Abstract
Costimulation of purified CD8+ T lymphocytes induces de novo expression of CD4, suggesting a previously unrecognized function for this molecule in the immune response. Here, we report that the CD4 molecule plays a direct role in CD8+ T cell function by modulating expression of IFN-γ and Fas ligand, two important CD8+ T cell effector molecules. CD4 expression also allows infection of CD8 cells by HIV, which results in down-regulation of the CD4 molecule and impairs the induction of IFN-γ, Fas ligand, and the cytotoxic responses of activated CD8+ T cells. Thus, the CD4 molecule plays a direct role in CD8 T cell function, and infection of these cells by HIV provides an additional reservoir for the virus and also may contribute to the immunodeficiency seen in HIV disease.
CD8+ cytotoxic T lymphocytes (CTL) have a major role in antiviral immunity, directly killing virally infected cells and producing antiviral cytokines. Activation of these cells requires interaction of the T cell receptor complex with antigenic peptide and major histocompatibility complex (MHC) class I molecules on antigen-presenting cells (APCs) followed by a second costimulatory signal (1). After activation, there is a coordinated expression of various cell surface molecules, many of which play a direct role in cytotoxic activity. We and others have shown that costimulation of CD8+ T cells from the peripheral blood results in the de novo expression of CD4, a molecule previously thought to be absent on this cell type at this stage of development (2–5). These CD8+CD4+ cells express higher levels of activation molecules than do costimulated CD8+ T cells lacking CD4 expression (2, 6). CD8+CD4+ T cells constitute ≈3–5% of the human peripheral blood lymphocyte pool (7–10). Certain conditions seem to influence CD8+CD4+ cell levels in humans, including infection with HIV (11), human T lymphotrophic virus-1 (12), Epstein–Barr virus (8), human herpesvirus 6 (13), and aging (10). CD8+CD4+ cells also have been observed in monkeys (14–17), and in mice, rats, swine, and chickens (reviewed in ref. 18). In mice, CD8+CD4+ cell levels increased after inoculation with reovirus or recombinant adenovirus (19, 20). In each species, the CD8+CD4+ populations usually displayed the phenotype of activated or previously activated T cells and, in the studies that assessed the composition of the CD8 dimer, were predominantly CD8αβ (versus CD8αα) cells (11, 15, 19). The presence of CD8+CD4+ cells in normal individuals and the increased representation of these cells in individuals with disease or increased antigenic stimulation suggest a role for this cell type in immunity.
The CD4 molecule has an important role in CD4+ T helper (Th) cell development and response to antigen, including functioning as an adhesion molecule, regulating cellular activation and gene expression, and serving as a chemotactic receptor (21–23). Its role as the primary receptor for HIV is well known (24, 25). The cytoplasmic tail of the CD4 molecule on Th cells is associated with the Src-family kinase Lck, through which it induces intracellular tyrosine phosphorylation and other signaling events (22, 23). We and others have shown that Lck is associated with CD4 in CD8+ T cells in a way similar to the association in Th cells, allowing signaling events to occur through this pathway (5, 6). We also have determined that CD4 can serve as a chemotactic receptor on CD8+ T cells, allowing cell migration in response to IL-16 (6). Inhibition of Lck signaling by specific chemical inhibitors ablates the ability of CD4 to direct this migration (6). Thus, CD4 expressed on CD8+ T cells is functionally capable of responding to an external signal, although its full role on this cell type is not yet known.
During the course of HIV infection, CD8+ CTL responses are unable to fully suppress viral replication (26). It is not clear how this occurs or whether HIV has a direct effect on CTL function. Direct infection of a target cell by HIV is known to impair the cell's ability to function properly through mechanisms such as cytotoxicity or apoptosis (27). In addition, HIV may inflict “collateral damage” on nearby uninfected cells by inducing dysfunction and/or cell death (27). HIV proviral DNA has been found in CD8+ T cells in vivo in the peripheral blood and pulmonary compartments (11, 28–35). Simian immunodeficiency virus proviral DNA also has been found in CD8+ T cells in the peripheral blood of infected macaques (16, 17), particularly in the CD8+CD4+ T cell subset (16). Infection of CD8+ T cells can occur via the CD4 molecule either through an immature CD4+ stage of T cell development (36, 37) or in a more mature cell after its induction by costimulation (3–5, 28). A recent study identified significant levels of HIV proviral DNA in mature CD8+ T cells through the use of ultra high purity polychromatic fluorescent cell sorting (FACS) procedures (28), further validating the presence of HIV in this population and quelling criticisms regarding sorting purity in earlier studies. However, the consequences of HIV or simian immunodeficiency virus infection in CD8+ T cells are unknown.
In this study, we investigated how the CD4 molecule on human CD8+ T cells plays a role in CD8+ cell function. We further examined the effects of HIV infection on these responses. Here, we show that ligation of the CD4 molecule on the surface of CD8+ T cells results in increased expression of IFN-γ and Fas ligand (FasL), two important effector molecules. Further, we found that CD4 plays a direct role in CTL activity, and that HIV infection down-regulates CD4 cell surface expression and perturbs the IFN-γ, FasL, and cytotoxic responses of CD8+ T cells. These findings identify a previously unrecognized role for the CD4 molecule in T cell-mediated responses and suggest that CD8+CD4+ cells are highly active effector cells. Furthermore, infection of activated CD8 cells via CD4 provides an additional reservoir for the virus, and may also provide an additional immune-escape mechanism by directly perturbing CD8+ T cell responses.
Methods
Analysis of Human Blood. Human whole blood was obtained in accordance with methods approved by the University of California, Los Angeles Institutional Review Board. Blood from HIV-negative donors was analyzed by flow cytometry within 4 h of withdrawal after lysis of red blood cells with ammonium chloride.
Flow Cytometry. Cells were analyzed by using mAbs to CD3, CD25, CD8, CD4, KC57 (anti-p24), CD69, CD71, CD45RA, CD45RO (Coulter), FasL (Pharmingen), or HLA-DR (Becton Dickinson) conjugated to FITC, phycoerythrin, perdinin chlorophyll protein, allophycocyanin, or biotin. Cells were analyzed by using a FACSCaliber flow cytometer and the cellquest (Becton Dickinson) software. Dot plot cursor settings were based on isotype control staining and samples were stained with single-color antibodies in the presence of isotype control antibodies (4). Intracellular HIV p24gag staining (KC57) was performed by permeabilizing cells with 0.2% Tween 20 after cell surface staining. Cellular apoptosis was assessed by annexin V and propidium iodide staining (Pharmingen) and staining with Mitotracker Deep Red 633 mitochondrial membrane staining (Molecular Probes).
Cell Culture. CD8+ T cells from fresh human leukopacks were purified by negative selection columns (purity >95%) and cultured in medium alone (unstimulated), anti-CD3 mAb alone, anti-CD28 mAb alone, or with anti-CD3 and anti-CD28 for 3 days as described in refs. 4 and 6. Cells then were analyzed by flow cytometry; gating was used to highlight the CD8+ population.
Microarray Analysis. Purified human CD8+ T cells were costimulated for 3 days and subsequently treated with medium alone, cross-linked (XL) CD4 mAb, or IL-16 (6) and cultured for 3 h before harvest. RNA was isolated (4) and gene expression analysis was performed by using U95A microarrays (Affymetrix, Santa Clara, CA) and the genespring program (Genespring, Palo Alto, CA). Genes were excluded from analysis if they did not have ≥1,200 arbitrary expression units at the time of their greatest levels of expression. Stimulation index was calculated as an increase or decrease over medium control. CD4 RNA expression was confirmed by RT-PCR (4).
Measurement of Cytokine Production. Three-day costimulated human CD8+ T cells were further stimulated for 24 h in triplicate with medium alone, medium with a XL-irrelevant mAb [anti-NK cell mAb (2B4)] (Pharmingen), XL-CD4 mAb, or IL-16, or XL-CD56 with or without 10 μM of the SRC-family kinase inhibitor 4-amino-5-(chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2) (6). IFN-γ production was measured by enzyme-linked immunospot (ELISPOT) using antihuman IFN-γ capture and detection antibodies (Pharmingen) (38, 39). Quantitation was performed with an Immunospot Series 1 Analyzer (CTL, Cleveland).
FasL Expression. Three-day costimulated human CD8+ T cells purified from normal, HIV-negative individuals were further stimulated for the indicated time period with medium, medium plus XL-CD4 mAb, or IL-16, or XL-CD56 with and without 10 μM PP2 (6). Cells were removed at the indicated times and stained with FasL-biotin, CD8-perdinin chlorophyll protein, and CD4 energy coupled dye (Clone SFCI12T4D11, Coulter) mAbs. Secondary staining with streptavidin-phycoerythrin was used to label the biotinylated mAb. A subset of cells was stained with mouse isotype controls. FasL expression was assessed by flow cytometry, gating on the CD8+ population. OKT4 does not inhibit binding of the SFCI12T4D11 mAb (data not shown).
Generation and Measurement of CTL Activity. To prepare effector, target, and stimulator cells for generation of allospecific CTL, various cell subsets were first isolated from fresh human peripheral blood leukocytes by magnetic activated cell sorting (Miltenyi Biotec, Auburn, CA). Dendritic cells, used to stimulate CD8 cells to react to alloantigen, were generated from peripheral blood monocytes cultured for 7 days in IL-4 and granulocyte–macrophage colony-stimulating factor as described in ref. 40. CD19+ B lymphocytes were isolated from the blood of the same donor as the dendritic cells and used as CTL target cells. These cells were viably frozen in RPMI medium 1640/10% DMSO until needed. Purified CD8+ T cells from a different donor were added to the dendritic cells as described and cultured for 7 days to generate alloantigen-specific CTL (4). Three days before use as target cells for CTL lysis, the CD19+ B cells were thawed and stimulated with XL anti-human IgM mAb (Coulter). These cells were then radioactively labeled by incubation with 350 μCi of sodium [51Cr]chromate for 1–2 h before exposure to allostimulated CD8 T cells. Cytotoxic activity of the effector CD8 cells was then assessed by quantitation of radioactivity in the supernatant by using a standard 51Cr release assay at various effector/target ratios. Each condition was measured in triplicate as described in ref. 41. In some 51Cr release assays, a mixture of anti-MHC class II mAbs (containing anti-HLA-DR, -DP, -DQ activity; 5 μg/ml, Pharmingen), soluble CD4 (100 ng/ml, NEN), anti-CD4 neutralizing antibody (5 μg/ml, R & D Systems), mouse Ig (5 μg/ml, Sigma), 50 μg of HIV-1NL4–3, 50 μg of HIV-1NFN-SX, or virus-free control cell supernatant was added to the cultures along with the CD8 cells, to determine any effects on lytic activity.
HIV Infection. Costimulated CD8+ T cells were infected with 10,000 infectious units per 106 cells of HIV-1BAL, HIV-1ADA-M, HIV-189.6, HIV-1IIIB, HIV-1NFN-SX, or HIV-1NL4–3, or mock infected by treatment with virus-free cell supernatant, depending on the experiment. Viral growth was measured by ELISA for HIV p24 as described in ref. 42.
Results
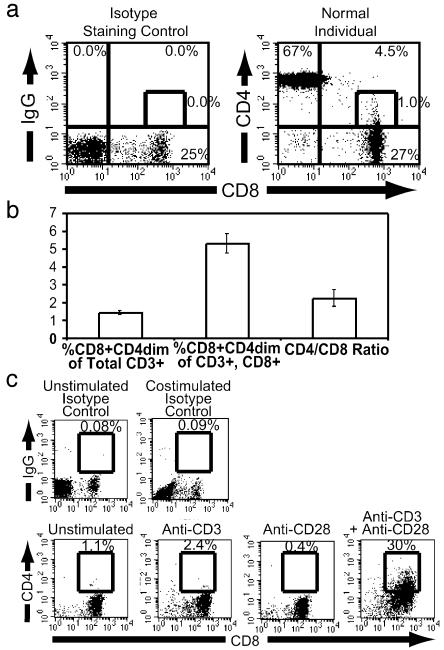
Induction of CD4 Expression by CD8+ T Cells. CD3+CD8+CD4dim cells constitute a significant and often overlooked subpopulation of CD8+ T cells in the peripheral blood (Fig. 1a) (7, 8, 10, 43). The CD8+CD4dim subset constitutes ≈5% of the CD3+CD8+ population (Fig. 1b). Phenotypic analysis reveals that the CD3+CD8+CD4dim subset is a highly activated population. These cells are αβ T cell receptor positive and express greater levels of the activation molecules CD25, CD69, HLA-DR, CD71, and CD45RO than do the CD3+CD8+CD4– population of mature T cells or thymocytes (2, 4) (data not shown). Thus, these cells are mature and are not thymocytes that have escaped the thymus into the peripheral blood. Consistent with this observation, purified human CD8+ T cells, when costimulated by anti-CD3 and anti-CD28 mAbs, substantially up-regulated CD4 expression, but not when stimulated by CD3 or CD28 alone (Fig. 1c) (3, 6). This finding suggests that the CD3+CD8+CD4dim subset of cells in the peripheral blood is a functionally active population of cells.
Fig. 1.
Identification of CD8+CD4dim T cells. (a) CD8 vs. CD4 expression on CD3+ peripheral blood lymphocytes from a healthy, HIV-negative individual. The single-color (energy coupled dye) isotype staining control identifying cells stained for CD8 and nonspecific mAb is identified (Left). CD3+ cells were identified by gating and CD4 versus CD8 expression is shown (Right). The percentage of cells in each quadrant is indicated at the outer edge; percentage CD8+CD4dim cells of entire population is given next to the indicated gate. (b) CD8+CD4dim cells in normal, HIV-negative individuals. The percentage of CD8+CD4dim cells of the total CD3+ population (left), the CD3+CD8+ population (center), and the ratio of CD4 to CD8 T cells (right) are given. (c) Costimulation of CD8+ T cells. CD4 and CD8 cell surface expression after 3 days of stimulation in the presence of medium, anti-CD3 alone, anti-CD28 alone, or anti-CD3 plus anti-CD28. The single-color (energy coupled dye) isotype controls are shown to indicate nonspecific background staining. The boxes are drawn to include CD8+CD4dim cells, and the numbers above the box denote the percentage of CD8 cells expressing CD4.
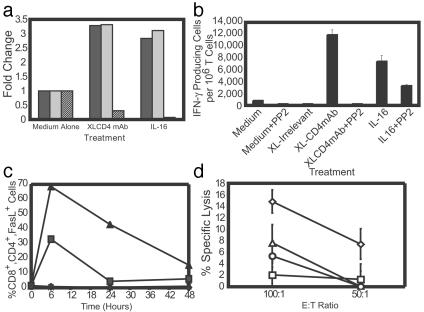
Functional Role of CD4 on CD8+ T Cells. Because CD4 plays a critical role in the activity of CD4+ Th and other hematopoietic cells, we investigated how the CD4 molecule on CD8+ T cells contributes to cell-mediated immunity. To determine the functional significance of CD4 on the surface of CD8+ T cells, we used microarray analysis to assess changes in gene expression profiles of costimulated human CD8+ T cells after ligation of CD4. Short-term ligation of CD4 either by XL anti-CD4 mAb or by the CD4-specific ligand IL-16 resulted in a >2.5-fold up-regulation of 127 genes and the >2.5-fold down-regulation of 159 genes. Specifically, RNA for CD4 was down-regulated, indicating that ligation rapidly shuts off expression of this molecule (Fig. 2a). Expression of IFN-γ and FasL, two genes critical for CD8+ T cell function (44, 45), was significantly up-regulated after CD4 ligation. We and others have shown that the signaling molecule p56lck (Lck) is functionally associated with the CD4 molecule in CD8+ T cells (5, 6). Treating cells with the src-family kinase inhibitor PP2, which inhibits signaling through Lck (46), abrogated the changes in IFN-γ, FasL, and CD4 RNA seen after ligation of CD4 (data not shown). These data strongly suggest that ligation of CD4 on CD8+ T cells has a role in CD8 T cell function and that CD4 mediates this activity through its primary signaling molecule, Lck.
Fig. 2.
IFN-γ and FasL expression after ligation of CD4 on costimulated CD8+ T cells. (a) Microarray analysis of IFN-γ (black bars), FasL (gray bars), and CD4 (striped bars) gene expression 3 h after CD4 ligation of costimulated CD8+ T cells. Ligation conditions are indicated. Data represent the average of two experiments. (b) IFN-γ production for 24 h after CD4 ligation with and without PP2 pretreatment, as measured by ELISPOT. XL-irrelevant refers to treatment with XL anti-NK cell (2B4) mAb (not reactive to markers expressed by CD8+ T cells). Data are expressed as the number of IFN-γ+ cells per million. The data are representative of four experiments. (c) Cell surface FasL expression after treatment of CD4 with medium (♦), XL CD4 mAb (▴), or XL-CD4 with PP2 treatment (▪). Mouse isotype staining controls are indicated by +. The data are representative of four independent experiments. (d) Effect of CD4 in the CTL response. Alloantigen-specific CTL were incubated with allogeneic CD19+ target cells at the indicated effector/target cell ratios in the presence of medium-plus mouse Ig (medium alone control) (⋄), a mixture of anti-MHC class II mAbs (▵), soluble CD4 (□), or neutralizing anti-CD4 mAb (○). The data are representative of three independent experiments.
To confirm that IFN-γ protein levels were increased after ligation of CD4, CD3/CD28 costimulated CD8+ T cells were further stimulated with XL anti-CD4 mAbs or with IL-16, and IFN-γ production was measured by ELISPOT (Fig. 2b). CD4 ligation increased the numbers of IFN-γ-producing cells, and treatment of cells with PP2 inhibited this effect (P ≤ 0.05). Similar results were observed when biotinylated anti-CD4 mAbs were cross-linked with streptavidin; thus, increased IFN-γ expression was not due to cross-linking of residual anti-CD3 and anti-CD28 mAbs from the original costimulation step (data not shown). Blocking the effect of IL-16 by prestaining cells with non-XL anti-CD4 mAb to inhibit IL-16 binding significantly decreased IL-16-mediated increases in IFN-γ (data not shown), indicating that IFN-γ activation occurs through IL-16 interaction with CD4. Further, ligation of CD56, another cell surface protein found on activated CD8+ T cells (47), did not induce IFN-γ production (not shown), indicating that this up-regulation is not simply a result of nonspecific receptor binding.
We next used flow cytometry to determine whether ligation of CD4 induces FasL expression on the surface of T cells. Cell surface FasL was up-regulated within 6 h of CD4 cross-linking, and treatment with PP2 inhibited this effect (Fig. 2c). IL-16 also up-regulated FasL, although to a lesser extent, and PP2 abrogated this effect (not shown). Blocking IL-16 interaction with CD4 by preexposing the cells to a non-XL anti-CD4 mAb inhibited FasL up-regulation (not shown). Ligation of CD56 did not result in up-regulation of FasL (not shown). Thus, our results indicate that the specific ligation and cross-linking of CD4 on CD8+ T cells results in Lck-mediated production of IFN-γ and FasL, suggesting that CD4 expression could directly influence CD8 cell effector function.
CD4 and Cytotoxicity. To examine the role of the CD4 molecule on CD8+ T cells in the CTL response, we generated alloantigen-reactive CD8+ cytotoxic T lymphocytes by stimulating purified human CD8+ T cells with allogeneic monocyte-derived dendritic cells (4). Cytotoxic function was assessed against activated B cells (which express MHC class I and class II target antigens, but do not express CD4) (data not shown) derived from the same donor as the monocyte-derived dendritic cell targets. Allogeneic dendritic cells, in the absence of CD4+ Th cells, induced cellular activation and CD4 expression on CD8+ T cells (4) (typically 22–50% of cells were CD4+) and generated CTL activity against alloantigen (Fig. 2d). Blocking the target cell-mediated ligation of CD4 by adding soluble CD4 (sCD4), or a non-XL anti-CD4 mAb that blocks MHC class II interaction, or a mixture of anti-MHC class II mAbs, which is known to inhibit the generation of a CD4+ T cell mixed leukocyte reaction, significantly inhibited the CTL response (P <0.05). Thus, the CD4 molecule present on CD8+ T cells plays a direct functional role in the primary CTL response against alloantigen.
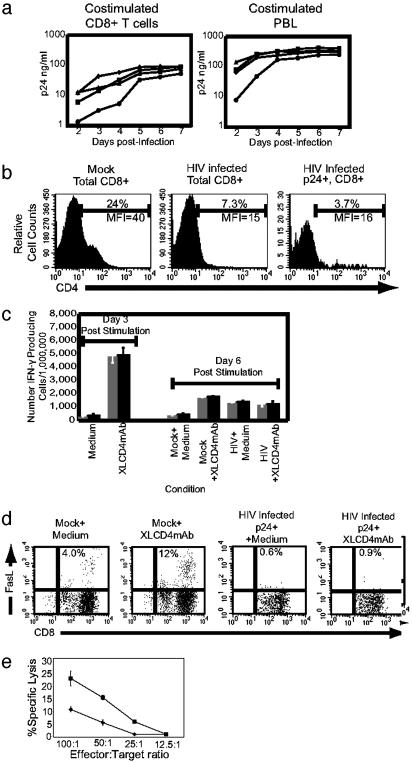
HIV Dysregulation of Cytotoxic T Cell Function. We then examined the effects of HIV infection on the CD8+CD4dim population of cells. CXCR4(X4)-tropic, CCR5(R5)-tropic, and X4/R5 dualtropic viruses are capable of replicating in vitro in purified costimulated CD8+ T cells, although not quite to the level seen in costimulated, unfractionated peripheral blood lymphocytes (Fig. 3a). We further examined HIV-infected CD8+ T cells for CD4 cell surface expression and found that the CD4 molecule was quickly down-regulated after infection by both X4-tropic (Fig. 3b) and R5-tropic (data not shown) strains of HIV-1. We observed no increases in apoptosis or necrosis as assessed by annexin V, propidium iodide staining, or changes in mitochondrial membrane potential 3 days after infection (data not shown). HIV gag protein was also found in viable CD8+CD4– cells (see below). Thus, loss of CD4 was because of down-regulation, not cell death. Further, infection with either an X4- or R5-tropic virus with a deletion in the nef ORF, resulted in similar amounts of down-regulation as the wild-type virus (data not shown). Thus, the observed down-regulation of CD4 probably occurs by nef-independent mechanisms.
Fig. 3.
Effects of HIV infection of CD8+ T cells. (a) HIV-1 replication in costimulated CD8+ T cells and peripheral blood leukocytes (PBL). Costimulated CD8+ T cells purified from PBL (Left) or whole PBL (Right) were infected with R5-tropic HIV-1BAL (▪), HIV-1ADA-M (♦), R5/X4 dual tropic HIV-189.6 (▴), or X4-tropic HIV-1IIIB (•) and p24gag production measured daily by ELISA. (b) HIV down-regulation of CD4 in CD8+ T cells. CD4 expression on costimulated CD8+ T cells from uninfected culture (Left), HIV-1NL4–3-infected cultures (Center), and HIVNL4–3 p24+ cells (Right). Percentages of CD4+ cells are above the indicated gate, and mean fluorescent intensities (MFI) of CD4 expression are below the gate. (c) HIV perturbation of the IFN-γ response. IFN-γ expression, measured by ELISPOT with or without ligation of CD4 by XL-CD4 for 3 days (left) or 6 days (right) after costimulation and mock treatment or infection with HIV-1NL4–3. Data are expressed as the number of IFN-γ+ cells per million. (d) Perturbation of FasL expression by HIV infection of CD8+ T cells. CD8 versus FasL expression of either mock-treated or HIV-1NL4–3-infected cells 3 days after infection, 6 h after treatment with either medium alone or with XL-CD4 mAb, as indicated. Percentages of CD8+FasL+ cells are indicated. (e) Impairment of CD8+ T cell lytic function by HIV. The percent lysis of allogeneic target cells at the specified effector/target cell ratios are indicated with mock-treated (▪) and HIV-infected (♦) cells.
We next investigated the functional consequences of HIV infection of CD8+ T cells. HIV replication requires several days, so cells were costimulated for 3 days and then infected for 3 additional days. To control these studies, uninfected CD8 cells were assessed in parallel. Compared with earlier studies (Fig. 2b), there is a diminished ability of uninfected control CD8+ cells to express IFN-γ in response to CD4 ligation 6 days after costimulation (Fig. 3c). Interestingly, IFN-γ was significantly up-regulated in HIV-infected CD8+ T cell cultures to levels similar to that of CD4-ligated control cells. XL anti-CD4 mAb did not further up-regulate IFN-γ production in infected cells. Most CD8+ T cells expressing intracellular gag protein displayed down-regulated CD4 expression similar to that observed in Fig. 3b (data not shown). In other studies, we observed that uninfected cells treated with XL anti-CD4 mAb 3 days after costimulation have levels of IFN-γ expression on day 6 similar to those of cells infected on day 3 (data not shown), suggesting that continual ligation of CD4 by HIV gp120 and subsequent down-regulation of CD4 produced the up-regulation of IFN-γ in infected cultures. Thus, our data further suggest that down-regulation of CD4 by HIV infection rendered these CD8+ T cells unable to modulate IFN-γ after treatment with XL anti-CD4 mAb.
FasL expression in HIV-infected CD8+ T cells was analyzed by flow cytometry. When subjected to CD4 cross-linking 6 days after costimulation, FasL expression in uninfected cells was up-regulated ≈3–4 fold (Fig. 3d). At the same time (6 days after costimulation, 3 days after infection), CD4 expression was significantly down-regulated in HIV-1 p24gag+ cells, rendering them unresponsive to XL mAb and thus unable to up-regulate FasL. Combined, these results indicate that in vitro HIV infection of CD8+CD4dim cells down-regulates CD4 expression and dysregulates IFN-γ and FasL responses associated with CD4 ligation.
To determine whether HIV infection of CD8+ T cells affects CD8+ T cell cytotoxic responses, we generated alloantigen-reactive CD8+ T cells similar to those described above. To allow cells to become activated and permissive for HIV infection, purified CD8+ T cells were cultured with allogeneic monocyte-derived dendritic cells for 5 days, when cells were incubated with or without HIV. Three days later (day 8), both infected and uninfected cells were assessed for their cytolytic ability. Cells expressing intracellular p24 exhibited down-regulated CD4 expression compared with uninfected cultures, similar to what we observed above, and no increase in apoptosis or necrosis was observed (not shown). CD8+ T cells from infected cultures demonstrated significantly reduced CTL responses compared with uninfected cultures (Fig. 3e). Thus, HIV infection of activated CD8+CD4dim T cells directly perturbs in vitro CD8+ CTL activity after the down-regulation of CD4 and the resulting alteration of IFN-γ and FasL expression.
Discussion
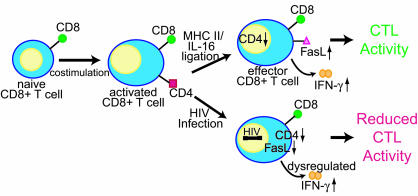
In this study, we have determined that the CD4 molecule has a direct functional role in CTL responses. Further, we have determined that ligation of the CD4 molecule found on activated CD8+ T cells modulates gene expression, particularly the expression of IFN-γ and FasL. Our data suggest that the ligation of CD4, either by MHC class II on an APC or target cell, or by secreted IL-16, modulates the CTL response by increasing IFN-γ and FasL expression (Fig. 4). It has been shown that ligation of CD4 on bona fide Th cells results in increased expression of IFN-γ (48, 49). IFN-γ may then have direct or indirect antiviral effects, including the up-regulation of Fas on infected cells, rendering these cells more susceptible to killing by CD8+ T cells expressing FasL (44, 45, 48, 49). CD4 ligation also has been determined to induce FasL expression on monocytes and to prime Th cells to express FasL after T cell receptor stimulation (49, 50). FasL on these cells has been demonstrated to subsequently induce apoptosis in cells expressing Fas.
Fig. 4.
Model of CD4 function on CD8+ T cells and the effects of HIV infection. Costimulation of naïve CD8+ T cells induces the expression of the CD4 molecule. Ligation of CD4 on CD8 cells by either MHC class II or IL-16 shuts off CD4 RNA expression and up-regulates expression of IFN-γ and FasL, which play a role in facilitating CTL activity. In HIV-infected individuals, CD4 expression allows infection of this cell type. HIV infection down-regulates CD4 cell surface expression, which prevents FasL induction and dysregulates IFN-γ production. This action has a detrimental effect on CTL activity, reducing the efficiency of the CD8+ T cell response against antigen.
Our results suggest that the CD4 molecule could influence several aspects of CD8+ T cell function. Our previous studies (6) indicate that CD4 on highly activated CD8+ T cells serves as a chemotactic receptor for IL-16, a proinflammatory cytokine produced by various cells, including Th and CD8+ T cells, eosinophils, bronchial epithelial cells, synovial fibroblasts, and mast cells (21). IL-16 production by these cells could attract activated CD8+ T cells bearing CD4 to sites of inflammation and viral replication. Together with data presented here, these events provide a potential mechanism through which CD4 directly modulates CD8+ CTL function.
We envision the following model for the role of CD4 in CD8 function: costimulation of an antigen naïve CD8+ T cell by an APC would induce CD4 expression. This likely occurs on the cell with the highest affinity T cell receptors coupled with the strongest costimulatory signal, because we have found that the greater the costimulatory signal, the greater the level of CD4 expressed (6). Thus, the higher-affinity naïve CD8+ T cells would be induced to express greater amounts of CD4. Although these cells remain MHC class I restricted by virtue of their T cell antigen receptor, CD4 expressed by these cells enhances the interaction with the APC by serving as an adhesion and a costimulatory molecule, interacting with MHC class II. Alternatively, the cell could then migrate in a CD4-dependent manner in response to IL-16 toward a site of inflammation. After the ligation of CD4 by either IL-16 or MHC class II, FasL and IFN-γ are induced. IFN-γ then elicits anti-microbial effects, including up-regulating Fas expression by the APC or by the infected cell. This effect renders the target cell susceptible to FasL-mediated attack and results in death of the target cell.
In HIV-infected individuals, the CTL response is generally ineffective at eliminating infection (26). There are many potential ways that HIV may escape the immune response. Our results suggest that infection of CD8+ T cells and constant ligation of CD4 by HIV gp120 on these cells might alter the CTL response in vivo through down-regulation of CD4 and dysregulation of IFN-γ and FasL. Through cytolysis or cellular dysregulation after infection, HIV may then cause loss of a high-affinity CTL shortly after costimulation of the naïve precursor. Infection of this cell before expansion could impact the clonal response to the specific peptide.
In HIV-infected individuals, proviral DNA has been found in a subset of the CD8+ T cell population in the peripheral blood and the pulmonary compartments (11, 28–35). A recent report indicated that the virus in the CD8+ T cell population represents a unique viral reservoir having a different drug resistance mutation pattern than virus found in other compartments in individuals undergoing antiretroviral therapy (33). This finding may represent unique viral growth kinetics and infection in this population. Virus that is found in the naïve CD8+ T cell population (29) is likely to have arisen through infection of a CD4+ progenitor cell that differentiated into a CD4-negative CD8+ T cell (36, 37). HIV and simian immunodeficiency virus proviral DNA that has been found in the more mature populations, particularly the CD8+CD4+ subset, likely entered the cell after costimulation and the induction of CD4 expression (11, 16, 17, 28). Further, our earlier studies suggest that CD4 expression is induced primarily on newly activated, previously antigen-naïve CD8+ T cells. Thus, it is unlikely that infected CD8+ T cells would differentiate to memory T cells. Overall, our studies indicate that CD4 expressed by the CD8 cell itself is required for optimal CD8 cell function. Our studies also suggest that if infection of these CD8+CD4+ T cells occurs in vivo, the function of these cells may be impaired, further perturbing the immune response to HIV and other antigens.
Acknowledgments
We thank Otto Yang, Beth Jamieson, Helen Brown, and Doug Nixon for critical review of the manuscript, and Steve Cole for technical advice and assistance. This work was supported by National Institutes of Health Grants AI57057, AI48392, and AI36059, and the University of California, Los Angeles, Center for AIDS Research (National Institutes of Health Grant AI28697).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, antigen-presenting cells; CTL, cytotoxic T lymphocytes; ELISPOT, enzyme-linked immunospot; FACS, fluorescent cell sorting; FasL, Fas ligand; PP2, 4-amino-5-(chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine; Th, T helper; XL, cross-linked.
References
- 1.Whitmire, J. K. & Ahmed, R. (2000) Curr. Opin. Immunol. 12, 448–455. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan, Y. B., Landay, A. L., Zack, J. A., Kitchen, S. G. & Al-Harthi, L. (2001) Immunology 103, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang, L. P., Riley, J. L., Carroll, R. G., June, C. H., Hoxie, J., Patterson, B. K., Ohshima, Y., Hodes, R. J. & Delespesse, G. (1998) J. Exp. Med. 187, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitchen, S. G., Korin, Y., Roth, M. D., Landay, A. & Zack, J. A. (1998) J. Virol. 72, 9054–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flamand, L., Crowley, R. W., Lusso, P., Colombini-Hatch, S., Margolis, D. M. & Gallo, R. C. (1998) Proc. Natl. Acad. Sci. USA 95, 3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitchen, S. G., LaForge, S., Patel, V. P., Kitchen, C. M., Miceli, M. C. & Zack, J. A. (2002) Blood 99, 207–212. [DOI] [PubMed] [Google Scholar]
- 7.Blue, M. L., Daley, J. F., Levine, H. & Schlossman, S. F. (1985) J. Immunol. 134, 2281–2286. [PubMed] [Google Scholar]
- 8.Ortolani, C., Forti, E., Radin, E., Cibin, R. & Cossarizza, A. (1993) Biochem. Biophys. Res. Commun. 191, 601–609. [DOI] [PubMed] [Google Scholar]
- 9.Tonutti, E., Sala, P., Feruglio, C., Yin, Z. & Colombatti, A. (1994) Clin. Immunol. Immunopathol. 73, 312–320. [DOI] [PubMed] [Google Scholar]
- 10.Laux, I., Khoshnan, A., Tindell, C., Bae, D., Zhu, X., June, C. H., Effros, R. B. & Nel, A. (2000) Clin. Immunol. 96, 187–197. [DOI] [PubMed] [Google Scholar]
- 11.Imlach, S., McBreen, S., Shirafuji, T., Leen, C., Bell, J. E. & Simmonds, P. (2001) J. Virol. 75, 11555–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macchi, B., Graziani, G., Zhang, J. & Mastino, A. (1993) Cell. Immunol. 149, 376–389. [DOI] [PubMed] [Google Scholar]
- 13.Lusso, P., De Maria, A., Malnati, M., Lori, F., DeRocco, S. E., Baseler, M. & Gallo, R. C. (1991) Nature 349, 533–535. [DOI] [PubMed] [Google Scholar]
- 14.Dykhuizen, M., Ceman, J., Mitchen, J., Zayas, M., MacDougall, A., Helgeland, J., Rakasz, E. & Pauza, C. D. (2000) Cytometry 40, 69–75. [DOI] [PubMed] [Google Scholar]
- 15.Akari, H., Terao, K., Murayama, Y., Nam, K. H. & Yoshikawa, Y. (1997) Int. Immunol. 9, 591–597. [DOI] [PubMed] [Google Scholar]
- 16.Khatissian, E., Monceaux, V., Cumont, M. C., Ho Tsong Fang, R., Estaquier, J. & Hurtrel, B. (2003) AIDS Res. Hum. Retroviruses 19, 267–274. [DOI] [PubMed] [Google Scholar]
- 17.Dean, G. A., Reubel, G. H. & Pedersen, N. C. (1996) J. Virol. 70, 5646–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuckermann, F. A. (1999) Vet. Immunol. Immunopathol. 72, 55–66. [DOI] [PubMed] [Google Scholar]
- 19.Hillemeyer, P., White, M. D. & Pascual, D. W. (2002) Cell. Immunol. 215, 173–185. [DOI] [PubMed] [Google Scholar]
- 20.Periwal, S. B. & Cebra, J. J. (1999) J. Virol. 73, 7633–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Center, D. M., Kornfeld, H., Ryan, T. C. & Cruikshank, W. W. (2000) Immunol. Today 21, 273–280. [DOI] [PubMed] [Google Scholar]
- 22.Konig, R. & Zhou, W. (2004) Curr. Issues Mol. Biol. 6, 1–15. [PubMed] [Google Scholar]
- 23.Ravichandran, K. S., Collins, T. L. & Burakoff, S. J. (1996) Curr. Top. Microbiol. Immunol. 205, 47–62. [DOI] [PubMed] [Google Scholar]
- 24.Dalgleish, A. G., Beverley, P. C., Clapham, P. R., Crawford, D. H., Greaves, M. F. & Weiss, R. A. (1984) Nature 312, 763–767. [DOI] [PubMed] [Google Scholar]
- 25.Klatzmann, D., Champagne, E., Chamaret, S., Gruest, J., Guetard, D., Hercend, T., Gluckman, J. C. & Montagnier, L. (1984) Nature 312, 767–768. [DOI] [PubMed] [Google Scholar]
- 26.Letvin, N. L. & Walker, B. D. (2003) Nat. Med. 9, 861–866. [DOI] [PubMed] [Google Scholar]
- 27.Stevenson, M. (2003) Nat. Med. 9, 853–860. [DOI] [PubMed] [Google Scholar]
- 28.Brenchley, J. M., Hill, B. J., Ambrozak, D. R., Price, D. A., Guenaga, F. J., Casazza, J. P., Kuruppu, J., Yazdani, J., Migueles, S. A., Connors, M., et al. (2004) J. Virol. 78, 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBreen, S., Imlach, S., Shirafuji, T., Scott, G. R., Leen, C., Bell, J. E. & Simmonds, P. (2001) J. Virol. 75, 4091–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livingstone, W. J., Moore, M., Innes, D., Bell, J. E. & Simmonds, P. (1996) Lancet 348, 649–654. [DOI] [PubMed] [Google Scholar]
- 31.Semenzato, G., Agostini, C., Ometto, L., Zambello, R., Trentin, L., Chieco-Bianchi, L. & De Rossi, A. (1995) Blood 85, 2308–2314. [PubMed] [Google Scholar]
- 32.Semenzato, G., Agostini, C., Chieco-Bianchi, L. & De Rossi, A. (1998) J. Leukocyte Biol. 64, 298–301. [DOI] [PubMed] [Google Scholar]
- 33.Potter, S. J., Dwyer, D. E. & Saksena, N. K. (2003) Virology 305, 339–352. [DOI] [PubMed] [Google Scholar]
- 34.Saha, K., Zhang, J. & Zerhouni, B. (2001) J. Acquired Immune Defic. Syndr. 26, 199–207. [DOI] [PubMed] [Google Scholar]
- 35.Saha, K., Zhang, J., Gupta, A., Dave, R., Yimen, M. & Zerhouni, B. (2001) Nat. Med. 7, 65–72. [DOI] [PubMed] [Google Scholar]
- 36.Kitchen, S. G., Uittenbogaart, C. H. & Zack, J. A. (1997) J. Virol. 71, 5713–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks, D. G., Kitchen, S. G., Kitchen, C. M., Scripture-Adams, D. D. & Zack, J. A. (2001) Nat. Med. 7, 459–464. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi, T., McGhee, J. R., Coffman, R. L., Beagley, K. W., Eldridge, J. H., Takatsu, K. & Kiyono, H. (1990) J. Immunol. Methods 128, 65–73. [DOI] [PubMed] [Google Scholar]
- 39.Murali-Krishna, K., Altman, J. D., Suresh, M., Sourdive, D. J., Zajac, A. J., Miller, J. D., Slansky, J. & Ahmed, R. (1998) Immunity 8, 177–187. [DOI] [PubMed] [Google Scholar]
- 40.Kiertscher, S. M. & Roth, M. D. (1996) J. Leukocyte Biol. 59, 208–218. [DOI] [PubMed] [Google Scholar]
- 41.Zarling, A. L., Johnson, J. G., Hoffman, R. W. & Lee, D. R. (1999) J. Immunol. 162, 5197–5204. [PubMed] [Google Scholar]
- 42.Zack, J. A., Arrigo, S. J., Weitsman, S. R., Go, A. S., Haislip, A. & Chen, I. S. (1990) Cell 61, 213–222. [DOI] [PubMed] [Google Scholar]
- 43.Zloza, A., Sullivan, Y. B., Connick, E., Landay, A. L. & Al-Harthi, L. (2003) Blood 102, 2156–2164. [DOI] [PubMed] [Google Scholar]
- 44.Siegel, R. M., Chan, F. K., Chun, H. J. & Lenardo, M. J. (2000) Nat. Immunol. 1, 469–474. [DOI] [PubMed] [Google Scholar]
- 45.Goodbourn, S., Didcock, L. & Randall, R. E. (2000) J. Gen. Virol. 81, 2341–2364. [DOI] [PubMed] [Google Scholar]
- 46.Hanke, J. H., Gardner, J. P., Dow, R. L., Changelian, P. S., Brissette, W. H., Weringer, E. J., Pollok, B. A. & Connelly, P. A. (1996) J. Biol. Chem. 271, 695–701. [DOI] [PubMed] [Google Scholar]
- 47.Pittet, M. J., Speiser, D. E., Valmori, D., Cerottini, J. C. & Romero, P. (2000) J. Immunol. 164, 1148–1152. [DOI] [PubMed] [Google Scholar]
- 48.Oyaizu, N., McCloskey, T. W., Than, S., Hu, R., Kalyanaraman, V. S. & Pahwa, S. (1994) Blood 84, 2622–2631. [PubMed] [Google Scholar]
- 49.Tateyama, M., Oyaizu, N., McCloskey, T. W., Than, S. & Pahwa, S. (2000) Blood 96, 195–202. [PubMed] [Google Scholar]
- 50.Oyaizu, N., Adachi, Y., Hashimoto, F., McCloskey, T. W., Hosaka, N., Kayagaki, N., Yagita, H. & Pahwa, S. (1997) J. Immunol. 158, 2456–2463. [PubMed] [Google Scholar]