Abstract
A compromised mitotic checkpoint, the primary mechanism for ensuring that each new cell receives one copy of every chromosome, has been implicated as a contributor to carcinogenesis. However, a checkpoint response is shown here to be essential for cell survival, including that of chromosomally instable colorectal cancer cells. Reducing the levels of the checkpoint proteins BubR1 or Mad2 in human cancer cells or inhibiting BubR1 kinase activity provokes apoptotic cell death within six divisions except when cytokinesis is also inhibited. Thus, suppression of mitotic checkpoint signaling is invariably lethal as the consequence of massive chromosome loss, findings that have implications for inhibiting proliferation of tumor cells.
Decreased strength of mitotic checkpoint signaling has been implicated as a driving force of carcinogenesis. Colorectal cancer cells that have an accelerated rate of chromosomal gains and losses [referred to as chromosome instability (CIN)] have a weaker mitotic checkpoint than similarly aggressive cancer cells with a stable chromosome content but which have microsatellite instability (MIN) from errors in DNA repair (1–3). The CIN phenotype has been associated with rare mutations in checkpoint genes (3–6) or decreased protein levels of checkpoint components (4, 6–8). Deletion in mice of one allele of Mad2, Bub3, or BubR1 compromises the mitotic checkpoint, yielding higher rates of chromosome missegregation and spontaneous (Mad2) or carcinogen-induced (Bub3 and BubR1) tumors (9–11). This finding has supported a causal relationship between a weakened checkpoint and carcinogenesis arising from CIN.
Absence of checkpoint components in mice is known to result in early developmental defects (10–14). To determine the short- and long-term effects of mitotic checkpoint inhibition on survival of individual cells, the critical checkpoint proteins BubR1 or Mad2 were reduced by plasmid-based expression of double-stranded small interfering RNAs (siRNAs) (15).
Materials and Methods
Plasmids. pSUPER-BubR1 and pSUPER-Mad2 were constructed as described (15) by using the sequences 5′-AGATCCTGGCTAACTGTTC-3′ and 5′-TACGGACTCACCTTGCTTG-3′, respectively. siRNA-resistant BubR1 (pcDNA3-myc-BubR1ΔsiRNA) was created by site-directed mutagenesis of bases 2823 (C to A) and 2826 (G to A) in pcDNA3-myc-BubR1 (a gift of S. Taylor, University of Manchester, Manchester, U.K.). BubR1ΔC and BubR1K795A alleles were created by site-directed mutagenesis of pcDNA3-myc-BubR1ΔsiRNA by inserting a T at position 1519 to create a premature STOP codon, or by mutating base pairs 2383–2384 to GC, respectively. pH2B-EYFP and pH2B-ECFP were created by inserting a fragment of H2B cDNA (a gift of K. Sullivan, The Scripps Research Institute, La Jolla, CA) into modified pEYFP or pECFP (Clontech). All constructs were verified by automated sequencing.
Cell Culture and Transfections. HeLa, YCA-2A3 (HeLa cells stably expressing EYFP-CENP-A), and T98G cells were grown in DMEM supplemented with 10% FBS and 50 μg/ml pen/strep (Invitrogen). SW480 and DLD-1 cells were grown in L-15 and Iscove's medium, respectively. Colcemid (KaryoMax, Invitrogen) was added to cells at a final concentration of 50 ng/ml and re-added every 2 days in experiments where treatment exceeded 2 days. Transfections were done by using Effectene (Qiagen, Valencia, CA).
Magnetic Activated Cell Sorting. Cells were transfected with pCMV-CD20 along with the various siRNA plasmids in a 1:10 ratio. Isolation of transfected cells was performed as described (16).
Antibodies and Immunoblotting. SDS/PAGE and Western blotting were standard. Antibodies used in this study were as follows: anti-BubR1 (5F9, a gift of S. Taylor), anti-CENP-E [Hpx1, (17)], anti-actin (N350, Amersham Pharmacia Biosciences), anti-cyclin B1 (GNS1), and anti-Mad2 (C19, Santa Cruz Biotechnology), anti-p85-PARP-1 (Promega), and anti-caspase-3 (Pharmingen).
In Vitro Kinase Assay. T98G cells were transfected with indicated siRNA plasmids for 8 hr, subjected to double 2 mM thymidine block, and released for 9.5 hr. Mitotic cells were collected and lysed in 50 mM Tris (pH 7.5), 200 mM NaCl, 1% Triton X-100, 1.5 mM MgCl2, and 5 mM EDTA, supplemented with protease and phosphatase inhibitors. The cleared supernatants were equalized for protein content, and BubR1 was immunoprecipitated with SBR1.1 antibody (a gift of S. Taylor) coupled to protein G-Sepharose beads for 2 hr at 4°C. Beads were washed twice with lysis buffer and twice with kinase buffer (50 mM Tris, pH 7.5/10 mM MgCl2/2 mM sodium vanadate). Phosphorylation reactions were performed with 25 μl of kinase mix (50 mM Tris, pH 7.5/10 mM MgCl2/1 mM DTT/50 μM ATP/250 μg/ml histone H1/2.5 μCi of [γ-32P]ATP) (1 Ci = 37 GBq) at 37°C for 30 min.
Immunofluorescence. Cells grown on poly-l-lysine-coated coverslips were washed once with PBS, fixed with 4% formaldehyde (Tousimis, Rockville, MD) for 10 min, extracted with 0.5% Triton X-100 for 5 min, and blocked in PBS containing 0.5% Tween 20 and 3% BSA (Sigma) for 1 hr. Coverslips were exposed to primary antibodies diluted in blocking buffer for 1 hr, and to secondary antibodies (Jackson ImmunoResearch) diluted 1:200 in blocking buffer for 1 hr in the dark. After each incubation, coverslips were washed extensively with PBS/0.5% Tween 20. Finally, coverslips were submerged in PBS containing 4′,6-diamidino-2-phenylindole (DAPI), washed once with PBS, and mounted by using ProLong antifade reagent (Molecular Probes). All treatments were performed at room temperature. Dilutions were as follows: anti-Mad2 (Covance, Princeton) 1:100, anti-BubR1 (5F9) 1:1,000, anti-CENP-E (Hpx1) 1:200, ACA (a gift of K. Sullivan) 1:1,000, and anti-active-caspase-3 (CM1, Idun Pharmaceuticals, San Diego) 1:200.
BrdUrd Incorporation Assay and Fluorescence-Activated Cell Sorter (FACS) Analysis. T98G cells were treated with 1 μM BrdUrd for 1 hr and analyzed by flow cytometry as described (16). For analysis of DNA content, all cells were collected, washed with PBS, and fixed overnight with 70% ethanol. Next, cells were washed with PBS and resuspended in PBS/propidium iodide/RNaseA.
Live Cell Microscopy. HeLa cells seeded on 35-mm glass-bottom dishes (MatTek, Ashland, MA) were transfected with pH2B-EYFP and the indicated siRNA plasmids in a ratio of 1:10. Forty-eight hours posttransfection, the medium was replaced with CO2-independent medium (GIBCO) supplemented with glutamine and 10% FBS. The dish was placed in a heat-controlled stage set to 37°C. Live cell images of H2B-EYFP and brightfield (to determine nuclear envelope breakdown and nuclear envelope reformation) were taken on a Nikon Eclipse 300 inverted microscope (Nikon) by using a 60XA/1.4 objective. Z-stack images were collected by a Photometrics COOLSNAP HQ camera (Roper Scientific, Tucson, AZ) and transferred to computer by metamorph software (Universal Imaging, Media, PA). Time-lapse sequences were captured with exposure times of 100 ms, at 2 × 2 binning and with interframe intervals of 2 min.
Chromosome Counts. YCA-2A3 cells were grown on poly-l-lysine-coated coverslips and transfected with pH2B-ECFP along with the various siRNA plasmids in a ratio of 1:10 for 48 hr, after which they were subjected to a double thymidine block. Fourteen hours after release from the block, the cells were fixed in 4% formaldehyde (Tousimis) and mounted. Z-stack images were collected by using a ×100 objective.
Colony Outgrowth Assay. Cells were transfected with the indicated siRNA plasmids and pBabe-Puro in a ratio of 10:1. Twenty-four hours posttransfection, cells were diluted 10-fold and grown in puromycin-containing medium for 9 days. Cells were fixed with methanol for 30 min at room temperature and stained with crystal violet.
Results and Discussion
Transient expression of BubR1 or Mad2 siRNA in the human cervical cancer cell line HeLa or the glioblastoma line T98G produced robust (≈90%), long-term (up to 6 days) depletion of BubR1 or Mad2 whereas control siRNA plasmids (mock) did not affect either BubR1 or Mad2 levels (Fig. 6 A and B, which is published as supporting information on the PNAS web site). Kinetochore integrity was not compromised by absence of BubR1 or Mad2. Although the ≈10% of BubR1 or Mad2 remaining after 48 hr was undetectable at HeLa cell kinetochores, the outer-kinetochore kinesin-like protein CENP-E and the antigens recognized by an anti-centromere antiserum (ACA) were present at levels similar to mock transfected cells (Fig. 6C), as were other checkpoint components including Cdc20, Mad1, and Bub1.
Both HeLa and T98 cells transfected with the mock siRNA plasmids activated and sustained mitotic checkpoint signaling after colcemid-mediated microtubule disassembly. By 16 hr, most had accumulated in mitosis with 4N DNA content and high levels of cyclin B1 and hyperphosphorylated BubR1 (Fig. 1A). Filming beginning at nuclear envelop disassembly of colcemid-treated HeLa cells expressing both a fluorescently tagged histone (histone H2B-YFP) and the mock siRNAs revealed that they entered mitosis normally and then remained arrested at prometaphase for as long as filming was continued (4 hr) (Movie 1, which is published as supporting information on the PNAS web site, and Fig. 1B). siRNA-mediated reduction of BubR1 or Mad2, however, yielded cells that, despite chronic microtubule disassembly, did not show mitotic arrest by any measure, entering and exiting mitosis without sister chromatid separation or cytokinesis (Movies 2 and 3, which are published as supporting information on the PNAS web site, and Fig. 1 B and C) and reduplicating their DNA in the subsequent S phase (as revealed by labeling with BrdUrd) (Fig. 1D). This treatment yielded a significant proportion of octaploid cells after 16 hr and cells with 16N DNA content after an additional 24 hr of continued microtubule disassembly (Fig. 1 D and E). Indistinguishable results were obtained when nocodazole or taxol was used to disrupt microtubule assembly or dynamics (data not shown).
Fig. 1.
Absence of mitotic checkpoint response in BubR1- or Mad2-depleted cells. (A) HeLa cells transfected with the indicated siRNA plasmids were treated with or without colcemid for 16 hr, and whole-cell lysates of the transfected population were immunoblotted for cyclin B1, BubR1, or actin. p-BubR1, phosphorylated BubR1. (B) Stills of time-lapse movies 1–3. Asterisks indicate nuclear envelope breakdown. (C) Time-lapse sequence of cells transfected with Mad2 siRNA plasmid and pH2B-EYFP. Arrows indicate the reassembled nuclear envelope. (D) T98G cells transfected with the indicated siRNA plasmids were treated with or without colcemid for 16 hr, and the entire population was analyzed for BrdUrd incorporation. S phase indicates the percentage of the cell population that is BrdUrd positive. (E) DNA content profiles of T98G cells that were transfected and treated as in D.(F) Stills of time-lapse movies 4–6. (G) HeLa cells were transfected with the indicated siRNA plasmids. Forty-eight hours posttransfection, transfected cells were isolated by magnetic activated cell sorting and replated onto coverslips. Twenty hours after replating, cells were fixed and DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI).
In the absence of microtubule poisons, the mammalian mitotic checkpoint is activated in every mitosis beginning at nuclear envelope disassembly, with each unattached kinetochore producing an inhibitor that prevents advance to anaphase (18). Silencing of the checkpoint occurs only after all kinetchores have attached to spindle microtubules (18). Filming of mitoses in HeLa cells with fluorescent histone-tagged chromosomes revealed that, after nuclear envelope disassembly, ≈33 min were required for complete chromosome attachment, with anaphase ensuing ≈18 min after alignment (Movie 4, which is published as supporting information on the PNAS web site, and Fig. 1F). In contrast, cells with reduced BubR1 or Mad2 entered anaphase ≈20 min after mitotic entry, with many unaligned chromosomes (Movies 5 and 6, which are published as supporting information on the PNAS web site, and Fig. 1F), producing cells that in the subsequent interphase displayed a variety of nuclear abnormalities (Fig. 1G). These effects were unlikely to be due to off-target events of the siRNAs because similar effects were seen with additional siRNAs (Fig. 7, which is published as supporting information on the PNAS web site). Thus, as suggested by antibody microinjection approaches (19, 20), BubR1 and Mad2 are each essential for the timing of normal mitosis and for the ability in such mitoses of arresting advance to anaphase until all chromosomes have attached.
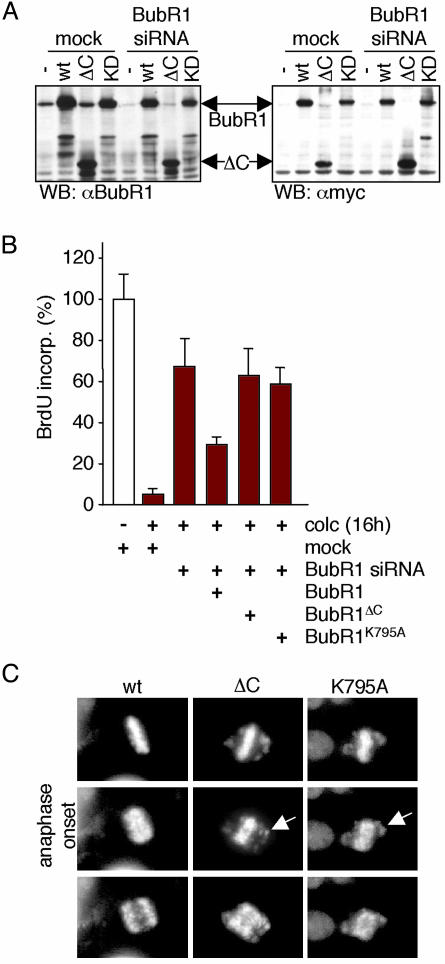
Checkpoint signaling was largely, but not completely, restored to BubR1-depleted cells by expression of WT BubR1 (Fig. 2B), but not comparable levels (Fig. 2 A) of kinase-deficient BubR1 [deleted in either the kinase domain (BubR1ΔC) or a kinase-inactive point mutant (BubR1K795A)], encoded by genes made siRNA-resistant by two silent base changes within the region targeted by the siRNA. The incomplete restoration even with WT BubR1 probably reflects the expected sensitivity of checkpoint signaling to optimal levels of BubR1 (18), including the lethality of high levels of kinase-active BubR1 (7). Live cell microscopy corroborated these results: siRNA-resistant WT BubR1 blocked anaphase entry in the presence of misaligned chromosomes in ≈65% of cells depleted for endogenous BubR1 whereas the kinase-deficient mutants never could (Fig. 2C). Thus, BubR1 kinase activity is essential for sustained checkpoint signaling in these human cancer cells.
Fig. 2.
BubR1 kinase activity is required for mitotic checkpoint signaling. (A) Total cellular lysates of T98G cells transfected with mock or BubR1 siRNA plasmid in combination with either empty vector or myc-tagged siRNA-resistant BubR1 mutants were immunoblotted for BubR1 or the myc epitope tag. KD, K795A. (B) T98G cells were transfected, treated, and analyzed as in Fig. 1D, with the addition of the various siRNA-resistant BubR1 alleles during transfection. Percentage BrdUrd positivity in the mock samples without colcemid was set to 100. (C) Stills of live cell microscopy of BubR1 siRNA HeLa cells transfected with pH2B-EYFP and the indicated siRNA-resistant BubR1 alleles. Forty-eight hours posttransfection, images of H2B-EYFP-positive cells were taken at 2-min intervals. Arrows indicate unaligned chromosomes during anaphase.
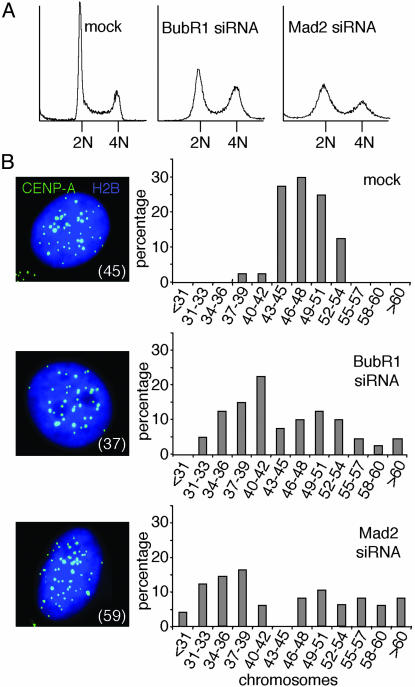
Analysis of HeLa cell DNA content 72 hr after introduction of BubR1 or Mad2 siRNA revealed extensive loss or gain of DNA content within two to three divisions, with a significant proportion of cells with DNA content that diverged considerably from the major 2N and 4N peaks observed in control cells (Fig. 3A). After depletion of Mad2 or BubR1 from HeLa cells stably expressing GFP-tagged CENP-A, a histone H3 variant found only at active centromeres (18), and after synchronizing those cells to be in G1 (with a double thymidine block and a subsequent 14-hr release), all centromeres within single cell nuclei were imaged and counted. Most mock-transfected cells had a range of 44–50 chromosomes (Fig. 3B). Cells lacking BubR1 or Mad2, however, displayed a much broader range of chromosome numbers (Fig. 3B), with severe chromosome loss appearing within two divisions in the absence of a functional mitotic checkpoint. Moreover, there did not seem to be equivalent gain and loss of chromosomes. Rather, within the first 2–3 divisions after lowering BubR1 or Mad2, there was a higher proportion of cells with <2N DNA content, almost certainly reflecting unattached or aberrantly (merotelically) attached (21) chromosomes lost from both daughter cells at cytokinesis (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
Mitosis with reduced BubR1 or Mad2 causes acute chromosome loss. (A) DNA content profiles of HeLa cells transfected with the indicated siRNA plasmids along with pBabe-Puro for 24 hr and grown in puromycin-containing medium for an additional 48 hr. Nontransfected cells were removed 14 hr before analysis. (B) Distribution of the amount of chromosomes within a G1 population of YCA-2A3 cells transfected with mock, BubR1 siRNA, or Mad2 siRNA. Images are Z-stack projections displaying all EYFP-CENP-A-containing centromeres in one plane of a transfected (H2B-ECFP-positive) cell. Number in parentheses indicates number of chromosomes.
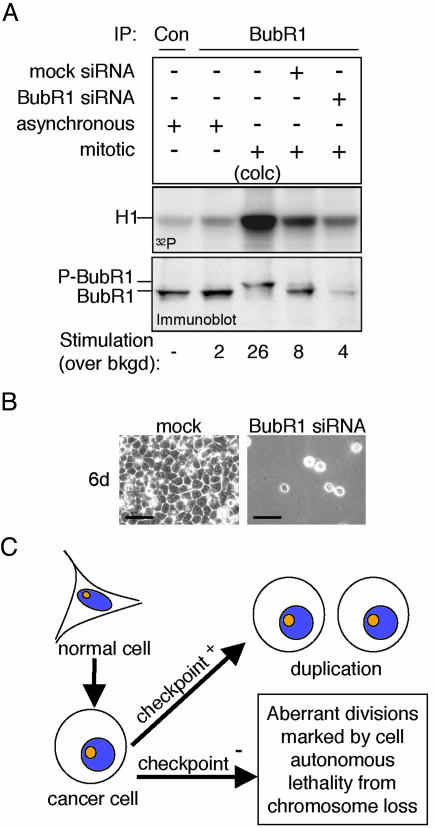
Colony outgrowth assays of HeLa cells with diminished levels of either BubR1 or Mad2 were performed by introducing siRNA-encoding plasmids together with a plasmid conferring puromycin resistance. Nontransfected cells were eliminated by continuous growth in puromycin-containing medium. After 9 days, surviving cells were stained with crystal violet, and colonies were counted. This analysis revealed that BubR1- or Mad2-depleted cells did not form colonies (Fig. 4A). Fluorescence-activated cell sorter (FACS, Becton Dickinson) analysis further showed a large increase in the proportion of cells containing less than a 2N amount of DNA, beginning as early as 4 days posttransfection (Fig. 4B). A similar result using HeLa cells treated with Mad2 siRNA oligonucleotides was recently reported (22). By 6 days, no BubR1 or Mad2 siRNA cells were viable. Because in control cells puromycin-related death occurred within the first 1–2 days of selection, death observed at 4–5 days in cells depleted of BubR1 or Mad2 must result from the absence of mitotic checkpoint signaling. Similar results were obtained with the glioblastoma cell line T98G (see Fig. 5B) and the osteosarcoma cell line U2OS (data not shown).
Fig. 4.
Loss of viability by inhibition of mitotic checkpoint signaling. (A) Colony outgrowth assay. Indicated is the number of colonies on each plate. (B) HeLa cells transfected with the indicated siRNAs along with pBabe-Puro were analyzed for DNA content after 4 or 5 days of growth in puromycin-containing medium. The extent of cell death is shown as percentage of cells with sub2N DNA content. (C) Colony outgrowth assay of DLD-1 and SW480 cells. Percentage of surviving colonies in mock transfected samples was set to 100. (D) HeLa cells transfected as in B were analyzed for DNA content (Upper) and morphology (Lower) after growth in puromycin- and colcemid-containing medium for an additional 3 or 6 days, respectively. (Bar = 50 μm.) (E) Whole-cell extracts of HeLa cells transfected with the indicated siRNA plasmids were immunoblotted for p85 poly-(ADP-ribose) polymerase-1 (PARP-1) protein cleavage product, caspase-3, or actin. Asynchronous and puromycin- and colcemid-treated cells were used as controls. (F) HeLa cells were transfected and selected as in B. Immunostaining of active caspase-3 was done 3 days posttransfection.
Fig. 5.
Loss of viability by partial inhibition of the essential BubR1 kinase. (A) Mitotic T98G cells, untransfected or transfected with mock or BubR1 siRNA plasmids, were analyzed for BubR1 protein levels (immunoblot) and BubR1 kinase activity (32P) by immunoprecipitation (IP). Asynchronous populations, a colcemid-treated population (10 hr; colc) and immunoprecipitation with control antibody (Con) were used as controls. (B) T98G cells were transfected as in Fig. 5A and grown for 6 days. (Bar = 50 μm.) (C) Proposed effects of mitotic checkpoint status on proliferation and survival of human cancer cells. Although a weakened mitotic checkpoint may contribute to carcinogenesis, tumor cells require checkpoint signaling for proliferation. More severe rates of chromosome loss by functionally inhibiting the mitotic checkpoint, however, are lethal.
Highly aneuploid CIN colorectal cancer cell lines have a considerably weaker checkpoint that cannot be sustained as long as in normal cells or in chromosomally stable MIN cancer cell lines (e.g., refs. 3 and 23). After siRNA-mediated reduction of BubR1 or Mad2, colony outgrowth assays on both CIN (SW480) and MIN (DLD-1) lines yielded few surviving colonies (Fig. 4C). Examination of BubR1 or Mad2 protein levels in these colonies revealed that all had escaped siRNA-mediated BubR1 or Mad2 suppression. Thus, survival of both CIN and MIN cancer cell lines depends on basal mitotic checkpoint signaling.
HeLa cells were also cultured continuously in colcemid after depletion of BubR1 or Mad2 with siRNA. Although these cells are mitotic checkpoint-deficient so that prevention of mitotic spindle assembly does not arrest cell cycle advance, it does block cytokinesis, which requires overlapping microtubules from the two spindle poles for the recruitment of components required for cleavage (24). Blocking division eliminated cell death of BubR1- or Mad2-depleted cells, producing instead giant cells and nuclei as a consequence of continued cycling (Fig. 4D). When cytokinesis was allowed, execution of a cell death pathway was provoked within two or three divisions in the majority of BubR1- or Mad2-depleted cells, including activation of caspase-3 (Fig. 4 E and F) as well as appearance of the p85 cleavage product of caspase-3-cleaved poly-(ADP-ribose) polymerase-1 (PARP-1) (Fig. 4E). Conversely, whereas chronic colcemid treatment of checkpoint-proficient cells (measured by the proportion of cells with sub-2N DNA content) triggered a cell-killing pathway after eventual mitotic exit, such death was nearly eliminated by reducing the expression of BubR1 or Mad2 (Fig. 4D). Thus, loss of viability of checkpoint-deficient cycling cells arises directly from rapid loss during cytokinesis of chromosomes encoding genes required for maintenance of viability of individual cells, presumably in a way similar to the bona fide apoptosis inducer puromycin, which acts as a stress by inhibiting general protein synthesis.
Considering the requirement of BubR1 kinase activity for checkpoint signaling (Fig. 2 B and C), remaining kinase activity was measured in immunoprecipitates from BubR1-depleted, mitotic cells obtained after release from synchronization at G1/S. As expected (18, 25), BubR1 kinase activity in mitotic cells was increased 4-fold over that of an asynchronously growing population (Fig. 5A). Despite a 5-fold reduction in BubR1 level and the lethal loss of a functional mitotic checkpoint (Fig. 5B), mitotic BubR1 kinase activity was reduced only 50% as compared with the parental cells with a functional checkpoint (Fig. 5A). Thus, as little as a 50% reduction in BubR1 kinase activity compromises the mitotic checkpoint sufficiently to eliminate cell viability. It should be noted that we cannot formally exclude the possibility that a kinase that associates with BubR1 is responsible for the observed H1 phosphorylation.
Thus, whereas an initial weakening of the mitotic checkpoint may enhance aspects of CIN-mediated tumorigenesis (3, 7, 9, 10), further weakening (or silencing) of checkpoint signaling is rapidly (within three divisions) and invariably lethal even to aggressive cancer cell lines (Fig. 5C). Therefore, manipulating the mitotic checkpoint to inhibit growth of both CIN and MIN tumor cells by designing drugs that target essential checkpoint functions, such as BubR1 kinase activity, could prove to be useful in treatment of certain cancers.
Supplementary Material
Acknowledgments
We thank Drs. J. Shah (Ludwig Institute for Cancer Research), R. Agami (Netherlands Cancer Institute, Amsterdam), K. Sullivan, S. Taylor, J. Carethers (Veterans Affairs Medical Center, University of California at San Diego), and K. Arden (Ludwig Institute for Cancer Research), for providing reagents and cell lines, and the Cleveland laboratory for helpful discussions. G.J.P.L.K. is supported by a fellowship from the Dutch Cancer Society (KWF Kankerbestrijding). D.R.F. is supported by a fellowship from the National Institutes of Health. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CIN, chromosome instability; MIN, microsatellite instability; siRNA, small interfering RNA.
References
- 1.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623–627. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643–649. [DOI] [PubMed] [Google Scholar]
- 3.Cahill, D. P., Lengauer, C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300–303. [DOI] [PubMed] [Google Scholar]
- 4.Shichiri, M., Yoshinaga, K., Hisatomi, H., Sugihara, K. & Hirata, Y. (2002) Cancer Res. 62, 13–17. [PubMed] [Google Scholar]
- 5.Ru, H. Y., Chen, R. L., Lu, W. C. & Chen, J. H. (2002) Oncogene 21, 4673–4679. [DOI] [PubMed] [Google Scholar]
- 6.Wang, X., Jin, D. Y., Ng, R. W., Feng, H., Wong, Y. C., Cheung, A. L. & Tsao, S. W. (2002) Cancer Res. 62, 1662–1668. [PubMed] [Google Scholar]
- 7.Shin, H.-J., Baek, K.-H., Jeon, A.-H., Park, M.-T., Lee, S.-J., Kang, C.-M., Lee, H.-S., Yoo, S.-H., Chung, D.-H., Sung, Y.-C., et al. (2003) Cancer Cell 4, 483–497. [DOI] [PubMed] [Google Scholar]
- 8.Wang, X., Jin, D. Y., Wong, Y. C., Cheung, A. L., Chun, A. C., Lo, A. K., Liu, Y. & Tsao, S. W. (2000) Carcinogenesis 21, 2293–2297. [DOI] [PubMed] [Google Scholar]
- 9.Michel, M. L., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. V. S. & Benezra, R. (2001) Nature 409, 355–359. [DOI] [PubMed] [Google Scholar]
- 10.Babu, J. R., Jeganathan, K. B., Baker, D. J., Wu, X., Kang-Decker, N. & Van Deursen, J. M. (2003) J. Cell Biol. 160, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai, W., Wang, Q., Liu, T., Swamy, M., Fang, Y., Xie, S., Mahmood, R., Yang, Y. M., Xu, M. & Rao, C. V. (2004) Cancer Res. 64, 440–445. [DOI] [PubMed] [Google Scholar]
- 12.Dobles, M., Liberal, V., Scott, M. L., Benezra, R. & Sorger, P. K. (2000) Cell 101, 635–645. [DOI] [PubMed] [Google Scholar]
- 13.Kalitsis, P., Earle, E., Fowler, K. J. & Choo, K. H. (2000) Genes Dev. 14, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putkey, F. R., Cramer, T., Morphew, M. K., Silk, A. D., Johnson, R. S., McIntosh, J. R. & Cleveland, D. W. (2002) Dev. Cell 3, 351–365. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 16.Medema, R. H., Kops, G. J., Bos, J. L. & Burgering, B. M. (2000) Nature 404, 782–787. [DOI] [PubMed] [Google Scholar]
- 17.Brown, K. D., Wood, K. W. & Cleveland, D. W. (1996) J. Cell Sci. 109, 961–969. [DOI] [PubMed] [Google Scholar]
- 18.Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- 19.Gorbsky, G. J., Chen, R. H. & Murray, A. W. (1998) J. Cell Biol. 141, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan, G. K., Jablonski, S. A., Sudakin, V., Hittle, J. C. & Yen, T. J. (1999) J. Cell Biol. 146, 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimini, D., Howell, B., Maddox, P., Khodjakov, A., Degrassi, F. & Salmon, E. D. (2001) J. Cell Biol. 153, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel, L., Diaz-Rodriguez, E., Narayan, G., Hernando, E., Murty, V. V. & Benezra, R. (2004) Proc. Natl. Acad. Sci. USA 101, 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tighe, A., Johnson, V. L., Albertella, M. & Taylor, S. S. (2001) EMBO Rep. 2, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maddox, A. S. & Oegema, K. (2003) Nat. Cell Biol. 5, 773–776. [DOI] [PubMed] [Google Scholar]
- 25.Weaver, B. A., Bonday, Z. Q., Putkey, F. R., Kops, G. J., Silk, A. D. & Cleveland, D. W. (2003) J. Cell Biol. 162, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.