Abstract
Overexpression of transforming growth factor β (TGF-β) has been shown to play pathogenic roles in progression of renal fibrosis, and the severity of tubulointerstitial fibrosis correlates better with renal function than the severity of glomerulosclerosis. Smad proteins are signaling transducers downstream from TGF-β receptors. Three families of Smad proteins have been identified: receptor-regulated Smad2 and Smad3, common partner Smad4, and inhibitory Smad7 (part of a negative-feedback loop). We investigated Smad-mediated TGF-β signaling pathway and regulatory mechanisms of inhibitory Smad7 in unilateral ureteral obstruction (UUO) kidneys in mice, a model of progressive tubulointerstitial fibrosis. Compared with sham-operated kidneys, the level of Smad7 protein, but not mRNA, decreased progressively in UUO kidneys, whereas immunoreactivity for nuclear phosphorylated Smad2 and Smad3 and renal fibrosis were inversely increased. Furthermore, we demonstrated that both the degradation and ubiquitination activity of Smad7 protein were increased markedly in UUO kidneys compared with sham-operated ones. We also found that both Smurf1 and Smurf2 (Smad ubiquitination regulatory factors), which are E3 ubiquitin ligases for Smad7, were increased and that they interacted with Smad7 in UUO kidneys. Our results suggest that the reduction of Smad7 protein resulting from enhanced ubiquitin-dependent degradation plays a pathogenic role in progression of tubulointerstitial fibrosis.
Keywords: transforming growth factor β, Smad proteins, tubulointerstitial fibrosis
Transforming growth factor β (TGF-β) is a multifunctional signaling protein that regulates cell cycle, apoptosis, differentiation, and extracellular matrix accumulation (1). TGF-β plays a significant role in the progression of renal fibrosis in clinical and experimental kidney diseases (2).
Smad proteins have been identified recently as important components of the TGF-β signaling pathway. TGF-β signals through the heteromeric complex of TGF-β type I receptor (TβRI) and TGF-β type II receptor (TβRII), which are transmembrane serine–threonine kinase receptors. Activation of the receptor complex occurs when type II receptor kinase transphosphorylates the glycine–serine domain of type I kinase. The activated type I kinase associates transiently with and also phosphorylates receptor-regulated Smad2 and Smad3 (R-Smads). When they are phosphorylated, R-Smads dissociate from the receptor, bind to Smad4, and then enter the nucleus. Smad7 is an intracellular antagonist for TGF-β signaling, and it is known to associate with activated TβRI and hinder the activation of R-Smads by preventing their interaction with activated TβRI and consequent phosphorylation (3, 4).
In human inflammatory bowel disease, it has been reported that Smad7 expression is increased and that TGF-β activity is down-regulated in augmented inflammation (5). However, decrease in Smad7 and increase in TGF-β activity were demonstrated in fibrotic tissues in human scleroderma and rat myocardial infarction (6, 7). It has been demonstrated also that overexpression of Smad7 induced by gene transfer inhibited Smad2 and Smad3 activation and tissue fibrosis in bleomycin-induced lung fibrosis and in rat obstructive nephropathy, in which increased expression of TGF-β has been shown to be involved in progression of fibrosis (8, 9). When these findings are considered together, it seems that the cellular level of Smad7 plays an important role in the regulation of Smad-mediated TGF-β signaling during progression of organ fibrosis in various diseases. However, the pathogenic implication of the intracellular Smad7 regulatory mechanism in the progression of organ fibrosis remains to be elucidated. Recent studies have demonstrated that the abundance of cellular proteins, including Smad7, is modulated dynamically by the ubiquitin–proteasome degradation pathway (10), in which Smurf1 and Smurf2 (Smad ubiquitination regulatory factors) are involved as E3 ubiquitin ligases for Smad7 (11, 12).
This study was designed to investigate Smad-mediated TGF-β signaling pathway and regulatory mechanisms of inhibitory Smad7 in mice with unilateral ureteral obstruction (UUO) kidneys, a model of kidney disease with progressive tubulointerstitial fibrosis. The results showed that UUO resulted in progressive tubulointerstitial fibrosis associated with increased expression of TGF-β1 and Smad7 mRNAs, activation of R-Smads, expression of Smurf1 and Smurf2, and a significant decrease in Smad7 protein level. We also demonstrated that both the degradation and ubiquitination activity of Smad7 protein were markedly enhanced in UUO kidney extracts and that the addition of proteasome inhibitor inhibited the degradation of Smad7 in UUO kidney extracts. Together, the results suggest that the reduction of Smad7 is caused by ubiquitin-dependent protein degradation (which could be mediated, at least in part, by Smurf proteins) and that such reduction plays an important role in the progression of tubulointerstitial fibrosis in UUO kidneys.
Materials and Methods
Experimental Animals and Design. Male C57BL6/J mice, weighing 20–25 g at the start of the experiment, were prepared. Ureteral obstruction was achieved by ligating the left ureter with 3-0 silk through a left lateral incision. Sham-operated animals (n = 9) were used as a control. Mice were killed at 3, 7, 14, or 28 days (for each group, n = 9) after operation, and obstructed kidneys were harvested and subjected to the studies described below. The experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of Hamamatsu University School of Medicine.
Histopathological and Immunohistochemical Analyses. Kidney tissues were fixed in 4% paraformaldehyde in PBS and embedded in paraffin. Tissue sections (3 μm thick) were stained with Masson's trichrome for histopathological analysis. Immunoreactivity for phosphorylated Smad2, Smad3, Smad7 proteins was determined by using a standard biotin–streptavidin–peroxidase method, as described (13, 14). The primary antibodies were rabbit anti-human phosphorylated Smad2 and Smad3 (Santa Cruz Biotechnology) and rabbit anti-human Smad7 (Santa Cruz Biotechnology). The secondary antibody was affinity-purified biotinylated donkey anti-rabbit IgG (Cortex Biochem, San Leandro, CA). Nuclei were counterstained lightly with hematoxylin. The immunoreactivity for Smad7 was scored as follows: 0.5, weak (<10% of cells in the sample were positive for the protein); 1, mild (≥10% to <30% of cells were positive); 2, moderate (≥30% to <50% of cells were positive); and 3, strong immunostaining (≥50% of cells were positive).
Immunoprecipitation and Immunoblot Analysis. Kidney tissues were dissolved in Triton X-100 lysis buffer (13–15). Equal amounts of proteins (40 μg) were loaded for SDS/PAGE, as described (13–15). The primary antibodies were rabbit anti-human Smad7 (Santa Cruz Biotechnology), goat anti-human Smurf1 (Santa Cruz Biotechnology), goat anti-human Smurf2 (Santa Cruz Biotechnology), and mouse anti-β-actin mAb (Sigma). We used β-actin as an internal control.
For immunoprecipitation, the renal lysates were preincubated with 2 μg of anti-Smad7 antibody and 30 μl of protein G–Sepharose beads for 3 h at 4°C. The resulting immunoprecipitates were washed thoroughly three times with cold lysis buffer, separated by SDS/PAGE, and immunoblotted with anti-Smurf1 and anti-Smurf2 antibodies. Quantification of the band intensity was performed by using image software (National Institutes of Health, Bethesda).
RNA Isolation and Quantitative Analysis of mRNA by Real-Time RT-PCR. Total RNA isolation, reverse transcription of the RNA, and all PCR experiments were performed as described (13). The PCR primer sequences were as follows: mouse TGF-β1, 5′-CCTGAGTGGCTGTCTTTTGACG-3′ (sense) and 5′-AGTGAGCGCTGAATCGAAAGC-3′ (antisense); mouse Smad7, 5′-TCGGACAGCTCAATTCGGAC-3′ (sense) and 5′-GGTAACTGCTGCGGTTGTAA-3′ (antisense); and mouse GAPDH, 5′-TGCACCACCAACTGCTTAG-3′ (sense) and 5′-GAGGCAGGGATGATGTTC-3′ (antisense). Data analysis was performed by using light cycler software (version 3.3.9; Roche). The ratios of TGF-β1 and Smad7 mRNAs to GAPDH mRNA were calculated in each sample.
Degradation Assay for Smad7. To investigate the degradation activity of Smad7 in the kidneys, we conducted an in vitro degradation assay for endogenous Smad7 in renal extracts collected from UUO kidneys on day 7 and renal extracts from sham-operated kidneys. The details of the method are given in ref. 15, in which we conducted the degradation assay of Smad2. In this study, we used anti-Smad7 antibody (Santa Cruz Biotechnology) instead of anti-Smad2 antibody for detection of Smad7. The degradation assay was performed also in the presence of 1× proteasome inhibitor mix (0.25 mM MG132/0.25 mM MG115).
Preparation of Flag-Tagged Smad7. HEK293 cells were grown in DMEM with 10% FBS at 37°C under a 95% air/5% CO2 atmosphere. pcDNA3-Flag-tagged mouse Smad7 was kindly provided by K. Miyazono (Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo). HEK293 cells were transfected with this expression vector by using FuGene 6 (Roche). As a control, pcDNA3 lacking Flag-tagged mouse Smad7 was used. The cells were lysed with Triton X-100 lysis buffer (13, 15) after incubation for 48 h. The cell lysates (1,000 μg/100 μl) were immunoprecipitated with 2 μg of anti-Flag M2 mAb (Sigma) and 30 μl of protein A–Sepharose beads (Bio-Rad) for 3 h at 4°C. The resulting immunoprecipitates were washed thoroughly four times with cold lysis buffer and then used for in vitro ubiquitination assays.
In Vitro Smad7-Ubiquitination Assay. To determine whether the levels of Smad7 ubiquitin-dependent degradation are up-regulated in UUO, we performed an in vitro assay of ubiquitination activity directed against exogenous Flag-tagged Smad7 protein. Renal extracts that were obtained from UUO and sham-operated kidneys and lysed in Triton X-100 lysis buffer were centrifuged for 8 h at 10,000 × g, and the resulting supernatants (S100) were used in the ubiquitination assay, as described (15, 16).
Statistical Analysis. All values are given as mean ± SEM. Differences between groups were examined for statistical significance by using ANOVA. When a significant difference was present, statistical analysis was performed further by using Scheffé's F test between the two groups. P values <0.05 indicate statistically significant difference.
Results
Progressive Fibrosis and Enhanced TGF-β Signaling in UUO Kidneys. Fig. 1A shows an example of progression of tubulointerstitial fibrosis, tubular atrophy, and interstitial mononuclear cell infiltration in UUO kidneys. These changes were associated with marked and time-dependent increase in the expression of TGF-β1 mRNA (4.8-fold, 5.0-fold, and 5.3-fold increases on days 7, 14, and 28, respectively, in obstructed kidneys compared with sham-operated kidneys; Fig. 1B).
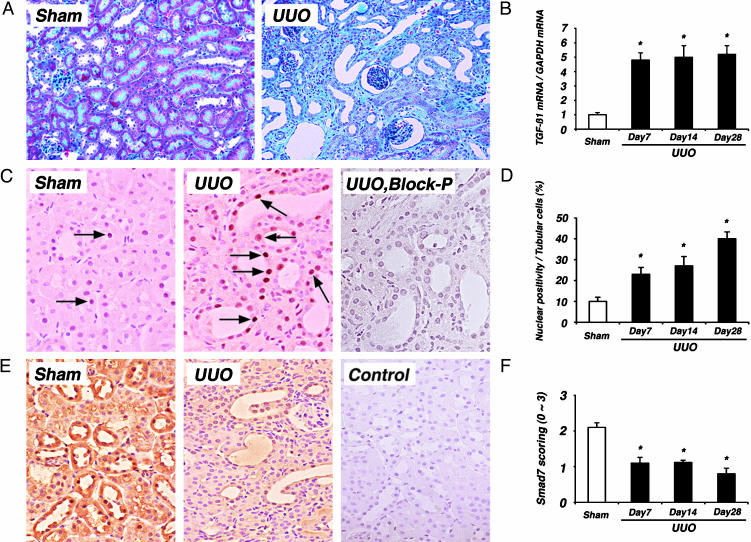
Fig. 1.
Histopathologic changes and TGF-β signaling components in UUO kidneys. (A) Masson's trichrome staining in sham-operated (Sham) and UUO kidney on day 28. Note the progressive development of tubulointerstitial fibrosis, tubular atrophy, and interstitial mononuclear cell infiltration in UUO kidneys. Images are shown at ×100 magnification. (B) Ratios of TGF-β1 mRNA/GAPDH mRNA in sham-operated kidneys (open bar) and UUO kidneys (filled bars) on days 7, 14, and 28. TGF-β1 mRNA expression was increased significantly in UUO kidneys compared with sham-operated kidneys (4.8-fold, 5.0-fold, and 5.3-fold increases on days 7, 14, and 28 in UUO, respectively, compared with sham-operated kidneys). (C) Immunostaining of phosphorylated Smad2 and Smad3 in sham-operated and UUO kidneys on day 14. Significantly increased nuclear immunostaining of phosphorylated Smad2 and Smad3 was noted in tubular and interstitial infiltrating mononuclear cells in UUO kidney, whereas no significant nuclear immunostaining was observed in serial sections of UUO kidney when antiphosphorylated Smad2 and Smad3 antibody was preincubated with blocking peptides in serial sections of UUO kidney (UUO, Block-P). Images are shown at ×400 magnification. (D) Nuclear positivity of phosphorylated Smad2 and Smad3 in tubular cells was increased significantly in UUO kidneys (filled bars) compared with sham-operated kidneys (open bar). (E) Immunostaining of Smad7 in sham-operated and UUO kidneys on day 14. Significant immunoreactivity for Smad7 was noted in tubular epithelial cells and peritubular capillary endothelial cells in sham-operated kidneys. However, immunoreactivity for Smad7 was markedly decreased in tubulointerstitial cells in UUO kidneys. No significant immunoreactivity was observed in serial sections of sham-operated kidney by using nonimmune rabbit IgG in place of anti-Smad7 antibody (Control). Images are shown at ×200 magnification. (F) Semiquantitative analysis of Smad7 staining in sham-operated (open bar) and UUO kidneys (filled bars). The immunoreactivity for Smad7 was significantly lower in UUO kidneys than in sham-operated kidneys. Data are given as mean ± SEM values of nine mice in each group. *, P < 0.05, compared with sham-operated kidneys.
TGF-β-mediated fibrosis is positively regulated by R-Smad2 and R-Smad3, and it is known that phosphorylated R-Smads translocate to the nucleus and, thereafter, regulate transcriptional responses of target genes (3). To determine the signaling activity of R-Smads, we investigated the nuclear localization of phosphorylated Smad2 and Smad3 by immunohistochemistry. Increased nuclear immunoreactivity for phosphorylated Smad2 and Smad3 was noted in tubular and interstitial infiltrating mononuclear cells in UUO kidneys (Fig. 1C). The nuclear positivity for phosphorylated Smad2 and Smad3 in tubular cells was significantly more in UUO kidneys than in sham-operated kidneys (Fig. 1D). Considering the current concept that phosphorylated R-Smads translocate to the nucleus to conduct TGF-β signaling from the cell surface TGF-β receptor complexes, our results suggest the enhancement of Smad-mediated TGF-β signaling in tubulointerstitial fibrotic lesions in UUO kidneys.
Decrease in Smad7 Protein, but Not mRNA, in UUO Kidneys. We examined the level of inhibitory Smad7, which is an intracellular antagonist for TGF-β signaling, by using Western blot analysis, immunohistochemistry, and quantitative RT-PCR. In sham-operated kidneys, Smad7 protein was expressed moderately and constantly throughout the experimental period. In contrast, Smad7 protein levels gradually but significantly decreased in a time-dependent fashion in UUO kidneys compared with sham-operated kidneys (Fig. 2 A and B). This observation was confirmed further by immunohistochemistry (Fig. 1 E and F). Significant immunoreactivity for Smad7 was noted in tubular epithelial cells and peritubular capillary endothelial cells in sham-operated kidneys (Fig. 1E). However, immunoreactivity for Smad7 was decreased markedly in tubulointerstitial cells in UUO kidneys.
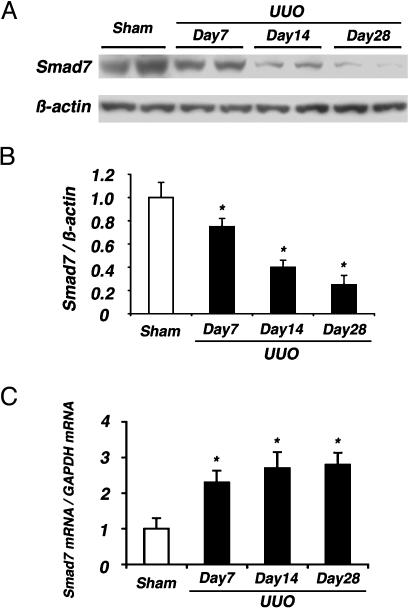
Fig. 2.
Smad7 expression in UUO kidneys. (A) Western blot analysis of Smad7 in renal extracts collected from UUO kidneys at indicated days after operation, showed a gradual reduction of Smad7 content in UUO kidneys compared with sham-operated kidneys. (B) Densitometric analysis of Smad7 protein levels detected by Western blot analysis. Significant decreases of Smad7 protein were noted in UUO kidneys (filled bars) compared with sham-operated kidneys (open bar). (C) Total RNA was subjected to real-time RT-PCR for measurement of Smad7 mRNA and GAPDH mRNA (internal control) levels. Data are ratios of Smad7 mRNA/GAPDH mRNA in sham-operated kidneys (open bar) and UUO kidneys (filled bars) on days 7, 14, and 28. In contrast to Smad7 protein, Smad7 mRNA expression was up-regulated significantly in UUO kidneys compared with sham-operated kidneys. Data are given as mean ± SEM values of nine mice in each group. *, P < 0.05, compared with sham-operated kidneys.
To clarify whether the reduction in Smad7 protein was the result of down-regulation of Smad7 mRNA, we evaluated the mRNA expression of Smad7 by real-time RT-PCR. In contrast to Smad7 protein, Smad7 mRNA was up-regulated in UUO kidneys compared with sham-operated kidneys (Fig. 2C). This result was consistent with reports (17) that Smad7 mRNA is induced rapidly by TGF-β in several cell lines. Based on these results, we speculated that the reduction of Smad7 protein in UUO kidneys was probably caused by enhanced protein degradation.
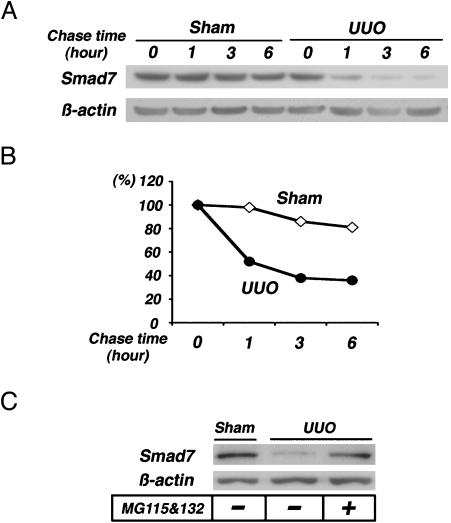
Enhanced Degradation of Endogenous Smad7 in UUO. To confirm the enhancement of Smad7 protein degradation in UUO kidneys, we subsequently subjected renal extracts to an in vitro degradation assay. Degradation of endogenous Smad7 progressed in renal extracts that were collected from UUO kidneys on day 7 (Fig. 3 A and B), when the levels of Smad7 protein were decreased (Fig. 2 A). In contrast, no significant reduction in Smad7 was noted in the renal extracts collected from sham-operated kidneys. Furthermore, the degradation of endogenous Smad7 was prevented by the addition of proteasome inhibitors (Fig. 3C). These results indicate the enhancement of proteasome-mediated degradation of Smad7 in UUO kidneys.
Fig. 3.
Smad7-degradation activity in UUO kidneys. (A) Renal extracts collected from sham-operated kidneys and UUO kidneys on day 7 were mixed with the ubiquitination mixture, incubated for 0–6 h at 37°C, and the reaction mixtures were subjected to immunoblot analysis to detect the degradation of endogenous Smad7 protein by using anti-Smad7 antibody. (B) The Smad7 protein signals in A were quantified by densitometry. The densitometric ratios were calculated relative to the densitometric values at time 0 in each group (⋄, sham; •, UUO). Degradation of endogenous Smad7 progressed in renal extracts collected from UUO mice. In contrast, no significant reduction in Smad7 was noted in renal extracts collected from sham-operated mice. (C) In vitro degradation assay of endogenous Smad7 was also performed for 6 h, with or without 1× proteasome inhibitor mixture (0.25 mM MG132/0.25 mM MG115). Enhanced degradation of Smad7 in UUO kidneys was prevented by the addition of the proteasome inhibitors.
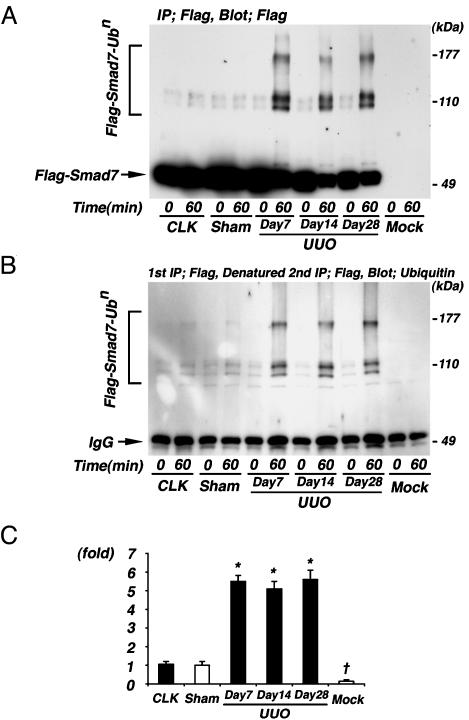
Enhanced Ubiquitination Activity Directed Against Exogenous Smad7 in UUO Kidneys. Recent studies (11, 12) demonstrated the degradation of Smad7 through the ubiquitin–proteasome pathway in cultured cells. To clarify whether the increased degradation of Smad7 in UUO involves the ubiquitin system, we investigated the in vitro ubiquitination activity directed against Smad7 in renal extracts by using Flag-tagged Smad7 protein, which was derived from HEK293 cells transfected with mouse Smad7 cDNA and then immunoprecipitated by anti-Flag M2 antibody as a substrate. As shown in Fig. 4A, by using anti-Flag M2 antibody, significant ladders were observed when Flag-tagged Smad7 protein was incubated with renal extracts collected from UUO kidneys. In contrast, no significant ladders were observed when Flag-tagged Smad7 protein was incubated with renal extracts collected from the contralateral kidneys of UUO mice and sham-operated kidneys. Also, no bands were observed when anti-Flag immunoprecipitate, which was derived from mock-transfected HEK293 cells as a negative control, was incubated with renal extracts collected from UUO kidneys on day 7. These results were confirmed also by detecting similar ladders by using an antiubiquitin antibody after the assay products were denatured in SDS to remove some Smad7-associated proteins and then subjected to re-immunoprecipitation by anti-Flag antibody (Fig. 4B). These results strongly suggested that the ladders were polyubiquitinated Smad7.
Fig. 4.
Ubiquitination assay of Smad7 in UUO kidneys. (A) Renal extracts collected from contralateral kidneys of UUO on day 7 (CLK), sham-operated kidneys on day 7 (Sham), and UUO kidneys on days 7, 14, and 28 were incubated with immunopurified Flag-tagged Smad7 protein for 60 min at 37°C. The assay products were subjected to immunoblot analysis using anti-Flag M2 antibody. Significant ladders of polyubiquitinated Smad7 were observed when Flag-tagged Smad7 protein was incubated with renal extracts collected from UUO kidneys but not from CLK and Sham. Furthermore, no bands were observed when anti-Flag immunoprecipitated protein, which was derived from mock-transfected HEK293 cells as a negative control, was incubated with renal extracts collected from UUO mice on day 7 (Mock). (B) To further purify the assay products, we next denatured the assay products in SDS, subjected to reimmunoprecipitation by anti-Flag antibody (2nd IP), and then immunoblotted by antiubiquitin antibody. The results shown in A were also confirmed by this experiment. (C) The densitometric ratios were calculated relative to the densitometric values in sham-operated kidneys. Densitometric analysis demonstrated that the increases in polyubiquitinated Smad7 after incubation with renal extracts collected from UUO kidneys at days 7–28 were 5- to 6-fold higher compared with sham-operated kidneys. CLK and Mock are as defined for A. Data are given as mean ± SEM values of nine mice in each group. *, P < 0.05, compared with sham-operated kidneys. †, P < 0.01, compared with sham-operated kidneys.
Densitometric analysis demonstrated 5- to 6-fold increases in polyubiquitination activity against Smad7 in the renal extracts collected from UUO kidneys at days 7, 14, and 28 compared with those from sham-operated kidneys (Fig. 4C).
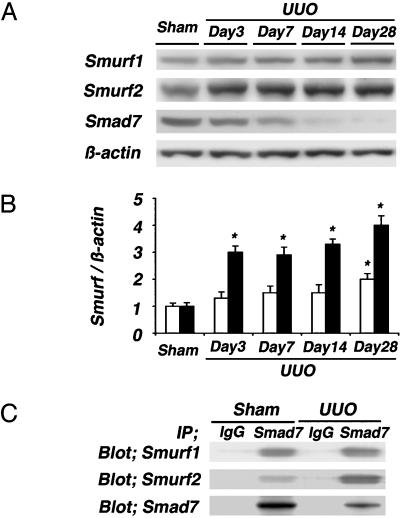
Increased Expression of Smurf1 and Smurf2 in UUO Kidneys. Both Smurf1 and Smurf2 are members of the HECT (homologous to E6–AP C terminus) family of E3 ubiquitin ligases for Smad7 (11, 12). Next, we investigated the levels of both Smurf1 and Smurf2 in UUO and sham-operated kidneys by using Western blot analysis. Smurf1 and Smurf2 were expressed weakly in renal extracts from sham-operated kidneys. In UUO kidneys, marked expression of Smurf2 was noted at day 3, which further increased thereafter in almost-inverse proportion to the levels of Smad7, whereas the increases in Smurf1 were milder than the increases in Smurf2 (Fig. 5 A and B). In addition, the expression levels of Smurf1 and Smurf2 mRNAs also increased and were almost proportionate to those of protein levels (see Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 5.
Immunoblot analyses of Smurf1 and Smurf2. (A) Western blot analyses of Smurf1 and Smurf2 were performed in sham-operated and UUO kidneys on days 3, 7, 14, and 28. (B) Densitometric analysis of the levels of Smurf1 and Smurf2 detected by Western blot analysis. In UUO kidneys, marked increases in Smurf2 (filled bars) were noted at day 3 and further enhanced thereafter in almost-inverse proportion to the levels of Smad7, whereas the increases in Smurf1 (open bars) were milder than the increases of Smurf2. (C) Renal extracts collected from sham-operated and UUO kidneys on day 7 were subjected to Smad7 immunoprecipitation (IP), followed by Smurf1 and Smurf2 immunoblotting (Blot). In UUO kidneys, both the Smad7–Smurf1 and Smad7–Smurf2 complexes were noted. The levels of Smurf2–Smad7 complex were relatively more than those of Smurf1–Smad7 complex. Representative data of three independent experiments are shown. Data are given as mean ± SEM values of nine mice in each group. *, P < 0.05, compared with sham-operated kidneys.
Finally, to investigate the interaction of Smurf1 and Smurf2 with Smad7 in kidney tissues, renal extracts collected from UUO kidneys on day 7 and sham-operated kidneys were immunoprecipitated by using anti-Smad7 antibody, followed by immunoblotting using anti-Smurf1 or anti-Smurf2 antibody (Fig. 5C). In UUO kidneys, both the complex composed of Smad7 and Smurf1 and that of Smad7 and Smurf2 were noted. The levels of the Smurf2–Smad7 complex were relatively more than the levels of the Smurf1–Smad7 complex. These data suggest that Smad7 ubiquitination could be promoted by both Smurf1 and Smurf2 in UUO kidneys, although the involvement of Smurf2 seems to be more evident than the involvement of Smurf1.
Discussion
Accumulating evidence indicates that TGF-β is a key mediator of the progression of renal fibrosis (2). Tubulointerstitial fibrosis is a final common feature of end-stage renal damage in various types of kidney diseases, and its severity correlates with renal prognosis (18). Therefore, elucidation of the pathogenic mechanisms involved in the progression of tubulointerstitial fibrosis is important for the design of a strategy to prevent progressive kidney diseases resulting in end-stage renal failure. The identification of Smad proteins has advanced our understanding of how TGF-β signals from the membrane receptors to the nucleus (3). Consistent with this concept, we have reported (13, 14) that TGF-β–Smad signaling system was up-regulated in rat progressive glomerulonephritis.
Recent studies (4) have identified several molecules that control the signal transduction of TGF-β, thereby regulating TGF-β activity. Smad7 is one of these molecules and is known to inhibit TGF-β-induced transcriptional responses (17). Smad7 associates with the activated TGF-β receptor and interferes with the activation of Smad2 and Smad3 by preventing their receptor interaction and phosphorylation. Therefore, it is conceivable that the cellular level of Smad7 is a major determinant of TGF-β responses, which could regulate the intensity and duration of TGF-β signals. Aberrant regulation of Smad7 activity could be implicated in the development of human diseases, such as inflammatory bowel diseases and scleroderma, in which inappropriate expression of TGF-β has been shown to play, at least in part, a causative role (5, 6). In chronic rat liver injury, the constitutive phosphorylation of Smad2 under a low level of Smad7 could be involved in the progression of liver fibrosis, whereas a transient induction of Smad7 terminated Smad2 phosphorylation in acute liver injury (19). However, controversy concerning the levels of Smad7 in kidney diseases remains; Uchida et al. (20) reported a decrease in glomerular expression of Smad7 in anti-Thy-1 nephritis, which is an experimental model of acute reversible mesangial proliferative glomerulonephritis in rats. In contrast, Ostendorf et al. (21) demonstrated increased Smad7 expression in a similar model. Therefore, the level of renal Smad7 remains to be determined and, to our knowledge, has not been investigated in a chronic model of kidney disease with progressive renal fibrosis.
The mechanisms that regulate the expression of Smad7 are not fully understood. Smad7 expression is strongly and rapidly induced by TGF-β (17), and the promoter region of the Smad7 gene contains a consensus Smad3–Smad4 binding element, the palindromic sequence of GTCTAGAC to which the Smad3–Smad4 complex binds (22). In the present study, the expression of Smad7 mRNA was increased and correlated with TGF-β1 expression, although the levels of Smad7 protein was decreased inversely in UUO kidneys.
Protein ubiquitination and subsequent proteasomal degradation is a common regulating mechanism (23). Several studies (11, 12, 24) have demonstrated that Smad proteins also undergo ubiquitin–proteasome-mediated degradation. Smurf1 was identified originally as a HECT (homologous to E6–AP C-terminus) type E3 ubiquitin ligase, which induces the ubiquitination and degradation of Smad1 and Smad5 in a signal-independent manner (25). Smurf2, which is structurally similar to Smurf1, also targets Smad1 for degradation (26). Subsequently, Smurf2 was shown to associate with Smad2 and to induce its ubiquitin-dependent degradation (24). Also, Smurf1 and Smurf2 interact with intranuclear Smad7 and induce nuclear export of Smad7. The Smurf–Smad7 complexes then associate with type I receptor for TGF-β and enhance its turnover (11, 12). These findings suggest that Smurf proteins can negatively regulate TGF-β signaling by targeting their positive signaling components for ubiquitin-dependent degradation. On the other hand, Bonni et al. (27) have demonstrated that Smurf2 binds to a transcriptional corepressor SnoN through activated Smad2 and, thereby, targets SnoN for ubiquitin-dependent degradation. This finding could indicate that Smurf2 positively regulates TGF-β signaling under other conditions. Together, the physiological roles of Smurf1 and Smurf2 remain to be clarified in various conditions in vivo.
In the present study, we demonstrated significant decrease in Smad7 protein and increase in ubiquitination and degradation activity of Smad7 protein in fibrotic kidneys, in which the expression levels of TGF-β1 and Smad7 mRNA, nuclear localization of phosphorylated Smad2 and Smad3, and expressions of Smurf1 and Smurf2 were increased. We demonstrated further that degradation of Smad7 in extracts from UUO kidneys was inhibited by the addition of proteasome inhibitor. These results confirmed that accelerated degradation of Smad7 by the ubiquitin–proteasome pathway, which could be mediated by Smurf proteins or other ligases, contributes to the reduction of Smad7 in fibrotic UUO kidneys. However, decreased translation of Smad7 may also contribute to the low Smad7 protein level, and this possibility cannot be ruled out in the present study. In addition, the present study could not indicate directly that the loss of Smad7 is a key contributory factor in the progression of renal fibrosis in UUO mice kidneys.
In conclusion, our study suggests that reduced levels of inhibitory Smad7 protein caused by ubiquitin-dependent degradation could be involved in the progression of renal fibrosis in mice UUO kidneys. Further studies are necessary to elucidate the pathophysiological implication of the ubiquitination and proteasomal degradation of Smad7 in the development of renal fibrosis.
Supplementary Material
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research 12671035 (to T.Y.) and a Grant-in Aid for the Center of Excellence from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: R-Smad, receptor-regulated Smad; TGF-β, transforming growth factor β; UUO, unilateral ureteral obstruction.
References
- 1.Sporn, M. B. & Roberts, A. B. (1992) J. Cell Biol. 119, 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Border, W. A. & Noble, N. A. (1994) N. Engl. J. Med. 331, 1286–1292. [DOI] [PubMed] [Google Scholar]
- 3.Massagué, J. (2000) Nat. Rev. Mol. Cell Biol. 1, 169–178. [DOI] [PubMed] [Google Scholar]
- 4.Wrana, J. L. (2000) Cell 100, 189–192. [DOI] [PubMed] [Google Scholar]
- 5.Monteleone, G., Kumberova, A., Croft, N. M., McKenzie, C., Steer, H. W. & MacDonald, T. T. (2001) J. Clin. Invest. 108, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, C., Zhu, S., Wang, T., Yoon, W., Li, Z., Alvarez, R. J., ten Dijke, P., White, B., Wigley, F. M. & Goldschmidt-Clermont, P. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3908–3913. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Wang, B., Hao, J., Jones, S. C., Yee, M. S., Roth, J. C. & Dixon, I. M. (2002) Am. J. Physiol. 282, H1685–H1696. [DOI] [PubMed] [Google Scholar]
- 8.Nakao, A., Fujii, M., Matsumura, R., Kumano, K., Saito, Y., Miyazono, K. & Iwamoto, I. (1999) J. Clin. Invest. 104, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan, H. Y., Mu, W., Tomita, N., Huang, X. R., Li, J. H., Zhu, H. J., Morishita, R. & Johnson, R. J. (2003) J. Am. Soc. Nephrol. 14, 1535–1548. [DOI] [PubMed] [Google Scholar]
- 10.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 11.Kavsak, P., Rasmussen, R. K., Causing, C. G., Bonni, S., Zhu, H., Thomsen, G. H. & Wrana, J. L. (2000) Mol. Cell 6, 1365–1375. [DOI] [PubMed] [Google Scholar]
- 12.Ebisawa, T., Fukuchi, M., Murakami, G., Chiba, T., Tanaka, K., Imamura, T. & Miyazono, K. (2001) J. Biol. Chem. 276, 12477–12480. [DOI] [PubMed] [Google Scholar]
- 13.Fukasawa, H., Yamamoto, T., Suzuki, H., Togawa, A., Ohashi, N., Fujigaki, Y., Uchida, C., Aoki, M., Hosono, M., Kitagawa, M. & Hishida, A. (2004) Kidney Int. 65, 63–74. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe, T., Yamamoto, T., Ikegaya, N., Fujigaki, Y., Suzuki, H., Togawa, A., Fukasawa, H., Nagase, M. & Hishida, A. (2002) J. Pathol. 198, 397–406. [DOI] [PubMed] [Google Scholar]
- 15.Togawa, A., Yamamoto, T., Suzuki, H., Fukasawa, H., Ohashi, N., Fujigaki, Y., Kitagawa, K., Hattori, T., Kitagawa, M. & Hishida, A. (2003) Am. J. Pathol. 163, 1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa, M., Hatakeyama, S., Shirane, M., Matsumoto, M., Ishida, N., Hattori, K., Nakamichi, I., Kikuchi, A. & Nakayama, K. (1999) EMBO J. 18, 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakao, A., Afrakhte, M., Moren, A., Nakayama, T., Christian, J. L., Heuchel, R., Itoh, S., Kawabata, M., Heldin, N. E., Heldin, C. H. & ten Dijke, P. (1997) Nature 389, 631–635. [DOI] [PubMed] [Google Scholar]
- 18.Eddy, A. A. (1996) J. Am. Soc. Nephrol. 7, 2495–2508. [DOI] [PubMed] [Google Scholar]
- 19.Tahashi, Y., Matsuzaki, K., Date, M., Yoshida, K., Furukawa, F., Sugano, Y., Matsushita, M., Himeno, Y., Inagaki, Y. & Inoue, K. (2002) Hepatology 35, 49–61. [DOI] [PubMed] [Google Scholar]
- 20.Uchida, K., Nitta, K., Kobayashi, H., Kawachi, H., Shimizu, F., Yumura, W. & Nihei, H. (2000) Mol. Cell Biol. Res. Commun. 4, 98–105. [DOI] [PubMed] [Google Scholar]
- 21.Ostendorf, T., Kunter, U., van Roeyen, C., Dooley, S., Janjic, N., Ruckman, J., Eitner, F. & Floege, J. (2002) J. Am. Soc. Nephrol. 13, 658–667. [DOI] [PubMed] [Google Scholar]
- 22.Nagarajan, R. P., Zhang, J., Li, W. & Chen, Y. (1999) J. Biol. Chem. 274, 33412–33418. [DOI] [PubMed] [Google Scholar]
- 23.Ciechanover, A., Orian, A. & Schwartz, A. L. (2000) BioEssays 22, 442–451. [DOI] [PubMed] [Google Scholar]
- 24.Lin, X., Liang, M. & Feng, X. H. (2000) J. Biol. Chem. 275, 36818–36822. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, H., Kavsak, P., Abdollah, S., Wrana, J. L. & Thomsen, G. H. (1999) Nature 400, 687–693. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y., Chang, C., Gehling, D. J., Hemmati-Brivanlou, A. & Derynck, R. (2001) Proc. Natl. Acad. Sci. USA 98, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonni, S., Wang, H. R., Causing, C. G., Kavsak, P., Stroschein, S. L., Luo, K. & Wrana, J. L. (2001) Nat. Cell Biol. 3, 587–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.