Abstract
Influenza virus infection is responsible for hundreds of thousands of deaths annually. Current vaccination strategies and antiviral drugs provide limited protection; therefore, new strategies are needed. RNA interference is an effective means of suppressing virus replication in vitro. Here we demonstrate that treatment with small interfering RNAs (siRNAs) specific for highly conserved regions of the nucleoprotein or acidic polymerase inhibits influenza A virus replication in vivo. Delivery of these siRNAs significantly reduced lung virus titers in infected mice and protected animals from lethal challenge. This protection was specific and not mediated by an antiviral IFN response. Moreover, influenza-specific siRNA treatment was broadly effective and protected animals against lethal challenge with highly pathogenic avian influenza A viruses of the H5 and H7 subtypes. These results indicate that RNA interference is promising for control of influenza virus infection, as well as other viral infections.
Influenza virus infection is a major public health problem, causing millions of cases of severe illness and as many as 500,000 deaths each year worldwide (1). Although inactivated vaccines are 60–80% effective against matched influenza strains (2), vaccination coverage is a problem worldwide. Moreover, this strategy provides no protection against unexpected strains, outbreaks such as the H5 and H7 avian influenza outbreaks in Hong Kong in 1997 and The Netherlands and Southeast Asia in 2003–2004, or pandemics. Currently, antiviral drugs are the best defense against these outbreaks, but they too provide only partial protection (3). New therapies to treat ongoing influenza infection are urgently needed, as well as new vaccination strategies inducing broader immunity (1, 4, 5).
RNA interference (RNAi) is an emerging technology that specifically inhibits gene expression. Small interfering RNAs (siRNAs), mediators of RNAi, are short (21–26 nt), double-stranded RNA duplexes that inhibit gene expression by inducing sequence-specific degradation of homologous mRNA (6). Many studies have shown that siRNA can significantly suppress gene expression when delivered into mammalian cells in vitro (7, 8). These findings raised the possibility that RNAi could inhibit viral gene expression and protect cells from viral infection. Subsequently, a number of studies demonstrated inhibition of replication of RNA viruses in vitro by RNAi (9–11), including HIV (12, 13), polio virus (14), hepatitis C virus (15, 16), West Nile virus (17), and influenza virus (17, 18). Moreover, a number of groups demonstrated effective silencing of both transgene and endogenous gene expression in vivo (19–25). Here we extend these studies to an animal model of virus infection and disease. We show that administration of influenza-specific siRNAs can decrease lung virus titers and protect mice from lethal challenge with a variety of influenza A viruses, including potential pandemic subtypes H5 and H7. This inhibition of influenza virus replication is specific, requiring homology between the siRNAs and gene targets, and is not the result of IFN induction by double-stranded RNA.
Materials and Methods
Mice. Female, 4- to 6-week-old BALB/cAnNCR mice were purchased from Division of Cancer Treatment, National Cancer Institute, Frederick, MD. Mice were challenged between 7 and 9 weeks of age. The institutions' animal care and use committees approved all protocols for all animal experiments.
Viruses. Influenza virus strains used were A/Puerto Rico/8/34 (PR/8, H1N1) (5), A/Hong Kong/156/97 (HK/156, H5N1) (26), A/NL/219/03 (Netherlands/219, H7N7) (27, 28), and A/Hong Kong/1073/99 (HK/1073, H9N2) (29). Virus stocks were propagated in the allantoic cavity of embryonated hen eggs at 34°C for 48–72 h (PR/8) or 37°C for 24 h (other viruses). All experiments with H5H1, H7N7, and H9N2 viruses were conducted under BSL-3+ containment.
siRNAs. siRNAs were purchased from Dharmacon Research (Lafayette, CO) as dried, 2′-deprotected, desalted duplexes and resuspended in AccuGENE PBS (BioWhittaker). Sequences used were as follows: GFP-949 (siGFP), sense 5′-GGCUACGUCCAGGAGCGCAUU-3′, antisense 5′-UUCCGAUGCAGGUCCUCGCGU-3′; nucleoprotein (NP)-1496 (siNP), sense 5′-GGAUCUUAUUUCUUCGGAGdTdT-3′, antisense 5′-dTdTCCUAGAAUAAAGAAGCCUC-3′; and acidic polymerase (PA)-2087 (siPA), sense 5′-GCAAUUGAGGAGUGCCUGAdTdT-3′, antisense 5′-dTdTCGUUAACUCCUCACGGACU-3′ [as reported by Ge et al. (18)].
siRNA Delivery and Virus Infection in Vivo. On day –1, siGFP, siNP, siPA, or combined siNP and siPA were diluted to 50 μg/ml (25 μg/ml of each with siNP and siPA combined) in PBS. Mice received 1 ml of diluted siRNA (3.78 nmol) or PBS through hydrodynamic i.v. injection as described in ref. 19. Sixteen to 24 h later (day 0), siRNA/oligofectamine complexes were prepared: oligofectamine (Invitrogen) was diluted 1:1 in PBS and incubated for 10 min at room temperature (RT), and siRNAs were diluted to 1 mg/ml in PBS. Diluted oligofectamine and siRNA complexes (or PBS for vehicle controls) were combined in a 3:2 ratio and incubated for 20 min at RT. In all experiments except that detailed in Table 1, challenge viruses were diluted to a final volume of 10 μl per challenge dose in PBS. For each mouse, 10 μl of diluted virus was combined with 50 μl of oligofectamine/siRNA. Sixty micro-liters of virus/oligofectamine/siRNA (1.51 nmol siRNA) was administered intranasally (i.n.) under anesthesia with isoflurane (PR/8 virus; Table 1 and Fig. 1), CO2 (PR/8 virus; Table 2 and Fig. 2), or ketamine/xylazine (1.98 and 0.198 mg per mouse, respectively; HK/156, NL/219, and HK/1073 viruses). For the experiment detailed in Table 1, viruses were diluted to a final volume of 50 μl per challenge dose in PBS. In this experiment, mice were anaesthetized with ketamine/xylazine, infected with the 50 μl PR/8 challenge dose i.n., and given the 50-μl i.n. siRNA dose (1.51 nmol) 20 min later while still under anesthesia. Virus challenge doses were as follows: 5 × 102 tissue culture 50% infective dose (TCID50) of PR/8 virus, 10 LD50 of HK/156 virus, 10 LD50 of NL/219 virus, and 106 egg 50% infective dose (eID50) of HK/1073 virus. Mice were killed day 2 postchallenge for analysis of lung virus titers or monitored for body weight and mortality until all animals had succumbed to infection or were recovering by body weight. Lungs for virus titer were homogenized in 3 ml of Leibovitz medium (Biofluids, Rockville, MD) and clarified by centrifugation. In the experiments detailed in Table 2, lung homogenates were titrated for virus infectivity by eID50 assay. For these assays, lungs were homogenized in 1 ml of sterile PBS and clarified by centrifugation.
Table 1. Influenza A-specific siRNA treatment inhibits influenza A/H1N1 virus replication in vivo.
| Treatment* | n | Mean lung virus titer† | P value‡ | Fold reduction§ |
|---|---|---|---|---|
| siGFP | 5 | 5.6 ± 0.4 | — | — |
| siNP | 5 | 3.8 ± 0.1 | <0.0001 | 63 |
| siPA | 4 | 4.6 ± 0.1 | <0.01 | 10 |
| siNP+siPA | 5 | 3.8 ± 0.2 | <0.0001 | 63 |
—, not applicable.
BALB/c mice were treated as indicated and challenged with 5 × 102 TCID50 of PR/8. Two days later, animals were sacrificed and lungs were collected for virus titer.
Expressed as log10 TCID50/ml ± SEM.
One-way ANOVA statistical analysis on log-transformed data, followed with comparison with control (siGFP) by Dunnett's method.
Compared with the siGFP-treated group.
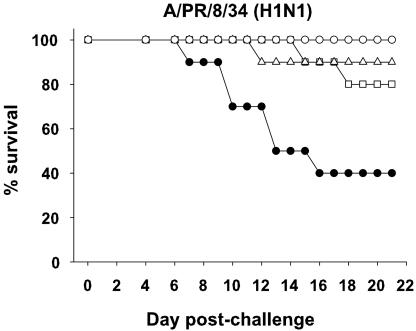
Fig. 1.
Influenza-specific siRNA treatment protects mice from lethal H1N1 virus challenge. BALB/c mice (10 per group) were treated with siGFP (filled circles), siNP (open squares), siPA (open triangles), or siNP+siPA (open circles), challenged with PR/8 virus, and monitored daily for mortality. Influenza siRNA groups all differ from siGFP controls (P < 0.05; log-rank).
Table 2. Influenza-specific siRNA treatment significantly decreases lung virus titers in mice challenged with H5, H7, and H9 avian influenza A viruses.
| Challenge virus* | Virus subtype | Treatment | Mean lung virus titer† | P value‡ | Fold reduction§ |
|---|---|---|---|---|---|
| PR/8 | H1N1 | siGFP | 6.0 ± 0.2 | — | — |
| siNP + siPA | 4.3 ± 0.6 | 0.0072 | 56 | ||
| HK/156 | H5N1 | siGFP | 7.6 ± 0.1 | — | — |
| siNP + siPA | 6.5 ± 0.4 | 0.0201 | 11 | ||
| NL/219 | H7N7 | siGFP | 7.7 ± 0.4 | — | — |
| siNP + siPA | 6.7 ± 0.2 | 0.0245 | 9 | ||
| HK/1073 | H9N2 | siGFP | 5.2 ± 0.2 | — | — |
| siNP + siPA | 3.9 ± 0.6 | 0.0022 | 21 |
—, not applicable.
BALB/c mice (n = 4) were treated and challenged as indicated. On day 2 postchallenge, animals were sacrificed and lungs were removed for virus titration by eID50.
Expressed as log10 eID50/ml ± SEM.
One-way ANOVA statistical analysis on log-transformed data, followed by comparison using Student's t test.
Compared with the siGFP-treated group.
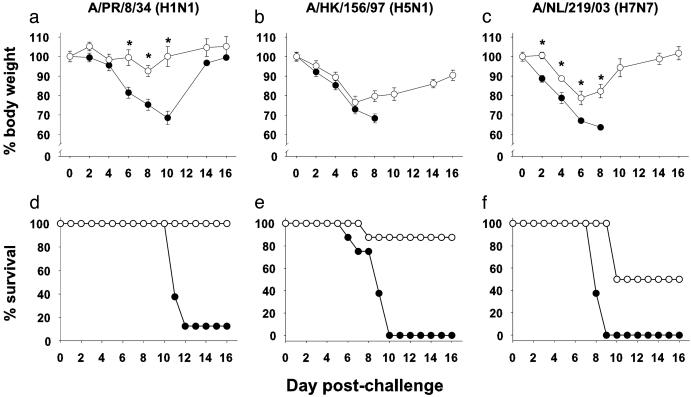
Fig. 2.
Influenza-specific siRNA treatment is broadly cross-reactive and protects mice against lethal challenge with highly pathogenic H5 and H7 avian influenza A viruses. BALB/c mice (eight per group) were treated with siGFP (filled circles) or siNP+siPA (open circles) and challenged with PR/8 virus (a and d), HK/156 virus (b and e), or NL/219 virus (c and f). The percent of initial body weight (a–c) and survival postchallenge (d–f) are shown. Error bars (a–c) depict the standard error of the mean. *, Groups differ for weight loss, P < 0.05 (Student's t test). The single surviving mouse from the PR/8-challenged, siGFP-treated group recovered fully. siNP+siPA groups differ from siGFP groups for survival [P < 0.002 by log-rank analysis for virus strains in d (PR/8), e (HK/156), and f (NL/219)].
Virus Quantitation. Madin–Darby canine kidney cells (MDCK) cells were cultured in OPTI-MEM I (Invitrogen). TCID50 assays were performed as described in ref. 30. Briefly, lung homogenates or tissue culture supernatants were assayed for virus infectivity on MDCK by endpoint dilution for cytopathic effect with a 10-fold dilution series. Titers are expressed as log10 TCID50/ml ± SEM. The detection limit of the assay is 1.5 log10 TCID50/ml. For the lung titer experiment detailed in Table 2, lung homogenates were titrated by eID50 assay as described in ref. 5. Briefly, lung homogenates were titrated in 10-day-old embryonated eggs in 10-fold steps from initial dilutions of 1:10, and positive eggs were identified by hemagglutination with allantoic fluid. Values are expressed as log10 eID50/ml ± SEM. The limit of virus detection is 1.2 log10 eID50/ml.
Statistical Analysis. Lung virus titers were compared by statistical analysis on log-transformed data, using one-way ANOVA followed by Dunnett's test for treatment versus control or Student's t test. Results for percent initial body weight were compared by using Student's t test. Comparison of survival was done by using log-rank test.
Results and Discussion
We first confirmed the results of Ge et al. (18), showing that pretreatment of MDCK cells with siRNAs specific for NP 1496–1514 (siNP) or PA 2087–2106 (siPA) (18) could inhibit influenza replication after infection in vitro. siNP-, siPA-, or siNP+siPA-pretreated MDCK cells were infected with A/PR/8 (PR/8) virus. A GFP-specific siRNA, siGFP (18), was used in all experiments to control for potential nonspecific siRNA effects. Virus titers were decreased for at least 48 h postinfection compared with untreated or control siRNA-treated cells (data not shown).
To assess whether RNAi could inhibit influenza virus replication in vivo, we used an established murine model of influenza infection. BALB/c mice were treated with influenza-specific or control siRNAs by using hydrodynamic i.v. delivery as described by Lewis et al. (19). Sixteen to 24 h later, mice were infected with PR/8 i.n. and also given a second dose of siRNA in a lipid carrier i.n. in the hope of improving effectiveness in the lungs. Two days postchallenge, lungs were removed and lung homogenates were assayed for virus. Virus titers were significantly reduced in the lungs of animals given siNP, siPA, or siNP+siPA compared with those given siGFP (Table 1). Lung virus titers in untreated, PBS plus delivery vehicle-treated, and siGFP-treated animals were identical (data not shown), showing that siGFP treatment did not affect virus replication. Additionally, the decreases in virus titer were not due to effects in the assay of influenza-specific siRNAs in the lung homogenates; lung samples from siRNA-treated, unchallenged animals did not inhibit detection of virus-containing samples in the TCID50 assay (data not shown).
Previous studies demonstrate that vaccines causing as little as 5- to 10-fold reduction in lung virus titers can protect against lethal influenza challenge (5, 30, 31). To determine whether the decreases in lung virus titers due to siRNA treatment were sufficient to protect animals from death, BALB/c mice were pretreated i.v. with siGFP, siNP, siPA, or siNP+siPA, followed 16–24 h later with lethal PR/8 challenge and a second i.n. dose of siRNA. By day 18 postchallenge (Fig. 1), 60% of animals given control siRNA had died, whereas mice given either siNP or siPA had significantly less mortality (20% and 10%, respectively). Strikingly, treatment with the combination of siNP+siPA resulted in 100% survival. In all cases, the protection provided by influenza-specific siRNA treatment was statistically significant.
Although a variety of studies testing virus-specific siRNAs in vitro found no induction of IFN-mediated antiviral responses (12, 14, 15, 18), it was recently reported by Sledz et al. (32) that siRNAs treatment could nonspecifically induce IFN-mediated innate immune responses. It was unlikely that IFN was mediating protection in our experiments, however, because animals treated with an identical amount of the control siGFP had significantly higher lung virus titers and significantly lower survival rates than animals treated with influenza-specific siRNAs (Table 1 and Fig. 1). Nonetheless, to verify that siNP+siPA was not inducing nonspecific antiviral responses, we tested serum and lung homogenates of mice treated with siRNAs in an IFN bioassay (33). We found no detectable IFN in an assay that readily detected IFN in the lungs of H3N2 virus-infected mice (data not shown). In addition, we compared the ability of the siRNA treatments to inhibit replication in vivo of PR/8 and influenza B/Ann Arbor/1/86 (B/AA). The influenza B/AA genome has only 52–67% homology with the siRNA target sequences. siNP+siPA treatment significantly decreased lung virus titers in PR/8-challenged animals, but not B/AA-challenged animals (data not shown), confirming that sequence homology between the siRNAs and the viral gene targets is necessary for suppression of virus replication.
Although influenza-specific RNAi provided potent protection against lethal challenge with PR/8, an H1N1 virus, it was unclear that it would protect against other influenza A subtypes because of the different kinetics and tissue tropism of these infections (34). The highly pathogenic avian influenza viruses that infected humans in the recent past were of particular interest because of their potential to unleash a pandemic; therefore, we tested the ability of siNP and siPA to inhibit replication of H5, H7, and H9 influenza subtypes. BALB/c mice were treated i.v. with siGFP or siNP+siPA on day –1. On the day of challenge (day 0), mice were given a second dose of siRNA in a lipid carrier i.n. and, at the same time, challenged with PR/8 virus (H1N1); HK/156 virus, an H5N1 isolate from the 1997 outbreak of avian influenza in Hong Kong; NL/219 virus, an H7N7 isolate from the 2003 outbreak of avian influenza in The Netherlands; or HK/1073 virus, an H9N2 isolate from a 1999 case of avian influenza in Hong Kong. Two days later, animals were killed and lungs were tested for virus titer by eID50 assay. As shown in Table 2, lung titers from HK/156- and NL/219-challenged mice were reduced 11- and 9-fold, respectively, whereas lung titers from PR/8- and HK/1073-challenged mice were more dramatically reduced (56- and 21-fold, respectively; all reductions were statistically significant). Thus, the siNP+siPA treatment could suppress replication of a broad spectrum of influenza subtypes, including highly pathogenic avian isolates. Additionally, the siRNA-mediated inhibition of HK/156 replication provides further evidence that IFN is not responsible for inhibition of virus replication, because H5 viruses are resistant to the antiviral effects of IFNs (35).
Certain isolates of avian influenza that have infected humans cause infections in mice that are rapidly lethal at low challenge doses (34). To test whether the inhibition of virus replication by RNAi was adequate for protection, BALB/c mice were treated i.v. with siGFP or siNP+siPA and treated again 16–24 h later and given a lethal challenge i.n. with PR/8, HK/156, or NL/219 virus. We did not include HK/1073 virus, because it is minimally lethal in mice (T.M.T., unpublished data). Animals treated with siNP+siPA and challenged with a lethal dose of PR/8 survived, whereas almost 90% of the siGFP-treated mice died (Fig. 2 a and d). Survival results for H5N1 virus were dramatic; influenza-specific RNAi protected seven of eight mice challenged with a dose of HK/156 virus that killed all of the control mice (Fig. 2 b and e). In the case of NL/219, protection against mortality although partial, was significant at a challenge dose lethal to all of the siGFP controls (Fig. 2 c and f). In addition, morbidity as indicated by weight loss was significantly reduced for PR/8 and NL/219.
Influenza NP and PA proteins are essential to viral replication, providing ideal targets for RNAi (18). Moreover, the NP and PA genes are highly conserved across subtypes of influenza A virus; therefore, siRNAs against these genes should inhibit most influenza A viruses. This hypothesis is supported by our results demonstrating specific inhibition of replication of H1, H5, H7, and H9 influenza A subtypes in vivo. With the recent publication characterizing NL/219 by Fouchier et al. (28), the sequences of the NP and PA genes are known for all of the viruses tested. The siRNA target sequences were identical in PR/8, HK/156, HK1073, and NL/219. However, there are naturally occurring influenza variants that have mismatches in the targeted regions. It will be important to test the ability of these siRNAs to inhibit replication of viruses lacking complete identity. Targeting multiple elements within the influenza genome decreases the likelihood of mismatches in all RNAi targets and could reduce the likelihood of development of siRNA-resistant virus escape variants (14).
To use siRNA as an in vivo therapeutic, it must be delivered efficiently to the appropriate tissue(s). We demonstrate that hydrodynamic i.v. delivery combined with i.n. delivery of siRNA can specifically inhibit virus replication in the site of infection. Intravenous delivery of siRNAs alone also provided significant protection, although some animals succumbed to infection, suggesting that i.n. delivery contributed to survival (data not shown). Concurrent with our report, Ge et al. (36) show that i.n. delivery of plasmids expressing influenza-specific siRNAs can significantly decrease lung virus titers in influenza-infected mice. Studies are underway to test alternative expression vectors, delivery vehicles, and routes of administration.
Although we have not studied the effect of siRNA treatment of established infection, Ge et al. (36) have shown reduction of virus replication in the lungs by siRNA given after infection. Our studies demonstrate that siRNA can be effective when given to animals before an otherwise lethal influenza infection. Although it will be important to test siRNA treatment in established infections, the data reported here suggest that this intervention would be useful during influenza outbreaks, and that siRNA could be given as a preventive in the face of a pandemic. Further development of this technology may provide an effective strategy for controlling influenza and other viral diseases.
Acknowledgments
We are especially grateful to Dr. Edward A. Havell (North Carolina State University, Raleigh) for his rapid and thorough help with IFN measurement and analysis. Additionally, we are grateful to Roger Brock (U.S. Department of Agriculture/Agricultural Research Service/Southeast Poultry Research Laboratory, Athens, GA) for his assistance with BSL3+ experiments and Julia Misplon (Laboratory of Immunology and Developmental Biology, Center for Biologics Evaluation and Research/Food and Drug Administration, Bethesda) for technical assistance and many useful discussions. We thank Dr. Ron A. M. Fouchier (Department of Virology and National Influenza Center, Erasmus Medical Center, Rotterdam) for providing the sequences of NL/219 NP and PA genes before their availability in GenBank, Carolyn Wilson for critical review of the manuscript, and Andrew Byrnes for critical review of the manuscript and assistance with statistical analysis.
Abbreviations: eID50, egg 50% infective dose; i.n., intranasal(ly); NP, nucleoprotein; PA, acidic polymerase; RNAi, RNA interference; siRNA, small interfering RNA; TCID50, tissue culture 50% infective dose.
References
- 1.World Health Organization (2004) Influenza: Report by the WHO Secretariat, World Health Assembly (WHO, Geneva), A56/23 3-17-2003.
- 2.Wright, P. F. & Webster, R. G. (2001) in Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Martin, M. A., Lamb, R. A., Roizman, B. & Straus, S. E. (Lippincott Williams & Wilkins, Philadelphia), pp. 1533–1579.
- 3.Nicholson, K. G., Aoki, F. Y., Osterhaus, A. E., Trottier, S., Carewicz, O., Mercier, C. H., Rode, A., Kinnersley, N. & Ward, P. (2000) Lancet 355, 1845–1850. [DOI] [PubMed] [Google Scholar]
- 4.Epstein, S. L. (2003) Expert Rev. Anti-infective Ther. 1, 89–100. [DOI] [PubMed] [Google Scholar]
- 5.Epstein, S. L., Tumpey, T. M., Misplon, J. A., Lo, C. Y., Cooper, L. A., Subbarao, K., Renshaw, M., Sambhara, S. & Katz, J. M. (2002) Emerg. Infect. Dis. 8, 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 7.McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 8.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457–467. [DOI] [PubMed] [Google Scholar]
- 9.Gitlin, L. & Andino, R. (2003) J. Virol. 77, 7159–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haasnoot, P. C. J., Cupac, D. & Berkhout, B. (2003) J. Biomed. Sci. 10, 607–616. [DOI] [PubMed] [Google Scholar]
- 11.Silva, J. M., Hammond, S. M. & Hannon, G. J. (2002) Trends Mol. Med. 8, 505–508. [DOI] [PubMed] [Google Scholar]
- 12.Jacque, J. M., Triques, K. & Stevenson, M. (2002) Nature 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, M. T., Coburn, G. A., McClure, M. O. & Cullen, B. R. (2003) J. Virol. 77, 11964–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitlin, L., Karelsky, S. & Andino, R. (2002) Nature 418, 430–434. [DOI] [PubMed] [Google Scholar]
- 15.Kapadia, S. B., Brideau-Andersen, A. & Chisari, F. V. (2003) Proc. Natl. Acad. Sci. USA 100, 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randall, G., Grakoui, A. & Rice, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCown, M., Diamond, M. S. & Pekosz, A. (2003) Virol. 313, 514–524. [DOI] [PubMed] [Google Scholar]
- 18.Ge, Q., McManus, M. T., Nguyen, T., Shen, C. H., Sharp, P. A., Eisen, H. N. & Chen, J. Z. (2003) Proc. Natl. Acad. Sci. USA 100, 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, D. L., Hagstrom, J. E., Loomis, A. G., Wolff, J. A. & Herweijer, H. (2002) Nat. Genet. 32, 107–108. [DOI] [PubMed] [Google Scholar]
- 20.McCaffrey, A. P., Meuse, L., Pham, T. T., Conklin, D. S., Hannon, G. J. & Kay, M. A. (2002) Nature 418, 38–39. [DOI] [PubMed] [Google Scholar]
- 21.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 22.McCaffrey, A. P., Nakai, H., Pandey, K., Huang, Z., Salazar, F. H., Xu, H., Wieland, S. F., Marion, P. L. & Kay, M. A. (2003) Nat. Biotechnol. 21, 639–644. [DOI] [PubMed] [Google Scholar]
- 23.Giladi, H., Ketzinel-Gilad, M., Rivkin, L., Felig, Y., Nussbaum, O. & Galun, E. (2003) Mol. Ther. 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 24.Song, E. W., Lee, S. K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J. S., Shankar, P. & Lieberman, J. (2003) Nat. Med. 9, 347–351. [DOI] [PubMed] [Google Scholar]
- 25.Sorensen, D. R., Leirdal, M. & Sioud, M. (2003) J. Mol. Biol. 327, 761–766. [DOI] [PubMed] [Google Scholar]
- 26.Subbarao, K., Klimov, A., Katz, J., Regnery, H., Lim, W., Hall, H., Perdue, M., Swayne, D., Bender, C., Huang, J., et al. (1998) Science 279, 393–396. [DOI] [PubMed] [Google Scholar]
- 27.Koopmans, M., Fouchier, R., Wilbrink, B., Meijer, A., Natrop, G., Osterhaus, A. D. M. E., van Steenbergen, J. E., du Ry van Beest Holle, M., Conyn van Spaendonck, M. A. E. & Bosman, A. (2003) Eurosurveill. Wkly. 7.
- 28.Fouchier, R. A., Schneeberger, P. M., Rozendaal, F. W., Broekman, J. M., Kemink, S. A., Munster, V., Kuiken, T., Rimmelzwaan, G. F., Schutten, M., Van Doornum, G. J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1356–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, X. H., Renshaw, M., Tumpey, T. M., Kelly, G. D., Hu-Primmer, J. & Katz, J. M. (2001) J. Virol. 75, 4896–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benton, K. A., Misplon, J. A., Lo, C.-Y., Brutkiewicz, R. R., Prasad, S. A. & Epstein, S. L. (2001) J. Immunol. 166, 7437–7445. [DOI] [PubMed] [Google Scholar]
- 31.Liang, S., Mozdzanowska, K., Palladino, G. & Gerhard, W. (1994) J. Immunol. 152, 1653–1661. [PubMed] [Google Scholar]
- 32.Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H. & Williams, B. R. (2003) Nat. Cell Biol. 5, 834–839. [DOI] [PubMed] [Google Scholar]
- 33.Havell, E. A. (1986) J. Infect. Dis. 153, 960–969. [DOI] [PubMed] [Google Scholar]
- 34.Katz, J. M., Lu, X. H., Tumpey, T. M., Smith, C. B., Shaw, M. W. & Subbarao, K. (2000) J. Virol. 74, 10807–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo, S. H., Hoffmann, E. & Webster, R. G. (2002) Nat. Med. 8, 950–954. [DOI] [PubMed] [Google Scholar]
- 36.Ge, Q., Filip, L., Bai, A., Nguyen, T., Eisen, H. N. & Chen, J. (2004) Proc. Natl. Acad. Sci. USA 101, 8676–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]