Abstract
Recent studies have delineated a large Nearctic Müllerian mimicry complex in Dasymutilla velvet ants. Psorthaspis spider wasps live in areas where this mimicry complex is found and are phenotypically similar to Dasymutilla. We tested the idea that Psorthaspis spider wasps are participating in the Dasymutilla mimicry complex and that they codiverged with Dasymutilla. We performed morphometric analyses and human perception tests, and tabulated distributional records to determine the fit of Psorthaspis to the Dasymutilla mimicry complex. We inferred a dated phylogeny using nuclear molecular markers (28S, elongation factor 1-alpha, long-wavelength rhodopsin and wingless) for Psorthaspis species and compared it to a dated phylogeny of Dasymutilla. We tested for codivergence between the two groups using two statistical analyses. Our results show that Psorthaspis spider wasps are morphologically similar to the Dasymutilla mimicry rings. In addition, our tests indicate that Psorthaspis and Dasymutilla codiverged to produce similar color patterns. This study expands the breadth of the Dasymutilla Müllerian mimicry complex and provides insights about how codivergence influenced the evolution of mimicry in these groups.
Introduction
Müllerian mimicry refers to the phenomenon in which sympatric, harmful species share a similar warning signal for mutual benefit against predation [1], [2]. This kind of mimicry has been well documented for several tropical groups, such as Heliconius butterflies [2]–[6] and poisonous Dendrobatidae and Mantellidae frogs [7]–[9]. Recently, a large Nearctic Müllerian mimicry complex was described in diurnally foraging Dasymutilla velvet ants (Hymenoptera: Mutillidae) [10]. These aposematic solitary wasps have wingless females that inflict a painful sting, which is effective as a defense against predators [10]. Although several Batesian mimics of velvet ants have been reported [11]–[15], the possibility that other harmful species might be Müllerian mimics of velvet ants has not been investigated.
Nine spider wasps in the genus Psorthaspis (Pompilidae) closely resemble velvet ant color patterns [16], and thus might be participating in the velvet ant mimicry complex. Because spider wasps are defended with a sting that invokes some of the most intense, instantaneous pain among stinging insects [17], and velvet ants and Psorthaspis spider wasps are attacked by some of the same predators (i.e., frogs, lizards and mammals) [18]–[22], Psorthaspis spider wasps and velvet ants could be Müllerian mimics of each other. However, the resemblance of Psorthaspis spider wasps to velvet ants, and the potential fit of both wasps to the same mimicry complex have never been quantified.
In the well-studied Heliconius Müllerian mimicry systems, codivergence, or the parallel divergence of ecologically associated, but unrelated, lineages, has been a major contributor to the development of numerous mimicry rings [23]. Codivergence has been proposed as some of the strongest evidence for coevolution [23]–[26]. Codivergence patterns alone, however, are not enough to demonstrate coevolution in the strict sense (i.e., evolution that occurs in populations of at least two species as the result of reciprocal selective influence) because selective pressures are often not measured between the two groups [23]. Although codivergence and the associated phenotypic convergence has been tested in some mimicry systems, investigations into the evolution of mimetic patterns in other systems, such as Psorthaspis spider wasps and velvet ants, have the potential to better illuminate the role of coevolution in the development of large Müllerian mimicry complexes.
Here, we investigate the phenotypic and phylogenetic similarities of Dasymutilla velvet ants and Psorthaspis spider wasps to address the following questions. 1) How well do Psorthaspis spider wasps fit in the described velvet ant mimicry rings? 2) Are the color pattern similarities between these wasp groups a result of codivergence?
Methods
Study system
Velvet ants and spider wasps are both classified as stinging wasps (Aculeata: Hymenoptera), and are both solitary parasitoids. Insect parasitoids are a special case of parasitic organisms because they ultimately kill their hosts during development [27]. Velvet ants are usually external parasitoids on the larvae or pupae of bees and solitary wasps. Their females are wingless, while males are typically winged and capable of flight [28]. There are more than 150 species of Dasymutilla velvet ants. Spider wasps (Pompilidae) are parasitoids of spiders. Both males and females are winged. There are 29 species of Psorthaspis spider wasps. These spider wasps use trapdoor spiders of the family Ctenizidae as hosts [29]. Even though the venom is primarily used to paralyze the host, the sting of both spider wasps and velvet ants also can be a deterrent to predation [10], [17].
Morphometric analysis of color patterns
We quantified the color patterns of Psorthaspis using digital images following the procedure described by Wilson et al. [10], with the exception of setal characters, as they are not comparable between velvet ants and spider wasps. Characters included the percent black of the metasoma, integument color, and non-black metasomal color measured in red, green and blue (RGB). The color pattern of all the Psorthaspis species putatively involved in the mimicry complex was studied. Because there is some degree of intraspecific variation in color and pattern characteristics in spider wasp species, a representative individual was selected for each Psorthaspis species, on which measurements were made. These representative individuals were selected after the examination of over 1,000 specimens from 15 insect museums from five countries. All area and percentage measurements were made using the program ImageJ (http://rsb.info.nih.gov/ij/). Morphological characters were analyzed together with the data from Wilson et al. [10] using resemblance matrices, nonmetric multidimensional scaling (NMDS) based on a Bray-Curtis distance matrix, and permutational multivariate analysis of variance (PERMANOVA, [30]) in R [31] using the adonis function in the vegan package. The data gathered for this analysis are available on Figshare.
Human perception of mimetic fidelity
Mimetic fidelity in Müllerian mimicry systems represents how well a given species matches a group of species (i.e., the mimicry ring). To measure mimetic fidelity of spider wasps involved in described Müllerian mimicry rings [10], we used methods outlined by Wilson et al. [32] for human perception tests. Even though many researchers are hesitant about the adequacy of using human rankings to establish mimetic fidelity, various studies have shown that human rankings are consistent with those of multivariate analyses of morphological data and avian response rankings [33]–[35]. Although human perception has been used mainly in systems where predators are birds [33], [36], other vertebrate predators like lizards have similar color vision to birds and humans [37], and may perceive prey them same way.
We presented slides showing an individual Psorthaspis species compared to all members of the velvet ant mimicry ring to which the species was most similar. Volunteers (N = 35) were directed to rank each Psorthaspis species on how well it fit into the associated mimicry ring. Rankings were based on a scale of 1 (very poor mimic) to 10 (excellent mimic). We included images of all the Psorthaspis species putatively included in the mimicry complex. All images were presented at magnifications such that all wasps had the same projected body length. Each slide was presented for 20 seconds following the protocols used by other similar studies [32], [33]. The mimetic fidelity of each spider wasp was estimated based on the mean score of a wasp compared to its assigned mimicry ring.
All volunteers participating in this study were students in lower division Biology courses at Utah State University–Tooele. Students were presented with a short presentation introducing the concepts of Batesian and Müllerian mimicry and were then given the option to participate in a survey designed to rank mimetic fidelity of wasps. If students agreed to participate, they were given a link to the website containing the survey. To our knowledge, the volunteers were not experts in insect identification. This effectively resulted in mimetic fidelity scores that were based on overall resemblance of a mimic to a mimicry ring rather than on preconceived ideas of what specific parts of a mimic should match the ring. All participants were over the age of 18, and no data relating to the volunteers were gathered. No approval from the university was requested for this research because no information about living individuals was collected (i.e., the research did not involve human subjects as per the Code of Federal Regulations 45 CFR part 46). Volunteers were simply used to gather information concerning morphological similarities between the insects involved in this study. Because of the need to protect the anonymity of our volunteers, no questions were asked regarding any physical characteristics that would affect ranking mimics and models (i.e., colorblindness). While this potentially could influence the reported mimetic fidelity scores, we think any influence of colorblindness would be minimal, due to the nature of aposematic signals in spider wasps and velvet ants. These warning signals primarily result from contrasting black and red or yellow patterns, which would still be visually distinct to colorblind individuals.
Estimation of geographical distribution
To determine the distribution of each of the Psorthaspis color patterns identified in this study we geo-referenced 1,032 Psorthaspis specimens, from all mimic species, from 13 natural history collections and downloaded data on geo-referenced Psorthaspis specimens in the Southwest Collections of Arthropods Network (SCAN) [38]. We manually plotted the collection localities of each species on a map using the software Google Earth 5.0 (http://earth.google.com) and estimated geographic distributions by drawing a line encompassing all of the collection localities. These estimated distributions were visually compared to the distributions of velvet ant mimicry rings published by Wilson et al. [10]. The data points used for this analysis are available on Figshare.
Molecular data and phylogenetic inference
We compiled a data set of four genes (28S, elongation factor 1-alpha, wingless, and long-wavelength rhodopsin) for 13 Psorthaspis species and one outgroup (Aporus idris), which were previously published by Rodriguez et al. [39]. Two of the putative mimic species, Psorthaspis nigriceps and Psorthaspis texana could not be included because of the lack of suitable molecular data. Sequences were aligned using Geneious Alignment in Geneious 5.4 [40], and manually refined. The model of molecular evolution used for each gene and by codon position was the same used by Rodriguez et al. [39] except for introns from long-wavelength rhodopsin, for which the model was determined in MrModelTest [41]. Single-gene phylogenies were estimated through a Bayesian framework implemented in MrBayes 3.2 [42] to check for potential conflict between gene trees. Single-gene matrices were then concatenated using Geneious 5.4 to produce a combined matrix, using the best partition scheme used by Rodriguez et al. [39], and an additional partition including long-wavelength rhodopsin introns with the model GTR+I+G. MCMC chains were run for 10,000,000 generations, with sampling every 1,000 generations. Effective sample size (ESS), burn-in, and graphical examination of chain convergence were examined in Tracer 1.5 [43].
A chronogram of Psorthaspis was inferred from the combined matrix in a Bayesian framework using BEAST 1.7.5 [44] under an uncorrelated lognormal relaxed-clock model [45], [46]. Substitution models were unlinked among partitions; the underlying clock and trees were linked. The crown-group node of all Psorthaspis was assigned a normal prior of mean = 12.9 Ma (SD = 10), based on results of Rodriguez et al. [39]. Two separate Markov Chain Monte Carlo (MCMC) searches were performed for 10,000,000 generations. Effective sample size (ESS) and graphical chain convergence were examined in Tracer 1.5. Independent runs were assembled with LogCombiner 1.7.5. and 10% of the generations were discarded as burn-in. Divergence time estimations of Dasymutilla were obtained from Williams [28].
Codivergence test
To determine if there was codivergence between Dasymutilla and Psorthaspis mimicry rings we performed two permutation analyses in R using the phylogenetic trees of both groups. First, an analysis that calculates the Pearson’s correlation coefficient [47] was implemented using the correlation between the distances of the two phylogenies. Second, we applied an analysis that calculates the ParaFitGlobal statistic [48], which uses transformed distances derived from the phylogenetic trees into matrices of principal coordinates. Both analyses test the null hypothesis that the two groups are evolving independently. We performed 100,000 simulations for both tests. Additionally, we constructed a tanglegram linking phenotypically similar species between the phylogenies of Dasymutilla and Psorthaspis. The tanglegram was created using the function “cophyloplot” from the Ape package in R. This function does not optimize the tanglegram and rather is just a visual representation of the shared branching events.
Results
Morphological results
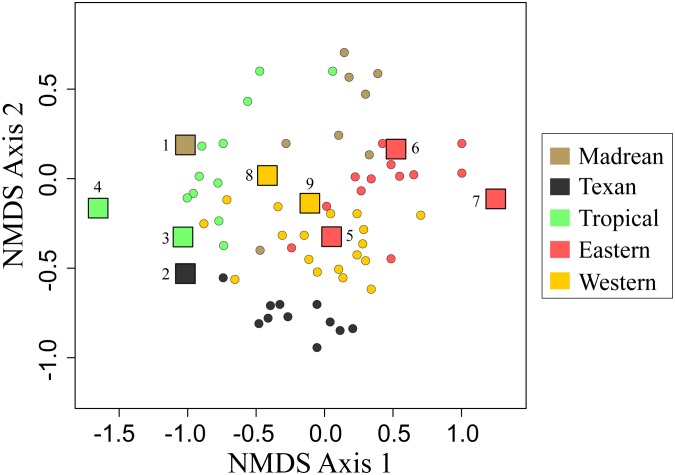
The NMDS and PERMANOVA analyses indicate that morphological traits of Psorthaspis spider wasps fall within Dasymutilla mimicry rings to which they were assigned a priori (Figures 1 and 2). The overall effect of the mimicry ring as a categorical variable was F = 22.503, R2 = 0.616, NMDS stress = 0.14, P<0.001. Despite the overall similarity, the plot of the NMDS and the stress value show that Psorthaspis often do not fit tightly with Dasymutilla in morphospace, but rather seem to fall out near the periphery of the velvet ant clusters. The sole exception was the Eastern mimicry ring, which fell within the middle of the velvet ant distribution (Figure 2).
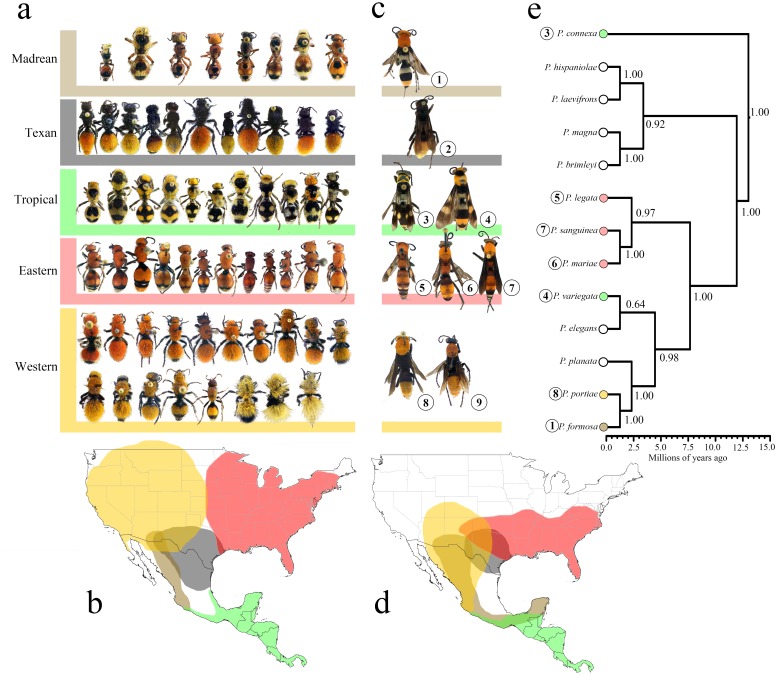
Figure 1. Psorthaspis spider wasp and velvet ant mimicry ring morphology and distribution, and Psorthaspis chronogram (a) Color patterns of the five velvet ant mimicry rings described by Wilson et al. (2012).
(b) Geographic distribution of the five velvet ant mimicry rings. (c) Color pattern of the nine Psorthaspis species placed next to their putative velvet ant mimicry rings. Numbers under each Psorthaspis species correspond to their positions on the phylogenetic tree and in Figure 2. Species number 2 [Psorthaspis texana] and number 9 [Psorthaspis nigriceps] did not yield usable DNA samples and was therefore not included in the phylogenetic analysis. (d) Geographic distributions of the Psorthaspis spider wasp mimicry rings. (e) Psorthaspis spider wasp chronogram. Bayesian posterior probabilities are displayed on nodes.
Figure 2. Morphological trait NMDS ordination plot of Psorthaspis spider wasps and the Dasymutilla mimicry rings to which they were assigned a priori.
Circles denote velvet ant data (from Wilson et al. 2012) and squares represent Psorthaspis data. Numbers represent Psorthaspis species numbered in Figure 1.
Mimetic fidelity reported by volunteers was more variable for spider wasps (Table 1) than for velvet ants [32]. Although some spider wasps received mimetic fidelity scores comparable to the velvet ants (e.g., the Tropical, Madrean and Eastern mimicry rings), others received much lower scores (e.g., the Western and Texan mimicry rings).
Table 1. Human perception tests of mimetic fidelity of Psorthaspis species reported by volunteers (N = 35).
| Spider wasp species | Average mimetic fidelity score | SD | Assigned mimicry ring |
| P. formosa | 4.60 | 2.19 | Madrean |
| P. texana | 4.71 | 3.18 | Texan |
| P. connexa | 8.74 | 1.52 | Tropical |
| P. variegata | 6.29 | 2.53 | Tropical |
| P. legata | 8.83 | 1.69 | Eastern |
| P. mariae | 6.74 | 2.17 | Eastern |
| P. sanguinea | 6.63 | 2.17 | Eastern |
| P. portiae | 5.26 | 2.13 | Western |
| P. nigriceps | 5.89 | 1.91 | Western |
Average mimetic fidelity of each spider wasp species indicates how well each species matches the velvet ant mimicry ring it was phenotypically and geographically most similar to. Scores are based on a scale of 1 (very poor mimic) to 10 (excellent mimic).
Geographical overlap between Psorthaspis and Dasymutilla mimicry rings
Distributions of Psorthaspis spider wasp and Dasymutilla velvet ant species putatively involved in the same mimicry rings are largely congruent (Figure 1). In general, Dasymutilla mimicry rings have a more widespread distribution than that of spider wasps, particularly in northern latitudes. Distributions of Psorthaspis mimicry rings show much greater overlap with each other than do those of Dasymutilla velvet ants (Figure 1). This is particularly apparent in the distribution of the Psorthaspis Madrean mimicry ring, which is geographically larger than the Madrean ring in Dasymutilla. Similarly, the Western Psorthaspis ring extends farther south than the Western Dasymutilla ring, resulting in a larger overlap between Psorthaspis Western and Madrean rings. In addition, the Texan Psorthaspis ring seems to be more restricted than its Dasymutilla counterpart (Figure 1).
Phylogenetic relationships, divergence times and codivergence results
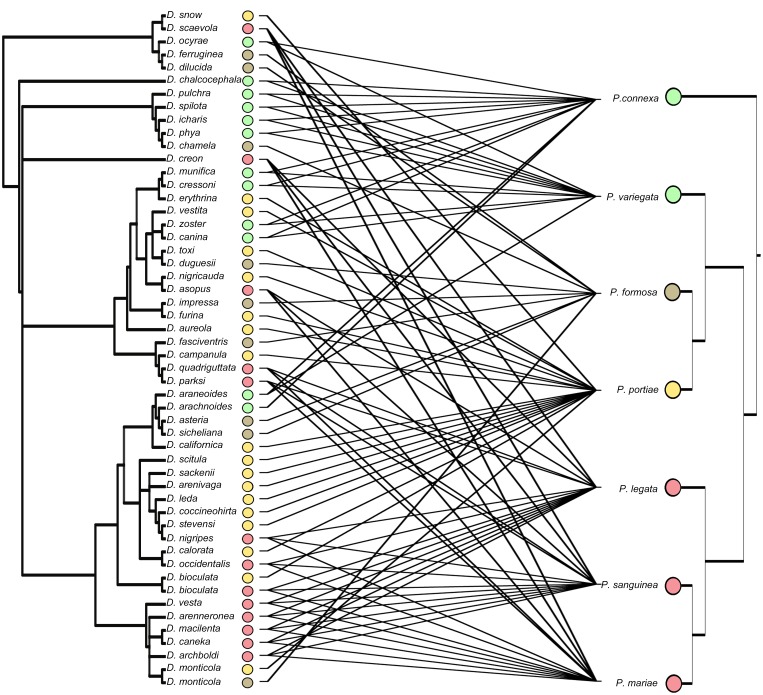
The phylogeny of Psorthaspis suggests that mimetic species do not compose a monophyletic group. Divergence time estimates suggest that the common ancestor of extant Psorthaspis species arose ca. 12.9 Ma (CI = 8.76,18.02). Because taxa composing the sister group to Psorthaspis (i. e. species of Allaporus) are non-mimics [39], it is probable that mimicry arose in Psorthaspis after it diverged from its sister group ca. 18.14 Ma (CI = 13.28,23.71). The origin of Dasymutilla was ca. 21 Ma (CI = 18,23), and the divergence from its sister group was 23 Ma (CI = 21,27) (Williams 2012); therefore, the origin of mimicry in Dasymutilla was likely 23 Ma or later. The codivergence tests suggest topological concordance between the lineages representing mimicry rings of Psorthaspis and Dasymutilla (Pearson’s p = 0.0027, ParaFitGlobal p = 0.047). The tanglegram of Psorthaspis and Dasymutilla, is somewhat complicated by the lack of order of mimetic color patterns in Dasymutilla. Even though at a first glance the phylogenies compared do not have obvious shared branching patterns (due partially to the random distribution of color characters on the velvet ant phylogeny [10]), statistical tests are often a more powerful way to detect correlation because, besides cospeciation, other types of events can be taking place, like independent speciation, and extinctions. Because of this, even host-parasite phylogenies are only rarely completely congruent [47].
Discussion
Fit of Psorthaspis to the velvet ant mimicry rings
Assessing the strength of the fit of mimics to their model is challenging; therefore, we used multiple lines of evidence to support our results. Results of the morphometric analyses and human perception tests indicate that Psorthaspis spider wasps likely participate in the Dasymutilla velvet ant mimicry complex, albeit with a lower mimetic fidelity than the velvet ant participants, which suggests some degree of imperfect mimicry. This lower fidelity of the spider wasps is not surprising, given the many morphological differences between the two groups (e.g., wings, setae, etc.). The lower mimetic fidelity might also be explained by the broad geographic overlap in some Psorthaspis mimicry rings. Such overlap between adjacent mimicry rings is correlated with lower mimetic fidelity in velvet ants [32], and likely accounts for lower mimetic fidelity in spider wasps as well.
Evidence for coevolution
While not tested directly in this study, our results suggest that coevolution played a role in the development of the large velvet ant and spider wasp mimicry complex. Several lines of evidence (i.e., morphological similarity, shared geographic distribution, codivergence) support this assertion. First, while it is not immediately evident from the topologies of the Dasymutilla and Psorthaspis phylogenies (Figure 3), statistical tests show evidence of codivergence between mimetic lineages of the two wasp families. This suggests that the evolution of mimicry between these wasp groups must have involved convergence at the genetic and phenotypic level, such as has been found for Neotropical butterflies [49], [50].
Figure 3. Tanglegram of Psorthaspis (left topology) and Dasymutilla (right topology).
Lines connect between members of the same mimicry rings in the two groups.
Molecular dating estimates suggest that Dasymutilla likely evolved approximately 5 Ma earlier than Psorthaspis, although there is some overlap in the CI estimates of the two groups. This would suggest that the similar color patterns of Psorthaspis spider wasps and Dasymutilla velvet ants likely are the result of codivergence (Figure 1). Interestingly, the low fidelity of spider wasp mimicry is not equal across all mimicry rings. For example, Psorthaspis participating in the Tropical mimicry ring received higher fidelity scores than many of the mimicry rings in higher latitudes (Table 1). This supports the hypothesis that tropical mimics converge on precise mimicry, whereas temperate mimics seem to converge on an “impressionistic” or more relaxed pattern [51]. It also supports the hypothesis that mimicry rings that are more isolated (have little geographic overlap with adjacent mimicry rings) tend to have higher mimetic fidelity because the ecological community is more uniform in coloration, which can lead stronger convergence on one color pattern [32]. The Tropical mimicry ring of Psorthaspis has the least amount of distributional overlap with other mimicry rings, which might explain their high mimetic fidelity.
Coevolution involves reciprocal selective pressures between two groups. While not tested directly, reciprocal selective pressures between Psorthaspis spider wasps and Dasymutilla velvet ants may indeed be taking place. These two wasp groups share predators [18]–[22], and while Dasymutilla velvet ants likely evolved aposematic coloration before Psorthaspis spider wasps, once spider wasps converged phenotypically, the aposematic signal of velvet ants would be strengthened because of the presence of harmful, aposematic co-mimics (spider wasps). Likewise, the spider wasp aposematic coloration would also be strengthened through the presence of their harmful aposematic co-mimics (velvet ants). Thus, both groups would be imposing coevolutionary selective pressures on each other.
Summary
We provide evidence that Psorthaspis spider wasps participate in velvet ant mimicry rings. Furthermore, we find evidence that the two groups codiverged to produce a similar color pattern. This study expands the breadth of the largest known North American Müllerian mimicry complex to include spider wasps as well as velvet ants. This large mimicry complex is an intriguing system that should be the focus of further investigations into the evolution of predator avoidance strategies in the temperate regions, the evolution of aposematic coloration, and the evolution of Müllerian mimicry involving unrelated taxa.
Acknowledgments
We would like to thank Joshua Jahner for reviewing the manuscript and students from Utah State University – Tooele for participating in the human perception tests. We would also like to thank several anonymous reviewers for their critical comments and suggestions to improve the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The molecular data presented in this manuscript have been published before. Data for human perception tests are included in the manuscript. Data for NMDS plots and distributional data points are available from Figshare using the following link: http://dx.doi.org/10.6084/m9.figshare.1210788.
Funding Statement
This work was supported by National Science Foundation (www.nsf.gov) award DEB-0743763 to JPP and CDvD, and Utah Agricultural Experiment Station (http://uaes.usu.edu/), Utah State UAES #8743, to JPP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Müller F (1879) Ituna and Thyridia; a remarkable case of mimicry in butterflies. Ecol Entomol 1879: 20–29. [Google Scholar]
- 2. Benson WW (1972) Natural selection for Müllerian mimicry in Heliconius erato in Costa Rica. Science 176: 936–939. [DOI] [PubMed] [Google Scholar]
- 3. Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC (1985) Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos Trans R Soc Lond B Biol Sci 308: 433–610. [Google Scholar]
- 4. Nijhout HF (1994) Developmental Perspectives on Evolution of Butterfly Mimicry. Bioscience 44: 148–157. [Google Scholar]
- 5. Flanagan NS, Tobler A, Davison A, Pybus OG, Kapan DD, et al. (2004) Historical demography of Müllerian mimicry in the neotropical Heliconius butterflies. Proc Natl Acad Sci U S A 101: 9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones RT, Poul YL, Whibley AC, Merot C, Ffrench-Constant RH, et al. (2013) Wing shape variation associated with mimicry in butterflies. Evolution 67: 2323–2334. [DOI] [PubMed] [Google Scholar]
- 7. Toledo LF, Haddad CFB (2009) Colors and some morphological traits as defensive mechanisms in anurans. Int J Zool 2009: 1–12. [Google Scholar]
- 8. Symula R, Schulte R, Summers K (2001) Molecular phylogenetic evidence for a mimetic radiation in Peruvian poison frogs supports a Müllerian mimicry hypothesis. Proc Biol Sci 268: 2415–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chouteau M, Summers K, Morales V, Angers B (2011) Advergence in Müllerian mimicry: the case of the poison dart frogs of Northern Peru revisited. Biol Lett 7: 796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson JS, Williams KA, Forister ML, von Dohlen CD, Pitts JP (2012) Repeated evolution in overlapping mimicry rings among North American velvet ants. Nat Commun 3: 1272. [DOI] [PubMed] [Google Scholar]
- 11. Acorn JH (1988) Mimetic tiger beetles and the puzzle of cicindelid coloration (Coleoptera: Cicindelidae). Coleopt Bull 42: 28–33. [Google Scholar]
- 12. Mawdsley JR (1994) Mimicry in Cleridae (Coleoptera). Coleopt Bull 48: 115–125. [Google Scholar]
- 13. Lanteri AA, Del Rio MG (2005) Taxonomy of the monotypic genus Trichaptus Pascoe (Coleoptera: Curculionidae: Entiminae), a potential weevil mimic of Mutillidae. Coleopt Bull 59: 47–54. [Google Scholar]
- 14. Edwards G (1984) Mimicry of velvet ants (Hymenoptera: Mutillidae) by jumping spiders (Araneae: Salticidae). Peckhamia 2: 46–49. [Google Scholar]
- 15. Nentwig W (1985) A mimicry complex between mutillid wasps (Hymenoptera, Mutillidae) and spiders (Araneae). Stud Neotrop Fauna Environ 20: 113–116. [Google Scholar]
- 16. Evans HE (1968) Studies on Neotropical Pompilidae (Hymenoptera). IV. Examples of dual sex limited mimicry in Chirodamus. Psyche 75: 1–22. [Google Scholar]
- 17. Schmidt JO (2004) Venom and the good life in tarantula hawks (Hymenoptera: Pompilidae): how to eat, not be eaten, and live long. J Kans Entomol Soc 77: 402–413. [Google Scholar]
- 18. Parker ML, Goldstein MI (2004) Diet of the Rio Grande leopard frog (Rana berlandieri) in Texas. J Herpetol 38: 127–130. [Google Scholar]
- 19. Cicek K, Mermer A (2007) Food composition of the marsh frog, Rana ridibunda Pallas, 1771, in Thrace. Turk Zool Derg 31: 83–90. [Google Scholar]
- 20. Best TL, Pfaffenberger GS (1987) Age and sexual variation in the diet of collared lizards (Crotaphytus collaris). Southwest Nat 32: 415–426. [Google Scholar]
- 21. Manley DG, Sherbrooke WC (2001) Predation on velvet ants (Hymenoptera: Mutillidae) by Texas horned lizards (Phrynosoma cornutum). Southwest Nat 46: 221–222. [Google Scholar]
- 22. Punzo F (2003) Observations on the diet composition of the grey shrew Notiosorex crawfordi (Insectivora) including interactions with large arthropods. Tex J Sci 55: 75–86. [Google Scholar]
- 23. Cuthill JH, Charleston M (2012) Phylogenetic codivergence supports coevolution of mimetic Heliconius butterflies. PLoS One 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futuyma DJ, Slatkin M (1983) Introduction. In: Futuyma DJ, Slatkin M, editors. Coevolution. Sunderland: Sinauer Associates Inc. pp. 1–13.
- 25.Gilbert LE (1983) Coevolution and mimicry. In: Futuyma DJ, Slatkin M, editors. Coevolution. Sunderland: Sinauer Associates Inc. pp. 263–281.
- 26.Page RDM (2003) Introduction. In: Page RDM, editor. Tangled trees: Phylogeny, cospeciation, and coevolution. Chicago: The University of Chicago Press. pp. 1–21.
- 27. Tschopp A, Riedel M, Kropf C, Nentwig W, Klopfstein S (2013) The evolution of host associations in the parasitic wasp genus Ichneumon (Hymenoptera: Ichneumonidae): convergent adaptations to host pupation sites. BMC Evol Biol 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams KA (2012) Systematics of Mutillidae (Hymenoptera) with special emphasis on Dasymutilla and their allies. Ph. D. Dissertation, Utah State University. Available: http://digitalcommons.usu.edu/etd/1200/. Accessed 10 June 2014.
- 29. Jenks GE (1938) Marvels of metamorphosis: a scientific “G-man” pursues rare trapdoor spider parasites for three years with a spade and a candid camera. Natl Geogr Mag 74: 807–828. [Google Scholar]
- 30.Anderson MJ (2005) PERMANOVA. Department of Statistics, University of Auckland, New Zealand.
- 31.R development core team (2010) R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing.
- 32. Wilson JS, Jahner JP, Williams KA, Forister ML (2013) Ecological and evolutionary processes drive the origin and maintenance of imperfect mimicry. PLoS One 8: e61610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Penney HD, Hassall C, Skevington JH, Abbott KR, Sherratt TN (2012) A comparative analysis of the evolution of imperfect mimicry. Nature 483: 461–U110. [DOI] [PubMed] [Google Scholar]
- 34. Dittrich W, Gilbert F, Green P, McGregor P, Grewcock D (1993) Imperfect mimicry - a pigeons perspective. Proc Biol Sci 251: 195–200. [Google Scholar]
- 35. Green PR, Gentle L, Peake TM, Scudamore RE, McGregor PK, et al. (1999) Conditioning pigeons to discriminate naturally lit insect specimens. Behav Processes 46: 97–102. [DOI] [PubMed] [Google Scholar]
- 36. Bain RS, Rashed A, Cowper VJ, Gilbert FS, Sherratt TN (2007) The key mimetic features of hoverflies through avian eyes. Proc Biol Sci 274: 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honkavaara J, Koivula M, Korpimaki E, Siitari H, Viitala J (2002) Ultraviolet vision and foraging in terrestrial vertebrates. Oikos 98: 505–511. [Google Scholar]
- 38.SCAN (2013) Southwest Collections of Arthropods Network. http//:symbiota1.acis.ufl.edu/scan/portal/index.php. Accessed 26 September 2013.
- 39.Rodriguez J, Pitts JP, von Dohlen CD (2014) Historical biogeography of the widespread spider wasp tribe Aporini (Hymenoptera: Pompilidae). J Biogeogr: In press.
- 40.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2011) Geneious v5.4. Available: http://www.geneious.com/.
- 41.Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Uppsala University: Evolutionary Biology Centre.
- 42. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 43.Rambaut A, Suchard MA, Xie D, Drummond AJ (2013) Tracer v1.5. Available: http://beast.bio.ed.ac.uk/Tracer.
- 44. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hommola K, Smith JE, Qiu Y, Gilks WR (2009) A Permutation Test of Host-Parasite Cospeciation. Mol Biol Evol 26: 1457–1468. [DOI] [PubMed] [Google Scholar]
- 48. Legendre P, Desdevises Y, Bazin E (2002) A statistical test for host-parasite coevolution. Syst Biol 51: 217–234. [DOI] [PubMed] [Google Scholar]
- 49. Hines HM, Counterman BA, Papa R, Albuquerque de Moura P, Cardoso MZ, et al. (2011) Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc Natl Acad Sci U S A 108: 19666–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reed RD, Papa R, Martin A, Hines HM, Counterman BA, et al. (2011) optix Drives the Repeated Convergent Evolution of Butterfly Wing Pattern Mimicry. Science 333: 1137–1141. [DOI] [PubMed] [Google Scholar]
- 51. Merrill RM, Jiggins CD (2009) Müllerian Mimicry: Sharing the Load Reduces the Legwork. Curr biol: CB 19: R687–R689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The molecular data presented in this manuscript have been published before. Data for human perception tests are included in the manuscript. Data for NMDS plots and distributional data points are available from Figshare using the following link: http://dx.doi.org/10.6084/m9.figshare.1210788.