Abstract
Background
Streptococcus suis is a major problem in the swine industry causing meningitis, arthritis and pericarditis in piglets. Pathogenesis of S. suis is poorly understood. We previously showed that introduction of a 3 kb genomic fragment from virulent serotype 2 strain 10 into a weakly virulent serotype 2 strain S735, generated a hypervirulent isolate. The 3 kb genomic fragment contained two complete open reading frames (ORF) in an operon-structure of which one ORF showed similarity to folylpolyglutamate synthetase, whereas the function of the second ORF could not be predicted based on database searches for protein similarity.
Results
In this study we demonstrate that introduction of orf2 from strain 10 into strain S735 is sufficient to dramatically increase the virulence of S735 in pigs. This increase in virulence could not be associated with changes in pro-inflammatory responses of porcine blood mononucleated cells in response to S. suis in vitro. Sequence analysis of the orf2-folC-operon of S. suis isolates 10 and S735 revealed an SNP in the −35 region of the putative promoter sequence of the operon, as well as several SNPs resulting in amino acid substitutions in the ORF2 protein. Transcript levels of orf2 and folC were significantly higher in the virulent strain 10 than in the weakly virulent strain S735 and in vitro mutagenesis of the orf2 promoter confirmed that this was due to a SNP in the predicted −35 region upstream of the orf2 promoter. In this study, we demonstrated that the stronger promoter was present in all virulent and highly virulent S. suis isolates included in our study. This highlights a correlation between high orf2 expression and virulence. Conversely, the weaker promoter was present in isolates known to be weakly pathogenic or non-pathogenic.
Conclusion
In summary, we demonstrate the importance of orf2 in the virulence of S. suis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-014-0264-9) contains supplementary material, which is available to authorized users.
Keywords: Streptococcus suis, Piglets, Virulence, Pathogenesis
Background
Streptococcus suis is a zoonotic pathogen that is ubiquitously present among swine populations in the pig industry. Thirty-three capsular serotypes have been described to date [1] of which serotypes 1, 2, 7, 9 and 14 are frequently isolated from diseased pigs in Europe [2]. Strain virulence differs between serotypes and even within a serotype: virulent, avirulent and weakly virulent isolates have been isolated based on the expression of virulence markers, muramidase released protein (MRP) and extracellular factor (EF) [3] and suilysin [4,5].
Nasopharyngeal carriage of S. suis in adult pigs is asymptomatic, whereas in young piglets this increases susceptibility to S. suis invasive disease, leading to meningitis, arthritis and serositis, and high rates of mortality [5]. In Western countries humans occupationally exposed to pigs or uncooked pork might also become infected by S. suis although the incidence is very low [6]. Invasive S. suis infection of humans gives similar clinical signs as in pigs; patients often suffer from remaining deafness after recovery [6]. In Southeast Asia however, S. suis is considered an emerging pathogen for humans, and is recognized as leading cause of bacterial meningitis [7-10]. In Southeast Asia, clinical signs of human infections with S. suis are reported to be more severe compared to other parts of the world, with patients developing toxic shock-like syndrome, sepsis and meningitis [8].
Previously, a hypervirulent S. suis isolate (S735-pCOM1-V[10]) was generated that causes severe toxic shock-like syndrome in piglets after infection resulting in death within 24 h post-infection [11]. S735-pCOM1-V[10] was selected from a library of clones generated in a weakly virulent serotype 2 isolate (S735), after transformation with plasmid DNA isolated from around 30,000 pooled clones carrying randomly cloned genomic DNA fragments from a virulent serotype 2 isolate (strain 10). Isolates with increased virulence were selected by infecting piglets with strain S735 containing the plasmid library of genomic fragments from strain 10. One prevalent clone isolated from the infected piglets contained a 3 kb genomic fragment from strain 10 designated V[10] and was demonstrated to be hypervirulent in subsequent animal experiments. V[10] contained an incomplete open reading frame (ORF), followed by two genes (orf2 and folC) in an operon structure as well as a second incomplete ORF [11]. Assuming that only the full-length ORFs could contribute to the hypervirulence of this isolate, we further characterized the orf2-folC-operon. The first ORF in the operon could not be annotated and was designated orf2, the second ORF in the operon showed homology to polyfolylpolyglutamate synthase (folC) [11]. This operon was present in all S. suis serotypes, including the parent strain S735. Strain S735 with low virulence, contained several single nucleotide polymorphisms (SNP) in orf2-folC and the non-coding regions compared to strain 10 [11].
In this study, we aimed to explain the increased virulence of the strain containing the orf2-folC operon. We demonstrate that overexpression of orf2 suffices to increase the virulence, and that a SNP in the predicted −35 region upstream of the promoter of the operon is strongly associated with virulence in S. suis isolates. Furthermore, the stronger promoter was shown to be present in all virulent or highly virulent S. suis isolates that were included in our study, highlighting a correlation between high orf2 expression and virulence.
Results
Overexpression of orf2 increases virulence of strain S735
Introduction of a 3 kb genomic fragment from virulent serotype 2 strain 10 increased the virulence of the weakly virulent serotype 2 strain S735 [11], creating a hypervirulent isolate (S735-pCOM1-V[10]). All pigs infected with S735-pCOM1-V[10] died within 1 day post infection (p.i.) and a high percentage of the pigs showed severe clinical signs of disease (Table 1), whereas nearly all pigs infected with the control strain S735-pCOM1 survived throughout the experiment. Clinical indices differed significantly (p ≤0.01) between pigs infected with S735-pCOM1-V[10] and S735-pCOM1 (Table 1). As a control we also tested the virulence of S735 transformed with a plasmid containing the homologous 3 kb fragment from strain S735 (S735-pCOM1-V[S735]). A high percentage of the pigs infected with S735-pCOM1-V[S735] survived throughout the experiment. In contrast pigs infected with S735-pCOM1-V[S735] showed significantly more specific clinical signs (p ≤0.01) than pigs infected with S735-pCOM1 (Table 1), although differences in clinical indices for fever and non-specific symptoms were not significantly different between the groups (p = 0.06). Thus the increased copy number of V[S735] in S735, due to introduction of plasmid pCOM1-V[S735] increased specific clinical signs of S. suis. Nevertheless, the specific and non-specific clinical signs due to porcine infection with S735-pCOM1-V[10] (p ≤0.01) were significantly increased compared to pigs infected with S735-pCOM1-V[S735], demonstrating that the introduction of V[10] in strain S735 increased the virulence more than introduction of V[S735]. This result indicated that hypervirulence of strain S735 pCOM-1-V[10] might be due to the different nucleotide polymorphisms in V[10] compared to V[S735].
Table 1.
Virulence of complemented S. suis strains in germfree piglets; all strains contained a plasmid (pCOM1) with or without insert
| Clinical index of the group | No. of pigs in which S. suis was isolated from | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | No. of pigs | Dose (CFU) | Mortality a (%) | Mean no. of days till death | Morbidity b (%) | Specific c symptoms | Non-specific d symptoms | Fever index e | CNS | Serosae g | Joints |
| S735-pCOM1-V[10] | 4 | 106 | 100 | 1 | 100 | 100** | 100** | 38* | 4 | 4 | 4 |
| S735-pCOM1-orf2[10] | 4 | 106 | 100 | 1 | 100 | 100** | 66** | 29 | 4 | 4 | 4 |
| S735-pCOM1-folC[10] | 4 | 106 | 0 | 11 | 0 | 4 | 21 | 1 | 0 | 0 | 0 |
| S735-pCOM1 | 4 | 106 | 0 | 11 | 0 | 0 | 21 | 5 | 0 | 0 | 0 |
| S735-pCOM1-V[10]f | 5 | 106 | 100 | 1 | 100 | 100** | 100** | 60* | 5 | 5 | 5 |
| S735-pCOM1-V[S735]f | 5 | 106 | 20 | 15 | 100 | 43** | 38 | 25 | 1 | 1 | 1 |
| S735-pCOM1f | 5 | 106 | 20 | 16 | 60 | 14 | 11 | 12 | 1 | 0 | 0 |
| T15-pCOM1-V[10] | 5 | 106 | 0 | 14 | 16 | 4 | 16 | 13 | 1 | 1 | 1 |
V[10]/V[S735]: original 3 kb fragment from strain 10 or strain S735 that was selected from library; orf2[10]: orf2 from V[10]; folC[10]: orf3 from V[10]encoding dihydrofolate synthase.
aPercentage of pigs that died due to infection or had to be killed for animal welfare reasons.
bPercentage of pigs with specific symptoms.
cPercentage of observations for the experimental group in which specific symptoms (ataxia, lameness of a least one joint and/or stillness) were observed.
dPercentage of observations for the experimental group in which non-specific symptoms (inappetite and/or depression) were observed.
ePercentage of observations for the experimental group of a body temperature of >40°C.
fPrevious experiments (Smith et al., 2001) were re-analyzed to allow for statistical comparison between experiments, this re-analysis required new stringent definitions of specific and aspecific symptoms as indicated in materials and methods.
*p ≤0.05 compared to S735-pCOM1.
**p ≤0.01 compared to S735-pCOM1.
gSerosae are defined as peritoneum, pericardium or pleura.
To determine whether V[10] alone could increase the virulence of an otherwise avirulent S. suis strain, pCOM1-V[10] was introduced into the avirulent serotype 2 strain T15 to generate strain T15 pCOM1-V[10]. All piglets infected with T15-pCOM1-V[10] survived throughout the experiment (14 d p.i.), some piglets did show mild clinical signs including fever and specific symptoms (Table 1). Although wild-type isolate T15 was not included into this study, clinical signs or fever have not been previously reported in piglets intranasally infected with this avirulent serotype 2 isolate [12,13]. Based on this results we tentatively concluded that the introduction of pCOM1-V[10] might increase the virulence of strain T15 slightly, although the effect was smaller than that observed in strain S735, and could also be due to intravenous inoculation. Obviously, the genetic background of a strain determines whether introduction of V[10] suffices to increase the virulence tremendously.
To determine if the complete orf2-folC-operon was required for the observed increase in virulence, both genes of the operon from strain 10 and its cognate promoter were introduced separately into strain S735 to generate strains S735-pCOM1-orf2[10] and S735-pCOM1-folC[10]. Virulence of these isolates was determined in an experimental infection in piglets, using S735-pCOM1-V[10] and S735-pCOM1 as controls. Table 1 shows that pigs infected with S735-pCOM1-V[10] or with S735-pCOM1-orf2[10] died within one day p.i. with severe clinical signs. Infected pigs developed toxic shock-like syndrome that was not observed using wild-type strain 10 in experimental infections, implying fragments V[10] and orf2[10] increased virulence of S735 yielding more virulent isolates than strain 10 [3]. Both specific and non-specific symptoms were significantly increased (p <0.01) in pigs infected with S735-pCOM1-V[10] or with S735-pCOM1-orf2[10] compared to S735-pCOM1 (Table 1). Bacteriological examination showed that CNS, serosae and joints were colonized by high CFU of S. suis. In contrast pigs infected with S735-pCOM1-folC[10] or S735-pCOM1 lived throughout the experiment (11 days p.i.) showing mild symptoms of infection, like fever. No significant differences in clinical outcome were observed between pigs infected with S735-pCOM1-folC[10] and with S735-pCOM1. This clearly demonstrates that introduction of folC[10] does not increase the virulence of strain S735, whereas introduction of V[10] and orf2[10] increased the virulence of strain S735. Therefore, we concluded that the observed increased virulence of S735-pCOM1-V[10] compared to S735-pCOM1 was attributed to introduction of orf2[10].
Innate immune response of porcine PBMCs
As clinical signs of S. suis strains S735-pCOM1-V[10] and S735-pCOM1-orf2[10] were severe, it was hypothesized that introduction of orf2 might exacerbate the innate inflammatory response to S. suis contributing to the host pathology and symptoms. To test this hypothesis, immune responses of porcine PBMCs to S735-pCOM1-orf2[10] and S735-pCOM1 were compared in vitro. Gene expression levels of innate immune genes of PBMCs were determined after incubation with S. suis isolates as a function of time (2, 4 and 6 h p.i.) using qPCR. Both S735-pCOM1 and S735-pCOM1-orf2[10] induced high gene expression of pro-inflammatory cytokine IL-1-β (170-fold) and chemokine IL-8 (130-fold) compared to controls whereas relatively low gene expression of pro-inflammatory IL-6 (15-fold), anti-inflammatory IL-10 (8-fold), and TNF-α (9-fold) and IFN-γ (4-fold) was induced by both isolates (Figure 1). The transcript levels peaked at 4 h p.i. for all genes tested except for IL-12 which was not expressed in response to S. suis during the incubation time of our experiment (Figure 1). There were no significant differences between the two isolates in the expression levels of any of the tested immune genes. These data suggest that the observed increased virulence of S735-pCOM1-orf2[10] in piglets compared to S735-pCOM1 is probably not due to differences in the host innate responses to this strain.
Figure 1.
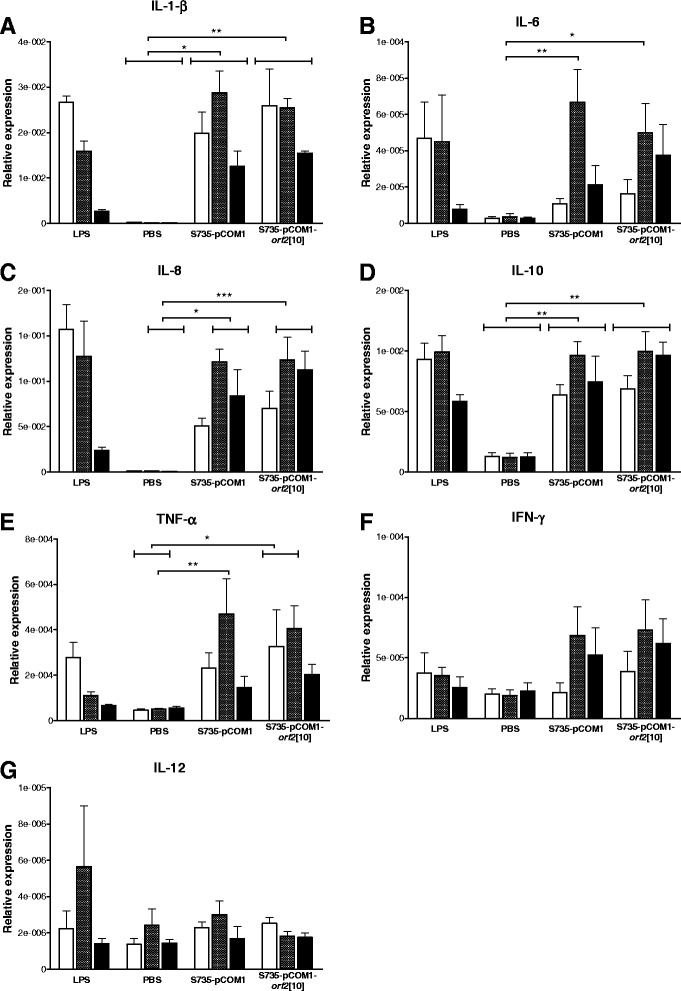
Innate immune response of porcine PBMCs to S. suis isolates. Porcine PBMCs were incubated with S. suis strains S735-pCOM1 and S735-pCOM1-orf2[10] at an MOI of 1. Gene expression of IL-1-β (panel A), IL-6 (panel B), IL-8 (panel C), IL-10 (panel D), TNF-α (panel E), IFN-γ (panel F), and IL-12 (panel G) was determined using qPCR after 2 h (white bars), 4 h (hatched bars) and 6 h (black bars) of stimulation. Relative expression was determined by expressing the amount of target gene relative to a housekeeping gene. LPS: lipopolysaccharide; PBS: phosphate buffered saline. Each bar represents two individual experiments each performed in duplo. Error bars represent standard error of the mean. Significance was determined by 2-way ANOVA analysis, only significant differences between PBS treatment, S735-pCOM1 and S735-pCOM1-orf2[10] are indicated, *p <0.05; **p <0.01; ***p <0.001.
A promoter SNP leads to differential expression of the orf2/folc operon
Sequence analysis of the putative promoter of orf2 revealed a difference at one nucleotide position in the −35 region of the putative promoter in strain 10 (TGGACA) compared to strain S735 (TGGTCA) [11]. The effect of this SNP on expression levels of orf2 and folC in strains 10 and S735 was determined using qPCR analysis. Significantly higher levels of expression of orf2 as well as folC were observed in strain 10 compared to strain S735 (Figure 2A). This clearly indicates that the SNP in the −35 region of the putative promoter affects the transcription of orf2 and folC. Thereby, it demonstrates that the identified SNP was indeed located in the promoter region co-transcribing orf2 and folC in an operon. Moreover, introduction of pCOM1-orf2[10] into S735 increased expression of orf2 31-fold compared to introduction of pCOM1, whereas introduction of pCOM1-orf2[S735] increased expression of orf2 only 5-fold (Figure 2B). As expected expression levels of folC were similar for both recombinant strains (Figure 2B). To confirm that the identified SNP in the −35 region of the promoter is responsible for the differences in transcription of orf2 in strains S735 and 10 the TGGTCA of orf2[S735] was mutated to TGGACA as found in the promoter of orf2[10] (yielding strain S735-pCOM1-orf2[S735][t488a]. Transcript levels of orf2 in S735-pCOM1-orf2[S735][t488a] were shown to be similar to that of strain S735-pCOM1-orf2[10] and four-fold higher than that of strain S735-pCOM1-orf2[S735] in different growth phases (Figure 2C). Both promoters are most active early in the growth phase of S. suis when grown in Todd Hewitt broth (Figure 2C). Together, these results clearly demonstrate that in strain 10, the promoter upstream of orf2-folC-operon is stronger than the promoter upstream of this operon in strain S735, due to an SNP in the −35 region.
Figure 2.
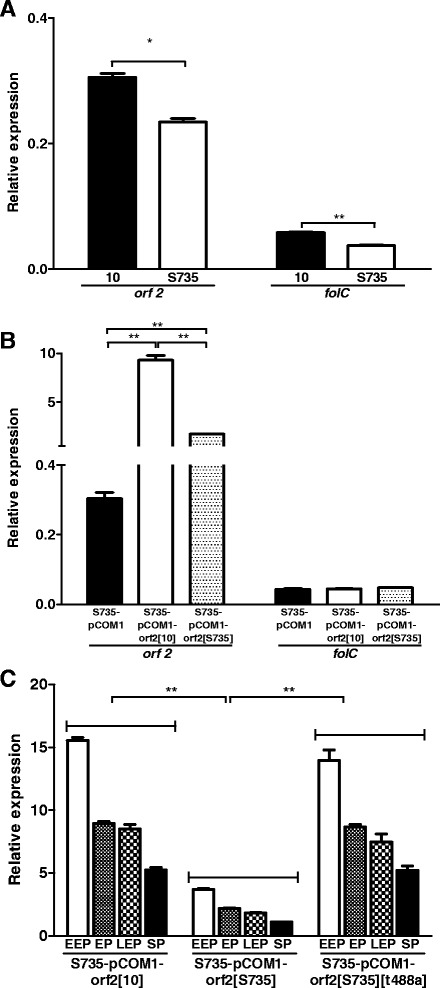
Expression levels of orf2 and folC in S. suis wild-type isolates and mutants. Expression level of orf2 and folC in S. suis wild-type isolates strain 10 (black bars) and S735 (white bars) grown exponentially in Todd Hewitt (panel A); and in strain S735 complemented with empty control plasmid pCOM1 (black bars), with orf2[10] (white bars) or with orf2[S735] (hatched bars) grown exponentially in Todd Hewitt (panel B). Expression level of orf2 in S735 complemented with orf2[10], orf2[S735] and orf2[S735][t488a] after growing in Todd Hewitt until early exponential phase (EEP) (white bars), exponential phase (EP) (small hatched bars), late exponential phase (LEP) (large hatched bars) and stationary phase (SP) (black bars) (panel C). Expression levels were determined using qPCR and expressed as relative expression to housekeeping gene recA. The experiments were performed in triplicate, error bars indicate standard error of the mean. Significance was determined by paired t-tests. *p <0.05; **p <0.01.
Sequence analysis of the-35 region of the orf2/folC promoter and orf2 sequence in different strains and serotypes of S. suis
To determine whether the SNP linked to increased expression of orf2-folC operon was associated with particular clonal types or serotypes of S. suis the promoter regions of a large collection of isolates were sequenced (Table 2 & Additional file 1: Table S1). All isolates used were recently characterized and typed by CGH and MLST [14]. Based on the sequence data obtained, isolates could be divided in two main groups (Table 2 & Additional file 1: Table S1). The strong −35 promoter region was exclusively found in serotype 1 and 2 isolates that belonged to CGH cluster A and MLST clonal complex 1 and that expressed the EF-protein. The SNP associated with lower promoter activity was found in serotype 7 and 9 isolates belonging to CGH group B (except for two), which are all negative for the expression of EF, as well as in weakly virulent isolates of serotype 2 belonging to CGH group A/Clonal Complex 1 (CC1) that were positive for the expression of the larger form of EF protein (EF*). There were two exceptions; serotype 7 isolate (C126), that belongs to CC1 but does not express the EF-protein contained the SNP linked to higher promoter activity and serotype 7 isolate (7711) which had a different −35 promoter sequence (TTGTCA) for which the promoter strength is undetermined. In conclusion, only CC1 isolates expressing EF protein (and 1 serotype 7 isolate) contain the SNP linked to strong promoter activity. As isolates of this combination of phenotype and genotype are strongly correlated with virulence [14,15], upstream of orf2-folC-operon is associated with virulent isolates of S. suis.
Table 2.
Sequence analysis of the −35 region of the orf2 / folC promoter among various S. suis isolates and serotypes 1
| Phenotype | −35 promoter sequence (5′- 3′) | ||||||
|---|---|---|---|---|---|---|---|
| Serotype | MRP 2 | EF 3 | CGH cluster 4 | Clonal complex | TGGACA | TGGTCA | TTGTCA |
| 1 | - | - | B | 13 | 1/1 | ||
| 1 | S | + | A | 1 | 4/4 | ||
| 2 | - | - | B | 16/29/147 | 6/6 | ||
| 2 | + | - | B | 28 | 1/1 | ||
| 2 | + | * | A | 1 | 7/7 | ||
| 2 | - | * | A | 1 | 1/1 | ||
| 2 | + | + | A | 1 | 9/9 | ||
| 7 | - | - | B | 29/1 | 1/85 | 6/8 | 1/8 |
| 9 | - | - | B | 16 | 2/2 | ||
| 9 | * | - | B | 16 | 6/6 | ||
| 9 | + | - | B | 16 | 1/1 | ||
1 S. suis isolates were described in de Greeff et al. [14].
2*indicates an higher molecular weight form of MRP; s indicates a lower molecular weight form of MRP.
3*indicates an higher molecular weight form of EF.
4All isolates were genotyped using Comparative Genome Hybridization (CGH) [14].
5This isolate belongs to clonal complex 1.
“-” indicates absence of the protein; “+” indicates presence of the protein.
Besides the correlation between promoter strength and virulence, we also looked for an association between amino acid sequence of ORF2 and virulence. Sequence analysis of the orf2-folC-operon of wild type strains 10 and S735 revealed several SNPs throughout the sequence [11]. A comparison of ORF2 protein sequences in different isolates revealed more heterogeneity. However, ORF2 protein sequence of all serotype 2 MRP+EF+ isolates were identical to the sequence of ORF2 in strain 10, besides some variation in the predicted start site of the proteins. Clustering of protein sequences revealed three groups of ORF2 sequences, indicated with cluster 1, 2 and 3 in Figure 3. Within cluster 1, two subclusters could be identified: cluster 1A and 1B. Cluster 1A contained strains with an ORF2 sequence that was identical to strain 10; these isolates seem to be associated with virulence. All of the characterized isolates within cluster 1A belong to CC1 and express EF protein. Furthermore, all isolates containing the stronger promoter belonged to this group. Cluster 1B contained two isolates, a Chinese serotype 7 isolate and a serotype 2 MRP−EF− isolate (89–1591), with respectively one and two amino acid substitutions in ORF2 compared to ORF2 of strain 10. The observation that 89–1591 clusters with a serotype 7 isolate, instead of with the other serotype 2 MRP−EF− isolates, adds to the speculation that strain 89–1591 is more similar to serotype 7 isolates than to other MRP−EF− isolates as was also suggested in our CGH study [14]. Within cluster 2, three subclusters could be identified: cluster 2B contained avirulent serotype 2 isolates that were MRP−EF−, cluster 2C contained weakly virulent MRP+EF− isolates including S735, and cluster 2A contained 1 serotype 3 isolate with unknown virulence. Within these subclusters amino acid sequences are identical, the subclusters differ at 13 (2A), 14 (2B) and 9 (2C) amino acid positions compared to strain 10. Cluster 3 contained serotype 9 isolates from different geographical locations, including the reference strain 5218. In conclusion, isolates that are associated with virulence have a strong promoter and an identical protein sequence of ORF2, whereas less virulent isolates have a weaker promoter and a different protein sequence. The relevance of the heterogenic amino acid composition of ORF2 is unknown, it could interfere with function or folding of ORF2.
Figure 3.
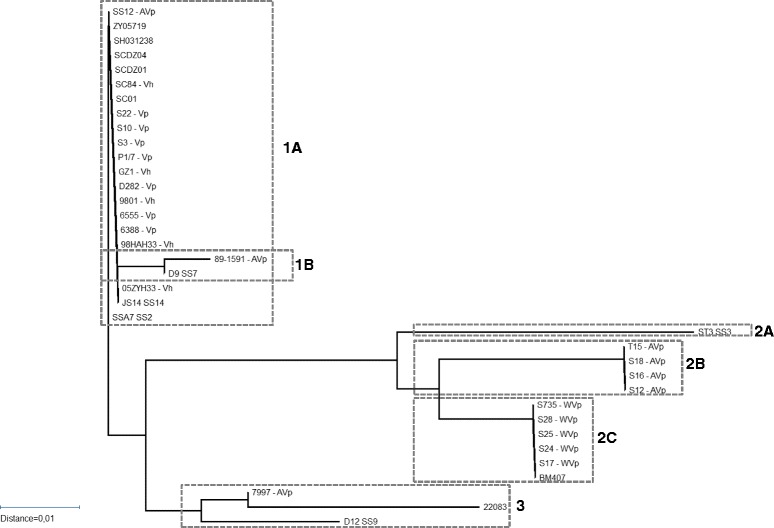
Alignment of Orf2 sequences of different S. suis isolates as available in the database. Phylogenetic tree of ORF2 sequences available in the database. Three different clusters are indicated in dotted boxes, sub-clusters are also indicated. Virulence of strains is indicated using V, AV, WV (virulent, weakly virulent, avirulent), as well as the host where virulence was determined indicatd with p or h (pigs, humans).
Discussion
In this study we showed that introduction of the orf2-gene (ssu0135 in P1/7) of the orf2-folC-operon from strain 10 increased the virulence of weakly virulent strain S735, thereby creating a hypervirulent isolate. The increased virulence of S735-pCOM1-V[10] was shown to be correlated with increased expression of orf2 due to the presence of a stronger promoter in strain 10 compared to strain S735. The promoter of the orf2-folC-operon differed at single nucleotide position close to the predicted −35 region, implicating that one SNP at a crucial position can increase the virulence of S. suis tremendously. The stronger promoter was only present in serotype 1 and 2 isolates belonging to MLST clonal complex 1 that express EF protein, suggesting an association between the strong promoter (and thus higher expression of orf2) and virulence.
Introduction of heterologous orf2[10] into weakly virulent S. suis strain S735 and subsequent infection of pigs with this strain resulted in unusually rapid development of toxic shock-like syndrome and severe clinical signs within 1 day p.i , which resulted in the pigs being euthanized for ethical reasons. Introduction of the homologous fragment containing the intact operon from strain S735 also increased the virulence of the resulting isolate S735-pCOM1-V[S735] significantly compared to the control isolate S735-pCOM1 but the virulence was significantly less that than that of strain S735 pCOM1-V[10]. The virulence of the isolates was correlated with gene expression level of orf2. The −35 sequence of the promoter region of the folC-orf2-operon in strain 10 (TGGACA) deviated from the σ70 consensus promoter (TTGACA) at 1 position, and induced higher expression of both orf2 and folC than the −35 sequence of the promoter region of the folC-orf2-operon in strain S735 (TGGTCA) that deviates from the σ70 consensus at 2 positions. Introduction of homologous V[S735] preceded by the weaker S735 promoter into strain S735, increased the expression level of orf2 5 fold and significantly increased the virulence of the isolate compared to the control S735-pCOM1. Hypervirulence, however, is only achieved when multiple copies of either V[10] or orf2[10] both preceded by the stronger promoter were present in strain S735 This demonstrates that increase of heterologous or homologous expression of orf2 in strain S735, increased the virulence of S735. A similar effect has been observed for the suilysin gene (sly), where 2 SNPs in the promoter region of sly were detected, and suggested to be associated with more virulent isolates [16]. In S. suis a slight increase of virulence occurred after introducing V[10] into avirulent strain T15 although the effect was less pronounced than in strain S735. This suggests that increased heterologous orf2[10] expression in strain T15 might increase the virulence, but the completely different genetic background of strain T15 [14,17] probably prohibited a more pronounced increase of virulence of this strain. In conclusion, increased heterologous or homologous expression of orf2 in either strain S735 or strain T15 was correlated with increased virulence.
Unfortunately, orf2 did not show homology to any known sequences in the database, so it was not possible to predict the function of orf2, and its role in S. suis pathogenesis. Based on hydrophobicity plots [11], ORF2 was predicted to be localized in the membrane, and could therefore interact with the host during infection. The host innate response to the hypervirulent isolate was studied in vitro in PBMCs. A strong pro-inflammatory immune response was induced in PBMCs that peaked 4 h p.i. both after incubation with the hypervirulent isolate as well as with the control isolate (S735-pCOM1). Similar pro-inflammatory host responses to S. suis were described for alveolar macrophages [18], brain microvascular cells [19], monocytes [20] and choroid plexus cells [21], suggesting the hypervirulent isolate induced a similar innate response to other S. suis isolates in vitro. This observation needs to be confirmed in vivo.
In post-mortem bacteriology of piglets infected with the hypervirulent isolate, the organs were found to be colonized by extremely large numbers of S. suis. This result suggests the hypervirulent isolate can multiply very fast in vivo, whereas in vitro the hypervirulent isolate S735-pCOM1-orf2[10] showed a slightly decreased growth rate compared to S735-pCOM1 (data not shown). It could have been expected that the high numbers of bacteria present within 24 h in organs of pigs infected with S735-pCOM1-orf2[10], resulted in a strong proinflammatory responses, that could ultimately result in the observed toxic shock-like syndrome. However, as in vitro measurements did not show any differences in host responses between S735-pCOM1-orf2[10] and S735-pCOM1 , it is more likely that introduction of orf2 into strain S735 is responsible for rapid growth and/or spreading of the bacteria in the host, and is not directly involved in a changed host response. This suggestion could be further substantiated by the in vitro expression studies that showed that orf2 expression is highest in the early exponential growth phase. Although the in vitro kinetics of gene expression do not necessarily reflect the in vivo kinetics, it is plausible that orf2 expression is regulated under in vivo conditions as well. This would suggest that the advantage of introduction of orf2 into strain S735 is inducible during the infection process, leading to more efficient breaking of host defence barriers, or to an increased growth rate as was already suggested by the large bacterial load. Recently, a gene homologous to ssu0135 was annotated as folate transporter folT in S. suis strain SC070731. This suggests that the whole V[10] operon is involved in folate metabolism. It is known that folate is essential for all living organism including bacteria. Our results would suggest that overexpression of genes involved in folate metabolism would be beneficial for growth and virulence in vivo.
In this study, we demonstrated that the stronger promoter was present in all virulent or highly virulent S. suis isolates that expressed EF protein that were included in our study, highlighting a correlation between high orf2 expression and virulence. Conversely, the weaker promoter was present in isolates known to be weakly pathogenic or non-pathogenic. There were some exceptions. A serotype 7 isolate (7711) was shown to have a different promoter that deviated from the σ70 consensus promoter at one position, like the strong promoter. Strength of this promoter is unknown. Another serotype 7 isolate (C126) contained the stronger promoter. This isolate belonged to MLST clonal complex 1, that strictly contains virulent S. suis isolates [15] and was isolated from the joints of a lame pig. Based on these observations, it could be speculated that this isolate is more virulent than other serotype 7 isolates. Finally, the serotype 1 reference strain (5482) contained the weak promoter despite being virulent. However, this isolate was already shown not to be a good representative of serotype 1 isolates [14].
Conclusion
Taken together, the difference in expression level of orf2 due to different promoter sequences could be responsible for the observed difference in virulence. However, we cannot exclude the possibility that the amino acid substitutions of ORF2 also affect function or effect of the ORF2 proteins of S. suis strains. In conclusion, the presence of a strong promoter in the −35 region of the orf2-folC-operon is associated with virulence. Further research on the biological function of orf2 may identify novel targets for vaccination or therapy of S. suis infections.
Methods
Bacterial strains and plasmids
S. suis isolates were grown in Todd-Hewitt broth (Oxoid, London, United Kingdom) and plated on Columbia blood base agar plates (Oxoid) containing 6% (vol/vol) horse blood. Escherichia coli was grown in Luria Broth and plated on Luria Broth containing 1.5% (wt/vol) agar. If required, erythomycin was added at 1 μg ml−1 for S. suis and at 200 μg ml−1 for E. coli. S. suis strain S735 complemented with a plasmid containing a 3 kb genomic fragment derived from strain 10 (S735-pCOM1-V[10]) and the other S. suis strains used in this study have been previously described [11,14] (Figure 4).
Figure 4.
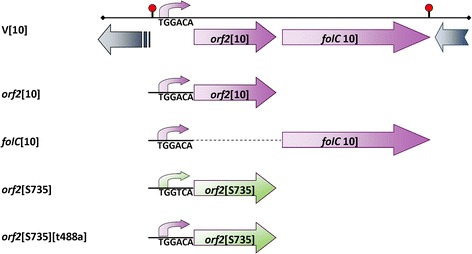
Constructs used in this study. V[10] depicts the clone that was identified using a complementation strategy containing two incomplete ORFs (greyish blue arrows) and a putative operon (purple) containing orf2[10] and folC[10] preceded by the putative promoter of the operon (for clarification only the sequence of the −35 region (TGGACA) of the putative promoter is depicted in the diagram). Constructs were made that contained either orf2[10] or folC[10] (purple) preceded by the putative −35 region of the putative promoter region of the operon (purple). A construct containing orf2 from strain S735 (green) with the −35 region of the putative S735 promoter (TGGTCA) (green) was made. The same construct was mutagenized to contain the −35 region of the putative promoter sequence of strain 10 (TGGACA) (purple) yielding orf2[735][t488a].
Complementation of S. suis strain S735
S735 was complemented with plasmid pCOM1 containing one of the two ORFs in the V[10] operon (i.e. orf2[10], or folC[10]) preceded by the putative promoter region of the operon from strain 10 or with plasmid pCOM1 containing orf2 and the cognate upstream promoter from strain S735 (orf2[S735]) (Figure 4). To construct these plasmids, primers with restriction sites were designed to amplify orf2[10] or orf2[S735] (comE1 – comE2), folC[10] (comE4 – comE6) or the promoter region of the operon (comE1 – comE3) (Table 3). The resulting PCR products orf2[10] and orf2[S735] were digested using restriction enzymes SacI and BamHI, cloned into pKUN19 [22], digested with the same restriction enzymes and subsequently cloned into pCOM1, yielding pCOM1-orf2[10] and pCOM1-orf2[S735], respectively. The PCR amplicon of folC[10] was digested using restriction enzymes SmaI and BamHI and cloned into pKUN19 cleaved with the same restriction enzymes. The PCR product comprising the promoter region of V[10] was cloned in front of folC[10] using restriction enzymes SacI and SmaI. Subsequently, the complete fragment of promoter V[10] – folC[10] was digested from pKUN19 using SacI and BamHI and cloned into pCOM1 digested with the same restriction enzymes, yielding pCOM1-folC[10]. To confirm that the fusion product of promoter – folC[10] was transcribed, in vitro transcription/translation was performed using 35S-methionine. A clear band of the molecular weight of FolC (46.8 kDa) was detected demonstrating that the fusion product could be expressed and translated. All plasmids were introduced into S. suis strain S735 by electroporation. In addition, pCOM1-V[10] was introduced into the avirulent serotype 2 strain T15 [3] by electroporation to yield T15-pCOM1-V[10].
Table 3.
Primer sequences
| Primer name | Sequence 5′-3′ | Target |
|---|---|---|
| comE1 | cgagctcggaagaattggttattgcgcgtg | orf2[10] – forward - SacI |
| comE2 | cgggatcccgggggatgacctgttgcttg | orf2[10] – reverse – BamHI |
| comE3 | tcccccgggggagtcgtgtgtattcgacagcgg | P-orf2-folC[10] – reverse – SmaI |
| comE4 | tcccccgggggacaagcaacaggtcatcccc | folC[10] – forward – SmaI |
| comE6 | cgggatcccggttgaatgcccggcaagcc | folC[10] – reverse - BamHI |
| IL-1-ß-fw | ggccgccaagatataactga | Porcine interleukin 1-ß |
| IL-1-ß-rev | ggacctctgggtatggctttc | Porcine interleukin 1-ß |
| IL-6-fw | gacaaagccaccacccctaa | Porcine interleukin 6 |
| IL-6-rev | ctcgttctgtgactgcagcttatc | Porcine interleukin 6 |
| IL-8-fw | ttcgatgccagtgcataaata | Porcine interleukin 8 |
| IL-8-rev | ctgtacaaccttctgcaccca | Porcine interleukin 8 |
| IL-10-fw | gagaaactagggagcccctttg | Porcine interleukin 10 |
| IL-10-rev | tggccacagctttcaagaatg | Porcine interleukin 10 |
| IL-12-fw | ggagtataagaagtacagagtgg | Porcine interleukin 12-p40 |
| IL-12-rev | gatgtccctgatgaagaagc | Porcine interleukin 12-p40 |
| TNF-α-fw | cgcccacgttgtagccaatgt | Porcine TNF-α |
| TNF-α-rev | cagatagtcgggcaggttgatctc | Porcine TNF-α |
| IFN-γ-fw | caaagccatcagtgaactcatca | Porcine interferon-γ |
| IFN-γ-rev | tctctggccttggaacatagtct | Porcine interferon-γ |
| Orf2-fw | ctacggctggttcttctatcgaa | S. suis orf2 |
| Orf2-rev | gcaatcggtgtcatgataaagg | S. suis orf2 |
| folC-fw | gtttgtccgtccatcggttt | S. suis polyfolylpolyglutamate synthase |
| Folc-rev | ctggtcggtcgcatagatga | S. suis polyfolylpolyglutamate synthase |
| RecA-fw | ggtttgcaggctcgtatgatg | S. suis recombinase A |
| RecA-rev | accaaacatgacaccgacttttt | S. suis recombinase A |
| t488a | gaaaggtatagtttttagcaagtggacaaaatatatagtgtgtgatacaat | Promoter orf2 |
| t488a_antisense | attgtatcacacactatatattttgtccacttgctaaaaactatacctttc | Promoter orf2 |
Bold characters in primers for com indicated the recognition site for restriction enzymes that was added to the primers.
Experimental infection
Experimental infection of caesarean derived germ-free piglets was performed as previously described [11]. Prior to infection, germ-free status of piglets was confirmed by plating tonsil swabs on Columbia agar plates containing 6% horse blood. Briefly, 4 or 5 one-week-old germ-free pigs were infected intravenously with 106 colony-forming units (CFU) of S. suis and then immediately orally administered 40 mg kg−1 body weight of erythomycin (erythomycin-stearate, Abbott B.V., Amstelveen, The Netherlands) twice a day to keep selective pressure on S. suis isolates harbouring the pCOM plasmids. Infected pigs were monitored twice daily for clinical signs and tonsil swabs collected for bacteriological analysis. Pigs were euthanized when clinical signs of arthritis, meningitis, or sepsis were observed after infection with S. suis. Tissue specimens of CNS, serosae and joints were collected during necropsy, homogenized and bacterial cell counts were determined by plating serial dilutions on Columbia agar plates containing 6% horse blood and 1 μg ml−1 of erythomycin. To be able to compare results from different animal experiments included in this manuscript, a uniform scoring of non-specific and specific symptoms was applied to all animal experiments. Non-specific symptoms included inappetite and depression that were scored 0 (none), 0.5 (mild inapptite/depression) or 1 (severe inapetite/depression). Specific symptoms included lameness, central nervous system (CNS) symptoms (locomotive disorders like cycling, or walking in circles; opistotonus; nystagmus), as well as raised hairs, arched back (kyphosis), and shivering, since these are all symptoms of sepsis or serositis. Based on these observation clinical indices were calculated by dividing the number of observations where either specific or non-specific symptoms were observed by the total number of observations for this parameter. This represents a percentage of observations where either specific or non-specific symptoms were observed. Fever was defined as a body temperature >40°C. ‘Mean number of days till death’ was used as a survival parameter. Although animals were euthanized after reaching humane end points (HEP), the time between inoculation and reaching HEPs is still indicative of severeness of infection. It is calculated by averaging the survival in days from inoculation until death.
All animal experiments were approved by the ethical committee of the Central Veterinary Institute of Wageningen UR, Lelystad, The Netherlands, in accordance with the Dutch law on animal experiments (#809.47126.04/00/01 & #870.47126.04/01/01).
Statistical analyses were performed on clinical indices of the groups (fever index, specific symptoms and non-specific symptoms) using a non-parametric Kruskal–Wallis test as there was no homogeneity of variance among groups. In subsequent analyses all groups were compared pairwise to the control group (S735-pCOM1) on all three parameters, using Mann–Whitney U tests. Differences were considered statistically significant at p <0.05. Calculations were performed using SPSS 19 (IBM, New York, USA).
Isolation and stimulation of porcine peripheral blood mononuclear cells
Blood from 3 – 4 week old specific pathogen free (SPF) pigs was aseptically collected and mixed with heparin (LEO Pharma, Breda, The Netherlands) to a final concentration of 5 international units (IU) ml−1 blood. Blood was subsequently diluted 1:1 with DPBS (Invitrogen, Carlsbad, CA, USA). Peripheral blood mononuclear cells (PBMCs) were isolated using Leucosep tubes (Greiner bio-one, Frickenhausen, Germany) according to manufacturer’s instructions. A cell suspension was generated by passage through a 100 μm cell strainer (BD Falcon, Bedford, MA, USA). PBMCs were washed once in DPBS containing 30 μg ml−1 penicillin, and once in RPMI 1640 (Invitrogen) supplemented with 2% of homologous serum (derived from the same donor pig as the PBMCs) and 30 μg ml−1 of penicillin. Cells (1 ml) were seeded at a concentration of 5.106 cells per well in 24-well tissue culture plates. After overnight incubation, cells were stimulated with 5.106 CFU exponentially growing S. suis cells (multiplicity of infection (MOI) = 1), with 1 μg of lipopolysaccharide as a positive control, or with DPBS as a negative control. Antibiotics were not washed away to increase stimulation of PBMCs as described by Wichgers Scheur [23]. At time point 0 (before stimulation) and after 2, 4, and 6 h of stimulation, the supernatant was removed from the PBMCs and RNA was isolated using Nucleospin RNA II kit (Machery Nagel, Düren, Germany) according to manufacturer’s instructions.
cDNA synthesis and quantitative PCR
RT-PCR
Two hundred ng of RNA was used to synthesize cDNA in a reaction containing 25 ng μl−1 random primers (Promega, Madison, WI, USA), 10 mM dNTPs (Promega), 10 mM DTT (Invitrogen), 40 U RNAsin (Promega) and SuperScriptII Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions.
qPCR
cDNA was diluted 20 times for qPCR analysis. Primers were designed using PrimerExpress software (Applied Biosystems, Foster City, CA, USA) (Table 3), except for the porcine reference genes that were ordered at GeNorm (Sequenom, San Diego, CA, USA). Each reaction contained 12.5 pmol forward primer, 12.5 pmol reverse primer and POWR SYBR Green PCR Master Mix (Applied Biosystems) according to manufacturer’s instructions. qPCR was performed using an ABI7500 (Applied Biosystems). GeNorm software (GeNorm) was used to determine the most stably expressed reference genes. Of the 5 candidate reference genes tested for porcine RNA, the transcript amounts of ppiA and gp1 were the least variable between the different samples. For S. suis recA was the least variable in expression of the 6 potential reference genes (phosphogelycerate dehydrogenase (pgd), acetyl coA acetyltransferase (aca), mutS, glutamate dehydrogenase (gdh), gyrase B) tested. Genorm combines expression data into a number, representing stability of expression, where 1 represents the most stabile gene. Stability numbers for S. suis ranged from 1.667 for gdh to 1.217 for recA. The level of expression of these reference genes was measured to control for variation in RNA-yield and RT-reaction conditions. In each qPCR run a standard curve was incorporated consisting of a vector containing a cloned PCR product of the target gene of that reaction. The standard curve consisted of seven 10-fold dilutions of the control vector. In this way both the expression level of the target gene and the expression levels of external reference genes could be calculated from a standard curve. For each reaction water was included in place of cDNA or template as a negative control. Analysis was performed using the ABI7500 Software (Applied Biosystems).
Sequence analysis
Sequence reactions were performed by Baseclear (Leiden, The Netherlands).
Site-directed mutagenesis
Site directed mutagenesis was achieved using the Quick-change II site-directed mutagenesis kit (Agilent Technologies, La Jolla, CA, USA) according to manufacturer’s instructions. PCR primers were designed with the accompanying software (Agilent Technologies) (Table 3). Using primers t448a and t488a_antisense the plasmid pCOM-orf2[S735] was amplified, introducing the desired mutation that changed the −35 region of the putative promotor region of the orf2-folC-operon of S735 from 5′-TGGTCA-3′ to 5′-TGGACA-3′ (Figure 4). The reaction mixture was digested using DpnI to inactivate the original template vector and subsequently transformed to XL-1-blue competent cells (Invitrogen). To exclude the possibility of introducing PCR errors into the vector backbone, the insert of the plasmid (orf2[S735]) was isolated from the template vector after digestion with restriction enzymes BamHI and SacI and cloned into pCOM1 digested with the same restriction enzymes. The resulting plasmid was introduced into S. suis isolate S735 by electroporation and transformants were selected on Columbia agar containing 1 μg ml−1 erythomycin, yielding S735-pCOM1-orf2[S735][t488a]. Sequencing was used to exclude presence of PCR errors in the final construct.
Acknowledgements
The authors would like to acknowledge the personnel of Animal Facilities at Central Veterinary Institute for performing the animal experiments described in this study.
Additional file
Promoter sequence for Streptococcus suis isolates of different serotypes and phenotypes.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AG performed in vitro experiments and drafted the paper; HB prepared the complemented isolates and assisted in animal experiments; JW advised on the study and helped to draft the manuscript; HS conceived of the study, and supervised the work. All authors read and approved the final manuscript.
Contributor Information
Astrid de Greeff, Email: astrid.degreeff@wur.nl.
Herma Buys, Email: herma.buys@wur.nl.
Jerry M Wells, Email: jerry.wells@wur.nl.
Hilde E Smith, Email: hilde.smith@wur.nl.
References
- 1.Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet Microbiol. 2005;107(1–2):63–69. doi: 10.1016/j.vetmic.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Wisselink HJ, Smith HE, Stockhofe-Zurwieden N, Peperkamp K, Vecht U. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet Microbiol. 2000;74(3):237–248. doi: 10.1016/S0378-1135(00)00188-7. [DOI] [PubMed] [Google Scholar]
- 3.Vecht U, Wisselink HJ, Jellema ML, Smith HE. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect Immun. 1991;59(9):3156–3162. doi: 10.1128/iai.59.9.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs AA, Loeffen PL, van den Berg AJ, Storm PK. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect Immun. 1994;62(5):1742–1748. doi: 10.1093/benz/9780199773787.article.b00034458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21(6):381–407. doi: 10.1023/A:1005870317757. [DOI] [PubMed] [Google Scholar]
- 6.Wertheim HF, Nghia HD, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48(5):617–625. doi: 10.1086/596763. [DOI] [PubMed] [Google Scholar]
- 7.Ye C, Zhu X, Jing H, Du H, Segura M, Zheng H, Kan B, Wang L, Bai X, Zhou Y, Cui Z, Zhang S, Jin D, Sun N, Luo X, Zhang J, Gong Z, Wang X, Wang L, Sun H, Li Z, Sun Q, Liu H, Dong B, Ke C, Yuan H, Wang H, Tian K, Wang Y, Gottschalk M, Xu J. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis. 2006;12(8):1203–1208. doi: 10.3201/eid1708.060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, Wang C, Feng Y, Yang W, Song H, Chen Z, Yu H, Pan X, Zhou X, Wang H, Wu B, Wang H, Zhao H, Lin Y, Yue J, Wu Z, He X, Gao F, Khan AH, Wang J, Zhao GP, Wang Y, Wang X, Chen Z, Gao GF: Streptococcal toxic shock syndrome caused byStreptococcus suisserotype 2.PLoS Med 2006, 3(5):e151. [DOI] [PMC free article] [PubMed]
- 9.Takamatsu D, Wongsawan K, Osaki M, Nishino H, Ishiji T, Tharavichitkul P, Khantawa B, Fongcom A, Takai S, Sekizaki T. Streptococcus suis in humans, Thailand. Emerg Infect Dis. 2008;14(1):181–183. doi: 10.3201/eid1401.070568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mai NT, Hoa NT, Nga TV, le Linh D, Chau TT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J, Nghia HD, Minh TN, Chau NV, de Jong MD, Chinh NT, Hien TT, Farrar J, Schultsz C. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46(5):659–667. doi: 10.1086/527385. [DOI] [PubMed] [Google Scholar]
- 11.Smith HE, Buijs H, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. Selection of virulence-associated determinants of Streptococcus suis serotype 2 by in vivo complementation. Infect Immun. 2001;69(3):1961–1966. doi: 10.1128/IAI.69.3.1961-1966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vecht U, Arends JP, van der Molen EJ, van Leengoed LA. Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am J Vet Res. 1989;50(7):1037–1043. [PubMed] [Google Scholar]
- 13.Vecht U, Wisselink HJ, van Dijk JE, Smith HE. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect Immun. 1992;60(2):550–556. doi: 10.1128/iai.60.2.550-556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Greeff A, Wisselink HJ, de Bree FM, Schultsz C, Baums CG, Thi HN, Stockhofe-Zurwieden N, Smith HE: Genetic diversity ofStreptococcus suisisolates as determined by comparative genome hybridization.BMC Microbiol 2011, 11:161. [DOI] [PMC free article] [PubMed]
- 15.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40(10):3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi D, Akeda Y, Nakayama T, Kerdsin A, Sano Y, Kanda T, Hamada S, Dejsirilert S, Oishi K. The contribution of suilysin to the pathogenesis of Streptococcus suis meningitis. J Infect Dis. 2014;209(10):1509–1519. doi: 10.1093/infdis/jit661. [DOI] [PubMed] [Google Scholar]
- 17.Smith HE, Rijnsburger M, Stockhofe-Zurwieden N, Wisselink HJ, Vecht U, Smits MA. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J Clin Microbiol. 1997;35(5):1049–1053. doi: 10.1128/jcm.35.5.1049-1053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Greeff A, Benga L, Wichgers Schreur PJ, Valentin-Weigand P, Rebel JM, Smith HE. Involvement of NF-kappaB and MAP-kinases in the transcriptional response of alveolar macrophages to Streptococcus suis. Vet Microbiol. 2010;141(1–2):59–67. doi: 10.1016/j.vetmic.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Vadeboncoeur N, Segura M, Al-Numani D, Vanier G, Gottschalk M. Pro-inflammatory cytokine and chemokine release by human brain microvascular endothelial cells stimulated by Streptococcus suis serotype 2. FEMS Immunol Med Microbiol. 2003;35(1):49–58. doi: 10.1111/j.1574-695X.2003.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 20.Segura M, Vanier G, Al-Numani D, Lacouture S, Olivier M, Gottschalk M. Proinflammatory cytokine and chemokine modulation by Streptococcus suis in a whole-blood culture system. FEMS Immunol Med Microbiol. 2006;47(1):92–106. doi: 10.1111/j.1574-695X.2006.00067.x. [DOI] [PubMed] [Google Scholar]
- 21.Schwerk C, Adam R, Borkowski J, Schneider H, Klenk M, Zink S, Quednau N, Schmidt N, Stump C, Sagar A, Spellerberg B, Tenenbaum T, Koczan D, Klein-Hitpass L, Schroten H. In vitro transcriptome analysis of porcine choroid plexus epithelial cells in response to Streptococcus suis: release of pro-inflammatory cytokines and chemokines. Microb Infect. 2011;13(11):953–962. doi: 10.1016/j.micinf.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Konings RN, Verhoeven EJ, Peeters BP. pKUN, vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- 23.Wichgers Schreur PJ, Rebel JM, Smits MA, van Putten JP, Smith HE: Lgt processing is an essential step inStreptococcus suislipoprotein mediated innate immune activation.PLoS ONE 2011, 6(7):e22299. [DOI] [PMC free article] [PubMed]


