Abstract
Pre-mRNA splicing of Pol II transcripts is executed in the mammalian cell nucleus within a huge (21 MDa) and highly dynamic RNP machine — the supraspliceosome. It is composed of four splicing active native spliceosomes, each resembling an in vitro assembled spliceosome, which are connected by the pre-mRNA. Supraspliceosomes harbor protein splicing factors and all the five-spliceosomal U snRNPs. Recent analysis of specific supraspliceosomes at defined splicing stages revealed that they harbor all five spliceosomal U snRNAs at all splicing stages. Supraspliceosomes harbor additional pre-mRNA processing components, such as the 5′-end and 3′-end processing components, and the RNA editing enzymes ADAR1 and ADAR2. The structure of the native spliceosome, at a resolution of 20 Å, was determined by cryo-EM. A unique spatial arrangement of the spliceosomal U snRNPs within the native spliceosome emerged from in-silico studies, localizing the five U snRNPs mostly within its large subunit, and sheltering the active core components deep within the spliceosomal cavity. The supraspliceosome provides a platform for coordinating the numerous processing steps that the pre-mRNA undergoes: 5′ and 3′-end processing activities, RNA editing, constitutive and alternative splicing, and processing of intronic microRNAs. It also harbors a quality control mechanism termed suppression of splicing (SOS) that, under normal growth conditions, suppresses splicing at abundant intronic latent 5′ splice sites in a reading frame-dependent fashion. Notably, changes in these regulatory processing activities are associated with human disease and cancer. These findings emphasize the supraspliceosome as a multi-task master regulator of pre-mRNA processing in the cell nucleus.
Keywords: Pre-mRNA splicing, Riponucleoproteins (RNPs), U snRNPs, Alternative splicing, Intronic microRNA biogenesis, Suppression of splicing
1. Introduction
Eukaryotic pre-mRNAs are transcribed by RNA Polymerase II (Pol II) and have to undergo several processing events before they can exit from the nucleus to the cytoplasm to encode for proteins. These processing events include 5′ end and 3′ end processing, editing and splicing. In the cell nucleus each mammalian pre-mRNA is assembled in a huge nuclear ribonucleoprotein (RNP) complex – the supraspliceosome – in which pre-mRNA processing most likely takes place.
Splicing of pre-mRNAs to remove the introns and ligate the exons is a two-step transesterification reaction that occurs within the highly dynamic splicing machine. The accuracy and efficiency of pre-mRNA splicing are attributed to a number of cis elements, such as the 5′ and 3′ splice sites (SS), a branch point, a polypyrimidine tract, and splicing enhancers and silencers. These are recognized by transfactors, which include the spliceosomal uridine-rich snRNPs (U1, U2, U4, U5 and U6 snRNPs) and several non-snRNP protein splicing factors, such as the Serine/Arginine (SR) rich protein family and hnRNP proteins [1].
The U snRNPs are central components of the spliceosome. They participate in splice-site recognition and have an essential function in splicing through cooperative RNA:RNA interactions between the snRNAs and the pre-mRNA [2–4]. Studies in vitro have shown that the assembly of the spliceosome occurs in a stepwise manner (reviewed in refs [2,4,5]), and involves an intricate series of interactions between the five major U snRNPs, as well as with a number of non-snRNP splicing factors, which are dynamically recruited to the spliceosome when an exogenous pre-mRNA is added to a nuclear extract. A number of intermediate complexes were identified during the assembly of the spliceosome in vitro [6]. First, U1 snRNP is bound to the 5′SS of the pre-mRNA together with various proteins generating complex E. Next, the entry of U2 snRNP defines the branch-point and leads to the formation of the A complex. Binding of the U4/U6.U5 tri-snRNP forms the pre-catalytic B complex, that harbors the five-spliceosomal U snRNPs. This step is followed by major rearrangements in RNA:RNA and RNA:protein interactions, destabilization of U1 and U4 snRNPs and formation of activated complex Bact. This is followed by generation of complex B*, which catalyzed the first step of splicing, generating complex C, which catalyzes the second step of splicing. Thus, the splicing active complex contains U2/U6.U5 snRNPs [1]. Studies of the structure and function of the in vitro assembled spliceosome have been summarized in updated reviews [1,7,8].
Here we focus on the structure and function of the splicing machine isolated from the nuclei of mammalian cells — the supraspliceosome (reviewed in ref. [9]). This machine is not only responsible for splicing and splicing regulation in the cell nucleus, but also responsible for all pre-mRNA processing activities mentioned above and more, as will be discussed here.
2. The supraspliceosome
When isolated from cell nuclei, under physiological conditions, mammalian pre-mRNAs are found packaged with all five spliceosomal U snRNPs [10–12], and splicing factors [9–15] in huge (21 MDa) [16] nuclear RNP particles — supraspliceosomes. Composition analyses including mass spectrometry (MS) analysis (Table 1) of the general population of supraspliceosomes isolated from cell nuclei [17], or of affinity purified supraspliceosomes assembled on a specific transcript [18], reveled the presence of over hundred proteins, and confirmed the presence of all required splicing factors in supraspliceosomes [17,18]. Supraspliceosomes are composed of four splicing active substructures – native spliceosomes – connected by the pre-mRNA [19]. Importantly, the entire repertoires of nuclear pre-mRNAs, appear to be assembled in splicing-active supraspliceosomes (Table 2b and reviewed in ref. [9]). A remarkable feature of supraspliceosomes is that they individually package a single pre-mRNA transcript each of different sizes and of different numbers of introns (Table 2a) into complexes of a unique size and hydrodynamic properties, indicating their universal nature [10,11,19–22]. The supraspliceosome structure provides a platform where numerous regulatory processing steps that the pre-mRNA undergoes occur [9], emphasizing the role of the supraspliceosome as a master regulator of pre-mRNA processing. The question arises why is the nuclear processing machine so complex? In the following sections we will detail the characteristic of the supraspliceosome with the aim of trying to elucidate this point.
Table 1.
Proteins associated with supraspliceosomes.
| Protein | Cells origin | References |
|---|---|---|
| snRNP proteins | Syrian Hamster; HeLa; TD40 | [17,18,94] |
| hnRNP proteins | Syrian Hamster; HeLa; TD40 | [17,18,94] |
| SR protein family | HeLa; 293T HEK; TD40 | [10,14,17,18] |
| Additional regulatory splicing factors | HeLa; 293T HEK; TD40 | [13–15,17,18] |
| hPRP19 complex | HeLa; DT40 | [17,18] |
| 3′-end processing components | HeLa; TD40 | [17,18,39] |
| Cap binding proteins | HeLa; 293T HEK; DT40 | [17,18,39] |
| A-to-I RNA editing enzymes | HeLa; 293T HEK; TD40 | [17,43,95,96] |
| Microprocessor components | HeLa | [64] |
| mRNA export and surveillance components | HeLa; TD40 | [17,18] |
| RNA metabolism | HeLa; TD40 | [17,18] |
Table 2.
Nuclear RNAs associated with supraspliceosomes.
| 2a. pre-mRNA/mRNAs individually associated with supraspliceosomes | ||
|---|---|---|
| Pre-mRNA/mRNA | Cells origin | References |
| Poly(A) + nuclear RNA | Syrian hamster | [21] |
| CADa | Syrian hamster | [97] |
| DHFRa | Syrian hamster | [21] |
| ß-Actina | Syrian hamster; HeLa | [94] |
| Histone H4a | Syrian hamster; HeLa | [21] |
| AdML minigenea | HeLa | [18] |
| SMNa | HeLa; HEK 293 | [33] |
| ADAR 2a | HeLa; HEK 293 | [33] |
| hnRNP A/Ba | HeLa; HEK 293 | [33] |
| Tau minigenea | HEK 293 | [14] |
| HTR2C minigenea | HEK 293 | [57] |
| 2b. U snRNAs associated with supraspliceosomes | ||
| U snRNA | Cell origin | References |
| U1 | HeLa; Syrian hamster; TD40 | [10–12,17–19,98] |
| U2 | HEK 293; HeLa; Syrian hamster; TD40 | [10–12,17–19,87,98] |
| U4 | HEK 293; HeLa Syrian hamster; TD40 |
[10–12,17–19,87] |
| U5 | HeLa; Syrian hamster; TD40 | [10–12,17–19] |
| U6 | HeLa; Syrian hamster; TD40 | [10–12,17–19,98] |
| 2c. Small non-coding RNAs associated with supraspliceosomesb | ||
| Small non-coding RNA | Cell origin | References |
| Initiator-tRNA | HEK 293 | [87] |
| Intronic pre-miRNAsc: | ||
| miR-762 (HTR2C)d | HeLa | [69] |
| miR-1912 (HTR2C)d | HeLa | [69] |
| miR-1264 (HTR2C)d | HeLa | [69] |
| miR-1298 (HTR2C)d | HeLa | [69] |
| miR-106b (MCM7)d | HeLa | [64] |
| miR-93 (MCM7)d | HeLa | [64] |
| miR-25 (MCM7)d | HeLa | [64] |
| miR-330 (EML2)d | HeLa | [64] |
Examples of pre-mRNA/mRNAs each individually associated with a supraspliceosome.
A temporary list of small non-coding RNAs associated with supraspliceosomes.
A list of published intronic pre-miRNAs found associated with supraspliceosomes.
Host gene of intronic miRNA.
2.1. Supraspliceosomes isolated from cell nuclei are assembled with all five spliceosomal U SnRNPs during the steps of the splicing reaction
The U snRNPs are central components of the spliceosome. Studies in vitro have shown that the assembly of the spliceosome occurs in a stepwise manner with changes in U snRNP composition (reviewed in refs. [2,4,5,8]). While a pre-catalytic complex involves the five spliceosomal U snRNPs, the splicing active complex contains U2/U6.U5 snRNPs [1].
In contrast to these findings, Northern blot analysis (Table 2b) revealed that the general population of supraspliceosomes harbors the five-spliceosomal U snRNPs [10,12,19]. Also, native spliceosomes, the subunits of the supraspliceosome, harbor all five spliceosomal U snRNAs [19]. These findings are consistent with the finding of a functional penta-snRNP in yeast [23].
The above studies represent averaged-characteristics of the supraspliceosome as they dealt with the steady state population of Pol II transcripts that differ in their number of introns and exons, and in their stage of the splicing reaction. We recently analyzed the U snRNP composition of affinity purified specific supraspliceosomes assembled on specific transcripts at defined functional states [18]. We used the Pseudomonas aeruginosa phage 7 (PP7) RNA binding site/recombinant PP7 coat protein (PP7CP) system [24], to affinity purify PP7-tagged splicing complexes [18]. A series of stable cell lines was prepared (Fig. 1A, B), each expressing a PP7-tagged wild type (WT), or mutant (Mut) AdML, having a mutation at the 3′SS that inhibits the second step of splicing, and having the PP7-tag either at the 3′ UTR or at the intron. We also prepared control cell lines lacking the PP7 tag. Specific affinity purification of the AdML assembled complexes from each of the cell lines expressing PP7-tagged AdML was achieved, and revealed that they were assembled in supraspliceosomes [18]. AdML-WT supraspliceosomes were found assembled on mature RNA, and AdML Mut supraspliceosomes were found assembled on pre-mRNA. Northern blot analysis using probes for the five spliceosomal U snRNAs revealed that all five spliceosomal U snRNAs are associated with each of the above affinity-purified AdML supraspliceosomes: AdML-WT-PP73′UTR supraspliceosomes that are assembled on mature mRNA (Fig. 1C, lane 2); as well as on AdML-Mut-PP73′UTR and AdML-Mut-PP7IVS supraspliceosomes that are assembled on AdML pre-mRNA (lanes 6 and 10, respectively) [18]. Very low levels of U snRNAs were found in the control samples that lack the PP7 tag (lanes 4, 8, and 12), with some non-specific binding of U1 snRNA, which is known to be in excess in the cell nucleus [25]. It can therefore be concluded that supraspliceosomes isolated from mammalian cell nuclei have all five spliceosomal U snRNAs associated with them at all splicing stages. These findings highlight the important role of large, pre-formed, complexes in pre-mRNA splicing in vivo.
Fig. 1.
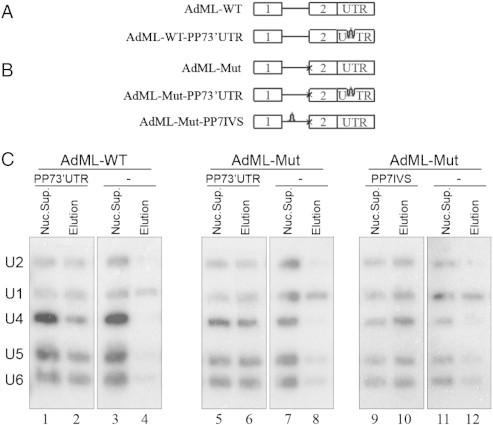
The five-spliceosomal U snRNAs are associated with in vivo assembled supraspliceosomes at all splicing stages. (A, B) Schemes of AdML minigenes, each expressed from a stable cell line from which specific supraspliceosomes at defined splicing stages were affinity purified using the PP7/PP7CP system [24]. Stable cell lines expressing the respective AdML transcripts lacking tag were used as controls. Open boxes represent exons, lines represent introns, and stem-loops represent the PP7 tag inserted either at the 3′UTR or the intron. (A) Constructs expressing AdML-WT. (B) Constructs expressing AdML-Mut, having a mutated 3′ splice site, designated “x”. (C) Northern blot analysis with probes directed against the five-spliceosomal U snRNAs was performed on RNA extracted from nuclear supernatants (Nuc. Sup.), and from affinity purified supraspliceosomes (Elution), prepared from HeLa cell-lines expressing the AdML constructs either with the PP7 tag or without it (−), as indicated. AdML-WT supraspliceosomes were found assembled on mature RNA, and AdML Mut supraspliceosomes were found assembled on pre-mRNA [18]. Left, the AdML-WT-PP73′UTR supraspliceosomes; middle, the AdML-Mut-PP73′UTR supraspliceosomes; Right, AdML-Mut-PP7IVS supraspliceosomes. The identity of the U snRNA probes is given on the left.
Adapted from Kotzer-Nevo et al. [18].
The apparent discrepancy between a stepwise assembly pathway of the spliceosome in vitro and the occurrence of a pre-formed splicing complex in vivo, has been explained by a “holospliceosome” model, in which the sequential complexes represent ordered modulations within the in vivo assembled spliceosome without the loss of components [4]. It has also been pointed out that such intermediate states in spliceosome assembly in vitro may not occur in vivo [23,26].
While the assembly pathways of supraspliceosomes in vivo occur in the nucleus co-transcriptionally, that of spliceosomes assembled in vitro occur when a full-length pre-mRNA is interacting with the spliceosomal components present in a nuclear extract. These two pathways may lead to different local minima in the respective free energy profiles, which may result in the assembly of slightly different complexes. Alternatively, the observed changes in composition between intermediate complexes assembled in vitro, which are not found in supraspliceosomes assembled in vivo, may be due to the lack of specific components in the in vitro assembled complexes, which might help keep the in vivo assembled complexes intact.
2.2. Supraspliceosome structure
The complexity and large size of the supraspliceosome make EM the method of choice for its structural analysis. 3-D image reconstruction of individual supraspliceosomes by automated electron tomography of negatively stained [20,27], and of frozen hydrated complexes [28] showed the supraspliceosome as forming a closed structure, 50 × 50 × 35 nm in dimensions, composed mainly of four similar native spliceosomes. Mass measurements by scanning transmission EM (STEM) showed that the supraspliceosome has a mass of 21 MDa, whereas each of the native spliceosomes has a mass of 4.8 MDa [16]. Notably, when the dynamics of U1 snRNPs within live cells was analyzed at the single-particle level [29], most U1 snRNPs were found bound to specific intranuclear sites, many of those presumably representing sites of pre-mRNA splicing. The dissociation kinetics from these sites reflects the involvement of U1 snRNP in numerous distinct interactions. Mobile U1 snRNPs moved with diffusion constants in the range of 0.5–8 μm2/s, values that are consistent with uncomplexed U1 snRNPs diffusing at a viscosity of 5 cPoise and U1 snRNPs moving in a largely restricted manner, and U1 snRNPs contained in large supramolecular assemblies such as spliceosomes or supraspliceosomes, diffusing in a hindered manner through the nucleoplasm [29].
The relatively uniform mass of the supraspliceosome (21.1 ± 1.6 MDa; n = 400) [16], likely reflects the presence of a general basic structure. Using a positive staining protocol, which allowed visualization of nucleic acids, we could show strands and loops of RNA emanating from positively stained supraspliceosomes [16] (see Fig. 3C), indicating that the RNA is loosely bound and therefore accessible for probing. Furthermore, fibers, presumably the pre-mRNA covered with proteins, through which the native spliceosomes are interconnected were observed by 2-D image restoration of ice-embedded particles [28]. These observations support our working hypothesis that the native spliceosomes are connected by the pre-mRNA. Within the supraspliceosome, the native spliceosomes are arranged such that their small subunits reside in its center. This configuration allows communication between the native spliceosomes [30].
Fig. 3.
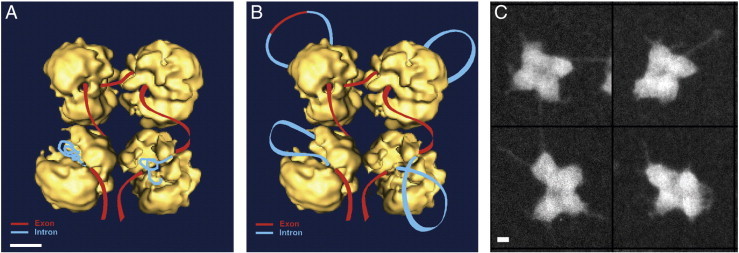
The supraspliceosome model. (A, B) Schematic models of the supraspliceosome in which the pre-mRNA (introns in blue, exons in red) is connecting four native spliceosomes. The supraspliceosome presents a platform onto which the exons can be aligned and splice junctions can be checked before splicing occurs. (A) The pre-mRNA that is not being processed is folded and protected within the cavities of the native spliceosome. (B, C) When a staining protocol that allows visualization of nucleic acids was used, RNA strands and loops bound with proteins were seen emanating from the supraspliceosomes [16]. (B) Under these conditions the RNA kept in the cavity is proposed to unfold and loop-out. In the looped-out scheme an alternative exon is depicted in the upper left corner. Adapted from Azubel et al. [19]. (C) STEM dark field images of supraspliceosomes [16] stained with a protocol that allows visualization of nucleic acids. Adapted from Muller et al. [16]. Bar represents 10 nm.
The four substructures of the supraspliceosome are interconnected in a flexible way and may thus adopt different angular settings, which impose a significant restriction on reaching high resolution in EM image analyses. We have therefore developed a methodology to isolate the native spliceosomes from supraspliceosomes, by specific cleavage of the general population of pre-mRNAs, while keeping the snRNAs within these substructures intact [19]. The 3-D structure of the native spliceosome was determined by the cryo-EM single particle technique at a resolution of 20 Å, and revealed an elongated globular particle composed of a large and a small subunit (Fig. 2A–C) [31]. The two subunits are interconnected to each other leaving a tunnel in between, which is large enough to allow the pre-mRNA to pass through (Fig. 2A). The other side of the native spliceosome exposes a cavity that could provide a place to transiently store the labile pre-mRNA and protect the part of the pre-mRNA that is not directly involved in the splicing reaction from non-specific degradation (Fig. 2B). Because RNA is denser than protein, high-density regions can provide some information about its internal localization. The large subunit was thus proposed as a suitable candidate to accommodate the five spliceosomal U snRNPs, as the high density regions were found there (Fig. 2C) [31].
Fig. 2.
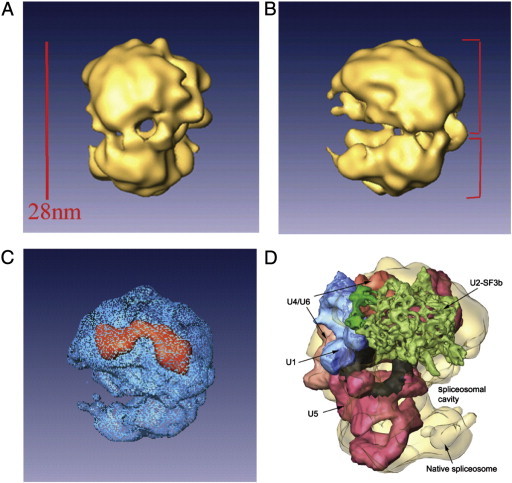
Structure of the native spliceosome.
(A, B) Two different views of the structure of the native spliceosome reconstructed at 20 Å resolution from cryo-images [31]. (C) The high-density mass region of the native spliceosome (red) represents the stable RNAs within the structure of the native spliceosome. The large subunit of the native spliceosome is thus a suitable candidate to harbor the five-spliceosomal U snRNPs. Adapted from Azubel et al. [31]. (D) A unique spatial arrangement of the U snRNPs within the native spliceosome emerges from in-silico studies [32]. The native spliceosome is transparent; U4/U6.U5 tri-snRNP is colored by functional regions, with U5 snRNP in pink and the region attributed to loop I in black [93]; U4/U6 in beige-orange; U2 snRNP subcomponent SF3b is in green; and U1 snRNP is blue. Adapted from Frankenstein et al. [32].
Support for this suggestion, comes from our recent analysis of the localization of U snRNPs within the native spliceosome, using a new computational procedure we designed to efficiently fit thousands of conformers into the spliceosome envelope. Despite the low-resolution limitations, a unique spatial arrangement of the spliceosomal U snRNPs within the native spliceosome emerged from our in-silico studies (Fig. 2D). Our model localizes the five U snRNPs mostly within the large subunit of the native spliceosome, requiring only minor conformation changes. The remaining free volume presumably accommodates additional spliceosomal components. The constituents of the active core of the spliceosome are juxtaposed, forming a continuous surface deep within the large spliceosomal cavity, which provides a sheltered environment for the splicing reaction [32].
3. The supraspliceosome model
In the proposed model (Fig. 3), the supraspliceosome represents a stand-alone complete macromolecular machine capable of performing splicing of every pre-mRNA — irrespective of its length or the number of introns it contains. It presents a closed-structure-model composed of four native spliceosomes connected by a single pre-mRNA. Critical experimental support of this model comes from experiments, which showed that cleavage of the pre-mRNA yielded functional native spliceosomes that could be reconstituted into supraspliceosomes by incubation with exogenously added pre-mRNAs [19,31]. The hypothesis that each supraspliceosome is assembled on a single pre-mRNA was verified by EM visualization of supraspliceosomes reconstituted from native spliceosomes and from pre-mRNA tagged at its 3′ end with one Nanogold particle (1.4 nm) [33]. The supraspliceosome structure provides a platform to juxtapose exons about to be spliced, and each of the four native spliceosomes, resembling an in vitro assembled spliceosome, can splice the intron wound around it (Fig. 3). In this configuration the supraspliceosome acts as a multiprocessor machine that can simultaneously splice four introns — not necessarily in a consecutive manner.
In this schematic model, each of the native spliceosomes is presented by the 20-Å resolution structure of the native spliceosome [31]. The small subunit of each native spliceosome, proposed to harbor non-snRNP components such as SR proteins and hnRNP proteins, is placed at the center of the supraspliceosome [30]. This configuration allows communication between the native spliceosomes, which is a crucial element for regulated alternative splicing and for quality control of the resulting mRNAs. This setting places the large subunit of each native spliceosome, where catalysis by the U snRNPs presumably takes place, in the periphery of the supraspliceosome. This localization of the U snRNPs within the native spliceosome was recently supported by our in-silico study showing unique localization of the U snRNPs within the native spliceosome [32]. Thus, the supraspliceosome presents a platform on which splice junctions could be checked prior to intron excision. An attractive feature of such a machine is that it allows rearrangement of splice junction combinations to select the appropriate ones. This way it comprises an important tool to ensure the fidelity of splicing and alternative splicing.
Splicing of a multi-intronic pre-mRNA can be facilitated by the translocation of the pre-mRNA through the complex in a ‘rolling model’ fashion. After processing four introns the RNA roles in to place a new subset of introns in the correct position for processing. Cotranscriptional splicing [34–36] may help explain how pre-mRNAs having an exceptionally large number of introns (e.g. dystrophin or CAD) can be spliced by the supraspliceosome in a rolling mode. At the other extreme, both reconstitution experiments with pre-mRNAs having only one intron [19], and affinity purification of supraspliceosomes assembled on PP7-tagged AdML transcripts with one intron [18], revealed that pre-mRNAs having less then four introns are also packaged in supraspliceosomes. Thus, the interactions of the RNA with the native spliceosomes per se are presumably sufficient to hold the structure together.
4. Supraspliceosomes function
4.1. Splicing
We have shown that both native spliceosomes and supraspliceosomes are active in splicing [19,31]. This was shown by complementing splicing activity of a micrococcal nuclease treated HeLa nuclear extract, which was inactive in splicing. We have further shown that this splicing activity could be attributed to the native spliceosomes and not to the U snRNAs or U snRNPs they contain. Support for the functional significance of supraspliceosomes was provided from a study showing that U2/U6 snRNA base pairing, which characterizes active spliceosomes assembled in vitro, was found in complexes sedimenting between 150-300S but not in 60S complexes [37]. These studies are in support of the supraspliceosome model, in which the native spliceosomes within the supraspliceosome are active in splicing.
4.2. Pre-mRNA processing activities: 5′ end, 3′ end processing and editing
Transcripts of Pol II have to undergo several processing steps before they can exit from the nucleus to the cytoplasm to encode for proteins. These activities include, in addition to splicing, 5′ end and 3′ end processing, and RNA editing. Since Pol II transcripts are assembled in supraspliceosomes from the transcription site until they reach the nuclear pore for export from the nucleus to the cytoplasm [38], we asked if components of the additional processing activities are present in supraspliceosome. We showed that the cap-binding proteins (CBPs), CBP20 and CBP80 are associated with supraspliceosomes (Table 1), and so are components of 3′ end processing [39]. These findings were confirmed my MS analyses [17,18].
The ADAR (adenosine deaminases acting on RNA) editing enzymes act on double stranded RNA and convert A-to-I. Because inosines are recognized by the splicing and translation machineries as guanosines, RNA editing in coding regions or in introns can change the codons, or splicing patterns, respectively [40–42]. The involvement of introns in editing, either as substrates for editing or as part of the complementary strand of the RNA duplex, suggested that A-to-I editing is likely to occur in supraspliceosomes. Accordingly, we have shown that the editing enzymes ADAR1 and ADAR2 are associated with supraspliceosomes (Table 1), and that they are enzymatically active [43]. These findings were confirmed my MS analyses [17,18].
4.3. Alternative splicing
Alternative splicing (AS) is a major source of the diversity of the human proteome and plays a major role in the regulation of gene expression. It is estimated that the majority of human genes undergo regulated AS [44–47]. Importantly, alterations in AS and misregulation of factors affecting AS were shown to be involved in several human diseases, including cancer [48–51].
Regulated splicing, as well as constitutive splicing, operates through the combinatorial interplay of positive and negative regulatory signals present in the pre-mRNA, which are recognized by trans-acting factors. The most studied of the latter are members of the hnRNP [52,53] and SR protein families [54–56]. The high fidelity of exon recognition is thus achieved by the combination of multiple weak protein:protein, protein:RNA and RNA:RNA interactions.
Because the entire repertoire of nuclear pre-mRNAs is individually found assembled in supraspliceosomes, we asked if the ability to perform alternative splicing is an integral part of the in vivo splicing machine. The tetrameric structure of the supraspliceosome, in which four native spliceosomes are connected by the pre-mRNA, suggests that such a multi-subunit structure was designed to coordinate multiple processing events, such as alternative splicing. In accordance with this suggestion, we have shown that the supraspliceosome contains splicing regulatory proteins as integral components (Table 1). The list includes the splicing regulators RBM4 and Wilm's tumor protein 1 (WT1) [13], the alternative splicing regulator ZRANB2 [15], and the splicing regulatory proteins belonging to the SR proteins family [10,39], and hnRNP G [14], which are predominantly found in supraspliceosomes, as well as additional splicing regulators identified by MS of affinity purified supraspliceosomes [18]. The association of regulatory splicing factors with supraspliceosomes is in support of their proposed role in splicing regulation and alternative splicing.
Importantly, the supraspliceosome was shown as the nuclear complex in which alternative splicing is regulated. First, hnRNP G was shown to affect the alternative splicing of the tau pre-mRNA transcript in supraspliceosomes [14]. Furthermore, both the constitutively and alternatively spliced mRNAs of the endogenous human pol II transcripts: hnRNP A/B, survival of motor neuron (SMN) and ADAR2 are predominantly found in supraspliceosomes. Notably, overexpression of SRSF5 (SRp40) enhanced exon 7 skipping in supraspliceosomes assembled on hnRNP A/B transcripts, while treatment with C6-ceramide was accompanied with exon 7 inclusion in hnRNP A/B supraspliceosomes [33].
Additional studied case is the regulation of alternative splicing of the serotonin receptor 2C (HTR2C) transcripts that occur in supraspliceosomes. A recent screen for substances that promote production of active HTR2C, through alternative splicing including exon 5b, identified pyrvinium pamoate as a drug promoting exon 5b inclusion [57]. We have shown that the HTR2C mRNA is found in supraspliceosomes. Importantly, addition of pyrvinium pamoate resulted in increase in exon 5b inclusion within the supraspliceosome fractions [57]. These findings support the proposed role of the supraspliceosome in splicing regulation and alternative splicing.
4.4. Intronic miRNA processing in supraspliceosomes
MicroRNAs (miRNAs) are small ~ 22 nt long molecules involved in the negative control of gene expression by binding mainly to the 3′UTR of target mRNA transcripts. Notably, a large fraction of miRNA genes are located in introns. The canonical biogenesis of intronic miRNAs from Pol II transcripts involves two main steps: First, cleavage of the primary (pri) miRNA transcript into a precursor (pre) miRNA stem-loop molecule of about 70–80 bases by the microprocessor in the nucleus. Second, after export to the cytoplasm, the pre-miRNA is cleaved by Dicer yielding mature miRNA, which loaded on the RNA Induced Silencing Complex (RISC) binds to its target gene [58–63].
Since a large proportion of miRNA genes are located in introns, we asked whether there is a cross-talk between their processing and pre-mRNA splicing within the in vivo pre-mRNA processing machine. In a recent study [64], we showed that the main microprocessor components, Drosha and DGCR8 are found in supraspliceosomes (Table 1). We further showed that inhibition of splicing increases miRNA expression, while knock-down of Drosha increases splicing. In order to analyze the processing of specific miRNAs, we focused on the miR-106b-25 cluster (miRNAs 106b, 93 and 25), which is conserved in mammals and is pro-oncogenic [65]. The miR-106b-25 cluster is harbored in intron 13 of MCM7 (minichromosome maintenance 7) [62,66], which plays a critical role in the G1/S phase transition, ensuring that all the genome is only replicated once at each cell cycle [65]. RT-PCR analysis revealed that pri-miR 25 is found in supraspliceosomes. Furthermore, deep sequencing of small RNA (< 200 nt) from supraspliceosomes revealed the presence of pre-miRNA 106b, pre-miRNA 93 and pre-miRNA 25 (Table 2c). This study showed that both the pri-miRNAs and the pre-miRNAs of the miR-106b-25 cluster are found in supraspliceosomes [64]. These findings bring further support to the cross-talk between the splicing and miRNA processing machines and to the hypothesis that the processing of the tested intronic miRNAs occurs in supraspliceosomes [64]. Earlier findings of association between certain splicing components and the microprocessor and pre-miRNAs [67,68] are in support of our findings.
An additional interplay between splicing and miRNA processing was unraveled when we identified a novel alternative 3′ splicing event in intron 13 of MCM7, between pre-miRNAs 93 and 25, which occurred at a highly conserved 3′SS sequence. Importantly, the new splice isoform incorporates the sequence of pre-miR-25 into exon 14 of MCM7. While both miRNA 25 and MCM7 mRNA can be generated from the same MCM7 pre-mRNA, when spliced at the normal SS, this is not the case when splicing occurs at the novel 3′SS. Splicing at this novel 3′SS excludes processing of pri-miRNA-25 into miR-25, affecting its biogenesis, and at the same time generates a new isoform of MCM7 mRNA, which encodes for a shortened protein that lacks the 3′ carboxy terminus. Thus, the expression of the novel splice isoform affects the levels of the different miRNAs of the cluster and also affects the splice isoforms of the hosting mRNA originating from the same transcript. This study shows a cross-talk and competition between splicing and miRNA biogenesis, which might also differentially affect the levels of clustered intronic miRNAs [64].
Among RNA sequences, found in deep sequencing of small RNAs from supraspliceosomes isolated from nuclei of HeLa cells, we identified sequences of pre-miRNAs of miR-764, miR-1260, miR-1912 and miR-1298 (Table 2c and Ref. [69]). These miRNAs are encoded in intron 2 of the serotonin receptor 2C (HTR2C), known to be exclusively expressed in neurons [70]. This observation led to the finding that HTR2C, which encodes for a G-protein coupled receptor, has an additional transcript coming from the 5′ untranslated region of the receptor, and this pre-mRNA as well as its hosted miRNAs are widely expressed in non-neuronal cell lines. This is an additional example of cross talk between the supraspliceosome and the processing of microRNA [69]. Furthermore, the presence of both pri-miRNAs and pre-miRNAs in supraspliceosomes [64,69] suggests that this nuclear processing step likely occurs in supraspliceosomes.
4.5. Stop codon-mediated suppression of splicing — SOS
A key step in pre-mRNA splicing involves the recognition and selection of a consensus sequence (AG/GTRAGT; in mammals, where R denotes purine and “/” denotes the splice junction) that defines the 5′SS [71]. An estimate based on alignment of EST data showed that splicing involving alternative 5′SSs account to about 8% of the total alternative splicing events that are conserved between the human and the mouse genomes [72]. This fraction, however, is much smaller than that expected from sequence alone. We found that over 70% of the introns within the coding regions of human genes harbor multiple sequences that comply with the 5′SS consensus (on average 9.19 sequences/intron [73]), but these sequences are not used for splicing (latent 5′SSs). Importantly, the intronic sequences upstream of almost all of these sites (> 98%) harbor at least one in-frame stop codon and therefore has the potential of introducing premature termination codons (PTCs) into the alternatively spliced isoform [73]. mRNAs that contain PTCs (nonsense mRNAs) might be toxic to cells as they code for truncated nonfunctional proteins that could have a deleterious dominant negative effect on the cell's metabolism [74–76].
The presence of such a large number of latent 5′SS raises the question: why splicing at latent 5′SSs (latent splicing) has not been detected under normal growth conditions? In principle, two scenarios can account for this phenomenon (Fig. 4A): (i) Splicing at latent 5′SSs does occur, but the nonsense mRNAs thus formed are rapidly and efficiently degraded, by any RNA surveillance mechanism (e.g., NMD) [74–78] to a level below detection; and (ii) Splicing events at intronic 5′SSs that are preceded by at least one stop codon in frame with the upstream exon are suppressed. We have tested the first scenario in a substantial number of independent experiments: It was thus shown that SOS was not affected (i.e. latent splicing was not elicited in the tested gene transcripts) upon abrogation or bypassing the NMD pathway by a number of ways: Inhibition of translation [79–81] and, in particular, inhibition of the pioneer round of translation, which had been shown to be essential for NMD [82], by expressing a dominant negative mutant of the translation initiation factor eIF2-α, did not elicit latent splicing [81]. Furthermore, RNAi of the NMD genes hUpf1 and hUpf2, or the expression of three mutants of hUpf1 that abrogated NMD [83], did not elicit latent splicing [80]. Because Upf1 is essential for mammalian NMD [84–86], these data do not fit a model that could attribute the lack of latent splicing to a rapid and complete degradation of latent mRNAs by NMD. We also ruled out degradation of latent mRNA by a yet unknown RNA degradation mechanism by showing that constructs in which we forced formation of latent mRNA, through its expression from a plasmid harboring the already spliced DNA, express latent mRNA at levels only slightly lower than the level of authentic mRNA expressed from a plasmid harboring the already spliced DNA at the authentic 5′SS [87]. On the other hand, the data summarized above, fit the second scenario and invoke a mechanism termed suppression of splicing (SOS) that suppresses splicing involving latent alternative 5′SSs whose usage could introduce an intronic stop codon into the resultant mRNA [88].
Fig. 4.
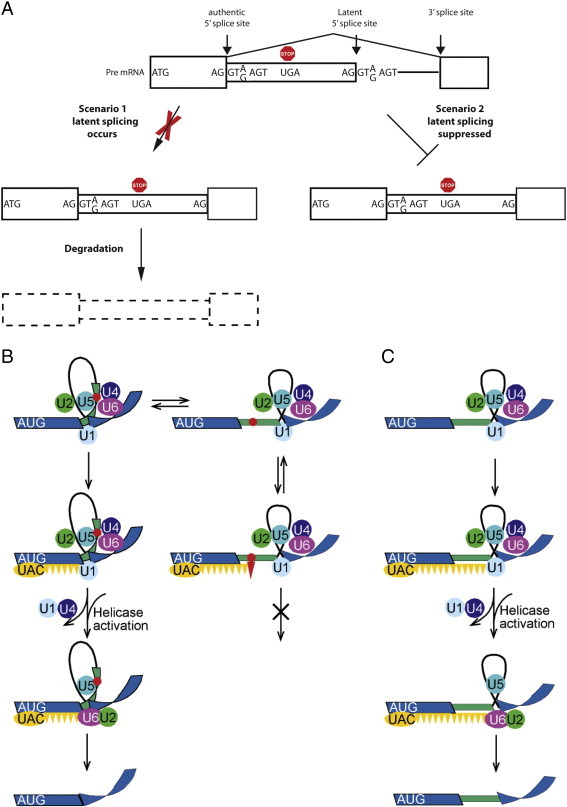
Suppression of splicing (SOS). (A) A scheme depicting the two scenarios that can account for lack of latent splicing under normal growth conditions, despite the abundance of latent 5′SS sequences in introns. As discussed, we have ruled out the 1st scenario [79–81,87]. Boxes, exons; narrow boxes, latent exons; lines, introns; red octagon, stop codon. (B, C) A schematic model for the quality control function of SOS. (B) Left scheme, splicing at the authentic 5′SS; right column, splicing at the latent 5′SS. (C) Splicing at the latent 5′SS after elimination of the in frame stop codon. Blue stripes, exons; black line, intron; green narrow stripe, latent exon; red octagon, stop codon; circles, U snRNPs; orange ellipse (UAC), hypothesized AUG-binding complex of initiator-tRNA; orange triangles, hypothesized triplet-binding proteins; red triangle, stop-codon-binding protein.
Adapted from Kamhi et al. [87].
Supporting evidence for the idea that PTC-harboring pre-mRNAs can be recognized in the cell nucleus, resulting in suppression of splicing has also been provided in a number of studies [89–92], including a recent study showing that un-spliced PTC-harboring transcripts are retained at nuclear transcription sites [92].
Latent 5′SSs are indeed legitimate and functional as has been shown by eliciting latent splicing in three different ways. First, by eliminating the stop codons (in several gene constructs) either by point mutations that converted the stop codons to sense codons or by frame shifting through the insertion (or deletion) of nucleotides upstream of the stop codons [79,80]. Second, by disrupting the reading frame through mutations made in the start ATG codon [81,87]. Third, by subjecting cells to stress conditions, such as heat-shock, hypoxia, or treatment with certain antibiotics [22,73,80,81], or in cancer cells, where activation of latent splicing was recently found in thousands of gene transcripts expressed in breast cancer cells and in gliomas [73].
It has been shown that AUG sequences are essential for SOS, while protein synthesis is not required. Mutations in the translation initiation codon (AUG) elicited latent splicing — even though the stop codons remained intact [81]. This finding indicated that maintenance of an open reading frame is required to sustain SOS, and suggested a role in SOS for the initiator-tRNA. Indeed, we have shown that this molecule may also act, in a manner that is independent of its role in protein translation, as a pre-mRNA splicing regulator. Specifically, we showed that activation of latent splicing, induced by mutations in the translation initiation AUG codon, could be suppressed by expressing initiator-tRNA constructs carrying anticodon mutations that compensate for the AUG mutations [87]. Importantly, the initiator-tRNA species proposed to participate in SOS reside in the cell nucleus, are found associated with supraspliceosomes (Table 2c), and appear not to be charged with an amino acid [87].
For the mechanism of SOS we have proposed a working model, based on what is known on the splicing mechanism and on our published observations, and termed it sense triplet-recognition mechanism that can be interrupted by stop codon-binding proteins (Fig. 4B). As its name implies, this mechanism is based on three elements. First, as already shown [87], the AUG sequence is recognized by the complementary anticodon (UAC) of the initiator-tRNA, probably in a complex with auxiliary proteins. This step is proposed to recruit the SOS mechanism and helps establish a register for the recognition of the reading frame. We next hypothesized that this step involves the cooperative polymerization of a protein(s) that bind triplets of nucleotides and in the lack of PTC constitutes the “approval” step of splice site selection that triggers the remodeling of the spliceosome to its functional state. The latter step is interfered in the presence of a PTC, perhaps through a competing interaction with a stop-codon-binding protein (e.g. a release factor-like protein). The unproductive complex may undergo a conformational change and revert to the productive splicing complex involving the authentic 5′SS, as indicated by the double arrows (see Fig. 4B).
The association of initiator-tRNA with the supraspliceosome suggests that the SOS mechanism, is likely incorporated as part of the splicing machine and may thus act as a quality control mechanism that scans along exons, avoiding the looped out introns and ensures the production of a translatable mRNA.
5. Summary and outlook
In the cell nucleus, mammalian transcripts of Pol II are assembled in supraspliceosomes (reviewed in ref [9]) and have to undergo several processing modifications before they can exit from the nucleus to the cytoplasm to encode for proteins. In addition to the 5′ end, and 3′ end processing, splicing and editing activities, most transcripts are multi-intronic and thus undergo alternative splicing. Furthermore, a large fraction of mammalian introns harbor small non-coding RNAs, (e.g. microRNAs), and thus their processing should be coordinated with the other pre-mRNA processing activities. As summarized here, all these processing activities: 5′ and 3′ end processing; RNA editing; constitutive splicing; alternative splicing; and intronic microRNA processing, occur in supraspliceosomes, the nuclear pre-mRNA processing machine.
Furthermore, since each transcript is subjected to a large number of processing activities, which are sometimes complementary, yet, sometimes competitive, the nuclear pre-mRNA processing machine should harbor also a quality control mechanism to coordinate all these processing activities and to ensure the production of a “meaningful” translatable mRNA. The SOS mechanism, which is part of the supraspliceosome provides for such quality control. The requirements for a quality control mechanism clarify also the question why the supraspliceosome is composed of multiple spliceosomes. A splicing machine composed of multi-spliceosomes enables coordinated processing of a number of spliceosomes (e.g. alternative splicing), so that the quality of the mRNA can be assessed before splicing, and only “correct combinations” of splice junction are approved. When “wrong combinations” of splice junctions are presented to the splicing machine, the quality control mechanism will not confirm such a combination and the unproductive complex may undergo a conformational change and revert to the productive splicing complex. The fact that the supraspliceosome is a closed structure in which the small subunits are placed in the center might explain why the supraspliceosome is composed of four subunits connected by the pre-mRNA. Because this structure enables close contact between the small subunits, which is likely required for coordinated processing of the pre-mRNA.
Introns of mammalian Pol II transcripts are long, and some of these intronic sequences encode for miRNAs, which are processed in supraspliceosomes, as discussed here. It is likely that these large intronic sequences may also host additional non-coding RNAs that are likely processed within supraspliceosomes. Thus further analyses of the supraspliceosome might help elucidate the processing of novel intronic small non-coding RNAs. Future structural and functional studies are required to decipher how all these processing activities are orchestrated within this giant nuclear pre-mRNA processing machine, and what changes and defects to this machine occur in disease.
Acknowledgments
We thank the US NIH, grant RO1 GM079549 (R.S. and J.S.), The Israel Science Foundation, grant No 624/14 (R.S.and J.S.) and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (J.S. and K.S.) for financial support.
References
- 1.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge C.B., Tuschl T.H., Sharp P.A. The RNA world. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 525–560. [Google Scholar]
- 3.Staley J.P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Brow D.A. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 5.Will C.L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 6.Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Galej W.P., Nguyen T.H., Newman A.J., Nagai K. Structural studies of the spliceosome: zooming into the heart of the machine. Curr Opin Struct Biol. 2014;25C:57–66. doi: 10.1016/j.sbi.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W.J., Moore M.J. The spliceosome: disorder and dynamics defined. Curr Opin Struct Biol. 2014;24:141–149. doi: 10.1016/j.sbi.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperling J., Azubel M., Sperling R. Structure and function of the pre-mRNA splicing machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Yitzhaki S., Miriami E., Sperling J., Sperling R. Phosphorylated Ser/Arg-rich proteins: limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc Natl Acad Sci U S A. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperling R., Sperling J. In: RNP particles, splicing and autoimmune diseases. Schenkel J., editor. Springer; 1998. pp. 29–47. [Google Scholar]
- 12.Miriami E., Angenitzki M., Sperling R., Sperling J. Magnesium cations are required for the association of U small nuclear ribonucleoproteins and SR proteins with pre-mRNA in 200 S large nuclear ribonucleoprotein particles. J Mol Biol. 1995;246:254–263. doi: 10.1006/jmbi.1994.0081. [DOI] [PubMed] [Google Scholar]
- 13.Markus M.A., Heinrich B., Raitskin O., Adams D.J., Mangs H., Goy C. WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp Cell Res. 2006;312:3379–3388. doi: 10.1016/j.yexcr.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich B., Zhang Z., Raitskin O., Hiller M., Benderska N., Hartmann A.M. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y.-H.J., Markus A.M., Mangs H.A., Raitskin O., Sperling R., Morris B.J. ZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptome. Mol Biol Rep. 2013;40:5381–5395. doi: 10.1007/s11033-013-2637-9. [DOI] [PubMed] [Google Scholar]
- 16.Müller S., Wolpensinger B., Angenitzki M., Engel A., Sperling J., Sperling R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J Mol Biol. 1998;283:383–394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y.I., Moore R.E., Ge H.Y., Young M.K., Lee T.D., Stevens S.W. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotzer-Nevo H., de Lima Alves F., Rappsilber J., Sperling J., Sperling R. Supraspliceosomes at defined functional states present portray the pre-assembled nature of the pre-mRNA processing machine in the cell nucleus. Int J Mol Sci. 2014;15:11637–11664. doi: 10.3390/ijms150711637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azubel M., Habib N., Sperling J., Sperling R. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J Mol Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 20.Sperling R., Koster A.J., Melamed-Bessudo C., Rubinstein A., Angenitzki M., Berkovitch-Yellin Z. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J Mol Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- 21.Spann P., Feinerman M., Sperling J., Sperling R. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc Natl Acad Sci U S A. 1989;86:466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miriami E., Sperling J., Sperling R. Heat shock affects 5′ splice site selection, cleavage and ligation of CAD pre-mRNA in hamster cells, but not its packaging in lnRNP particles. Nucleic Acids Res. 1994;22:3084–3091. doi: 10.1093/nar/22.15.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens S.W., Ryan D.E., Ge H.Y., Moore R.E., Young M.K., Lee T.D. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 24.Hogg J.R., Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baserga S.J., Steitz J.A. The RNA world. In: Gesteland R.F., Atkins J.F., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1993. pp. 359–381. [Google Scholar]
- 26.Nilsen T.W. The spliceosome: no assembly required? Mol Cell. 2002;9:8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- 27.Medalia O., Koster A.J., Tocilj A., Angenitzki M., Sperling J., Berkovitch Y.Z. Automated electron tomography of large nuclear RNP (lnRNP) particles — the naturally assembled complexes of precursor messenger RNA and splicing factors. J Struct Biol. 1997;120:228–236. doi: 10.1006/jsbi.1997.3926. [DOI] [PubMed] [Google Scholar]
- 28.Medalia O., Typke D., Hegerl R., Angenitzki M., Sperling J., Sperling R. Cryoelectron microscopy and cryoelectron tomography of the nuclear pre-mRNA processing machine. J Struct Biol. 2002;138:74–84. doi: 10.1016/s1047-8477(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 29.Grunwald D., Spottke B., Buschmann V., Kubitscheck U. Intranuclear binding kinetics and mobility of single native U1 snRNP particles in living cells. Mol Biol Cell. 2006;17:5017–5027. doi: 10.1091/mbc.E06-06-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen-Krausz S., Sperling R., Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J Mol Biol. 2007;368:319–327. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 31.Azubel M., Wolf S.G., Sperling J., Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell. 2004;15:833–839. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 32.Frankenstein Z., Sperling J., Sperling R., Eisenstein M. A unique spatial arrangement of the snRNPs within the native spliceosome emerges from In silico studies. Structure. 2012;20:1097–1106. doi: 10.1016/j.str.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebbag-Sznajder N., Raitskin O., Angenitzki M., Sato T.A., Sperling J., Sperling R. Regulation of alternative splicing within the supraspliceosome. J Struct Biol. 2012;177:152–159. doi: 10.1016/j.jsb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neugebauer K.M. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 35.Kornblihtt A.R. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 36.de Almeida S.F., Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 37.Wassarman D.A., Steitz J.A. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in > 150S complexes in HeLa cell extracts: Implications for the spliceosome assembly pathway. Proc Natl Acad Sci U S A. 1993;90:7139–7143. doi: 10.1073/pnas.90.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iborra F.J., Jackson D.A., Cook P.R. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J Cell Sci. 1998;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- 39.Raitskin O., Angenitzki M., Sperling J., Sperling R. Large nuclear RNP particles-the nuclear pre-mRNA processing machine. J Struct Biol. 2002;140:123–130. doi: 10.1016/s1047-8477(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 40.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79(79):321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savva Y.A., Rieder L.E., Reenan R.A. The ADAR protein family. Genome Biol. 2012;13 doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raitskin O., Cho D.S., Sperling J., Nishikura K., Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc Natl Acad Sci U S A. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Dov C., Hartmann B., Lundgren J., Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- 46.Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coelho M.B., Smith C.W. Regulation of alternative pre-mRNA splicing. Methods Mol Biol. 2014;1126:55–82. doi: 10.1007/978-1-62703-980-2_5. [DOI] [PubMed] [Google Scholar]
- 48.Srebrow A., Kornblihtt A.R. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 49.Pajares M.J., Ezponda T., Catena R., Calvo A., Pio R., Montuenga L.M. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- 50.Ward A.J., Cooper T.A. The pathobiology of splicing. J Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irimia M., Blencowe B.J. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 53.Busch A., Hertel K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin S., Fu X.D. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 55.Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long J.C., Caceres J.F. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 57.Shen M., Bellaousov S., Hiller M., de La Grange P., Creame T.P., Malina O. Pyrvinium pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Res. 2013;41:3819–3832. doi: 10.1093/nar/gkt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 59.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y.K., Kim V.N. Processing of intronic microRNAs. Embo J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shomron N., Golan D., Hornstein E. An evolutionary perspective of animal microRNAs and their targets. J Biomed Biotechnol. 2009;2009:594738. doi: 10.1155/2009/594738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agranat-Tamir L., Shomron N., Sperling J., Sperling R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucl Acids Res. 2014;42:4640–4651. doi: 10.1093/nar/gkt1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrocca F., Vecchione A., Croce C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 66.Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 68.Kataoka N., Fujita M., Ohno M. Functional association of the microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z., Falaleeva M., Agranat-Tamur L., Pages A.P., Eyras E.E., Sperling J. The 5′ untranslated region of the serotonin receptor 2C pre-mRNA generates miRNAs and is expressed in non-neuronal cells. Exp Brain Res. 2013;230:387–394. doi: 10.1007/s00221-013-3458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pompeiano M., Palacios J.M., Mengod G. Distribution of the serotonin 5-HT2 receptor family mRNAs: comparison between 5-HT2A and 5-HT2C receptors. Brain Res. 1994;23:163–178. doi: 10.1016/0169-328x(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 71.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugnet C.W., Kent W.J., Ares M., Jr., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput. 2004;66–77 doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 73.Nevo Y., Kamhi E., Jacob-Hirsch J., Amariglio N., Rechavi G., Sperling J. Genome-wide activation of latent donor splice sites in stress and disease. Nucleic Acids Res. 2012;40:10980–10994. doi: 10.1093/nar/gks834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Popp M.W., Maquat L.E. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schweingruber C., Rufener S.C., Zund D., Yamashita A., Muhlemann O. Nonsense-mediated mRNA decay — mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta Gene Regul Mech. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Behm-Ansmant I., Kashima I., Rehwinkel J., Sauliere J., Wittkopp N., Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 77.Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 78.Nicholson P., Yepiskoposyan H., Metze S., Zamudio Orozco R., Kleinschmidt N., Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li B., Wachtel C., Miriami E., Yahalom G., Friedlander G., Sharon G. Stop codons affect 5′ splice site selection by surveillance of splicing. Proc Natl Acad Sci U S A. 2002;99:5277–5282. doi: 10.1073/pnas.082095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wachtel C., Li B., Sperling J., Sperling R. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA. 2004;10:1740–1750. doi: 10.1261/rna.7480804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamhi E., Yahalom G., Kass G., Hacham Y., Sperling R., Sperling J. AUG sequences are required to sustain nonsense-codon-mediated suppression of splicing. Nucleic Acids Res. 2006;34:3421–3433. doi: 10.1093/nar/gkl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiu S.Y., Lejeune F., Ranganathan A.C., Maquat L.E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mendell J.T., ap Rhys C.M.J., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 84.Gehring N.H., Kunz J.B., Neu-Yilik G., Breit S., Viegas M.H., Hentze M.W. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 85.Chan W.K., Huang L., Gudikote J.P., Chang Y.F., Imam J.S., MacLean J.A., II An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsuda D., Hosoda N., Kim Y.K., Maquat L.E. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 87.Kamhi E., Raitskin O., Sperling R., Sperling J. A potential role for initiator-tRNA in pre-mRNA splicing regulation. Proc Natl Acad Sci U S A. 2010;107:11319–11324. doi: 10.1073/pnas.0911561107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sperling J., Sperling R. Nuclear surveillance of RNA polymerase II transcripts. RNA Biol. 2008;5:220–224. doi: 10.4161/rna.7162. [DOI] [PubMed] [Google Scholar]
- 89.Mühlemann O., Mock-Casagrande C.S., Wang J., Li S., Custodio N., Carmo-Fonseca M. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol Cell. 2001;8:33–44. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- 90.Aoufouchi S., Yelamos J., Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell. 1996;85:415–422. doi: 10.1016/s0092-8674(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 91.Gersappe A., Burger L., Pintel D.J. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J Biol Chem. 1999;274:22452–22458. doi: 10.1074/jbc.274.32.22452. [DOI] [PubMed] [Google Scholar]
- 92.de Turris V., Nicholson P., Orozco R.Z., Singer R.H., Mühlemann O. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA. 2011;17:2094–2107. doi: 10.1261/rna.02918111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sander B., Golas M.M., Makarov E.M., Brahms H., Kastner B., Luhrmann R. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 94.Sperling R., Sperling J. In: The eukaryotic nucleus, molecular biochemistry and macromolecular assemblies. Strauss P.R., Wilson S.H., editors. Vol. 2. Telford Press; Caldwell, NJ: 1990. pp. 453–476. [Google Scholar]
- 95.Agranat L., Raitskin O., Sperling J., Sperling R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc Natl Acad Sci U S A. 2008;105:5028–5033. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agranat L., Sperling J., Sperling R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010;7:253–262. doi: 10.4161/rna.7.2.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sperling R., Sperling J., Levine A.D., Spann P., Stark G.R., Kornberg R.D. Abundant nuclear ribonucleoprotein form of CAD RNA. Mol Cell Biol. 1985;5:569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sperling R., Spann P., Offen D., Sperling J. U1, U2 and U6 small nuclear ribonucleoprtoeins (snRNPs) are associated with large nuclear RNP particles containing transcripts of an amplified gene in vivo. Proc Natl Acad Sci U S A. 1986;83:6721–6725. doi: 10.1073/pnas.83.18.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]


