 44 amplification cycles of 95° for 15 sec, 56° for 30 sec, 72° for 30 sec, and ending with a melting curve analysis to monitor for potential nonspecific products. The quantification cycle values (crossing point, Cp) were calculated with LightCycler 480 software (v. 1.5.0) according to the second derivative maximum algorithm.
The gene dosage of each studied gene was calculated from Cp values and subsequently normalized to the gene dosage of the single copy gene EF1a based on the equation: R = [2Cp gene/2Cp EF1a]−1 (Cawthon 2002). Finally, the relative gene dosage ratio (r) between sexes was calculated for each gene as r = Rmale/Rfemale. Since ACA has the XX/XY sex determination system, we expected a relative gene dosage ratio (r) of 0.5 for X-linked genes missing on the Y chromosome and 1.0 for autosomal or pseudoautosomal genes.
Results
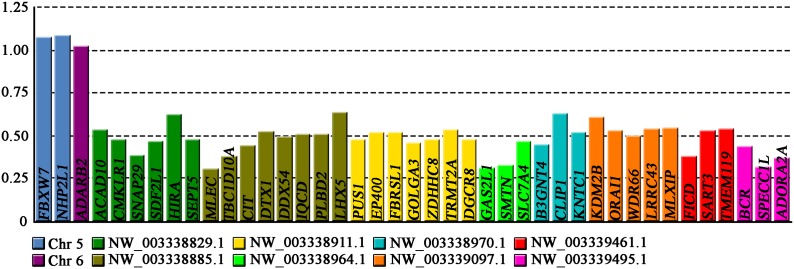
The relative gene dosage ratios between males and females were estimated by qPCR for two genes from chromosome 5 (contig No. NC_014780), one gene from chromosome 6 (NC_014781), six genes from the known X-linked region of ACA (NW_003338829), and 32 putative X-linked genes assigned to seven unanchored scaffolds (see Figure 1, Table 1, Table 2, and Table S1). In all cases, the values from the gene EF1a were used for normalization.
Figure 1.
Male-to-female relative gene dosage ratios for genes tested by quantitative polymerase chain reaction in A. carolinensis. Value 1.0 is expected for autosomal or pseudoautosomal genes, while value 0.5 is consistent with X-specific position.
Table 2. List of contigs from the ACA genome project shown to be X-linked.
| ACA X-Linked Contigs | Contig Size, bp | Number of Genes | Studied Genes by qPCR | Percentage of Studied Genes per Contig |
|---|---|---|---|---|
| NC_014783 | 3 271 537 | 58 | 5a,b | 9 |
| NW_003338829 | 1 779 868 | 48 | 9a,b,c | 19 |
| NW_003338885 | 1 258 094 | 45 | 8 | 18 |
| NW_003338911 | 1 083 274 | 17 | 7 | 41 |
| NW_003338964 | 831 895 | 37 | 4a,b | 11 |
| NW_003338970 | 834 740 | 19 | 6a,b | 32 |
| NW_003339097 | 526 944 | 11 | 5 | 45 |
| NW_003339461 | 147 151 | 5 | 3 | 60 |
| NW_003339495 | 117 443 | 10 | 3 | 30 |
Presented are the contig size, gene content, and number of genes tested by qPCR per contig. ACA, Anolis carolinensis; qPCR, quantitative polymerase chain reaction.
Data from Rovatsos et al. 2004a.
Data from Rovatsos et al. 2014b.
Data from Gamble et al. 2014.
Our qPCR results confirmed the autosomal position of the genes located in chromosomes 5 and 6 of ACA. As expected for autosomal or pseudoautosomal genes, their relative gene dosage ratios were very close to, and did not differ significantly from, the expected value of 1.0 (t-test, P = 0.08). In addition, the six genes from the known X-linked region of ACA had male to female gene dosage ratios ranging from 0.38 to 0.62 (mean 0.50), which did not differ significantly from the expected value for X-linkage of 0.5 (t-test, P = 0.90). The 32 putative X-linked genes demonstrated ratios varying from 0.31 to 0.64 (mean 0.48), which also did not vary significantly from the expected 0.5 (t-test, P = 0.16). Thus, this leads to the conclusion that these genes are localized to the X chromosome of ACA and are absent from Y. We detected no autosomal or pseudoautosomal genes.
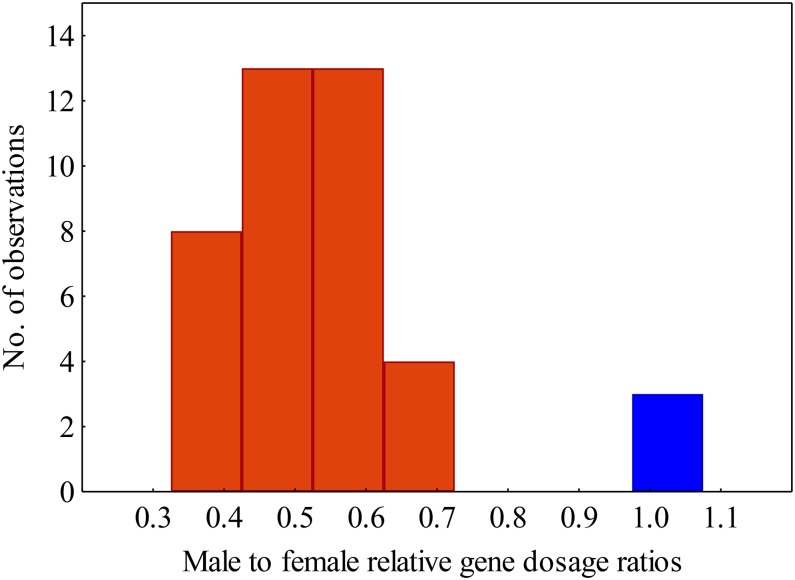
Our qPCR approach identified X-specific genes robustly, since the relative gene dosage ratios for the genes located on autosomes (chromosomes 5 and 6) differ significantly from those of the X-linked genes (analysis of variance, P = 0.004). The two sets are thus clearly distinguishable, without overlap or intermediate values (Figure 2).
Figure 2.
Distribution of male-to-female relative gene dosage ratios in the autosomal (blue) and X-specific (red) genes.
Discussion
The first X-linked genes in ACA had been discovered during the whole-genome sequencing project (Alföldi et al. 2011) by in situ hybridization of 11 BACs containing DNA fragments from two contigs (NC_014783 and NW_003338829). Those two contigs include 58 and 48 genes, respectively. Subsequent studies had tested five genes (TMEM132D, CCDC92, ATP2A2, PEBP1, and MED13L) from the NC_014783 and three genes (PPM1F, PI4KA, and CLTCL1) from the NW_003338829 contig (Gamble et al. 2014; Rovatsos et al. 2014a,b) by qPCR for relative gene dosage between sexes, thereby verifying their X-linkage in ACA. Four more X-specific genes (GAL3ST1, ZCCHC8, CUX2, and SH2B3) from the scaffolds NW_003338964 and NW_003338970 were discovered by qPCR (Rovatsos et al. 2014b). These genes were chosen because they have orthologs linked to GGA15 as the previously known ACAX genes. This work provided the first direct evidence that additional X-linked genes exist on unanchored scaffolds. In the present study, we verified that these two unanchored scaffolds indeed contain X-specific genes by testing six additional genes located within them using qPCR (Table 1). Furthermore, we demonstrate that an additional 26 genes located on five unanchored scaffolds are also part of the ACA X chromosome (Table 1). Using qPCR, we tested 11–60% of the gene content per scaffold (Table 2). Assuming that all genes included on those scaffolds are localized to the ACA X chromosome, we can increase the number of known ACA X-linked genes from 106 (Alföldi et al. 2011; Gamble et al. 2014; Gamble and Zarkower 2014; Rovatsos et al. 2014a,b) to 250. According to the Galgal 4.0 assembly, the GGA15 chromosome has a size of 12.66 Mbp with approximately 430 genes, which suggests that ACAX may contain more genes than the 250 identified here.
Despite intensive study of the ACA sex chromosomes (Alföldi et al. 2011; Gamble et al. 2014; Rovatsos et al. 2014a,b) only limited data about the gene content of the Y chromosome is available. Gamble and Zarkower (2014) identified a partial sequence from the Y-specific gene RTDR1Y using restriction site-associated DNA sequencing. In addition, the identification of many ACA X-specific genes localized to distinct contigs (Table 1 and Table 2) indicates that the ACA Y-chromosome is highly degenerated and that it had lost much of its original content during evolution.
In the current study, we detected no gene with an ortholog at GGA15 having a relative gene dosage ratio between male and female consistent with an autosomal or pseudoautosomal position. Using a similar qPCR approach, Gamble and Zarkower (2014) had tested two ACA genes (RTDR1 and GNA2) located in a short contig composed from only three genes for relative gene dosage between sexes. RTDR1 displayed relative gene dosage ratios between male and female consistent with an X-specific position. GNA2 yielded equal ratios between sexes, thus suggesting that this gene has a gametolog on the Y chromosome (Gamble and Zarkower 2014). Data from the Genomicus database (http://www.genomicus.biologie.ens.fr/genomicus-75.02/cgi-bin/search.pl; Louis et al. 2013) indicate, however, that some genes from the GGA15 chromosome are localized to autosomes in ACA (e.g., ACACB on ACA chromosome 1 or SFI1 on ACA chromosome 3). This probably is due to translocations that occurred during the 250 million years of divergence between ACA and GGA. The test for putative pseudoautosomal position of GNA2 requires further experimental work.
All studied genes were X-specific and not present on the Y chromosome in ACA. We did not detect any evidence for autosomal or pseudoautosomal position of the genes with orthologs linked to GGA15. It is therefore possible that the pseudoautosomal region in ACA is small or absent. Alternatively, a pseudoautosomal region in ACA could be homologous to a chromosome other than GGA15 and, as such, it could not be determined using our approach. These hypotheses should be tested by, for instance, observing the behavior of sex chromosomes during male meiosis.
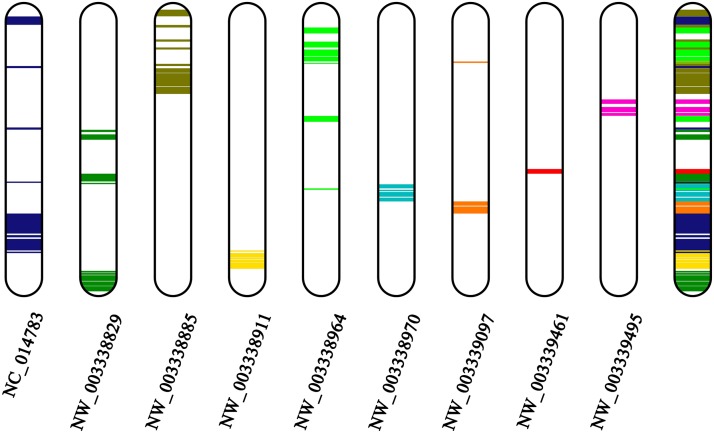
Comparison of the topology between the X-linked ACA contigs and GGA 15 (Figure 3), as illustrated by the Genomicus database (http://www.genomicus.biologie.ens.fr/genomicus-75.02/cgi-bin/search.pl; Louis et al. 2013), shows that despite high conservation of the chromosome, several intrachromosomal rearrangements, such as inversions, probably occurred at this chromosome after the divergence of ACA and GGA from their common ancestor. Similarly, no interchromosomal but numerous intrachromosomal rearrangements have been documented in the microchromosomes of chicken, turkey, and zebra finch (Lithgow et al. 2014). Several pericentromeric inversions have been revealed in the chromosomal pairs 1–4 of ACA by in situ hybridization with BACs, but their functional importance remains unclear (Alföldi et al. 2011). We should keep in mind, however, a recent counterexample based on fluorescent in situ hybridization mapping of 11 markers whereby a high level of synteny was revealed between ACA chromosome 6 and its homologous chromosome in snakes (Z in colubroid snakes) without any detectable large-scale chromosomal rearrangements (Vicoso et al. 2013).
Figure 3.
Homology between nine A. carolinensis contigs and the Gallus gallus chromosome 15. The homology was predicted by the Genomicus database (http://www.genomicus.biologie.ens.fr/genomicus-75.02/cgi-bin/search.pl).
Reptiles (excluding their inner avian group) usually are considered a group with rapid turnover of sex-determining mechanisms (Sarre et al. 2004; Organ and Janes 2008). In general, they do indeed exhibit large variability in sex-determining systems (Valenzuela and Lance 2004; Pokorná and Kratochvíl 2009; Gamble 2010). It has been suggested that poikilotherms possess more frequent turnovers of sex chromosomes than do homoiotherms, whose effective thermoregulation can prevent the emergence of sex reversals induced by environmental temperature. Despite their species richness, wide distribution, and enormous ecological and morphologic variability, however, iguanas possess great stability of sex chromosomes (Rovatsos et al. 2014b) that is comparable with the well-documented cases of birds (ca 120 Mya according to Mank and Ellegren 2007) and therian mammals (ca 166 Mya based on Veyrunes et al. 2008). Although such stability of sex chromosomes has been reported for several poikilothermic amniotes, such as within the turtle family Trionychidae (ca. 95 Mya) (Badenhorst et al. 2013) or within colubroid snakes (Matsubara et al. 2006; Vicoso et al. 2013), where the split between the studied families Viperidae and Colubridae occurred, ca. 36 Mya according to Wiens et al. (2006), but even up to 75 Mya according to Wüster et al. (2008), to the best of our knowledge, iguanian lizards possess the oldest sex chromosomes currently known among amniotic poikilothermic vertebrates. By reporting the large list of X-specific genes in ACA, our recent contribution enables future comparative study on the evolution of sex chromosomes in iguanas. For example, applying the qPCR approach within lineages derived from basal splitting of iguanas may reveal whether highly differentiated X and Y chromosomes described in ACA evolved via a stepwise series of suppressions of recombination along the iguana Y chromosome, i.e., whether some “evolutionary strata” found in mammals and birds (Lahn and Page 1999; Handley et al. 2004; Nam and Ellegren 2008) could be observed also in iguanas.
Supplementary Material
Acknowledgments
We express our gratitude to František Marec and Romana Stopková for their valuable guidance on qPCR and to Jan Červenka for animal handling and care. Deborah Charlesworth and an anonymous referee provided valuable comments. This project was funded by the Czech Science Foundation (GAČR 506/10/0718) and the Grant Agency of Charles University (GAUK 591712). This paper represents part 11 of our series “Evolution of sex determining systems in lizards.”
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014084/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Alföldi J., Di Palma F., Grabherr M., Williams C., Kong L., et al. , 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477: 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst D., Stanyon R., Engstrom T., Valenzuela N., 2013. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 21: 137–147. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D., Bouskila A., Mishmar D., 2013. The first chameleon transcriptome: comparative genomic analysis of the OXPHOS system reveals loss of COX8 in Iguanian lizards. Genome Biol. Evol. 5: 1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D. J., et al. , 2013. GenBank. Nucleic Acids Res. 41: D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia R., Flores-Villela O., Bezerra A. M., Muñoz A., Gornung E., 2013. Pattern of chromosomal changes in “beta” Anolis (Norops group) (Squamata: Polychrotidae) depicted by an ancestral state analysis. Zool. Stud. 52: 60. [Google Scholar]
- Castoe T. A., de Koning A. P. J., Hall K. T., Yokoyama K. D., Gu W., et al. , 2011. Sequencing the genome of the Burmese python (Python molurus bivittatus) as a model for studying extreme adaptations in snakes. Genome Biol. 12: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. M., 2002. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Valle L., Nardi A., Bonazza G., Zuccal C., Emera D., et al. , 2010. Forty keratin-associated β-proteins (β-keratins) form the hard layers of scales, claws, and adhesive pads in the green anole lizard, Anolis carolinensis. J. Exp. Zool. B Mol. Dev. Evol. 314: 11–32. [DOI] [PubMed] [Google Scholar]
- Eckalbar W. L., Lasku E., Infante C. R., Elsey R. M., Markov G. J., et al. , 2012. Somitogenesis in the anole lizard and alligator reveals evolutionary convergence and divergence in the amniote segmentation clock. Dev. Biol. 363: 308–319. [DOI] [PubMed] [Google Scholar]
- Eckalbar W. L., Hutchins E. D., Markov G. J., Allen A. N., Corneveaux J. J., et al. , 2013. Genome reannotation of the lizard Anolis carolinensis based on 14 adult and embryonic deep transcriptomes. BMC Genomics 14: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T., Quinn A. E., Miura I., Sarre S. D., Georges A., et al. , 2005. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 13: 763–776. [DOI] [PubMed] [Google Scholar]
- Ezaz T., Azad B., O’Meally D., Young M. J., Matsubara K., et al. , 2013. Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genomics 14: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith M. A., Trifonov V., 2007. Mammalian karyotype evolution. Nat. Rev. Genet. 8: 950–962. [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Billis K., et al. , 2014. Ensembl 2014. Nucleic Acids Res. 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. K., Edwards S. V., Ponting C. P., 2011. The Anolis lizard genome: an amniote genome without isochores. Genome Biol. Evol. 3: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T., 2010. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex Dev. 4: 88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T., Zarkower D., 2014. Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol. Ecol. Resour. 14: 902–913. [DOI] [PubMed] [Google Scholar]
- Gamble T., Geneva A. J., Glor R. E., Zarkower D., 2014. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68: 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M., Mathur A., Khan M. A., Majumdar S. S., Rai U., 2013. Transcriptome analysis of spermatogenically regressed, recrudescent and active phase testis of seasonally breeding wall lizards Hemidactylus flaviviridis. PLoS One 8: e58276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. K., Robertson L. B. W., Tempest H. G., Skinner B. M., 2007. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 117: 64–77. [DOI] [PubMed] [Google Scholar]
- Handley L. L., Ceplitis H., Ellegren H., 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. A., Wade J., 2010. Behavioural display systems across nine Anolis lizard species: sexual dimorphisms in structure and function. Proc. Biol. Sci. 277: 1711–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. A., Cohen R. E., Vandecar J. R., Wade J., 2011. Relationships among reproductive morphology, behavior, and testosterone in a natural population of green anole lizards. Physiol. Behav. 104: 437–445. [DOI] [PubMed] [Google Scholar]
- Kolbe J. J., Revell L. J., Szekely B., Brodii E. D., III, Losos J. B., 2011. Convergent evolution of phenotypic integration and its alignment with morphological diversification in Caribbean Anolis ecomorphs. Evolution 65: 3608–3624. [DOI] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K., Mori A. D., Kaynak B. L., Cebra-Thomas J., Sukonnik T., et al. , 2009. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 461: 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Lithgow P. E., O’Connor R., Smith D., Fonseka G., Al Mutery A., et al. , 2014. Novel tools for characterising inter and intra chromosomal rearrangements in avian microchromosomes. Chromosome Res. 22: 85–97. [DOI] [PubMed] [Google Scholar]
- Losos J. B., 2009. Lizards in an Evolutionary Tree: Ecology and Adaptive Radiation of Anoles. University of California Press, Berkeley. [Google Scholar]
- Losos J. B., Jackman T. R., Larson A., de Quieroz K., Rodríguez-Schettino L., 1998. Contingency and determinism in replicated adaptive radiations of island lizards. Science 279: 2115–2118. [DOI] [PubMed] [Google Scholar]
- Louis A., Muffato M., Roest Crollius H., 2013. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41: D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., Ellegren H., 2007. Parallel divergence and degradation of the avian W sex chromosome. Trends Ecol. Evol. 22: 389–391. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Tarui H., Toriba M., Yamada K., Nishida-Umehara C., et al. , 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 103: 18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Nishida-Umehara C., Tarui H., Kuroiwa A., Yamada K., et al. , 2005. Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 13: 601–615. [DOI] [PubMed] [Google Scholar]
- Matthey R., 1931. Chromosomes de reptiles. Sauriens, ophidiens, chéloniens. L’évolution de la formule chromosomiale chez les sauriens. Rev. Suisse Zool. 38: 117–186. [Google Scholar]
- Miller H. C., Biggs P. J., Voelckel C., Nelson N. J., 2012. De novo sequence assembly and characterisation of a partial transcriptome for an evolutionarily distinct reptile, the tuatara (Sphenodon punctatus). BMC Genomics 13: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B. F., Thompson M. B., 2011. A review of the evolution of viviparity in squamate reptiles: the past, present and future role of molecular biology and genomics. J. Comp. Physiol. B 181: 575–594. [DOI] [PubMed] [Google Scholar]
- Nam K., Ellegren H., 2008. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180: 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P., Sýkorová M., Šíchová J., Kůta V., Dalíková M., et al. , 2013. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc. Natl. Acad. Sci. USA 110: 6931–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P. A., Basta H., Floumanhaft M., McClure M. A., Boissinot S., 2009. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol. Biol. Evol. 26: 1811–1822. [DOI] [PubMed] [Google Scholar]
- Organ C. L., Janes D. E., 2008. Evolution of sex chromosomes in Sauropsida. Integr. Comp. Biol. 48: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O., Nishihara H., Okada N., 2009. The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene 441: 111–118. [DOI] [PubMed] [Google Scholar]
- Pokorná M., Kratochvíl L., 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156: 168–183. [Google Scholar]
- Pokorná M., Giovannotti M., Kratochvíl L., Kasai F., Trifonov V. A., et al. , 2011. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma 120: 455–468. [DOI] [PubMed] [Google Scholar]
- Pokorná M., Giovannotti M., Kratochvíl L., Caputo V., Olmo E., et al. , 2012. Conservation of chromosomes syntenic with avian autosomes in squamate reptiles revealed by comparative chromosome painting. Chromosoma 121: 409–418. [DOI] [PubMed] [Google Scholar]
- Rovatsos M., Altmanová M., Pokorná M., Kratochvíl L., 2014a Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution 68: 2079–2085. [DOI] [PubMed] [Google Scholar]
- Rovatsos M., Pokorná M., Altmanová M., Kratochvíl L., 2014b Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10: 20131093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarre S. D., Georges A., Quinn A., 2004. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. BioEssays 26: 639–645. [DOI] [PubMed] [Google Scholar]
- Skinner B. M., Griffin D. K., 2012. Intrachromosomal rearrangements in avian genome evolution: evidence for regions prone to breakpoints. Heredity 108: 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K., Nishida C., Matsubara K., Uno Y., Thongpan A., et al. , 2009. Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res. 17: 975–986. [DOI] [PubMed] [Google Scholar]
- Srikulnath K., Uno Y., Nishida C., Matsuda Y., 2013. Karyotype evolution in monitor lizards: cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 21: 805–819. [DOI] [PubMed] [Google Scholar]
- St John J. A., Braun E. L., Isberg S. R., Miles L. G., Chong A. Y., et al. , 2012. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 13: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y., Nishida C., Tarui H., Ishishita S., Takagi C., et al. , 2012. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7: e53027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N., Lance V. A., 2004. Temperature Dependent Sex Determination in Vertebrates. Smithsonian Books, Washington, DC. [Google Scholar]
- Veyrunes F., Waters P. D., Miethke P., Rens W., McMillan D., et al. , 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Emerson J. J., Zektser Y., Mahajan S., Bachtrog D., 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11: e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viets B. E., Ewert M. A., Talent L. G., Nelson C. E., 1994. Sex-determining mechanisms in squamate reptiles. J. Exp. Zool. 270: 45–56. [Google Scholar]
- Völker M., Backström N., Skinner B. M., Langley E. J., Ellegren H., et al. , 2010. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 20: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk F. J., Casewell N. R., Henkel C. V., Heimberg A. M., Jansen H. J., et al. , 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 110: 20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Pascual-Anaya J., Zadissa A., Li W., Niimura Y., et al. , 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 45: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens J. J., Brandley M. C., Reeder T. W., 2006. Why does a trait evolve multiple times within a clade? Repeated evolution of snakelike body form in squamate reptiles. Evolution 60: 123–141. [PubMed] [Google Scholar]
- Wu Q., Wei Z., Yang Z., Wang T., Ren L., et al. , 2010. Phylogeny, genomic organization and expression of lambda and kappa immunoglobulin light chain genes in a reptile, Anolis carolinensis. Dev. Comp. Immunol. 34: 579–589. [DOI] [PubMed] [Google Scholar]
- Wüster W., Peppin L., Pook C. E., Walker D. E., 2008. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogenet. Evol. 49: 445–459. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., et al. , 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. J., O’Meally D., Sarre S. D., Georges A., Ezaz T., 2013. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 21: 361–374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.